Abstract
Background and Aim
Chronic hepatitis B virus (HBV) infection is a major cause of hepatocellular carcinoma (HCC). Circulating cell-free DNA (cfDNA) methylation of tumor suppressor genes are emerging potential biomarkers in HCC. We aimed to evaluate the cfDNA methylation status of RASSF1 and CDKN2AIP genes in patients with liver cirrhosis (LC) with or without HCC caused by HBV.
Materials and Methods
A total of 47 patients with HBV cirrhosis were included in the study. Patients were divided into two groups: HCC and LC (HCC+LC, n=22) and HBV cirrhosis only (LC, n=25). cfDNA was isolated from the plasma samples of the patients. Methylation analysis was performed for RASSF1 and CDKN2AIP genes.
Results
Mean methylation percentage of CDKN2AIP gene was 0.001±0.004% in the HCC+LC group and 0.008±0.004 % in the LC only group. The mean methylation percentage of RASSF1 gene was 5.1±16.1% in the HCC+LC group and 9.7±25.9% in the LC only group. The methylation rate of CDKN2AIP was significantly lower in the HCC+LC group (p=0.027). A positive correlation was found with the absence of cfDNA methylation of CDKN2AIP gene in the presence of HCC (R=0.667, p=0.018).
Conclusion
cfDNA methylation of CDKN2AIP and RASSF1 genes may provide important diagnostic information regarding the development of HCC in the setting of HBV cirrhosis.
Keywords: Cirrhosis, hepatitis B virus, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver and the fourth most common cause of cancer-related mortality worldwide.[1,2] Chronic hepatitis B virus (HBV) infection is the most common etiology of HCC.[1] HBV is an oncogenic virus that can be integrated into the host genome and affect the expression of tumor suppressor genes and oncogenes.[3,4]
According to phylogenic analyses, there are ten HBV genotypes (A–J).[5] Genotypes C and D have a higher risk of progression to cirrhosis and HCC, and their response to conventional treatment is usually less favorable than genotypes A and B.[6,7] Predominant HBV genotype in Turkiye is genotype D.[8–10]
Chronic liver injury induced by various etiologies, including infectious, metabolic, autoimmune, chronic cholestatic, and toxic insults leading to liver cirrhosis (LC) or chronic HBV without LC, increases the risk of HCC.[1] Patients with cirrhosis of all etiologies and high-risk patients with HBV are recommended to undergo surveillance for the HCC.[11] HCC surveillance is crucial to detect the presence of an early tumor, and liver ultrasonography (US) with or without serum alpha-fetoprotein (AFP) levels every 6 months is one of the recommended practice guidelines.[12] In a recent meta-analysis, US was found to have a sensitivity of 45% in detecting early-stage HCC, and the combination of US with AFP increased the sensitivity to 63%.[13] However, US is highly operator dependent and provides a limited evaluation in patients with cirrhosis and obesity.[14]
Several novel serum biomarkers have been studied for the early detection of HCC.[10,11,13] The circulating cell-free DNA (cfDNA) analysis, also called “liquid biopsy,” in serum or plasma samples is one of the methods that have been studied in the diagnosis of HCC.[15] The primary source of cfDNA is the degradation of apoptotic and necrotic tumor cells and the release of tumor DNA particles.[16] This technique is minimally invasive and has the potential to screen, diagnose, and evaluate the disease progression. DNA methylation is an epigenetic mechanism that influences gene activities by regulating gene expression.[17] RASSF1A and CDKN2A (p16) are tumor suppressor genes, and their hypermethylation has been observed in HCC caused by HBV, significantly higher as compared with chronic HBV.[15,18] RASSF1A methylation has also been shown to provide better sensitivity and specificity than AFP in HCC caused by HBV.[19] Identification of tumor suppressor genes that are methylated early in the tumor development, along with the analyses of the cfDNA methylation of select genes, may be a useful tool for screening patients at risk for HCC.[19]
Materials and Methods
Patient Selection
Patients were recruited for the study from the clinics of gastroenterology and hepatology, nuclear medicine, and interventional radiology of our institution between October 2018 and December 2019. Patients over the age of 18 years who have cirrhosis caused by HBV with or without HCC were included in the study. Patients with malignancies other than HCC and patients with severe systemic illnesses were excluded. The study was approved by the Institutional Review Board of Istanbul University, Istanbul School of Medicine. Written and verbal informed consent was obtained from all patients.
Collection and Storage of Specimens
The blood samples were taken into tubes (2 mL) with K3 EDTA, centrifuged twice at 4°C at 1350 RCF for 10 min in 4 h and then aliquoted to 1 mL in 1.5 mL Eppendorf tube in a sterile cabinet. Plasma samples were stored at –80°C until the day of analyses.[20,21] After the sample collection process, nucleic acids (cfDNA) in each sample were extracted using the QIAGEN Cell-Free isolation kit (QIAamp Circulating Nucleic Acid Kit Cat No/ID: 55114, Qiagen, UK), according to the manufacturer’s instructions. cfDNA was extracted from 1 mL plasma, and 50 µL of elution buffer was added.
The concentrations and purities of the obtained cfDNA samples were measured with a MaestroNano Micro-Volume spectrophotometer. To perform the methylation analysis, the DNA concentration of each sample was adjusted to 25 ng/µL. The nucleic acid digestion procedures required for methylation analyses were performed following the protocol of Qiagen-EpiTect Methyl II DNA Restriction Kit (SA Bioscience, cat no.: 335452). After digestion, determination of the methylation status of RASSF1 and CDKN2AIP genes was performed using the protocol of Qiagen – EpiTect Methyl II (Qiagen–EpiTect Methyl II PCR Assay for 2 Genes Using 1 DNA Sample, Cat No./ID: 335002, Qiagen, UK). Real-time PCR was used for methylation analysis. Real-time PCR studies were performed using Agilent brand Real-Time PCR device (Agilent Technologies, CA, USA).
Statistical Analysis
IBM SPSS Statistics 22.0 for Windows was used for statistical analysis. Student’s t-test was performed when appropriate, and p<0.05 was considered significant. Pearson’s correlation analysis was performed.
Results
Forty-seven HBsAg-positive patients with HBV were included in the study and divided into two groups: 22 patients (46.8%) with HCC+LC and 25 patients (53.2%) with LC only. HCC+LC group was older than LC only group (64.0±10.01 vs 53.9±10.6 years, p=0.003). Male gender was predominant in HCC+LC group; however, it was not statistically significant between the two groups (81.8% vs 60%, p=0.106, respectively), as shown in Table 1. Mean AST, ALT, ALP, and GGT were significantly higher in the HCC+LC group compared with LC only group (50.43±295.3 IU/L vs 30.9±12.9 IU/L, p=0.018; 74±107.1 IU/L vs 22.7±8.2 IU/L, p=0.031; 164.05±128.2 IU/L vs 86.6±32.6 IU/L, p=0.016; and 94.52±124.2 IU/L vs 35.0±29.9 IU/L, p=0.028, respectively). Mean AFP was higher in HCC+LC group than in the LC only group, although it was not statistically significant (234.0±505 ng/mL vs 4.04±2.94 ng/mL, p=0.085, respectively).
Table 1.
Clinical and laboratory features of study patients
| HCC+LC (n=22) | LC (n=25) | p | |||||
|---|---|---|---|---|---|---|---|
| n | % | Mean±SD | n | % | Mean±SD | ||
| Gender | 0.106 | ||||||
| Male | 4 | 10 | |||||
| Female | 18 | 15 | |||||
| Age | 64.0±10.01 | 53.92±10.6 | 0.003 | ||||
| Child | 0.605 | ||||||
| A | 76.2 | 64 | |||||
| B | 4.8 | 28 | |||||
| C | 19 | 8 | |||||
| Barcelona | |||||||
| A | 61.9 | NA | NA | ||||
| B | 14.3 | NA | NA | ||||
| C | 9.5 | NA | NA | ||||
| D | 14.3 | NA | NA | ||||
| AST (IU/L) | 150.43±295.3 | 30.9±12.9 | 0.018 | ||||
| ALT (IU/L) | 74±107.1 | 22.7±8.2 | 0.031 | ||||
| ALP (IU/L) | 164.05±128.2 | 86.6±32.6 | 0.016 | ||||
| GGT (IU/L) | 94.52±124.2 | 35.0±29.9 | 0.028 | ||||
| AFP (ng/mL, range) | 234±505.1 (1.2-1651) | 4.04±2.94 (1.2-11) | 0.085 | ||||
| Cell-free DNA (ng/mL) | 27.89±8.68 | 27.82±7.75 | 0.48 | ||||
| Methylation of RASSF1 | 5.1±16.1 | 9.7±25.9 | 0.431 | ||||
| Present | 27.3 | 36 | 0.027 | ||||
| Absent | 72.7 | 64 | |||||
| Methylation of CDKN2AIP | 0.001±0.004 | 0.008±0.004 | |||||
| Present | 4.5 | 20 | |||||
| Absent | 95.5 | 80 | |||||
HCC: Hepatocellular carcinoma; LC: Liver cirrhosis; AST: Aspartate aminotransferase; ALT: Alanine aminotransferase; AFP: Alpha-fetoprotein; ALP: Alkaline phosphatase; GGT: Gamma-glutamyl transferase.
In the LC only group, 64% had Child-Pugh class A, 28% had Child-Pugh class B, and 8% had Child-Pugh class C cirrhosis. In HCC+LC group, 76.2% had Child-Pugh class A, 4.8% had Child-Pugh class B, and 19% had Child-Pugh class C cirrhosis. Barcelona classification of patients with HCC was 61.9% A, 14.3% B, 9.5% C, and 14.3% D.[22]
cfDNA levels were similar in both groups (p>0.05). The mean methylation percentage of CDKN2AIP gene was 0.001±0.004% in the HCC+LC group and 0.008±0.004% in the LC only group. No methylation was detected in CDKN2AIP gene in 80% of the patients in the LC only group and in 95.5% of patients in the HCC+LC group (Fig. 1). The mean methylation percentage of RASSF1 gene was 9.7 ± 25.9% in LC only group and 5.1±16.1 in the HCC+LC group. No methylation was detected for RASSF1 gene in 64% of the patients in LC only group and 72.7% of the patients in the HCC+LC group. Although the methylation rates of CDKN2AIP and RASSF1 genes were not statistically different in the two groups, the presence of methylation of CDKN2AIP was significantly lower in the HCC+LC group (p=0.027) compared with the LC only group. Correlation analysis was performed, which revealed a positive correlation with the absence of methylation of CDKN2AIP gene in the presence of HCC (R=0.667, p=0.018).
Figure 1.
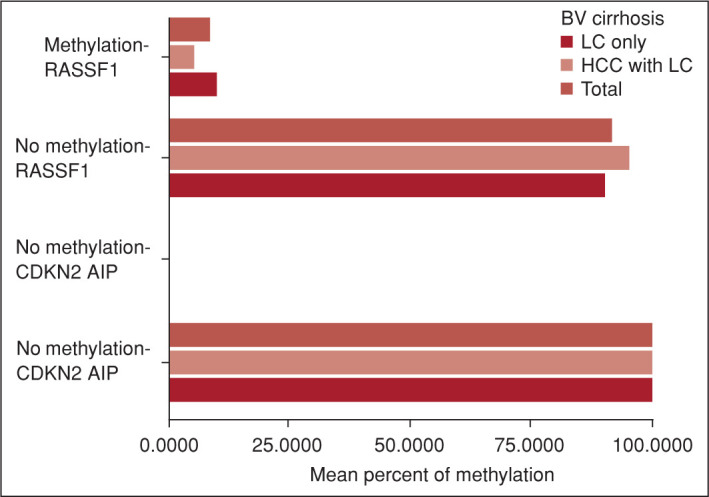
Percent cell-free methylation status of RASSF1 and CDKN2AIP genes. Cell-free methylation of CDKN2AIP gene was absent in 95.5% of patients in the HCC+LC group and in 80% of the patients in the LC only group. Cell-free methylation of RASSF1 gene was absent in 72.7% of patients in the HCC+LC group and in 64% of patients in LC only group.
HCC: Hepatocellular carcinoma; LC: Liver cirrhosis.
Discussion
The discovery of circulating DNA fragments, or cfDNA, in plasma dates back to 1948.[23] Healthy subjects have been shown to have an average of 30 ng/mL (0–100 ng/mL) cfDNA in their plasma.[24]
cfDNA has been studied extensively in several fields such as cancer research because of the genetic and epigenetic information provided by the cfDNA.[25] It is hypothesized that the cells that underwent apoptosis and necrosis are the primary sources of the cfDNA in the circulatory system.[26]
DNA methylation is an epigenetic modification affecting gene expression.[27] The formation of 5-methylcytosine by covalently binding a methyl group to the C5 position of the cytosine residues in the CpG dinucleotide sequences in differentiated cells is the basic epigenetic tag.[28] Although methylation formation generally occurs in CpG sequences, it can also be observed in non-CpG sequences; however, the importance of this occurrence is not fully understood.[17] DNA methylation is effective in critical cellular events such as regulation of gene expression, inactivation of X chromosome, and silencing of retroviral element.[17] 5-Methylcytosine, which is formed as a result of DNA methylation in promoter regions, generally suppresses gene expression by affecting the recognition and binding of transcription factors.[29] Gene methylation regulates immune responses by changing the expression of inflammatory cytokines.[30] Tumor suppressor gene methylations play an essential role in carcinogenesis.[31]
HBV-infected cells contain covalently closed circular DNA (cccDNA) in the nucleus.[32] Both episomal and integrated HBV DNA can be methylated in hepatocytes.[31] Methylation of viral DNA plays a vital role in viral replication and protein expression.[31] There are differences in methylation status between different HBV genotypes that show specific geographical distribution.[33] Previously, the presence of HBV cccDNA methylation and its role in HBV replication were reported from the studies in Asia, where genotypes B and C are dominant. It showed that this methylation determines the suitability of HBV replication,[34,35] whereas genotype D is the predominant HBV genotype in Turkiye.[8–10]
In a study conducted on liver biopsy samples of HBV-infected patients in France, where genotypes A and D are common, methylation in the HBV genome was reported as a rare occurrence.[36] High mutation rates observed in HBV DNA replication may affect the methylation by altering the C5 position in CpG dinucleotides, which is the only place where methylation can occur in mammalian cells.[33,37]
RASSF1A and CDKN2A are tumor suppressor genes. Hypermethylation of these genes in HCC has been shown mainly in HCC developed in the background of chronic HBV infection.[15] RASSF1A gene product is a folded protein and is responsible for cell signaling.[38] DNA hypermethylation occurs as a result of the effects of various proteins and viruses on DNA.[38] CDKN2AIP is a protein that consists of 580 amino acids belonging to the CARF family that regulates damage responses through signaling pathways related to cell proliferation, apoptosis, and aging.[39]
In several studies, it has been reported that methylation of CDKN2AIP gene is altered.[40,41] However, consistent results regarding the relationship between CDKN2AIP gene and the risk of HCC has not been shown.[42] A meta-analysis conducted by Zhou et al.[43] included 59 studies and concluded an enhanced promotor methylation rate of CDKN2A gene in the HCC and concluded that CDKN2A might be a tool for triage. However, in this meta-analysis, no distinction was made regarding the etiology of HCC; mainly tissue biopsies were analyzed, and study patients were from Japan, China, Korea, Germany, Italy, Spain, and the United States.[43] In our study on the role of cfDNA methylations of RASSF1 and CDKN2AIP genes in the diagnosis of HBV-related LC and HCC, we found that cfDNA levels in the group with HCC and LC were similar to those with LC only. However, methylation rates in CDKN2AIP gene were significantly lower in the HCC and LC group than in the LC only group.
Our study has a few limitations: small sample size and the lack of cases with chronic HBV (without LC or HCC) as a control group. Studies to assess and validate the utility of cfDNA methylations may provide important clues in the diagnosis of HCC and may aid in stratifying high-risk patients with HBV cirrhosis.
Acknowledgement
The test kits were supplied unconditionally with the support of Gilead Sciences Turkey.
Footnotes
How to cite this article: Telli P, Ozturk NB, Hakan MT, Cavus B, Cifcibasi Ormeci A, Yakut A, et al. Cell-free methylation of RASSF1 and CDKN2AIP genes in the diagnosis of hepatocellular carcinoma associated with hepatitis B virus cirrhosis. Hepatology Forum 2022; 3(3):77–81.
Ethics Committee Approval
The Istanbul University Clinical Research Ethics Committee granted approval for this study (date: 29.03.2019, number: 2019/442).
Peer-review
Externally peer-reviewed.
Author Contributions
Concept – PT, NBO, MTH, IY, FA; Design – PT, NBO, MTH, BC, ACO, AY, VS, ZI, AP, EGI, KD, FB, SK, IY, FA; Supervision – KD, FB, SK, IY, FA; Fundings – NBO, IY, FA; Materials – PT, MTO, BC, ACO, AY, VS, ZI, AP, EGI, KD, FB, SK, FA; Data Collection and/or Processing – PT, NBO, MTH, BC, ACO, AY, VS, ZI, AP, EGI; Analysis and/or Interpretation – PT, NBO, MTO, BC, BC, ACO, AY, VS, ZI, AP, EGI, KD, SK, IY, FA; Literature Search – PT, NBO, FA; Writing – PT, NBO, IY, FA; Critical Reviews – PT, NBO, MTH, BC, ACO, AY, VS, ZI, AP, EGI, KD, FB, SK, IY, FA.
Conflict of Interest
The authors have no conflict of interest to declare.
Financial Disclosure
The authors declared that this study has received no financial support.
References
- 1.Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019 Oct;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fung J, Lai CL, Yuen MF. Hepatitis B and C virus-related carcinogenesis. Clin Microbiol Infect. 2009;15(11):964–970. doi: 10.1111/j.1469-0691.2009.03035.x. [DOI] [PubMed] [Google Scholar]
- 4.Yang JD, Kim WR, Coelho R, Mettler TA, Benson JT, Sanderson SO, et al. Cirrhosis is present in most patients with hepatitis B and hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2011;9(1):64–70. doi: 10.1016/j.cgh.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, et al. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype. J Virol. 2009;83(20):10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erhardt A, Blondin D, Hauck K, Sagir A, Kohnle T, Heintges T, et al. Response to interferon alfa is hepatitis B virus genotype dependent: genotype A is more sensitive to interferon than genotype D. Gut. 2005;54(7):1009–1013. doi: 10.1136/gut.2004.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonneveld MJ, Rijckborst V, Cakaloglu Y, Simon K, Heathcote EJ, Tabak F, et al. Durable hepatitis B surface antigen decline in hepatitis B e antigen-positive chronic hepatitis B patients treated with pegylated interferon-α2b: relation to response and HBV genotype. Antivir Ther. 2012;17(1):9–17. doi: 10.3851/IMP1887. [DOI] [PubMed] [Google Scholar]
- 8.Akyuz F, Ciftci S, Keskin F, Cakiris A, Pinarbasi B, Baran B, et al. Ultra-deep pyrosequencing of precore/core promoter mutations in patients with genotype d chronic hepatitis B. J Gastroenterol Hepatol Res. 2014;3(4):1030–1034. [Google Scholar]
- 9.Leblebicioglu H, Eroglu C, Members of the Hepatitis Study Group Acute hepatitis B virus infection in Turkey: epidemiology and genotype distribution. Clin Microbiol Infect. 2004;10(6):537–541. doi: 10.1111/j.1469-0691.2004.00871.x. [DOI] [PubMed] [Google Scholar]
- 10.Yalcin K, Degertekin H, Bahcecioglu IH, Demir A, Aladag M, Yildirim B, et al. Hepatitis B virus genotype D prevails in patients with persistently elevated or normal ALT levels in Turkey. Infection. 2004 Feb;32(1):24–29. doi: 10.1007/s15010-004-3010-7. [DOI] [PubMed] [Google Scholar]
- 11.Harris PS, Hansen RM, Gray ME, Massoud OI, McGuire BM, Shoreibah MG. Hepatocellular carcinoma surveillance: An evidence-based approach. World J Gastroenterol. 2019;25(13):1550–1559. doi: 10.3748/wjg.v25.i13.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68(3):526–549. doi: 10.1016/j.jhep.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: A meta-analysis. Gastroenterology. 2018;154(6):1706–1718.e1. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parikh ND, Mehta AS, Singal AG, Block T, Marrero JA, Lok AS. Biomarkers for the Early Detection of Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev. 2020;29(12):2495–2503. doi: 10.1158/1055-9965.EPI-20-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pezzuto F, Buonaguro L, Buonaguro FM, Tornesello ML. The Role of Circulating Free DNA and MicroRNA in Non-Invasive Diagnosis of HBV-and HCV-Related Hepatocellular Carcinoma. Int J Mol Sci. 2018;19(4):1007. doi: 10.3390/ijms19041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butt AN, Swaminathan R. Overview of circulating nucleic acids in plasma/serum. Ann N Y Acad Sci. 2008;1137:236–242. doi: 10.1196/annals.1448.002. [DOI] [PubMed] [Google Scholar]
- 17.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv X, Ye G, Zhang X, Huang T. p16 Methylation was associated with the development, age, hepatic viruses infection of hepatocellular carcinoma, and p16 expression had a poor survival: A systematic meta-analysis (PRISMA) Medicine (Baltimore) 2017;96(38):e8106. doi: 10.1097/MD.0000000000008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mocan T, Simão AL, Castro RE, Rodrigues CMP, Słomka A, Wang B, et al. Liquid biopsies in hepatocellular carcinoma: are we winning? J Clin Med. 2020;9(5):1541. doi: 10.3390/jcm9051541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barták BK, Kalmár A, Galamb O, Wichmann B, Nagy ZB, Tulassay Z, et al. Blood collection and cell-free DNA Isolation methods influence the sensitivity of liquid biopsy analysis for colorectal cancer detection. Pathol Oncol Res. 2019;25(3):915–923. doi: 10.1007/s12253-018-0382-z. [DOI] [PubMed] [Google Scholar]
- 21.Gyanchandani R, Kvam E, Heller R, Finehout E, Smith N, Kota K, et al. Whole genome amplification of cell-free DNA enables detection of circulating tumor DNA mutations from fingerstick capillary blood. Sci Rep. 2018 Nov 23;8(1):17313. doi: 10.1038/s41598-018-35470-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llovet JM, Fuster J, Bruix J, Barcelona-Clínic Liver Cancer Group The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl. 2004;10(2 Suppl 1):S115–S120. doi: 10.1002/lt.20034. [DOI] [PubMed] [Google Scholar]
- 23.Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): Clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 2016;14(10):898–908. doi: 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]
- 24.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta. 2007;1775(1):181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 26.Breitbach S, Tug S, Simon P. Circulating cell-free DNA: an up-coming molecular marker in exercise physiology. Sports Med. 2012;42(7):565–586. doi: 10.2165/11631380-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 27.Choy MK, Movassagh M, Goh HG, Bennett MR, Down TA, Foo RS. Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated. BMC Genomics. 2010;11:519. doi: 10.1186/1471-2164-11-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 29.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010;105(1):4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 30.White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO-T cells. J Immunol. 2002;168(6):2820–2827. doi: 10.4049/jimmunol.168.6.2820. [DOI] [PubMed] [Google Scholar]
- 31.Vivekanandan P, Thomas D, Torbenson M. Hepatitis B viral DNA is methylated in liver tissues. J Viral Hepat. 2008;15(2):103–107. doi: 10.1111/j.1365-2893.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 32.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51(3):581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Li C, Zhang Y, Zhu H, Kang Y, Liu H, et al. Comparative analysis of CpG islands among HBV genotypes. PLoS One. 2013;8(2):e56711. doi: 10.1371/journal.pone.0056711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y, Li Y, Mu S, Zhang J, Yan Z. Evidence that methylation of hepatitis B virus covalently closed circular DNA in liver tissues of patients with chronic hepatitis B modulates HBV replication. J Med Virol. 2009;81(7):1177–1183. doi: 10.1002/jmv.21525. [DOI] [PubMed] [Google Scholar]
- 35.Kim JW, Lee SH, Park YS, Hwang JH, Jeong SH, Kim N, et al. Replicative activity of hepatitis B virus is negatively associated with methylation of covalently closed circular DNA in advanced hepatitis B virus infection. Intervirology. 2011;54(6):316–325. doi: 10.1159/000321450. [DOI] [PubMed] [Google Scholar]
- 36.Kaur P, Paliwal A, Durantel D, Hainaut P, Scoazec JY, Zoulim F, et al. DNA methylation of hepatitis B virus (HBV) genome associated with the development of hepatocellular carcinoma and occult HBV infection. J Infect Dis. 2010;202(5):700–704. doi: 10.1086/655398. [DOI] [PubMed] [Google Scholar]
- 37.Chotiyaputta W, Lok AS. Hepatitis B virus variants. Nat Rev Gastroenterol Hepatol. 2009;6(8):453–462. doi: 10.1038/nrgastro.2009.107. [DOI] [PubMed] [Google Scholar]
- 38.García-Gutiérrez L, McKenna S, Kolch W, Matallanas D. RASSF1A tumour suppressor: target the network for effective cancer therapy. Cancers (Basel) 2020;12(1):229. doi: 10.3390/cancers12010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miki TS, Richter H, Rüegger S, Großhans H. PAXT-1 promotes XRN2 activity by stabilizing it through a conserved domain. Mol Cell. 2014;53(2):351–360. doi: 10.1016/j.molcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Tannapfel A, Busse C, Weinans L, Benicke M, Katalinic A, Geissler F, et al. INK4a-ARF alterations and p53 mutations in hepatocellular carcinomas. Oncogene. 2001;20(48):7104–7109. doi: 10.1038/sj.onc.1204902. [DOI] [PubMed] [Google Scholar]
- 41.Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics. 2011;12(2):130–137. doi: 10.2174/138920211795564359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zang JJ, Xie F, Xu JF, Qin YY, Shen RX, Yang JM, et al. P16 gene hypermethylation and hepatocellular carcinoma: a systematic review and meta-analysis. World J Gastroenterol. 2011;17(25):3043–3048. doi: 10.3748/wjg.v17.i25.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Wang XB, Qiu XP, Zhang S, Wang C, Zheng F. CDKN2A promoter methylation and hepatocellular carcinoma risk: A meta-analysis. Clin Res Hepatol Gastroenterol. 2018;42(6):529–541. doi: 10.1016/j.clinre.2017.07.003. [DOI] [PubMed] [Google Scholar]


