Abstract
The pBHR1 plasmid is a derivative of the small (2.6-kb), mobilizable broad-host-range plasmid pBBR1, which was isolated from the gram-negative bacterium Bordetella bronchiseptica (R. Antoine and C. Locht, Mol. Microbiol. 6:1785–1799, 1992). Plasmid pBBR1 consists of two functional cassettes and presents sequence similarities with the transfer origins of several plasmids and mobilizable transposons from gram-positive bacteria. We show that the Mob protein specifically recognizes a 52-bp sequence which contains, in addition to the transfer origin, the promoter of the mob gene. We demonstrate that this gene is autoregulated. The binding of the Mob protein to the 52-bp sequence could thus allow the formation of a protein-DNA complex with a double function: relaxosome formation and mob gene regulation. We show that the Mob protein is a relaxase, and we located the nic site position in vitro. After sequence alignment, the position of the nic site of pBBR1 corresponds with those of the nick sites of the Bacteroides mobilizable transposon Tn4555 and the streptococcal plasmid pMV158. The oriT of the latter is characteristic of a family of mobilizable plasmids that are found in gram-positive bacteria and that replicate by the rolling-circle mechanism. Plasmid pBBR1 thus appears to be a new member of this group, even though it resides in gram-negative bacteria and does not replicate via a rolling-circle mechanism. In addition, we identified two amino acids of the Mob protein necessary for its activity, and we discuss their involvement in the mobilization mechanism.
Bacteria are omnipresent and have an exceptional ability to adapt to environmental changes. Plasmids play a crucial role in bacterial evolution and adaptation by mediating the horizontal exchange of genetic material via conjugation. This genetic exchange is possible between different bacterial species (via broad-host-range plasmids) and even between bacteria and eukaryotic cells (yeasts and plant cells) (5, 20, 24, 32). Conjugation involves unidirectional transfer of a single DNA strand, with 5′–3′ polarity, from a donor to a recipient cell. DNA transfer is initiated by a plasmid-encoded protein, a DNA relaxase, which cleaves the phosphodiester bond of a specific dinucleotide (the nick site) within the origin of transfer (oriT) (for a review see references 39 and 54). Conjugative plasmids possess the genes necessary for DNA transfer, whereas some plasmids, called mobilizable plasmids, possess their own transfer origin and encode a relaxase but need the help of a conjugative plasmid for their transfer from a donor to a recipient. The small (2.6-kb) broad-host-range plasmid pBBR1 was first isolated from the gram-negative bacterium Bordetella bronchiseptica and sequenced (2). It contains enough genetic information to be mobilized by IncP plasmids, to display a medium copy number, and to be stably maintained in all gram-negative bacteria tested to date. No phenotypic trait which might represent a selective advantage has been found, however. Plasmid pBBR1 is compatible with all plasmids tested. It consists of two functional cassettes (the replication and mobilization regions), as is common in small plasmids from gram-positive bacteria (23). Sequence similarities have been found with the transfer origins (also called recombination site A [RSA]) of several plasmids and mobilizable transposons from gram-positive bacteria (2, 10). In plasmids from gram-positive bacteria, the RSA is known as the specific site necessary for mobilization and recombination mediated by a Mob/Pre protein. Recombination events at this site result in cointegrate formation, but the site is not involved in plasmid maintenance (14, 40). Plasmid transfer from gram-positive to gram-negative bacteria is usually considered a rare event. The origin of the RSA of the pBBR1 plasmid thus seems enigmatic. The plasmid has been used frequently to design cloning vectors (2, 13, 25, 34), but involvement of the RSA of pBBR1 in mobilization between two gram-negative bacteria has never been demonstrated. The similarity between pBBR1 and some plasmids of gram-positive bacteria has led us to precisely examine its mobilization function. The DNA sequence of the mob gene of pBBR1 predicts a protein of 329 amino acids (molecular weight, 36,707). Here we demonstrate that this Mob protein is a relaxase binding specifically to the transfer origin (RSA) in order to nick the DNA and regulate its own synthesis. In addition, we identify two amino acids of this protein (aspartate 120 and glutamate 121) that are necessary for its mobilization activity.
MATERIALS AND METHODS
Media.
Rich Luria-Bertani (LB) broth (30) was the growth medium used. Antibiotic concentrations were as follows: 100 μg of ampicillin/ml, 15 μg of chloramphenicol/ml, 15 μg of tetracycline/ml, 50 or 1,000 μg of kanamycin/ml, 20 μg of nalidixic acid/ml, and 10 μg of gentamicin/ml.
Conjugation.
Overnight cultures of donor and recipient strains were mixed on LB plates (26). After overnight incubation at 30°C, the recipients, donors, and transconjugants were resuspended in 10 mM MgSO4 and titrated on LB plates supplemented with the appropriate antibiotics. For matings involving pETMob, pETMob-GFP, or pKKMob, the mixed bacteria were incubated on LB plates supplemented with isopropyl-β-d-thiogalactoside (IPTG) (usually 0.5 mM).
PCR amplifications and cloning.
Cloning manipulations were done according to the procedure of Sambrook et al. (41). The mob gene was amplified from pBBR1CM DNA by means of primers CS12 (5′-GAGGAATTCATGGCGGCATACGCGATC-3′) and CS13 (5′-GTGAAGCTTCAGGGCCTCGTGATACGCC-3′) containing, respectively, the EcoRI and the HindIII recognition sequence (italicized). The amplified fragment cleaved by these enzymes was cloned into the same sites in vectors pKK223-3 (Pharmacia) and pET21a(+) (Novagen), yielding pKKMob and pETMob, respectively. The gfp (green fluorescent protein) gene was amplified from DNA of the pGREEN Lantern-1 plasmid (Life Technologies) using primers 5′-GCAGCGCGCAAGCAAGGGCGAGGAAC-3′ and 5′-CATAAGCTTTCACTTGTACAGCTCG-3′, containing respectively, the BssHII and the HindIII recognition sequence (italicized). The amplified fragment cleaved by these enzymes was cloned into the same sites in pETMob, yielding pETMob-GFP. To obtain pETMobhis, the mob gene was amplified by PCR using primers CS12 and CS13b (5′-GCGAAGCTTTGATAATAATGGTTTCTTAG-3′). The amplified fragment was cleaved with the EcoRI and HindIII restriction enzymes and cloned into the corresponding sites of the pET21a(+) vector, generating a fusion between the mob gene and a sequence coding for six histidines under the control of the T7 promoter. Plasmids pJL52bp and poriT52bp containing the oriT were constructed by cloning the oligonucleotide 5′-AGCTTCCACTCAATGCTTGAGTATACTCACTAGACTTTGCTTCGCAAAGTCGTGACCTGCA-3′ and its complementary strand into the HindIII and PstI restriction sites of the pJL207 vector (27) and the pKilper2 vector (12). These oligonucleotides contain extremities compatible with the protruding ends of the restriction enzymes used. Plasmid poriT18bp was constructed by deleting a 34-bp fragment of poriT52bp with the enzymes Bst1107 and EcoRV. Plasmid poriT34bp was constructed by the same method, using the Bst1107, HindIII, and Klenow enzymes. PCR amplification from pBBR1CM DNA with primers CS4 (5′-TTGTCCACGGGCCGAGCG-3′) and CS7 (5′-CGAAGACGAAAGGGCCTC-3′) and cloning of the amplified fragment into the pCRBlunt vector (Invitrogen) resulted in pMob3. The amplified fragment contains the mob gene and the 327 bp upstream from this gene. Plasmid p2oriT was constructed by cloning this 327-bp fragment into poriT52bp. The 327-bp fragment was amplified by PCR, using CS4 and CS9 (5′-GGCGTGCTTGAGACTGGC-3′), and cloned into the pCRBlunt vector. The resulting plasmid was digested with Ecl136II and PstI. The 391-bp fragment formed was cloned into poriT52bp digested with StuI and PstI. The resulting plasmid, p2oriT, contains two oriTs in opposite directions. Plasmids poriTA and poriTC were constructed with primer CS7 and either primer NicA (5′-TCACGACTTTGCGAAGCAAAGTCTAGTGAATA-3′) or primer NicC (5′-TCACGACTTTGCGAAGCAAAGTCTAGTGAGCA-3′), respectively. Each amplified fragment was cloned into the TOPO-XL vector (Invitrogen).
Regulation and β-galactosidase assays.
Overnight cultures of TOP10F− (Invitrogen) strains containing the appropriate plasmids were diluted 50-fold and grown for 2 h at 37°C in LB medium supplemented with antibiotics. When plasmid pKKMob was used, 0.5 mM IPTG was added at the beginning of growth. β-Galactosidase activity units were determined according to the work of Miller (30). Each result is the mean from three independent experiments ± the standard deviation.
Protein expression and extraction.
Overnight cultures of BL21(DE3, pLys) carrying pETMob, pETMob-GFP, pETMobhis, or pETLacZ (induction control plasmid with an insert encoding β-galactosidase tagged by six histidines [Novagen]) were washed with fresh LB broth. Of this suspension, 300 μl was inoculated into 15 ml of LB medium containing 200 μg of ampicillin/ml. The cultures were grown at 37°C to an optical density at 600 nm of 0.8 and were then washed once more with fresh LB broth. Next, the cells were grown for 30 min at 37°C in LB medium supplemented with 500 μg of ampicillin/ml and 1 mM IPTG, after which 100 μg of rifampin/ml was added and the cultures were incubated for 90 more minutes. Protein extracts were prepared according to the work of Chaconas et al. (7). Briefly, the harvested cells were suspended in 100 μl of solution containing 50 mM Tris-HCl and 10% sucrose (pH 8) and were then frozen in liquid nitrogen for a few seconds. After thawing on ice, the bacteria were lysed by addition of 0.8 μl of a solution containing 0.2 mM dithiothreitol (DTT), 6.4 μl of 0.5 M EDTA, 6.4 μl of lysozyme (10 mg/ml), and 13 μl of Complete (Boehringer-Roche) and incubation on ice for 20 min, followed by addition of 1.6 μl of Brij 35 and incubation for 40 min. The lysed cells were centrifuged (at 22,000 × g for 20 min at 4°C). The supernatant was collected and stored at −80°C until use. Samples of each crude extract were mixed with an equal volume of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled for 5 min, and subjected to SDS-PAGE on 10% gels. The gels were stained with 0.5% Coomassie brilliant blue in 25% methanol–10% acetic acid.
DNA labeling and DNA mobility shift assay.
HindIII-XbaI DNA fragments containing different parts of the oriT were obtained by digestion of 9 μg of poriT52bp, poriT18bp, or poriT34bp. The fragments were labeled by addition of DNA polymerase I Klenow fragment, dATP, dTTP, dGTP (1 nmol each), and [α-32P]dCTP (60 μCi, 20 pmol [Amersham]) and incubation at 37°C for 30 min. The labeled DNA was electrophoresed on 10% polyacrylamide–0.5× Tris-borate-EDTA (TBE) gels. The DNA band was visualized by autoradiography, excised, and eluted overnight with 200 μl of Tris-EDTA buffer at 4°C. DNA binding reactions were performed on ice for 30 min by incubating 2 μl of purified labeled fragment (1 ng of DNA), 1 μl of protein extract, and nonspecific competitor DNA (salmon sperm DNA in varying quantities) in binding buffer (50 mM HEPES KOH [pH 7.8] 150 mM KCl, 60% glycerol, 25 mM MgCl2, 2.5 mM DTT, 0.5 mM EDTA). The final volume of the reaction mixture was 50 μl. The reaction was stopped by adding 5 μl of stop solution (0.25% bromophenol blue, 0.25% xylene cyanol, and 40% sucrose) before loading onto a 10% polyacrylamide–0.5x TBE gel. Electrophoresis was carried out on ice at 150 V for 2 h. The gels were vacuum dried and exposed to Kodak Biomax film.
Protein purification.
Protein extracts of BL21(DE3, pLys, pETMobhis) were diluted twofold with binding buffer (30 mM Tris-HCl [pH 8], 100 mM NaCl, 10% glycerol, 0.5× Triton X-100) and incubated with 500 μl of His-resin (Talon Metal Affinity Resin; Clontech). After centrifugation (for 5 min at 16,000 × g) the supernatant was removed and analyzed by SDS-PAGE (10% acrylamide gels). The resin was washed once with 1.5 ml of binding buffer and once with 1.5 ml of washing buffer (binding buffer plus 10 mM imidazole). The Mob(His6) protein was eluted with 150 μl of elution buffer (binding buffer plus 50 to 125 mM imidazole).
In vitro cleavage of supercoiled DNA and mapping of the nick position.
Supercoiled plasmid DNA was isolated by column purification according to the manufacturer's instructions (Qiagen; plasmid maxi kit). Plasmid DNA (1 μg) was incubated with Mob protein (≅400 ng) for 30 min at 30°C in buffer A (25 mM Tris-HCl [pH 7.6], 0.1 mM EDTA, 15 mM MgCl2, 10% glycerol, 1 mM DTT; final volume, 30 μl). The reactions were stopped by addition of 3 μl of a solution containing 10% SDS, 15 μl of 0.5 M EDTA (pH 8), and proteinase K (100 μg/ml). The mixtures were loaded onto Tris-agarose-EDTA (TAE) gels (0.9% agarose), and electrophoresed (at 120 V for 2 h), and the gels were stained with ethidium bromide. Bands corresponding to the relaxed form (and to the linear form for p2oriT) were cut out of the gel, recovered using spin columns (Supelco), and purified with a Qiagen column (Qiaquick nucleotide removal kit). The position of the nick site was determined by sequencing with an automatic sequencer (ABI310; Perkin-Elmer), using the M13 reverse, M13 forward, or CS9 primer for p2oriT according to the manufacturer's instructions.
Isolation of Mob(His6)-DNA complexes.
Supercoiled DNA (1 μg of p2oriT or 1 μg of the same vector [pKilpcr2] containing a 1,900-bp fragment without the oriT sequence) was incubated with Mob(His6) (≅400 ng) under standard conditions (in a final volume of 30 μl). The reactions were stopped by addition of 15 μl of 0.5 M EDTA (pH 8) and 3 μl of 10% SDS. The mixtures were incubated at 37°C for 4 min, KCl was added (to a final concentration of 0.25 M), and the incubation was continued at 0°C for 10 min. The precipitate was collected by centrifugation (for 2 min at 16,000 × g), washed with 1 ml of cold buffer B (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM KCl), and resuspended in 0.5 ml of a solution containing 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 100 mM NaCl, 10 mM MgCl2, and 10 μg of tRNA. The complexes were precipitated with 2 volumes of ethanol (100%), washed with 1 volume of 70% ethanol, and dissolved in 15 μl of 10 mM Tris-HCl (pH 7.5)–1 mM EDTA (TE buffer). The supernatant-containing free DNA was collected. A 0.5-ml portion of TE buffer was added, and the DNA was precipitated with 2 volumes of ethanol (100%), washed twice with 70% ethanol, and dissolved in 15 μl of TE buffer. The samples were analyzed by electrophoresis on 0.9% agarose gels after incubation with proteinase K to remove attached proteins.
Site-directed mutagenesis.
The site-directed mutations were introduced by PCR by overlap extension using the method described by Ho et al. (21). For each mutation, pBBR1CM DNA was used as the template in two sequential PCRs: first, primer CS7 and a primer containing the mutation were used to amplify the C-terminal part of the mob gene; then primer CS4 and a primer complementary to that containing the mutation amplified the N-terminal part of the mob gene and the 327-bp sequence upstream from this gene. Both amplified fragments were agarose gel purified and used as templates for a third PCR amplification with primers CS4 and CS7, thus reconstituting the mob gene. The amplified fragment was cloned into the TOPO-XL vector. Mutations were chosen so as to introduce a restriction site into the mob gene. In each resulting plasmid, the presence of the mutation was checked using this restriction site and by sequencing. The primers used and the corresponding mutations are listed in Table 4.
TABLE 4.
Site-directed mutagenesis of the mob gene
| Mutation(s)a | Oligonucleotideb | Restriction site introducedc | Mobilization frequencyd |
|---|---|---|---|
| Wild type | 10−1 | ||
| Y4F | 5′-GGCATGGCGGCATTCGCGATC-3′ | NruI | 10−1 |
| Y4L | 5′-ATGGCGGCACTAGCGATCATG-3′ | MaeI | 10−1 |
| Y26G | 5′-CAAGCACGCCGGCCGCGAGCG-3′ | NaeI | 10−1 |
| Y75L | 5′-CGGTCGAGCTCGTCATGACG-3′ | SacI | 10−1 |
| Y107L | 5′-CGGACAAGCTTGGGGCGGATCG-3′ | HindIII | 10−1 |
| Y185L | 5′-GGCGTTCCTCGAGGCCCTGG-3′ | AvaI | 10−1 |
| Y208L | 5′-CACGCGCCGGCGCACCGCAG-3′ | NaeI | 10−1 |
| Y241L | 5′-GCAGGGGCTCGAGCCTGCC-3′ | AvaI | 10−1 |
| F94L F95L | 5′-GGCGGCGCTCCTCGAGAAGG-3′ | AvaI | 10−1 |
| D120L | 5′-CGTCTCGAGACCAGCCCGCACATGAC-3′ | AvaI | <10−8 |
| E121G | 5′-CGTGACGGTACCAGCCCGCACATGAC-3′ | Asp718 | <10−8 |
| D120L E121G | 5′-CGTTTAGGTACCAGCCCGCACATGAC-3′ | Asp718 | <10−8 |
| D120L complemented by pBBR1CM | 10−1 |
The site-directed mutations are indicated: all the tyrosines (Y) of the pBHR1 mob gene were mutated to leucine (L) or glycine (G) by means of modified oligonucleotides.
Mutated codons are boldfaced.
Each mutation introduced a restriction site in the mob gene. Restriction and sequencing were used to check for the presence of the mutation in the donor strain and transconjugants.
For the mobilization experiments, Nals S17-1 bacteria containing one of the plasmids to be tested were used as donors and the Nalr F− strain XA106 was used as a recipient. Matings were performed at 30°C on LB plates as previously described. The mobilization frequency was calculated as the ratio of the number of transconjugants to the number of donors. Each value is an average from three independent experiments. The limit of detection of transconjugants was estimated at 10−8.
RESULTS
Mobilization frequencies of different pBBR1 derivatives.
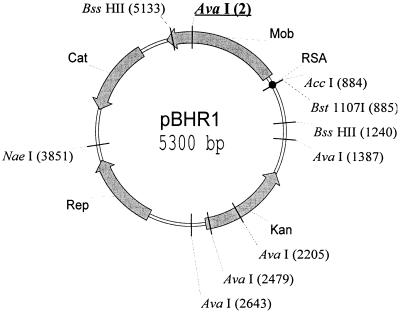
Different vectors (pBBR1CM, pBBR1MCS-2, pBBR1MCS-4, and pBBR1MCS-5) (2, 25) were derived from pBBR1 by insertion of “resistance cassettes” into the 3′ end of the mob gene. Thus, the Mob protein produced by each of these derivatives differs from its parent. We determined the mobilization efficiencies of these plasmids using the RP4 transfer system (Table 1). Except for that of pBBR122, the Mob proteins of the pBBR1 derivatives were found to remain active despite replacement of the 7-amino-acid C-terminal sequence by different sequences of various lengths (Table 1). A comparison of the plasmid DNA sequences revealed that one of the AvaI sites present in the mob gene of pBBR1CM and in Kovach's derivatives is lost in pBBR122. This probably occurred by “filling in” during introduction of the kanamycin resistance cassette of pBBR122. Such filling in introduces a frameshift mutation in the C-terminal end of the mob gene (the last 63 amino acids are modified). To determine whether modifications of these 63 C-terminal amino acids inactivate the Mob protein, we constructed a pBBR122 derivative called pBHR1: the 1,411-bp BssHII fragment of pBBR122 was replaced with the homologous 1,407-bp fragment of the Mob+ plasmid pBBR1CM (Fig. 1). Plasmid pBHR1 could be efficiently mobilized. We used plasmid pBHR1 to test the mobilization host range of pBBR1 derivatives. We observed that it was efficiently mobilized from E. coli by the RP4 plasmid into 11 different species of gram-negative bacteria (Table 2). We detected low or no mobilization, however, into four species: Erwinia herbicola, Acinetobacter sp. strain AC58, Proteus mirabilis NCTC5887, and Proteus vulgaris OX19. Such negative results may reflect an inability to mobilize the plasmid or nonoperation of the replication or resistance function. Our results thus show (i) that the 7 last amino acids of the Mob protein are not necessary for mobilization and (ii) that loss of mobilization in the pBBR122 plasmid is due to a frameshift mutation in the mob gene and not to the transfer origin.
TABLE 1.
C-terminal sequences of Mob proteins from different pBBR1 derivatives and their corresponding mobilization frequenciesa
| Plasmid (selective marker)b | C-terminal sequencec | Mobilization frequencyd |
|---|---|---|
| 322 329 | ||
| ↓ ↓ | ||
| pBBR1 (none) | .ERGRDRGGYSR | ND |
| pBBR1CM (KAN, CHL) | .ERGRAHFPEKCHLTSKKPLLS | 8 × 10−2 |
| pBBR1MCS-2 (KAN) | .ERGRAHFPEKCHLG | 1 × 10−1 |
| pBBR1MCS-4 (AMP) | .ERGRAHFPEKCHLDFGHEILKKDLHLDPFKLKMKF | 8 × 10−2 |
| pBBR1MCS-5 (GEN) | .ERGRAHFPEKCHLAAL | 3 × 10−2 |
| pBBR122 (KAN, CHL) | Frameshift mutation | 7 × 10−7 |
| pBBR122 (complementation by pMOB3) | 1 × 10−2 | |
| pBHR1 (KAN, CHL) | .ERGRAHFPEKCHLTSKKPLLS | 1 × 10−1 |
For the mobilization experiments, Nals S17-1 bacteria (45) containing one of the plasmids to be tested were used as donors and the Nalr F− strain XA106 (from our laboratory collection) was used as a recipient. Matings were performed at 30°C on LB plates as described previously (26). After overnight incubation, the transconjugants were selected at 30°C on LB plates supplemented with nalidixic acid (20 μg/ml) and the antibiotic allowing selection of the bacteria with the mobilizable plasmid (kanamycin at 50 μg/ml, ampicillin at 100 μg/ml, gentamicin at 10 μg/ml, or chloramphenicol at 15 μg/ml).
KAN, kanamycin; CHL, chloramphenicol; AMP, ampicillin; GEN, gentamicin. Plasmid pMOB3 contains a functional mob gene and the 327-bp sequence upstream from this gene (oriT), cloned in the pCRBlunt vector.
The C-terminal sequence of each Mob protein is represented; the coordinates are positions in the pBBR1CM Mob protein. Amino acids modified in the Mob protein of the pBBR1 derivative are boldfaced.
Calculated as the ratio of the number of transconjugants to the number of recipients. ND, not determined.
FIG. 1.
Map of the pBHR1 broad-host-range vector. pBHR1 was obtained by replacement of the 1,411-bp BssHII fragment of the Mob− vector pBBR122 (Mobitec, Göttingen, Germany) with the 1,407-bp homologous fragment of the Mob+ plasmid pBBR1CM (2). This construct was shown to be mobilizable by the IncP plasmid into different gram-negative strains (see Table 1). The AvaI site (underlined at 2 bp) is the site that was mutated in pBBR122. The rep and mob genes previously reported by Antoine and Locht (2) to be essential to plasmid replication and mobilization are indicated. Cat and Kan, the genes conferring resistance to chloramphenicol and kanamycin, respectively.
TABLE 2.
Frequencies of mobilization of pBHR1 into different bacteriaa
| Recipient | Transfer frequencyb:
|
|
|---|---|---|
| Per donor | Per recipient | |
| Escherichia coli XA106F− | 3 × 10−2 | 1 × 10−1 |
| Ralstonia eutropha AE110 | 2 × 10−1 | 3 × 10−1 |
| Erwinia chrysanthemi A1240 | 1 × 10−2 | 1 × 10−1 |
| Klebsiella pneumoniae KAY2026 Nalr | 2 × 10−3 | 1 × 10−1 |
| Agrobacterium tumefaciens C58 Nalr | 3 × 10−2 | 2 × 10−2 |
| Salmonella enterica serovar Typhimurium MA767 Nalr | 2 × 10−2 | 1 × 10−2 |
| Pseudomonas stutzeri ATCC 17588 Nalr | 4 × 10−2 | 9 × 10−3 |
| Pseudomonas aeruginosa PAO362 Nalr | 9 × 10−3 | 7 × 10−3 |
| Azospirullum brasilense 7000 Nalr | 2 × 10−3 | 6 × 10−3 |
| Pseudomonas aeruginosa PAO531 | 8 × 10−3 | 4 × 10−3 |
| Serratia marcescens HY(y− Ψ−) Nalr | 2 × 10−3 | 1 × 10−3 |
| Erwinia herbicola RH6101 Nalr | 4 × 10−4 | 5 × 10−6 |
| Acinetobacter sp. strain AC58 Nalr | ≤2 × 10−6 | ≤9 × 10−7 |
| Proteus mirabilis NCTC5887 Nalr | ≤4 × 10−7 | ≤6 × 10−7 |
| Proteus vulgaris OX19 Nalr | ≤8 × 10−6 | ≤8 × 10−7 |
The Nals E. coli strain S17-1 containing an integrated RP4 plasmid and the Kanr pBHR1 plasmid was mated at 30°C on LB plates with various Nalr Kans bacteria as described by Lejeune et al. (26). After overnight incubation, the recipient, donor, and transconjugants were titrated at 30°C on LB medium supplemented with the appropriate antibiotics (nalidixic acid at 20 μg/ml; kanamycin at 50 or 1,000 μg/ml according to the recipient used).
Mobilization frequencies were determined by dividing the number of transconjugants (Kanr Nalr) by the number of recipients (Nalr) or donors (Kanr − Kanr Nalr).
Regulation of the mob gene.
Sequence analyses have revealed a putative promoter of the mob gene (2). To examine the regulation of this gene, we cloned a 52-bp sequence (coordinates 867 through 918) containing the putative promoter into the pJL207 vector (27) upstream from the lacZ gene, creating plasmid pJL52bp. To express the mob gene under the control of an inducible promoter, we introduced the mob gene into the pKK223-3 vector under the control of Ptac (the resulting plasmid was called pKKMob). The ability of the Mob protein produced from pKKMob to perform its function was tested by mating an E. coli S17-1 (Nals) strain containing the RP4 plasmid in its chromosome, the pKKMob plasmid, and the mini-pBHR1 plasmid (a Mob− oriT+ pBHR1 derivative [48]) with an E. coli XA106F− (Nalr) strain. As a negative control, we used a donor strain in which pKK223-3 was substituted for pKKMob. The frequency of mobilization of the mini-pBHR1 plasmid increased with the IPTG concentration used to induce Mob production from Ptac (frequency range, 10−2 without IPTG to 7 × 10−1 with 500 μM IPTG). No mobilization was observed with the negative control, pKK223-3. The mobilization observed in the absence of IPTG suggests that a small amount of Mob protein is enough to promote mobilization of the mini-pBHR1 plasmid.
Regulation of the mob gene was studied by measuring β-galactosidase activity in Δlac bacteria containing plasmid pJL52bp. As a control, we used bacteria containing the pJL207 vector: the background was 5 ± 4 Miller units. In the absence of pKKMob, β-galactosidase production from the fusion gene on pJL52bp was 185 ± 10 Miller units, but in the presence of pKKMob, the measured activity was reduced to 8 ± 4 units. We conclude that (i) the 52-bp sequence contains the promoter and (ii) the mob gene is regulated by the mob product.
The transfer origin.
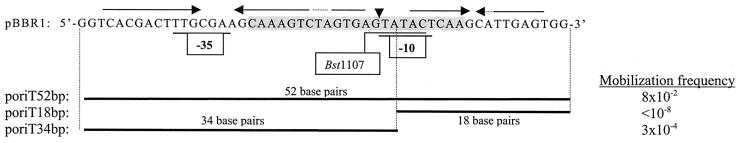
The region containing the putative promoter of the mob gene shows sequence similarity to the transfer origins (RSA) of plasmids and mobilizable transposons isolated from gram-positive bacteria (2, 10). This region may thus have a double function: to regulate the mob gene and to act as a transfer origin. To test this, we cloned the 52-bp sequence containing the RSA and the promoter (Fig. 2) into the pKilPCR2 vector (12), obtaining plasmid poriT52bp. This plasmid was tested for mobilization (Fig. 2). Mobilization was observed only in the presence of a plasmid containing the mob gene. This shows that the transfer origin (oriT) is contained in the 52-bp sequence and confirms that the Mob protein is necessary for mobilization. The sequence was divided into two parts by Bst1107 restriction (Fig. 2). The 3′ fragment (18 bp) and the 5′ fragment (34 bp) formed each contained an inverted repeat. Separate plasmids carrying the 3′ or 5′ fragment were constructed and called, respectively, poriT18bp and poriT34bp (Fig. 2). Plasmid poriT34bp was mobilized by the IncP plasmid pSL2T (47) in the presence of pBBR1CM, but no mobilization of poriT18bp was observed under these conditions. These results show that the transfer origin is contained in the 34-bp 5′ part of the RSA region. Yet the mobilization frequency of poriT34bp was lower than that of poriT52bp. This may be due to the fact that poriT34bp lacks 8 bp of the 23-bp RSA region (Fig. 2).
FIG. 2.
The 52-bp sequence containing the oriT and the promoter of the mob gene (coordinates 918 to 867). Horizontal arrows indicate inverted repeats. The vertical arrow indicates the position of the nick site. Boxes marked −10 and −35, proposed regions for the promoter of the mob gene. The 52-bp sequence was cloned in vector pKilpCR2, yielding poriT52bp. The Bst1107 site used to construct poriT18bp and poriT34bp is indicated. Mobilization frequencies were determined by matings between Nals B462 donors (4) containing pBBR1CM (Mob donor), the conjugative IncP plasmid pSL2T, and the poriT plasmid to be tested and Nalr B462 recipients. Shaded sequence, RSA region as defined for the reference plasmid pMV158.
The Mob protein binds specifically to the oriT.
To test the ability of the Mob protein to bind to the oriT region, we performed electrophoretic mobility shift assays with crude extracts of bacteria overproducing the Mob protein.
(i) Overproduction of the Mob protein.
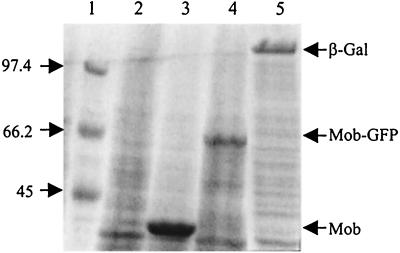
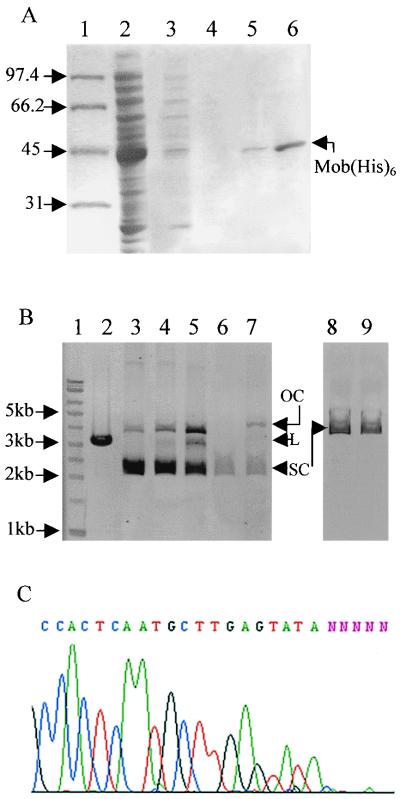
The mob gene of pBHR1 was amplified by PCR, and the amplified fragment was cloned into the pET21a(+) expression vector (Novagen). The resulting plasmid, pETMob, carries the mob gene under the control of the T7 promoter. In order to visualize production of the Mob protein in vivo, the green fluorescent protein (GFP) gene was added at the BssHII site located at the end of the mob gene (the resulting plasmid was called pETMob-GFP). Production of the chimeric Mob-GFP protein was visualized with a fluorescence microscope (data not shown). In E. coli strain BL21(DE3, pLys) containing the T7 RNA polymerase gene under the control of the lac promoter, expression from plasmids pETMob and pETMob-GFP led upon IPTG induction to high levels of the Mob and Mob-GFP proteins, respectively. These products were detectable by PAGE and Coomassie blue staining (Fig. 3). The positions of the bands observed on the polyacrylamide gels confirmed that the overproduced proteins were the 39.5-kDa Mob protein and the 62.7-kDa chimeric Mob-GFP protein.
FIG. 3.
Overproduction of the Mob protein. One microliter of crude extract of BL21(DE3, pLys) containing the appropriate plasmid was electrophoresed on SDS–10% polyacrylamide gels. The gels were stained with Coomassie brilliant blue. Lane 1, molecular mass marker (masses in kilodaltons are shown on the left); lane 2, 1 μl of crude extract of 15 ml of BL21(DE3, pLys, pETMob) without induction; lane 3, 1 μl of crude extract of 15 ml of BL21(DE3, pLys, pETMob) with induction; lane 4, 1 μl of crude extract of 15 ml of BL21(DE3, pLys, pETMob-GFP); lane 5, 1 μl of crude extract of 15 ml of BL21(DE3, pLys, pETLacZ).
The activities of the overproduced proteins were tested by mating an E. coli BL21 strain containing the overproducing vector, the mini-pBHR1 plasmid, and the IncP plasmid pSL2T with the Nalr E. coli strain DH5α. As a negative control, we used plasmid pETlacZ, (Novagen), producing β-galactosidase instead of pETMob or pETMob-GFP. As expected, the mini-pBHR1 plasmid was mobilized only in the presence of the vector producing the Mob protein (pETMob) or the Mob-GFP protein (pETMob-GFP) (data not shown).
(ii) Gel mobility shift assay.
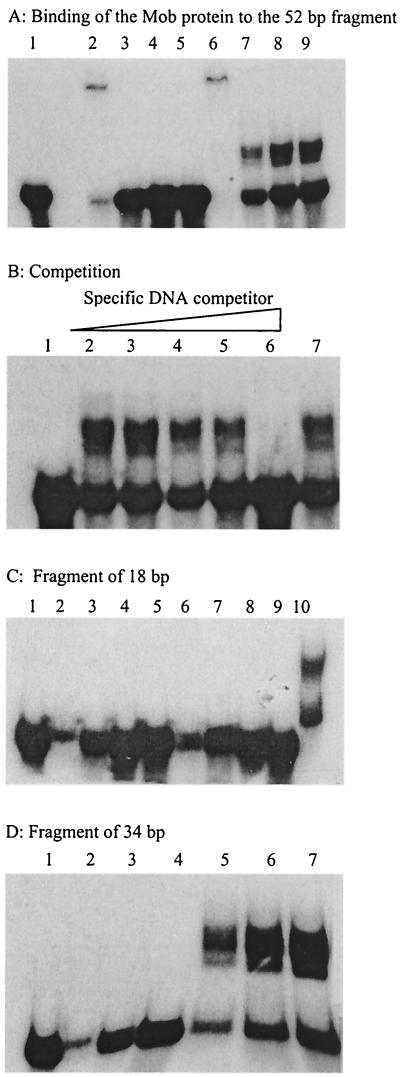
The 52-bp oriT fragment was labeled and incubated on ice in binding buffer with crude extracts of bacteria overproducing the Mob protein (see Materials and Methods). As a negative control, we used a crude extract of bacteria overproducing β-galactosidase. The reaction mixtures were electrophoresed in 10% polyacrylamide gels. In the absence of a nonspecific competitor DNA (salmon sperm DNA), migration of the labeled fragment was completely shifted, owing to nonspecific binding of proteins contained in the crude extracts (Fig. 4A, lanes 2 and 6). When nonspecific competitor DNA was added at increasing concentrations, migration of the oriT DNA fragment was shifted only in the presence of crude extracts containing the Mob protein (Fig. 4A, lanes 7 to 9). This shows that the Mob protein can bind to linear double-stranded DNA. To test the specificity of this binding, we incubated the labeled oriT fragment with crude extracts containing the Mob protein and increasing concentrations of unlabeled oriT DNA (Fig. 4B). A 500-fold excess of this specific competitor completely inhibited the reaction (lane 6), whereas a 500-fold excess of another DNA did not affect binding of the Mob protein (lane 7). This demonstrates that the Mob protein binds specifically to the 52-bp oriT fragment in the absence of any other transfer protein. Since the oriT is contained in the 34-bp 5′ fragment of the RSA (see above), we tested the capacity of the Mob protein to bind to this fragment and to the 18-bp 3′ fragment of the RSA by use of gel mobility shift assays. The Mob protein was found to bind only to the 34-bp 5′ fragment containing the oriT (Fig. 4C and D).
FIG. 4.
The Mob protein binds to the oriT of pBBR1. One nanogram of a DNA fragment labeled at the 5′ terminus with [α-32P]dCTP was incubated with crude protein extract. Lane 1, labeled fragment without addition of protein. (A) Fragment of 52 bp. Lanes 2 to 5, addition of 1 μl of crude extract of BL21(DE3, pLys, pETLacZ) and 0, 0.25, 0.5, and 1 μg of nonspecific competitor DNA (salmon sperm DNA), respectively; lanes 6 to 9, 1 μl of crude extract of BL21(DE3, pLys, pETMob) and 0, 0.25, 0.5, and 1 μg of salmon sperm DNA, respectively. (B) Fragment of 52 bp. Lanes 2 to 6, 1 μl of crude extract of BL21(DE3, pLys, pETMob) plus 1 μg of salmon sperm DNA and 0, 1, 10, 100, and 500 ng of specific competitor DNA (unlabeled oriT fragment), respectively; lane 7, same as lane 2 plus 500 ng of nonspecific DNA from plasmid. (C) Fragment of 18 bp. Lanes 2 to 9, same as panel A. Lane 10, positive control with the 52-bp fragment. (D) Fragment of 34 bp. Lanes 2 to 4, addition of 1 μl of crude extract of BL21(DE3, pLys, pETLacZ) and 0.25, 0.5, and 1 μg of nonspecific competitor DNA (salmon sperm DNA), respectively; lanes 5 to 7, 1 μl of crude extract of BL21(DE3, pLys, pETMob) and 0.25, 0.5, and 1 μg of salmon sperm DNA, respectively.
Position of the nick site.
To identify the position of the nick site, we tried to extract nicked DNA using the method of Clewell and Helinski (9) and to perform runoff experiments as in Zechner et al. (53). These experiments were unsuccessful. The Mob protein was thus purified, and the position of the nick site was determined in vitro.
(i) Purification of the Mob protein.
The Mob protein was tagged with six histidines at its carboxy terminus and purified as described in Materials and Methods. The purification steps were monitored by SDS-PAGE and Coomassie blue staining (Fig. 5A). The ability of the Mob(His6) protein to function was tested and confirmed in vivo as described for the Mob protein produced from the pETMob plasmid.
FIG. 5.
In vitro cleavage of supercoiled DNA by the purified Mob protein. (A) The Mob protein was tagged with six histidines at its carboxy-terminal end and purified. The proteins were separated in an SDS–10% polyacrylamide gel. Lanes: 1, protein standards (molecular sizes are given in kilodaltons); 2, 1 μl of crude extract of 15 ml of induced BL21(DE3, pLys, pETMobhis); 3, 25 μl of supernatant after the first wash of the His-resin; 4, 25 μl of supernatant after the second wash of the His-resin; 5, elution with 250 μl of buffer containing 50 mM imidazole (7.5 μl on the gel); 6, elution with 250 μl of buffer containing 125 mM imidazole (7.5 μl on the gel). (B) In vitro cleavage of supercoiled DNA. Plasmid DNA profiles after electrophoresis in a 0.9% agarose gel are shown. Lanes: 1, DNA molecular size marker (with sizes given in kilobases on the left); 2, restriction by PstI of the p2oriT plasmid DNA containing two oriTs in opposite directions (the linear plasmid has a size of 3,143 bp); 3, supercoiled p2oriT DNA (1 μg); 4, incubation of 1 μg of p2oriT DNA with the purified Mob protein (≅200 ng); 5, incubation of 1 μg of p2oriT DNA with the purified Mob protein (≅400 ng); 6, supercoiled poriT52bp DNA containing one oriT (0.5 μg); 7, incubation of 0.5 μg of poriT52bp DNA with Mob(His6) (≅200 ng); 8, supercoiled DNA of the control plasmid which contains no oriT (0.8 μg); 9, incubation of 0.8 μg of the control plasmid DNA with the purified Mob protein (≅200 ng). Note the increase of relaxed DNA (OC, open circle) in lanes 4, 5, and 7 and the partial linearization of p2oriT due to the cleavage of both oriTs in lane 5 (L, linear DNA; SC, supercoiled DNA). (C) Position of the nick site. Shown is an electropherogram of the sequence of the relaxed p2oriT DNA purified from the agarose gel using an automatic sequencer.
(ii) In vitro cleavage of supercoiled DNA by the Mob protein.
Purified Mob(His6) was incubated with supercoiled poriT52bp DNA in buffer containing Mg2+. (Mg2+ is the only cofactor required for all types of relaxase-mediated cleaving-joining reactions [39].) In order to scale up production of the relaxed plasmids, we constructed a plasmid (p2oriT) containing two oriTs and incubated this DNA with the Mob(His6) protein. The reactions were stopped with EDTA, followed by incubation with proteinase K and SDS. The relaxed plasmids were then visualized by agarose gel electrophoresis (Fig. 5B). In the p2oriT plasmid, the two oriTs are in opposite directions and very close to each other (225 bp separate them). Cleavage of both oriTs creates a discontinuity in each strand and can linearize the plasmid. Partial linearization of p2oriT DNA was indeed observed, showing double cleavage of this plasmid (Fig. 5B, lane 5). Supercoiled DNA of the plasmid lacking the pBBR1 oriT region was not cleaved by the Mob protein. This shows the substrate specificity of the protein (Fig. 5B, lane 9). In conclusion, the Mob protein of pBBR1 is a relaxase and, in the absence of any other transfer protein, can cleave supercoiled DNA containing the pBBR1 oriT region.
(iii) Mapping of the nick position.
To map the site of strand discontinuity within the nicked plasmid DNA, we purified the relaxed and linearized forms from the agarose gel and sequenced them as indicated in Materials and Methods. In one strand of the oriT sequence (the Mob coding strand), we detected an interruption (Fig. 5C) that appeared neither in the complementary strand nor in the untreated DNA (data not shown). It was located between a T nucleotide (at position 886 on the complementary strand of pBBR1) and a G nucleotide (position 887). To confirm this result in vivo, we mutated these nucleotides separately (T to C and G to A). After PCR amplification with modified primers, the amplified fragments containing the mutated oriT and the mob gene were cloned into the TOPO-XL vector (Invitrogen). Mobilization frequencies were determined for both constructs, called poriTC and poriTA (Table 3). No mobilization was detectable in matings between plasmid-harboring S17-1 as the donor and XA106F− as the recipient (Table 3, first two rows). Because the mutations were in the promoter sequence, we made sure that the loss of mobilization was not due to nonexpression of the mob gene. This was done in experiments showing that both constructs are nonmobilizable even in the presence of a mob gene expressed in trans from plasmid pBBR1CM (Table 3, fifth and sixth rows) and by quantifying the mobilization of pJL52bp (containing the wild-type oriT) by poriTA and poriTC. The mobilization frequencies of pJL52bp showed that expression of the mob gene is reduced in poriTA and poriTC but not totally abolished (Table 3, last three rows). These results are consistent with the view that the nick site lies between nucleotides T at position 886 and G at position 887 of pBBR1.
TABLE 3.
Mobilization frequencies of the mutated transfer originsa
| Plasmid testedb | Mob-producing plasmid in trans | Mobilization frequency of the plasmid testedc |
|---|---|---|
| poriTA | <10−8 | |
| poriTC | <10−8 | |
| pBBR1CM | 1 × 10−1 | |
| pBBR1MCS2 | 1 × 10−1 | |
| poriTA | pBBR1CM | <10−8 |
| poriTC | pBBR1CM | <10−8 |
| pJL52bp | poriTA | 9 × 10−5 |
| pJL52bp | poriTC | 1 × 10−3 |
| pJL52bp | pBBR1MCS2 | 8 × 10−2 |
For the mobilization experiments, Nals S17-1 bacteria containing the plasmids to be tested were used as donors and the Nalr F− strain XA106 (from our laboratory collection) was used as a recipient. After overnight incubation at 30°C, the transconjugants were selected at 37°C on LB plates supplemented with nalidixic acid (20 μg/ml) and the antibiotic allowing selection of bacteria with the mobilizable plasmid (kanamycin at 50 μg/ml or chloramphenicol at 15 μg/ml).
The poriTA and poriTC plasmids contain the mutated oriT and the wild-type mob gene. Because the mutations were in the promoter sequence, the pBBR1CM plasmid was used to produce the Mob protein in trans, and the pJL52bp plasmid containing the wild-type oriT was used to test expression of the mob gene from poriTA or poriTC. The pBBR1MCS2 plasmid is a kanamycin-resistant derivative of pBBR1 and was used as a positive control.
Calculated as the ratio of the number of transconjugants to the number of donors. Each value is an average from three independent experiments. The limit of detection of transconjugants was estimated at 10−8.
The bond between Mob and the oriT is resistant to SDS treatment.
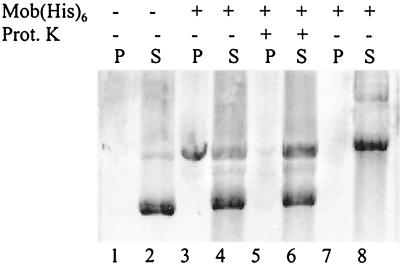
In all systems encoded by self-transmissible and mobilizable plasmids studied so far, the DNA cleavage reaction involves strand transfer with formation of a covalent DNA-relaxase bond (6, 54). To test whether this is true of the nicked DNA strand and the Mob relaxase, we used a method developed to isolate SDS-resistant protein-DNA complexes (49). The method is based on selective precipitation of DNA-protein complexes by KCl in the presence of SDS. It has been used to show tight binding between relaxases and oriTs (17, 29). Purified Mob(His6) protein was incubated with supercoiled p2oriT DNA as described above, and the reactions were terminated by addition of EDTA and SDS. KCl was added with or without prior proteinase K digestion. The precipitates and supernatants were analyzed by agarose gel electrophoresis (Fig. 6). In the absence of Mob(His6), this procedure precipitated only background levels of DNA and most of the DNA was recovered in the supernatant as a supercoiled form (Fig. 6, lanes 1 and 2). In the presence of Mob(His6), a substantial amount of nicked DNA and a small amount of linearized DNA were precipitated (lanes 3 and 4). When proteinase K was added after the cleavage reaction, only background levels of DNA were found in the precipitate (lanes 5 and 6). The DNA of a plasmid lacking the pBBR1 oriT region was not precipitated by the Mob protein, indicating that binding of the protein is specific (lanes 7 and 8). We conclude that a specific tight bond exists between Mob(His6) and the oriT. This bond is SDS resistant and probably covalent, as observed for other relaxases and oriTs (6, 39, 54).
FIG. 6.
After nicking, the relaxase remains tightly associated with its target DNA. The Mob(His6)-DNA complexes were precipitated with KCl (0.25 M) in the presence of SDS (1%). The DNA recovered in the precipitate (P) and supernatant (S) was analyzed by agarose gel electrophoresis. Supercoiled DNA (1 μg of p2oriT DNA [lanes 1 to 6] or control DNA lacking the oriT [lanes 7 and 8]) was incubated in the presence (+) or absence (−) of Mob(His6) protein (≅400 ng). The reaction was terminated by addition of EDTA (0.16 M) and SDS (1%) followed (+) or not followed (−) by incubation with proteinase K.
Site-directed mutagenesis of the putative Mob catalytic residue.
It is currently believed that the nicked double-stranded DNA produced by the action of a relaxase is covalently bound to the hydroxyl group of a specific tyrosyl residue of the protein (6, 15). The previously characterized conjugative relaxases RP4 TraI (36), RSF1010 MobA (44), and Ti VirD2 (51) each contain one catalytic tyrosine near the N terminus of the protein. Recently it was shown that the R388 TrwC relaxase carries two active-site tyrosyl residues (15). In all cases, mutation of one tyrosine to another residue results in a decreased transfer frequency. By PCR amplification, we separately mutated each of the seven tyrosines of the pBHR1 Mob protein to another residue, using modified primers (Table 4). The amplified fragments containing the oriT and a mutated mob gene were cloned separately into the TOPO-XL vector (Invitrogen). The mobilization frequencies of the resulting plasmids were determined. Surprisingly, no mutation of any tyrosine residue produced a change in the mobilization frequency (Table 4). As previously shown, the pBBR1 oriT sequence looks like the RSA of several plasmids (e.g., pMV158, pT181, and pG12) and a mobilizable transposon (Tn4451) from gram-positive bacteria (2, 10, 33). We also found sequence similarities to other plasmids from gram-positive bacteria (e.g., pTA1015, pIP823, and pUH1) and to two transposons (Tn5520 and Tn4555) and two plasmids (pFL1 and pZM2) from gram-negative bacteria (a total of 35 sequences were found using the Psi-Blast program [1]) (3, 8, 19, 29a, 31, 46, 50). These similarities are all located in the RSA and the amino-terminal half of the pBBR1 Mob protein. On the basis of sequence alignments, we identified two very well conserved clusters: two phenylalanine residues (F94 and F95) and an aspartate and a glutamate residue (D120 and E121). These amino acids were mutated separately or together as described above. As shown in Table 4, no effect was observed with the mob gene containing the mutated phenylalanines (F94L and F95L), but the D120L and E121G mutations completely abolished mobilization. We conclude that the aspartate 120 and glutamate 121 residues play a major part in the mobilization activity of Mob.
DISCUSSION
The cryptic plasmid pBBR1 has several interesting properties: its broad-host-range replication and mobilization, small size (2.6 kb), medium copy number, and similarity to plasmids from gram-positive bacteria. One of the fundamental steps in bacterial conjugation is the formation of the relaxosome (a protein-DNA complex that forms at the oriT) and the introduction of a nick at the transfer origin (oriT). In the case of pBBR1, our gel mobility shift data show that the Mob protein specifically recognizes a 52-bp sequence in the absence of any other transfer protein. We have shown that this sequence contains, in addition to the oriT, the promoter of the mob gene. We have further shown that this gene is autoregulated. Binding of the Mob protein to the 52-bp sequence may thus allow formation of a protein-DNA complex with a double function: relaxosome formation and mob gene regulation. The 52-bp sequence contains two inverted repeats. By deletion, we reduced the sequence necessary for mobilization and binding of the Mob protein to a 34-bp sequence containing one inverted repeat. In the case of pMV158, it was shown recently that the binding domain of the MobM protein spans 28 nucleotides and also includes an inverted repeat (16).
In our mobility shift assays, two shifted bands were distinguishable on the gels. This could be indicative of Mob protein multimerization either before or during binding. Alternatively, it could reflect a concentration-dependent increase in binding to additional regions within the oriT fragment. In an overlay assay using Mob and Mob(His6) protein, we found that the Mob protein has affinity for itself in vitro and that this interaction does not require the oriT or any other transfer protein (data not shown). This supports the idea that the two bands of the mobility shift assays reflect multimerization of the Mob protein.
We have located the nick site position in vitro between nucleotides T at position 886 and G at position 887 (of the complementary strand of pBBR1) through the action of overproduced Mob(His6) protein. Cleavage of supercoiled DNA was shown to be specific to the oriT sequence and independent of ATP and of other plasmid-encoded proteins, as shown for several other relaxases such as R388 TrwC (28) and pMV158 MobM (17). This is not the case for TraI of RP4 (35), MobA of RSF1010 (43), or VirD2 of Ti (37, 42), where the action of other plasmid-encoded proteins is required for cleavage of supercoiled DNA. We have confirmed the nick site position in vivo by introducing point mutations of the nucleotides T at position 886 and G at position 887. Each mutation of these nucleotides (T to C and G to A) totally abolishes mobilization of a plasmid containing the mutated oriT. Although both mutations are located in the mob promoter, neither totally prevents synthesis of the Mob protein.
As previously shown, the pBBR1 oriT sequence looks like the RSA of several plasmids and mobilizable transposons from gram-positive bacteria (2, 10, 33). We have also found sequence similarities with other plasmids and transposons from gram-positive and gram-negative bacteria, and surprisingly, with a gene of the pUH1 plasmid described as a γ-glutamyltranspeptidase gene (19) (a total of 35 sequences were found). The sequence similarities are all in the RSA and the amino-terminal half of the proteins. It is noteworthy that after sequence alignment, the position of the nick site of pBBR1 corresponds with those of the nick sites of the Bacteroides mobilizable transposon Tn4555 (46) and the streptococcal pMV158 plasmid. The oriT of the latter is characteristic of a family of mobilizable plasmids that are found in gram-positive bacteria and that replicate by the rolling-circle mechanism (17). Plasmid pBBR1 thus appears to be a new member of this group, even though it resides in gram-negative bacteria and does not replicate via a rolling-circle mechanism (2).
The conjugative mechanisms required for transfer of plasmids in gram-positive bacteria are not yet well understood. It would appear, however, that such mechanisms follow the same general principles as those reported for well-characterized systems such as the F, IncP, and IncW plasmids. In these systems, relaxase catalyzes a transesterification reaction resulting in a nicked double-stranded DNA molecule with its 5′ end covalently bound to the active tyrosine of the protein (6). Pansegrau et al. (38) propose that removal by a histidine of a proton from the aromatic hydroxyl group of the active tyrosine could result in an efficient nucleophile. The tyrosyl oxygen could attack the phosphodiester bond at the nick site. We have shown by KCl precipitation in the presence of SDS that after nicking, the Mob protein remains tightly associated with plasmid DNA containing the oriT. This strong association of Mob with its target DNA probably reflects a covalent bond. We have mutated all the tyrosines of the Mob protein, and surprisingly, none of the mutations results in a decreased mobilization frequency. On the basis of our sequence alignment, however, we identify two conserved clusters: two phenylalanines (F94 and F95) and the motif UHXDE (where U represents a hydrophobic residue and X represents any amino acid). Mutating the phenylalanines (F94L F95L) has no effect on the mobilization frequency, but mutating the aspartate (D120L) and/or glutamate (E121G) completely abolishes mobilization. These results show that aspartate 120 and glutamate 121 ensure an essential function of the Mob protein. It has been shown previously that mutating aspartic acids 128 and 130 of the VirD2 T-DNA transfer protein leads to a loss of the activity of the protein (52), whereas mutating the corresponding aspartates of the RP4 TraI relaxase has no effect on TraI activity (38). In the case of VirD2, the protein being linked to the 5′ end of the nicked DNA by its tyrosine 29, it was proposed that the aspartate region provides a magnesium-binding site (22, 38, 51). Aspartate 120 and glutamate 121 of the pBBR1 Mob protein might also provide such a site. The fact that no single replacement of a tyrosine with another amino acid alters pBBR1 Mob function could mean that more than one tyrosine is involved in the Mob activity, as shown for the bacteriophage φX174 gene A protein (18) and the R388 TrwC relaxase (15). Two tyrosines could alternate in DNA cleavage. Another interpretation could be that the aspartate and/or the glutamate is directly involved in the covalent bond: the oxygen of one of these amino acids could attack the phosphodiester bond at the nick site. We are currently testing this hypothesis. It would also be interesting to test the effects of mutations of the corresponding aspartate and glutamate residues in other pMV158 family relaxases.
ACKNOWLEDGMENTS
We thank C. Locht for sending the pBBR1CM plasmid and for communicating unpublished results. We are grateful to M. E. Kovach for providing plasmids pBBR1MCS-2, pBBR1MCS-4, and pBBR1MCS-5. We thank N. Mine and L. Wacheul for technical assistance.
This work was supported by grants from the Fonds National de la Recherche Scientifique, the Fonds de la Recherche Scientifique Médicale, the Actions de la Recherche Concertée, the Foundation Van Buuren, the European Union (MECBAD, BIO4980099), and the INTERREG program. C.Y.S. is an Aspirant of the Fonds National de la Recherche Scientifique.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine R, Locht C. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from Gram-positive organisms. Mol Microbiol. 1992;6:1785–1799. doi: 10.1111/j.1365-2958.1992.tb01351.x. [DOI] [PubMed] [Google Scholar]
- 3.Ashiuchi M, Zakaria M M, Sakaguchi Y, Yagi T. Sequence analysis of a cryptic plasmid from Flavobacterium sp. KP1, a psychrophilic bacterium. FEMS Microbiol Lett. 1999;170:243–249. doi: 10.1111/j.1574-6968.1999.tb13380.x. [DOI] [PubMed] [Google Scholar]
- 4.Bernard P. Positive selection of recombinant DNA by CcdB. BioTechniques. 1996;21:320–323. doi: 10.2144/96212pf01. [DOI] [PubMed] [Google Scholar]
- 5.Bravo-Angel A M, Gloeckler V, Hohn B, Tinland B. Bacterial conjugation protein MobA mediates integration of complex DNA structures into plant cells. J Bacteriol. 1999;181:5758–5765. doi: 10.1128/jb.181.18.5758-5765.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrd D R, Matson S W. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- 7.Chaconas G, Gloor G, Miller J L. Amplification and purification of the bacteriophage Mu encoded B transposition protein. J Biol Chem. 1985;260:2662–2669. [PubMed] [Google Scholar]
- 8.Charpentier E, Gerbaud G, Courvalin P. Conjugative mobilization of the rolling-circle plasmid pIP823 from Listeria monocytogenes BM4293 among gram-positive and gram-negative bacteria. J Bacteriol. 1999;181:3368–3374. doi: 10.1128/jb.181.11.3368-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clewell D B, Helinski D R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc Natl Acad Sci USA. 1969;62:1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crellin P K, Rood J I. Tn4451 from Clostridium perfringens is a mobilizable transposon that encodes the functional Mob protein, TnpZ. Mol Microbiol. 1998;27:631–642. doi: 10.1046/j.1365-2958.1998.00712.x. [DOI] [PubMed] [Google Scholar]
- 11.Disqué-Kochem C, Dreiseikelmann B. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J Bacteriol. 1997;179:6133–6137. doi: 10.1128/jb.179.19.6133-6137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabant P, Dreze P L, Van Reeth T, Szpirer J, Szpirer C. Bifunctional lacZα-ccdB genes for selective cloning of PCR products. BioTechniques. 1997;23:938–941. doi: 10.2144/97235pf01. [DOI] [PubMed] [Google Scholar]
- 13.Gabant P, Szpirer C Y, Couturier M, Faelen M. Direct selection cloning vectors adapted to the genetic analysis of Gram-negative bacteria and their plasmids. Gene. 1998;207:87–92. doi: 10.1016/s0378-1119(97)00610-0. [DOI] [PubMed] [Google Scholar]
- 14.Gennaro M L, Kornblum J, Novick R. A site-specific recombination function in Staphylococcus aureus plasmids. J Bacteriol. 1987;169:2601–2610. doi: 10.1128/jb.169.6.2601-2610.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandoso G, Avila P, Cayón A, Hernando M A, Llosa M, de la Cruz F. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J Mol Biol. 2000;295:1163–1172. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- 16.Grohmann E, Guzmán L M, Espinosa M. Mobilisation of the streptococcal plasmid pMV158: interactions of MobM protein with its cognate oriT DNA region. Mol Gen Genet. 1999;261:707–715. doi: 10.1007/s004380050014. [DOI] [PubMed] [Google Scholar]
- 17.Guzmán L M, Espinosa M. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J Mol Biol. 1997;266:688–702. doi: 10.1006/jmbi.1996.0824. [DOI] [PubMed] [Google Scholar]
- 18.Hanai R, Wang J C. The mechanism of sequence-specific DNA cleavage and strand transfer by φX174 gene A protein. J Biol Chem. 1993;268:23830–23836. [PubMed] [Google Scholar]
- 19.Hara T, Nagatomo S, Ogata S, Ueda S. The DNA sequence of the γ-glutamyltranspeptidase gene of Bacillus subtilis (natto) plasmid pUH1. Appl Microbiol Biotechnol. 1992;37:211–215. doi: 10.1007/BF00178173. [DOI] [PubMed] [Google Scholar]
- 20.Heinemann J A, Sprague G F. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 21.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 22.IIyina T V, Koonin E. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Josson K, Soetaert P, Michiels F, Joos H, Mahillon J. Lactobacillus hilgardii plasmid pLAB1000 consists of two functional cassettes commonly found in other gram-positive organisms. J Bacteriol. 1990;172:3089–3099. doi: 10.1128/jb.172.6.3089-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kado C I. Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 25.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 26.Lejeune P, Mergeay M, Van Gijsegem F, Faelen M, Gerits J, Toussaint A. Chromosome transfer and R-prime plasmid formation mediated by plasmid pULB113 (RP4::mini-Mu) in Alcaligenes eutrophus CH34 and Pseudomonas fluorescens 6.2. J Bacteriol. 1983;155:1015–1026. doi: 10.1128/jb.155.3.1015-1026.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Light J, Molin S. The sites of action of the two copy number control functions of plasmid R1. Mol Gen Genet. 1982;187:486–493. doi: 10.1007/BF00332633. [DOI] [PubMed] [Google Scholar]
- 28.Llosa M, Grandoso G, de la Cruz F. Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J Mol Biol. 1995;246:54–62. doi: 10.1006/jmbi.1994.0065. [DOI] [PubMed] [Google Scholar]
- 29.Matson S W, Nelson W C, Morton B S. Characterization of the reaction product of the oriT nicking reaction catalyzed by Escherichia coli DNA helicase I. J Bacteriol. 1993;175:2599–2606. doi: 10.1128/jb.175.9.2599-2606.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Meijer W J, Venema G, Bron S. Characterization of single strand origins of cryptic rolling-circle plasmids from Bacillus subtilis. Nucleic Acids Res. 1995;23:612–619. doi: 10.1093/nar/23.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 31.Misawa N, Nakamura K. The nucleotide sequence of the 2.7-kb pair plasmid of Zymomonas mobilis ATCC 10988. J Biotechnol. 1989;12:67–70. [Google Scholar]
- 32.Nester E W, Kosuge T. Plasmids specifying plant hyperplasias. Annu Rev Microbiol. 1981;35:531–565. doi: 10.1146/annurev.mi.35.100181.002531. [DOI] [PubMed] [Google Scholar]
- 33.Ogata K, Sekizaki T, Aminov R I, Tajima K, Nakamura M, Nagamine T, Matsui H, Benno Y. A small cryptic plasmid from Ruminobacter amylophilus NIAH-3 possesses functional mobilization properties. FEMS Microbiol Lett. 1999;181:41–48. doi: 10.1111/j.1574-6968.1999.tb08824.x. [DOI] [PubMed] [Google Scholar]
- 34.Ouahrani-Bettache S, Porte F, Teyssier J, Liautard J P, Köhler S. pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. BioTechniques. 1999;26:620–622. doi: 10.2144/99264bm05. [DOI] [PubMed] [Google Scholar]
- 35.Pansegrau W, Balzer D, Kruft V, Lurz R, Lanka E. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci USA. 1990;87:6555–6559. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pansegrau W, Schröder W, Lanka E. Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc Natl Acad Sci USA. 1993;90:2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pansegrau W, Schröder W, Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J Biol Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 39.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 40.Priebe S D, Lacks S A. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J Bacteriol. 1989;171:4778–4784. doi: 10.1128/jb.171.9.4778-4784.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Scheiffele P, Pansegrau W, Lanka E. Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J Biol Chem. 1995;270:1269–1276. doi: 10.1074/jbc.270.3.1269. [DOI] [PubMed] [Google Scholar]
- 43.Scherzinger E, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherzinger E, Kruft V, Otto S. Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 1993;217:929–938. doi: 10.1111/j.1432-1033.1993.tb18323.x. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Priefer U, Pühler A. A broad-host-range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 46.Smith C J, Parker A C. The transfer origin for Bacteroides mobilizable transposon Tn4555 is related to a plasmid family from gram-positive bacteria. J Bacteriol. 1998;180:435–439. doi: 10.1128/jb.180.2.435-439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szpirer C Y, Top E, Couturier M, Mergeay M. Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology. 1999;145:3321–3329. doi: 10.1099/00221287-145-12-3321. [DOI] [PubMed] [Google Scholar]
- 48.Szpirer C Y, Faelen M, Couturier M. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol Microbiol. 2000;37:1283–1292. doi: 10.1046/j.1365-2958.2000.02077.x. [DOI] [PubMed] [Google Scholar]
- 49.Trask D K, Didonato J A, Muller M T. Rapid detection and isolation of covalent DNA/protein complexes: application to topoisomerase I and II. EMBO J. 1984;3:671–676. doi: 10.1002/j.1460-2075.1984.tb01865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vedantam G, Novicki T J, Hecht D W. Bacteroides fragilis transfer factor Tn5520: the smallest bacterial mobilizable transposon containing single integrase and mobilization genes that function in Escherichia coli. J Bacteriol. 1999;181:2564–2571. doi: 10.1128/jb.181.8.2564-2571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogel A M, Das A. Mutational analysis of Agrobacterium tumefaciens virD2: tyrosine 29 is essential for endonuclease activity. J Bacteriol. 1992;174:303–308. doi: 10.1128/jb.174.1.303-308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel A M, Yoon J, Das A. Mutational analysis of a conserved motif of Agrobacterium tumefaciens VirD2. Nucleic Acids Res. 1995;23:4087–4091. doi: 10.1093/nar/23.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zechner E, Prüger H, Grohmann E, Espinosa M, Högenauer G. Specific cleavage of chromosomal and plasmid DNA strands in Gram-positive and Gram-negative bacteria can be detected with nucleotide resolution. Proc Natl Acad Sci USA. 1997;94:7435–7440. doi: 10.1073/pnas.94.14.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zechner E L, de La Cruz F, Eisenbrandt R, Grahn A M, Koraimann G, Lanka E, Muth G, Pansegrau W, Thomas C M, Wilkins B M, Zatyka M. Conjugative-DNA transfer processes. In: Thomas C M, editor. The horizontal gene pool: bacterial plasmids and gene spread. Amsterdam, The Netherlands: Harwood Academic Publishers; 2000. pp. 87–174. [Google Scholar]