Abstract
The limited knowledge of genomic diversity and functional genes associated with the traits of soybean varieties has resulted in slow progress in breeding. In this study, we sequenced the genomes of 250 soybean landraces and cultivars from China, America, and Europe, and investigated their population structure, genetic diversity and architecture, and the selective sweep regions of these accessions. Five novel agronomically important genes were identified, and the effects of functional mutations in respective genes were examined. The candidate genes GSTT1, GL3, and GSTL3 associated with the isoflavone content, CKX3 associated with yield traits, and CYP85A2 associated with both architecture and yield traits were found. The phenotype–gene network analysis revealed that hub nodes play a crucial role in complex phenotypic associations. This study describes novel agronomic trait-associated genes and a complex genetic network, providing a valuable resource for future soybean molecular breeding.
Keywords: Agronomic trait, GWAS, Network, Resequence, Soybean
Introduction
Soybean Glycine max [L.] Merr. is one of the most important crops worldwide, serving as a vegetable oil and protein source for human and livestock feed. Soybean originated in China, and its wild species (G. soja Sieb. & Zucc.) was domesticated in approximately 3000 BC before being introduced into Korea and Japan about 3000 years later. It was brought to Europe and North America in the 18th century and cultivated globally since the 19th century [1].
With the rapid development of modern molecular biology and high-throughput sequencing technologies, whole-genome resequencing and genome-wide association studies (GWAS) have become common methods to study population genetic diversity and locate phenotype-related quantitative trait loci (QTLs) or genes. These methods have significantly improved our knowledge of crop genomes and selective breeding. In recent years, an increasing number of reports have been published on the domestication and improvement of soybean at the genome-wide level. These include genes and genetic networks related to agronomic traits and functions of soybean [2], [3], [4]. However, our knowledge of the soybean genome and its functional genes is still limited compared to rice and maize [5], [6], [7], which is attributed to the diversity of soybean varieties and their complex genetic backgrounds. Therefore, a large number of soybean varieties need to be further explored at the genomic level, particularly in relation to molecular traits associated with edible quality, ideal plant architecture, and the underlying genetic network of high-yielding varieties.
In this study, 250 soybean accessions were collected from the core Northeast China soybean germplasm pool, consisting of 134 accessions of landraces and cultivars from Northeast and Northwest China and 116 accessions from European and North American cultivars. The genomes of most of these accessions were not sequenced previously. The high-depth whole-genome resequencing and comprehensive analyses of these 250 soybean accessions were performed. The resulting dataset revealed valuable information on soybean genome structure, novel genes associated with important agronomic traits, and genetic networks. These genetic resources provide unique references for further exploring molecular breeding and evolution in soybean.
Results
Genotyping of 250 diverse soybean accessions by genome resequencing
High-depth whole-genome resequencing was performed on 250 soybean accessions, including 51 landraces and 83 cultivars originating from provinces in Northeast China (i.e., Heilongjiang, Jilin, Liaoning, and Inner Mongolia) and Northwest China (i.e., Xinjiang, Ningxia, and Gansu), as well as 116 cultivars originating from Europe and North America (Table S1). Approximately 10 gigabyte (GB) / 3 tera-base-pair (Tb) of pair-end reads were obtained. The maximum sequencing depth of a single accession was 22.5×, with an average depth of 11×. After filtering out the raw sequencing data (see Materials and methods), the remaining high-quality cleaned data were compared with the soybean reference genome G. max v2.0 [8]. The effective mapping rates ranged from 74.8% to 87.6%, while the genome coverages ranged from 94.8% to 97.0% (Table S1). The high mapping rates and high coverages guaranteed that the sequencing data are reliable and high-quality.
A total of 6,333,721 single-nucleotide polymorphisms (SNPs) and 2,565,797 insertions and deletions (InDels) were detected through standard variation detection, genotype filtering, and imputation steps (see Materials and methods). This included 244,360 SNPs and 62,714 InDels located in exon regions. The ratio of nonsynonymous to synonymous SNP substitutions was 1.37. Furthermore, we found 4,311,814 SNPs with a minor allele frequency (MAF) larger than 0.05 (Figure S1; Table S3). In summary, more than 6 megabyte (MB) high-density and high-quality genotype data were obtained from 250 soybean accessions with a density of 1 SNP every 15 bases.
Population structure analysis of 250 soybean landraces and cultivars
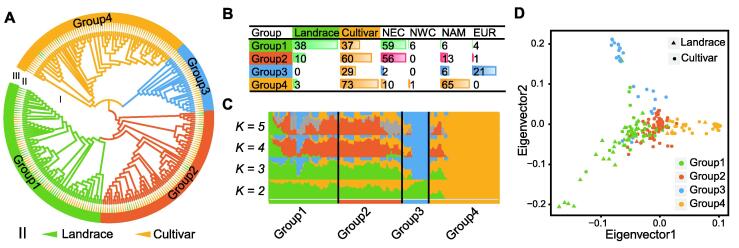
Using the 6 MB SNP genotype dataset, a phylogenetic tree was constructed using the neighbor-joining (NJ) method. As a result, the 250 soybean accessions were classified into 4 groups (Figure 1A). Group 1 comprised 65 Chinese, 4 European, and 6 American varieties; Group 2 comprised 56 Chinese, 1 European, and 13 American varieties; Group 3 comprised 21 European, 2 Chinese, and 6 American cultivars; Group 4 comprised 65 American cultivars and 11 Chinese varieties (Figure 1B). A Bayesian clustering algorithm based on a mixed model was used to estimate the proportion of ancestors in each accession. When K = 2, the main ancestor component (yellow) of Group 4 was split, indicating that Group 4 had the highest level of selection. When K = 3, the main ancestor component (blue) of Group 3 was split, indicating that Group 3 had the second level of selection. However, when K = 4 and K = 5, Groups 1 and 2 exhibited complex differentiated mixed ancestor components, indicating the higher genetic diversities and lower selection levels in Groups 1 and 2 (Figure 1C; Table S3).
Figure 1.
Population structure analysis of 250 soybean landraces and cultivars
A. Phylogenetic tree constructed for all soybean accessions. Groups 1–4 are shown with different colors, landraces are labeled with green triangles, and cultivars are labeled with yellow triangles. B. Statistics of the geographic origin for each subpopulation. C. Mixed ancestor analysis for soybean subpopulations. Each color represents an ancestral component. K from 2 to 5 is set to trace different ancestral components. D. Principal component analysis plot of the first two eigenvectors for all soybean accessions. Landraces and cultivars are shown with different shapes, while groups are shown with different colors. NEC, Northeast China; NWC, Northwest China; NAM, North America; EUR, Europe.
The results of the principal component analysis (PCA) were consistent with those of the phylogenetic tree. Three groups, Groups 1, 3, and 4, radiated away from Group 2 within the rectangular coordinate system projected using eigenvector 1 and eigenvector 2 data on the x-axis and y-axis, respectively. Concurrently, the distribution of varieties in the four groups showed continuity, indicating that the varieties located in different groups also had genetic similarities (Figure 1D).
These results indicated that the group classification of the 250 soybean accessions was closely related to their geographical distribution. That is, varieties with similar geographical distribution had similar genetic backgrounds. Generally speaking, the group classification was also related to the level of domestication. Landraces had a lower level of domestication, while cultivars had higher levels of domestication. Varieties with similar domestication levels tended to have a higher similarity in genetic backgrounds. However, still, differences were found in the geographical distribution and domestication level among breeds with similar genetic backgrounds, indicating that gene exchange might have occurred between accessions of different groups. This observation reflected the complexity of soybean domestication history.
Genetic diversity and selective sweep analysis for soybean subpopulations
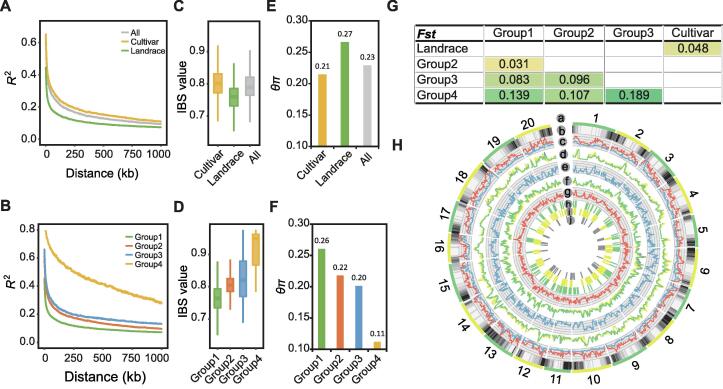
We next performed a linkage disequilibrium (LD) analysis. The results showed that the overall LD decay distance was more than 100 kb, and the LD decay distance of the landraces was smaller than that of the cultivars (Figure 2A). Further LD decay analysis of the four groups showed that the LD decay distance in Group 1 was the smallest, followed by Groups 2 and 3, while Group 4 had the largest LD decay distance (Figure 2B). In addition, the LD levels varied for different chromosomes or different regions across one chromosome. An identical-by-state (IBS) analysis, which reflects the degree of relatedness among individuals by calculating the consistency of all genetic markers, revealed that the average IBS value of landraces was less than that of cultivars (Figure 2C). The IBS values of Groups 1–4 followed the same trends as the LD decay distances. In particular, the IBS values of Group 1 were the lowest, and those of Group 4 were the highest among all groups (Figure 2D). θπ values reflect the genetic diversity within a population by calculating the number of different sites between any two sequences or individuals within a population. Fst is a calculation used to measure the differentiation and genetic distance between two populations. θπ values were calculated for landraces, cultivars, all accessions, and Groups 1–4. Fst values were calculated between landraces and cultivars and between the four groups. The results showed that a population with a higher LD decay distance or higher IBS values was correlated with a smaller θπ (Figure 2E and F), indicating an opposite pattern to those of the LD decay distances and IBS values. The lowest Fst value was for Group 1 versus Group 2, while the highest Fst value was for Group 3 versus Group 4. Also, the Fst value of Group 2 versus Group 3 was higher than that of Group 1 versus Group 3 (Figure 2G). The Fst value of Group 2 versus Group 4 was smaller than that of Group 1 versus Group 4. In addition, the results of our allele frequency distribution (AFD) analysis, as an alternative population similarity measurement, were consistent with the Fst results (Figure S2). When combined with the population structure and geographical distribution information, the results of our population diversity analysis inferred that the European and American soybean varieties might have originated from different Chinese ancestors before undergoing independent selection. The results indicated that European cultivars and the Chinese landrace group (Group 1) had a more recent common ancestor, while North American cultivars and the Chinese cultivar group (Group 2) had a more recent common ancestor.
Figure 2.
Genetic diversity analysis and putative selective regions of soybean subpopulations
A. LD decay plots for landraces (green), cultivars (yellow), and all soybean accessions (gray). B. LD decay plots for soybean subpopulations. C. IBS value distribution for landraces (green), cultivars (yellow), and all soybean accessions (gray). D. IBS value distribution for soybean subpopulations. E. Comparison of θπ values for landraces (green), cultivars (yellow), and all soybean accessions (gray). F. Comparison of θπ values for soybean subpopulations. G. Comparison of Fst values between landraces and cultivars and between subpopulations. H. Landscape of soybean genetic diversity across the whole genome. (a) Chromosomes. (b) Density of genes. (c) Density of SNPs (red) and InDels (blue). (d) LD value distribution for landraces (green), cultivars (yellow), and all accessions (gray). (e) Fst value distribution of landraces versus cultivars. (f) θπ value distribution for landraces (green), cultivars (yellow), and all accessions (gray). (g) Tajima’s D value distribution of all accessions. (h) Putative selective sweep regions detected by Tajima’s D combined with θπ. (i) Putative selective sweep regions detected by Fst combined with θπ ratios. (j) ROH region larger than 300 kb. LD, linkage disequilibrium; IBS, identical-by-state; SNP, single-nucleotide polymorphism; InDel, insertion and deletion; ROH, runs of homozygosity.
Tajima’s D (based on a neutral test), θπ (based on genetic diversity within a population), and Fst (based on genetic diversity between two populations) are highly effective tools that can screen selective sweep signals across a genome [9]. These methods were combined in pairs for mining potential selective sweep regions in the soybean genome that might have undergone artificial selection. One pair was Tajima’s D combined with θπ for the whole population. Another pair was Fst combined with θπ ratios between two subpopulations/landraces/cultivars. A sliding window method was used to calculate the values of Tajima’s D, θπ, and Fst in each window across the whole genome (Figure 2H), and the top 5% significant windows were selected as potential selective sweep regions (Figure S3A and B). A total of 148 and 222 potential selective sweep regions were screened by the aforementioned two combined methods, and they covered 36.09 Mb and 88.15 Mb genome regions, respectively (Table S4). These potential selective sweep regions covered 9128 genes, accounting for approximately one-sixth of all soybean genes. A total of 1876 genes were screened by both methods (Figure S3C). Runs of homozygosity (ROH) regions, continuous homozygous chromosome regions in a genome, may be related to domestication or artificial selection [10]. Through an ROH analysis, 71 ROH regions larger than 300 kb were obtained from all 250 accessions, with a total length of 27.84 Mb. The longest ROH region up to 911 kb was located at the beginning of chromosome 10 (Table S5). Furthermore, 3397 genes were located in these ROH regions, 924 of which were also located in the potential selected sweep regions (Figure S3D).
Identification of significantly associated loci and genes through GWAS of 50 agronomic traits
A total of 50 agronomic traits were measured in 250 soybean accessions from three geographic locations for three years and then integrated using the best linear unbiased prediction (BLUP). The 50 traits included traits related to architecture (15), color (5), isoflavone (1), oil (4), protein (18), and yield (7), and were classified into six categories (Table S6). Pearson correlation coefficients for traits were calculated to compare within and between categories, revealing that traits within the same category were more strongly correlated than traits in different categories. Specifically, strong positive or negative correlations were found between almost all traits within the protein-related, oil-related, and yield-related categories. For example, the linoleic acid content was positively correlated with the linolenic acid content but negatively correlated with the oleic acid content; stem intension was negatively correlated with lodging (Figure S4). Some traits were evenly distributed, while others were ranked (Figures S5–S54).
GWAS was performed on 4,311,814 SNPs with MAF > 0.05, using the mixed linear model (MLM) method for the aforementioned 50 agronomic traits. For each trait, a clump-based method [11] was used, and a significantly associated locus (SAL) in a chromosome region was defined with a substantial amount of SNPs associated with a specific trait. A total of 203 SALs were detected for 43 traits (Figures S5–S54; Table S7). Since each SAL may contain dozens of genes, a functional mutation-based haplotype test was used for further mining of the most reliable candidate trait-associated genes [12]. In particular, only the nonsynonymous SNPs, frameshift InDels, and mutations within a gene that happened on a start or stop codon, splice sites, or transcription start sites were considered as effective functional mutations. These mutations were used to classify each gene into different haplotypes, and the phenotypic differences of the accessions belonging to each haplotype were subsequently tested. A gene with significant phenotypic differences was defined as a significantly associated gene (SAG), and 3165 SAGs were thus screened for 43 traits. These SAGs included some QTLs or genes that were previously identified, such as the flower color-related chr13:16551728–19506795, pubescence color-related chr6:16930159–19168772, seed coat luster-related chr15:8910798–10281804, palmitic acid content-related chr5:879095–1682551 [4], isoflavone content-related chr5:38880530–39142565 [13], plant height-related Dt1 [4], and oil content-related FAD2 and SAT1 [14]. These SAGs also contained genes that were identified for the first time in soybean, such as the isoflavone content-related GL3 and glutathione S-transferase (GST) L3 (GSTL3), the yield trait-related cytokinin oxidase/dehydrogenase 3 (CKX3), and the architecture and yield trait-related CYP85A2 (Table 1).
Table 1.
Functional variants of representative significant associated genes
| Variant ID | Chromosome | Positon | Reference | Alternative | Variant type | Gene ID | Gene symbol |
|---|---|---|---|---|---|---|---|
| c5s38936266 | 5 | 38,936,266 | C | T | Nonsynonymous SNV | GLYMA_05G206900 | GSTT1a |
| c5s38940717 | 5 | 38,940,717 | C | T | Nonsynonymous SNV | GLYMA_05G207000 | GSTT1b |
| c5s39035509 | 5 | 39,035,509 | G | C | Nonsynonymous SNV | GLYMA_05G208300 | GL3 |
| c5s39036346 | 5 | 39,036,346 | T | C | Nonsynonymous SNV | GLYMA_05G208300 | GL3 |
| c13s24804891 | 13 | 24,804,891 | C | T | Nonsynonymous SNV | GLYMA_13G135600 | GSTL3 |
| c13s24805363 | 13 | 24,805,363 | A | T | Splicing SNV | GLYMA_13G135600 | GSTL3 |
| c17s4143663 | 17 | 4,143,663 | C | T | Nonsynonymous SNV | GLYMA_17G054500 | CKX3 |
| c17s4143832 | 17 | 4,143,832 | T | C | Nonsynonymous SNV | GLYMA_17G054500 | CKX3 |
| c17s4146922 | 17 | 4,146,922 | G | T | Nonsynonymous SNV | GLYMA_17G054500 | CKX3 |
| c17s4151713 | 17 | 4,151,713 | C | A | Nonsynonymous SNV | GLYMA_17G054600 | CKX4 |
| c17s4151752 | 17 | 4,151,752 | T | C | Nonsynonymous SNV | GLYMA_17G054600 | CKX4 |
| c18s55526062 | 18 | 55,526,062 | C | T | Nonsynonymous SNV | GLYMA_18G272300 | CYP85A2 |
Note: SNV, single nucleotide variant.
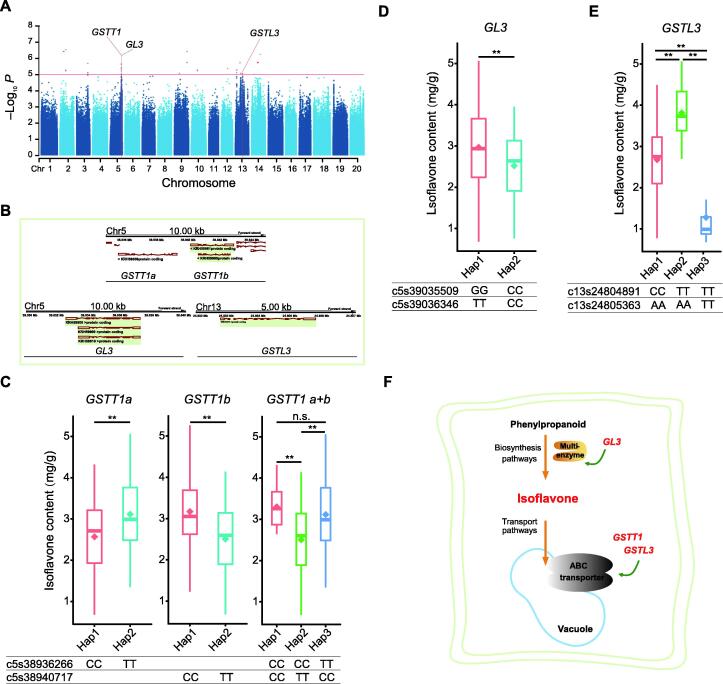
Association of GSTT1, GL3, and GSTL3 with the isoflavone content
The isoflavone content is an important quality-related trait in soybean, but its molecular mechanism is still unclear. This study identified four SALs related to the isoflavone content, namely chr3:38590023–38728718, chr5:3888053–39142565, chr13:18342836–18541809, and chr5:24726091–24852447. Only one SAL, chr5:24726091–24852447, overlapped with a previously reported QTL that contained a GST gene GST theta 1 (GSTT1) [13]. All the other SALs were newly identified. Furthermore, 48 genes were located within these SALs (Table S8), and three genes (GSTT1, GL3, and GSTL3) might be related to the isoflavone content (Figure 3A and B). Two functional mutations were present at c5s38936266 and c5s38940717, forming two haplotypes for GSTT1a and GSTT1b, respectively. For each GSTT1 gene, soybean accessions with a different haplotype were related to significantly different isoflavone contents. Since GSTT1a and GSTT1b were approximately only 1 kb apart from each other in the same genome region, the two genes were considered to be one in the subsequent analysis. Three haplotypes were formed by two functional mutations when the two GSTT1 genes were analyzed as one. Haplotype 1 versus Haplotype 2 and Haplotype 2 versus Haplotype 3, showed significant differences in the isoflavone content, while Haplotype 1 versus Haplotype 3 showed no significant difference (analyzed using Tukey’s test). This finding suggested that GSTT1a was associated with the isoflavone content due to its linkage with GSTT1b. However, c5s38936266 did not contribute to the difference in the isoflavone content. Thus, only c5s38940717 on GSTT1b was associated with the isoflavone content (Figure 3C). Two functional mutations, c5s39035509 and c5s39036346, producing two haplotypes in GL3, were associated with different isoflavone contents in soybean accessions (Figure 3D). Also, another GST gene, GSTL3, was identified, which was located on chromosome 13. Two functional mutations within GSTL3 produced three haplotypes, and significant associations between the different haplotypes and the isoflavone contents were detected for each comparison (Figure 3E). Based on these results, a schematic diagram of the roles of these three candidate genes was drawn according to their biological functions. The diagram indicates that GL3 regulates isoflavone synthesis, while GSTT1 and GSTL3 participate in isoflavone transport (Figure 3F).
Figure 3.
Three genes associated withsoybean isoflavone content identified by GWAS
A. Manhattan plot and candidate genes for the soybean isoflavone content. B. Chromosome location and transcript structure of the candidate genes. C. Soybean isoflavone content distribution for the haplotypes of GSTT1. D. Soybean isoflavone content distribution for the haplotypes of GL3. E. Soybean isoflavone content distribution for the haplotypes of GSTL3. F. Diagram of soybean isoflavone synthesis and transport and the roles of candidate genes detected by GWAS. *, P < 0.05; **, P < 0.01; n.s., not significant. GWAS, genome-wide association study; Hap, haplotype.
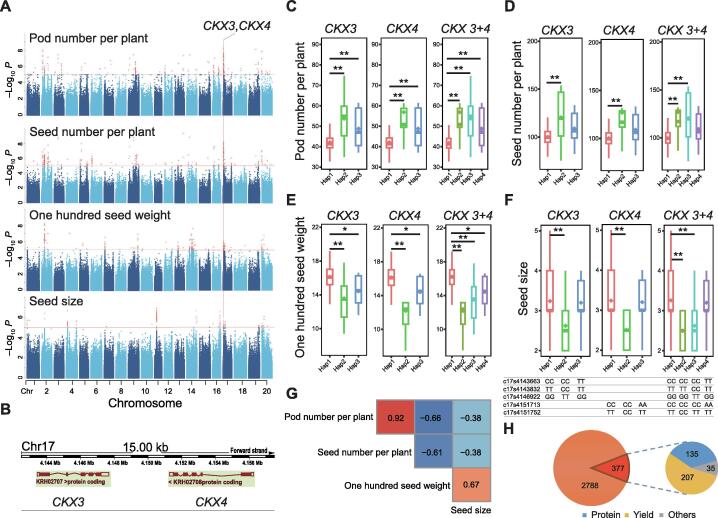
Association of CKX3 with yield-related traits and its location in an artificial selection region
Four yield-related traits (pod number per plant, seed number per plant, one hundred seed weight, and seed size) have a common SAL located in the region between ~ 4.0 Mb and ~ 4.2 Mb on chromosome 17 (Figure 4A). Further analyses revealed that this SAL contained two tandem repeat CKX genes named CKX3 and CKX4, approximately 15 kb apart from each other (Figure 4B).
Figure 4.
CKX3 was associated with soybean yield-related traits
A. Manhattan plot of four yield-related traits (pod number per plant, seed number per plant, one hundred seed weight, and seed size) and the candidate CKX genes. B. Chromosome location and transcript structure of CKX3 and CKX4. C. Pod number per plant distribution for the haplotypes of the CKX genes. D. Seed number per plant distribution for the haplotypes of the CKX genes. E. One hundred seed weight distribution for the haplotypes of the CKX genes. F. Seed size distribution for the haplotypes of the CKX genes. G. Phenotype correlation of four yield-related traits. H. Statistics of the SAGs located in selective sweep regions and their distributions for different trait categories. *, P < 0.05; **, P < 0.01.
The relationship between functional mutations of CKX3 and CKX4 was further analyzed. Three and two nonsynonymous SNPs were located in CKX3 and CKX4, respectively. As these two genes were only approximately 15 kb apart from each other in the same genomic region, the two genes were analyzed separately as well as combined as one in relation to their association with haplotypes and traits (Figure 4C–F). The results showed that the functional mutations could form either three haplotypes for each individual genes or four haplotypes for the CKX 3+4 combined. For all comparisons in all traits, Haplotype 1 always showed significant differences compared with the other haplotypes. The relationship between different haplotypes in terms of pod or seed number per plant showed a consistent trend, while that in terms of one hundred seed weight or seed size showed a consistent but opposite trend. A phenotypic correlation was found between pod number per plant and seed number per plant (0.92) as well as between one hundred seed weight and seed size (0.67) (Figure 4G). Furthermore, we observed that CKX3 and CKX4 were located in different strands of the same chromosome, suggesting that they were more likely to have independent functions. However, the expression of CKX4 was not detected in the subsequent qRT-PCR validation. Thus, only CKX3 was regarded as a real candidate gene, while the role of CKX4 needs further investigation.
When the soybean accession information of each haplotype for the four yield-related traits was compared, most accessions with the Haplotype 1 genotype had dominant traits (lower pod or seed numbers and larger seeds and seed weights) and were more associated with cultivars. The other haplotypes were mainly landrace-specific haplotypes, and their accessions all belonged to Group 1. CKX3 was also located in a strong selective sweep region. This indicated that the functional mutation sites in CKX3 experienced strong directed artificial selection, resulting in genotype differences and affecting yield-related traits. Furthermore, all SAGs and selective sweep regions for all traits were compared, revealing that approximately 12% of the SAGs were located in the selected sweep regions, which experienced artificial selection (Table S9). Of all the SAGs located in selective sweep regions, about 55% were related to yield traits, 36% were related to protein traits, and less than 10% were related to other traits (Figure 4H).
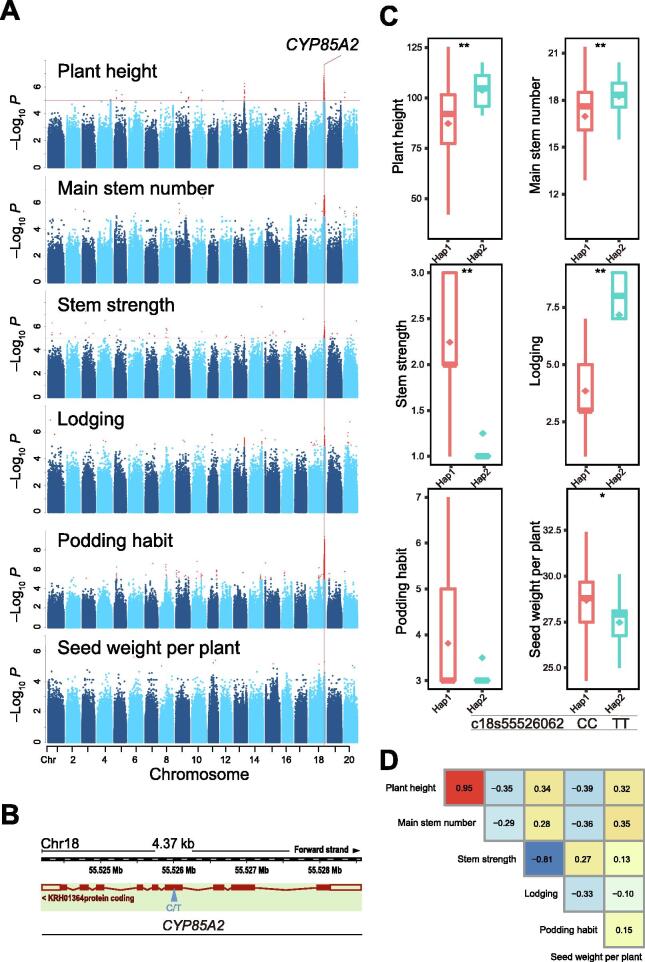
CYP85A2 was associated with both architecture- and yield-related traits
One SAL was located on chromosome 18, which was associated with six traits, including plant height, main stem number, stem strength, lodging, podding habit, and seed weight per plant. Interestingly, these traits included both architecture- and yield-related traits. A cytochrome P450 family gene named CYP85A2 was located within a 4.37 kb region of this SAL (Figure 5A and B). The association of CPY85A2 with the architecture- and yield-related traits in soybean was a novel finding. We also observed that a nonsynonymous mutation site c18s55526062 was involved in producing two haplotypes. Haplotype 1 with a CC genotype had a dwarf plant height, a low main stem node number, a high stem strength, and a low lodging rate. When plants produced mostly limited or semi-limited pods, their seed weight per plant was also found to increase (Figure 5C). Phenotypically, plant height was positively correlated with main stem node number (0.95), while stem strength and lodging were negatively correlated (–0.81), showing a trend consistent with the genotype (Figure 5D). The CC genotype of c18s55526062 is a dominant genotype, which is useful when designing an ideal plant type and increasing soybean yield.
Figure 5.
CYP85A2 was associated with soybean architecture and yield-related traits
A. Manhattan plot of six architecture- and yield-related traits (plant height, main stem number, stem strength, lodging, podding habit, and seed weight per plant) and the candidate gene CYP85A2. B. Chromosome location and transcript structure of CYP85A2.C. Trait distribution for the haplotypes of CYP85A2. D. Phenotype correlation of the six traits. *, P < 0.05; **, P < 0.01.
Construction of a complex phenotype–gene network using different phenotypes coupled with hub gene modules
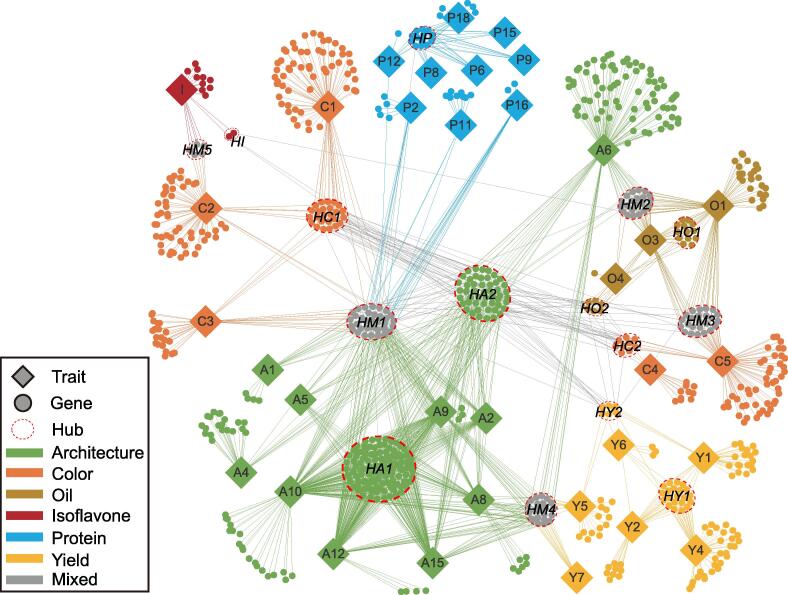
Based on the in-depth exploration of the GWAS results, one trait was found to be associated with multiple genes and vice versa. At the same time, a complex network between various phenotypes and genes was also found due to the widespread protein-level interactions between genes. To explore this further, a functional mutation-based haplotype test was performed to screen SAGs in all SALs for all traits. Then, a phenotype–gene network was constructed, including 34 traits and 853 SAGs (Figure 6). At the trait level, they were divided into six categories, namely architecture, color, oil, isoflavone, protein, and yield. At the gene level, besides the six categories, a mixed category emerged in which genes associated with more than one trait category. Traits in the same category were closely linked within the entire network. However, some trait categories were also linked with each other, such as yield, oil, protein, and color, and they were all closely linked to architecture through common SAGs. This suggested subtle relationships between architecture and other trait categories. In this genetic network, 6 trait categories were linked through 15 hub nodes containing 367 genes (Table S10). The largest hub was Hub Architecture 1 (HA1). The genes in this hub were associated with only two or more architecture-related traits. Unlike HA1, the genes in the HA2 node were not only associated with two or more architecture-related traits but also had protein interactions with other genes. Multiple yield-related traits were associated with the Hub Yield 1 (HY1) node, containing CKX3, while the Hub Mixed 4 (HM4) node, containing CYP85A2, was connected with architecture- and yield-related traits.
Figure 6.
Phenotype–gene association/interaction network for six trait categories in soybean
Traits are represented as solid rhombuses, genes as solid circles, and hubs as hollow ellipses. Six trait categories, their associated genes, and the links between them are colored accordingly; genes associated with more than one trait category are colored in gray. Genes with protein–protein interactions are linked with gray lines. HA, Hub Architecture; HY, Hub Yield; HM, Hub Mixed; HI, Hub Isoflavone; HC, Hub Color; HO, Hub Oil; HP, Hub Protein.
Discussion
In this study, 250 representative landrace and cultivar soybean accessions were deeply sequenced. It is novel in evaluating the genetic structure of European soybean varieties through population genetics and GWAS analyses. Novel candidate genes related to seed isoflavone content, yield, and architecture traits were identified. Moreover, a soybean phenotype–gene interaction network was constructed, and evidence of the improvement in soybean yield-related traits at the molecular level was found.
A total of ~ 3 Tb pair-end reads and 6 MB SNPs were obtained. The maximum sequencing depth of a single accession was 22.5×, with an average depth of 11× (higher than those in previous soybean resequencing studies [2], [3], [4]). Moreover, 84% of the accessions were sequenced for the first time, providing new data for soybean genome research. Previous soybean research mainly focused on varieties from Asia and North America but not from Europe [3], [4]. This study completed the resequencing of 26 European accessions and, for the first time, outlined a breeding history of the European soybean. We found that European soybean cultivars had higher genetic diversities and lower breeding levels than North American cultivars. Both European and American soybean cultivars might have been introduced from different ancestors in China. This theory was based on the following findings: a small population difference between European cultivars and Chinese landraces and between American cultivars and Chinese cultivars, but a large population difference between American and European cultivars (Figure 2G, Figure S2). The findings were consistent with the current hypothesis that soybean originated in China; they showed that ancestral components from the area of origin were the most complex. This study showed that the heterozygosity rates of most accessions were less than 0.2, except for four accessions with higher heterozygosity caused by their complex ancestral compositions (Table S3). Further combination analysis of the selective sweep and GWAS revealed that the artificial selection of soybean at the phenotypic level was consistent with that at the genomic level. Genomic regions associated with yield and quality traits were more likely to experience artificial selection. This might reflect the yield- and quality-directed artificial selection of soybean breeding at the genetic level. Furthermore, we found evidence of functional mutations under artificial selection for a candidate gene CKX3 related to multiple yield traits. This study provides valuable information for marker-assisted selection, which is vital for the improvement of soybean breeding.
Isoflavone is a secondary metabolite produced via phenylpropane metabolic pathways in higher plants. Isoflavone is associated with plant stress resistance, defense against microbial and insect infection, promotion of rhizobium chemotaxis, and development of rhizome and nitrogen fixation in plants. It also provides health benefits to humans, such as reducing the incidence of cancer and cardiovascular diseases and regulating the immune response [15]. Therefore, increasing the seed isoflavone content of soybean can improve its nutritional and health benefits. However, few genome-wide studies have investigated the molecular mechanism regulating the soybean isoflavone content. Isoflavone is synthesized in the cytoplasm, but it cannot be accumulated in the cytoplasm due to its cell cytotoxicity and must be continuously transported to vacuoles for storage. Therefore, the isoflavone content mainly depends on two factors: synthesis efficiency and transport efficiency [16]. The transcription factor GL3 is a bHLH family member that can form the MYB–bHLH–WD40 complex with two other transcription factors (MYB and WD40) to jointly regulate the synthesis of flavonoids and anthocyanin in plants [17]. GST can bind with glutathione to form an ABC transporter to transport and catalyze the entry of flavonoids into vacuoles for accumulation [16]. In this study, four novel genes associated with the isoflavone content were identified. These genes included GL3, which participated in regulating multi-enzyme systems from the phenylpropanoid pathway to the isoflavone biosynthesis pathway, and two GST genes GSTT1 and GSTL3, which facilitated the transport of isoflavone from the cytoplasm to vacuoles (Figure 3F). In addition, many other genes were observed in the SALs, such as cation/H+ exchanger 20 (CAX20), pyrophosphorylase 4 (PPa4), actin-depolymerizing factor 7 (ADF7), and four genes encoding a mitochondrial substrate carrier family protein, a myosin heavy chain-related protein, an ATP synthase alpha/beta family protein, and a protein kinase superfamily protein, respectively (Table S8); they were all related to isoflavone transport. This over-representation of transport-related genes further suggested that the accumulation of soybean isoflavone was related to its transport to the vacuole. In conclusion, the soybean isoflavone content is not determined merely by one or several genes or loci but by a multiple gene system involved in synthesis, regulation, transport, and storage.
Other novel candidate genes, such as CKX3, were associated with multiple yield-related traits. This study also found for the first time that CYP85A2 was associated with multiple architecture- and yield-related traits in soybean. Cytokinin promotes cell division and plant growth, and CKX is one of the key enzymes in cytokinin metabolism. A functional variation in the CKX gene may affect the cytokinin metabolism, thus affecting grain yield and related traits. A number of studies on Arabidopsis thaliana, rice, and other crops have shown that mutations or reduced expression levels of the CKX family genes are related to a decrease in seed setting rate and an increase in seed weight [18], [19]. CYP85A2 is involved in the brassinosteroid (BR) biosynthesis pathway in A. thaliana and converts 6-deoxocastasterone into castasterone, which is followed by the conversion of castasterone into brassinolide [20]. BRs are broad-spectrum plant growth regulators playing an important role in plant growth and development, as well as in biological and abiotic stress responses [21]. Mutations in CYP85A2 led to increased production of the dwarf phenotype [22], and overexpression of the CYP85A family gene resulted in increases in BR content, biomass, plant height, plant fresh weight, and fruit yield [23]. These results showed that CKX3 and CYP85A2 may affect soybean yield- and architecture-related traits through different molecular mechanisms. The potential effect of functional mutations in these genes on phenotypes was further confirmed by the haplotype tests. However, further functional verification of these genes is necessary to verify whether these candidate genes and functional mutations are the true cause of phenotypic differences. Multiple methods, such as construction of isolated populations, transgene, gene knockout, gene editing, and expression verification, can be used for this purpose. In this study, expression verification was performed in seedlings with different haplotypes/phenotypes for six genes GL3, GSTL3, GSTT1b, CKX3, CKX4, and CYP85A2. The results showed that, except for CKX4 (no expression was detected), the other five genes were differentially expressed in seedlings with different haplotypes/phenotypes. The expression levels of GL3, GSTL3, and GSTT1b were significantly higher in the seedlings with high isoflavone content than those with low isoflavone content (P < 0.05, t-test). The expression level of CKX3 was significantly higher in the seedlings with high-yield phenotype than those with low-yield phenotype (P < 0.05, t-test; Figure S55).
The highest goal of plant breeding is to aggregate many desired traits into a single genome. Breeders need to select and improve multiple related traits simultaneously. However, since multiple traits are interrelated, it is possible that when screening for a favorable trait, an unfavorable one is also selected. Understanding the genetic network behind different traits can help breeders increase breeding efficiency. Although soybean genetic networks for multiple agronomic traits have been established at the locus level [4], a new phenotype–gene network including 34 traits and 853 genes was built in this study. This network reflects the relationships between phenotypes and genes more directly than the previous phenotype–SAL network and is more conducive to discoverying important candidate genes. For example, the Hub Mixed 1 (HM1) node is associated with two or more trait types (architecture, color, or protein). Specifically, the HBT gene in the HM1 node is associated with six architecture-related traits (branch number, main stem number, plant height, stem strength, lodging, and podding habit) and four protein-related traits (phenylalanine content, isoleucine content, tyrosine content, and glycine content). The HBT gene belongs to the CDC27b gene family and is involved in cell cycle regulation, which is related to cell development and division [24]. Therefore, soybean architecture is likely affected by HBT; however, its relationship with the amino acid content is unclear. Leaf shape is known to affect photosynthesis efficiency, followed by carbohydrate accumulation and, consequently, oil accumulation; in our network, the HM2 node, containing FAD2, also relates oil content to leaf shape [25]. Oil-related traits and seed coat luster-related traits have experienced parallel selection during bean domestication [26]; the HM3 node also connects oil-related traits and seed coat luster-related traits. Anthocyanin synthesis and isoflavone synthesis share part of their metabolic pathways, and the HM5 node connects color-related traits and isoflavone content as well. This phenotype–gene network may surpass the previous phenotype–SAL network in terms of candidate gene selection, which is also beneficial to polymerization breeding programs. For example, breeders can achieve polymerization breeding by directly selecting a favorable gene (such as CYP85A2) in hub HM4, which is related to both yield and architecture traits and eliminates the confusion of other adverse genes located in the same SAL. Furthermore, the architecture-related traits, which centrally connect various other trait categories, have the most extensive connectivity. In other words, numerous relationships exist between architecture-related traits and other trait categories in the phenotype–gene network (Figure 6), suggesting that some candidate genes related to architecture traits may also be related to other trait types. This may provide theoretical support and practical guidance for parallel selection breeding and promote “ideotype” breeding in soybean. The next step is to conduct more in-depth functional investigations on genes with a potential application value, such as CKX3 and CYP85A2. This would help promote the design and breeding process of soybean varieties with a higher yield and quality. Overall, the present study is conducive to promoting soybean genome functional research and genomic breeding.
Materials and methods
Plant materials and phenotyping
A total of 250 soybean accessions were analyzed in this study, which was provided by the National Crop Germplasm Resources Platform, Institute of Crop Genetics, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences. All materials were planted and phenotyped at three locations: the Gongzhuling experimental site in the Jilin Academy of Agricultural Sciences, China (north latitude 43.51°, east longitude 124.80°), the Harbin experimental site in the Heilongjiang Academy of Agricultural Sciences, China (north latitude 45.68°, east longitude 126.61°), and the Chifeng experimental site in the Agricultural Science Institute in Inner Mongolia, China (north latitude 42.27°, east longitude 118.90°) in late April of 2008, 2009, and 2010, respectively. The grain protein content was measured using the Kjeldahl method from the National Food Safety Standard GB5009.5-2010, China (GB/T 5009.5-2010 National food safety standard—Determination of protein in foods), while the grain fatty acid content was determined using the Soxhlet extraction method from the National Food Safety Standard GB/T5512-2008, China (GB/T 5512-2008 Inspect of grain and oilseeds—Determination of crude fat content in grain). The amino acid content was determined using high-performance liquid chromatography (HPLC; Catalog No. S433D, Seckam, Germany) following a previous amino acid determination method from the National Food Safety Standard GB/T 18246-2000, China (GB/T 18246-2000 Determination of amino acids in feeds). The grain isoflavone content was determined using HPLC following the National Food Safety Standard GB/T23788-2009, China (GB/T 23788-2009 Determination of soybean isoflavone in health-care food-High-performance liquid chromatography). Finally, the phenotypic data were integrated by the BLUP method using R [27] to remove environmental effects and obtain stable genetic phenotypes. The seeds were planted in CLC-BIV-M/CLC404-TV (MMM, Germany) at 20 °C (with 12 h day/12 h night) and relative humidity of 60%–80% till the six-leaf stage (about 2-week-old). Two-week-old seedlings (24 °C, 12 h day/12 h night cycle) were used for qRT-PCR validation.
DNA preparation and sequencing
The genomic DNA for all soybean accessions was extracted from soybean leaves after 3 weeks of growth. DNA was extracted using the cetyltrimethylammonium bromide method [28]. The library for each accession was constructed with an insert size of approximately 500 bp following the manufacturer’s protocols (Illumina, CA). All soybean accessions were sequenced, and paired-end 150 bp reads were produced using an Illumina NovaSeq 6000 sequencer at the BerryGenomics Company (http://www.berrygenomics.com/; Beijing, China).
Total RNA extraction, cDNA synthesis, and qRT-PCR analysis
Total RNA was isolated from each sample using TRIzol reagent (Invitrogen, Nottingham, UK) following the manufacturer’s protocols. The purified RNA was stored at −80 °C until subsequent analyses. The first-strand cDNA synthesis was performed using M-MLV reverse transcriptase following the manufacturer’s protocols (TaKaRa, Shiga, Japan). qRT-PCR was performed using an SYBR Premix Ex Taq Kit (TaKaRa) and a real-time PCR machine (Catolog No. CFX96, Bio-Rad, CA) following the manufacturer’s protocols. The procedure used for qRT-PCR was 95 °C for 10 min, followed by 38 cycles of 15 s at 95 °C and 60 s at 61–62 °C. β-actin was used as the reference gene for analyzing the relative expression patterns of mRNA. The reactions were carried out with three biological replicates, with at least two technical replicates for each sample. The data were analyzed as previously described [29]. Finally, the data of three biological replicates were presented as mean ± SD.
Mapping, variant calling, and annotation
Raw paired-end resequencing reads were first cleaned by removing reads with adaptors, reads of low quality, and reads with “N”s. The high-quality clean reads were subsequently mapped to the soybean reference genome (Williams 82 assembly v2.1) with BWA [30]. Statistical analyses of mapping rate and genomic coverage of clean reads were performed using in-house scripts. The Speedseq pipeline [31] was used for SNP and InDel calling, and VCFtools [32] was used for genotype filtering. Missing genotypes were imputed and phased through a localized haplotype clustering algorithm implemented using Beagle v3.0 [33]. Variant annotation was performed using ANNOVAR [34] against the soybean gene model set v2.1.42. After annotation, SNPs and InDels were categorized into exonic, intronic, intergenic, splicing, 5′-UTRs, 3′-UTRs, upstream, and downstream. Exonic SNPs were further categorized into synonymous, nonsynonymous, stop gain, and stop loss. Exonic InDels were further categorized into frameshift, non-frameshift, stop gain, and stop loss.
Population structure analysis
Approximately 6 MB SNPs from the 250 soybean accessions were concatenated for constructing a phylogenetic tree. The phylogenetic tree was constructed with MegaCC using an NJ algorithm with a pairwise gap deletion method for 100 bootstrap replications [35]. The output was displayed using the iTOL [36] web tool. With the whole-genome genotype as input, a PCA was performed using flashPCA [37], and the first two eigenvectors were plotted. A population admixture analysis with K = 2 to K = 5 was performed to infer the admixture of ancestors using fastSTRUCTURE [38].
Genetic diversity analysis
Genetic diversity analysis was performed using the scripts provided by the SR4R database [39]. LD analyses for each subpopulation were performed using PLINK [40] by calculating the correlation coefficient (r2) of any two SNP pairs in one chromosome. An LD decay plot was drawn using the average r2 value for the distance from 0 to 1000 kb. Pairwise IBS calculations were also performed using PLINK, and a distance matrix was generated for each subpopulation. Population genetic diversities were measured using VCFtools [32] by calculating θπ and Fst. θπ was used to measure the genetic diversity of each subpopulation, while Fst, plus the AFD plot (which was generated by in-house scripts), was used to measure genetic diversity between subpopulations. In addition, sliding window calculations of r2, θπ, Fst, and Tajima’s D values were performed for genome-wide displays of soybean genetic diversities with a 100 kb window and a 10 kb step.
Selective sweep analysis
Two methods were used to detect selective sweep regions across the soybean genome: Tajima’s D combined with θπ and Fst combined with θπ ratios. First, a genome-wide sliding window calculation of θπ, Fst, and Tajima’s D (with a 100 kb window and a 10 kb step) was performed on landraces, cultivars, and the whole population, respectively. Second, the top 5% of the Tajima’s D and θπ windows for the whole population were selected. In addition, the top 5% of the Fst and θπ ratio windows for the landraces versus cultivars were also selected. Third, the selected windows using these two methods were merged to form the final selective sweep regions. ROH analyses for each accession were performed using PLINK [40] by setting the minimum ROH length to 300 kb.
GWAS and detection of SALs
The association analysis for each trait on each SNP with a MAF larger than 0.05 was performed using a single-locus MLM implemented in GEMMA [41] (which corrects for confounding effects due to the population structure and the relatedness matrix). The GWAS results were displayed using a Manhattan plot and a QQ-plot created with the R package CMplot [42]. A clump-based method implemented in PLINK [40] was used to reduce a false peak and detect real SALs. The P-value cutoff was set to 1 × 10−5 to first uncover significantly associated SNPs. Following this, for each significantly associated SNP, the region was regarded as a potential SAL if more than 10 SNPs within a 100 kb distance had P values smaller than 1 × 10−4. Finally, all overlapping SALs were merged to generate final SAL sets, and the SNP with the smallest P value in a SAL was defined as a peak.
Detection of SAGs
Usually, tens of genes are present in a SAL; hence, it is difficult to determine which genes are truly associated with traits. An improved functional mutation-based haplotype test method was used in this study for SAG discovery in SALs. As most variants within a gene are nonfunctional, the amino acid sequence and function of the gene do not change. Only a few variants have the potential to change the amino acid sequence of a gene, such as nonsynonymous SNPs, frameshift InDels, and variants in splicing sites, promoter regions, start codons, and stop codons. These combined functional mutations can only produce two or three different gene haplotypes. It is possible to test the relationship between gene haplotypes and traits. If they are significantly associated, then the gene is also most likely associated with the trait, which is how a SAG is defined. In this study, Welch’s test was used for a two-group haplotype test, and a Tukey’s test was used for a multiple-group haplotype test to detect SAGs. The functional annotation of SAGs was directly retrieved from SoyBase [43].
Network construction
Of all the genes located in the SALs, the most significant SAGs with P < 1 × 10−5, and their corresponding traits, were retained to build the phenotype–gene network for soybean. Protein–protein interaction information for soybean was retrieved from the STRING database [44] and mapped to the soybean genes using BLAST [45]. The construction, visualization, and exploration of the network were performed using Cytoscape [46].
Code availability
The bioinformatics analysis scripts used in this study can be downloaded through https://github.com/yjthu/GPB_250SoyReseq.
Data availability
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive [47] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: CRA002552), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa. The variation data are publicly accessible at the Genome Variation Map database (GVM: GVM000076), and are publicly accessible at https://ngdc.cncb.ac.cn/gvm [48].
CRediT author statement
Chunming Yang: Resources, Investigation, Validation. Jun Yan: Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Shuqin Jiang: Formal analysis. Xia Li: Investigation, Validation. Haowei Min: Conceptualization, Supervision, Formal analysis, Writing - review & editing. Xiangfeng Wang: Conceptualization, Supervision, Writing - review & editing. Dongyun Hao: Conceptualization, Supervision, Writing - review & editing. All authors have read and approved the final manuscript.
Acknowledgments
Competing interests
The authors declare no competing financial interests.
Acknowledgments
This study was supported by grants from the Agricultural Science and Technology Innovation Project, Jilin Province, China (Grant No. CXGC2017ZY027), the Program of Accurate Identification and Display of Soybean Germplasm, China [Grant Nos. NB08-2130315-(25-31)-06, NB07-2130315-(25-30)-06, NB06-070401-(22-27)-05, and NB2010-2130315-25-05], and the Chinese Universities Scientific Fund. We are grateful to Prof. Lijuan Qiu for her agreement of using the 250 soybean accessions from her laboratory at the China Academy of Agricultural Sciences. We appreciate Dr. Zhangxiong Liu of China Academy of Agricultural Sciences for the technical guidance in soybean phenotypic characterization. We also thank Yunshan Wei (Inner Mongolia Academy of Agriculture & Animal Husbandry Sciences, China) and Shuhong Wei and Qiang Wang (Heilongjiang Academy of Agricultural Sciences, China) for partial phenotypic characterization of the soybean population used in this study.
Handled by Peng Cui
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2021.02.009.
Contributor Information
Haowei Min, Email: biotrust_st@163.com.
Xiangfeng Wang, Email: xwang@cau.edu.cn.
Dongyun Hao, Email: dyhao@cjaas.com.
Supplementary material
The following are the Supplementary data to this article:
The density distribution of SNPs on chromosomes.
Allele frequency distribution between soybean subpopulations.
Selective sweep analysis for 250 soybean accessions. A. Selective sweep analysis by Tajima’ D combined with θπ. B. Selective sweep analysis by Fst combined with θπ ratios. Red dots present the top 5% of selected windows. C. Venn diagram of genes screened by two selective sweep analysis methods. D. Venn diagram of genes screened by two selective sweep analysis methods and ROH analysis.
Phenotype correlations between 50 soybean traits.
GWAS of pod height at bottom using MLM. A. Density distribution of pod height at bottom. B. Manhattan plots for pod height at bottom. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pod height at bottom. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of effective branch number using MLM. A. Density distribution of effective branch number. B. Manhattan plots for effective branch number. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for effective branch number. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pubescence density using MLM. A. Density distribution of pubescence density. B. Manhattan plots for pubescence density. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pubescence density. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of defollation using MLM. A. Density distribution of defollation. B. Manhattan plots for defollation. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for defollation. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of inflorenscence length using MLM. A. Density distribution of inflorenscence length. B. Manhattan plots for inflorenscence length. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for inflorenscence length. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leaf shape using MLM. A. Density distribution of leaf shape. B. Manhattan plots for leaf shape. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leaf shape. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leaflet size using MLM. A. Density distribution of leaflet size. B. Manhattan plots for leaflet size. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leaflet size. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of lodging using MLM. A. Density distribution of lodging. B. Manhattan plots for lodging. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for lodging. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of the number of nodes on the main stem using MLM. A. Density distribution of the number of nodes on the main stem. B. Manhattan plots for the number of nodes on the main stem. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for the number of nodes on the main stem. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of plant height using MLM. A. Density distribution of plant height. B. Manhattan plots for plant height. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for plant height. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of plant type using MLM. A. Density distribution of plant type. B. Manhattan plots for plant type. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for plant type. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of stem termination using MLM. A. Density distribution of podding habit. B. Manhattan plots for stem termination. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for stem termination. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed crack using MLM. A. Density distribution of seed crack. B. Manhattan plots for seed crack. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed crack. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of stem diameter using MLM. A. Density distribution of stem diameter. B. Manhattan plots for stem diameter. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for stem diameter. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of stem intension using MLM. A. Density distribution of stem intension. B. Manhattan plots for stem intension. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for stem intension. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pubescence color using MLM. A. Density distribution of pubescence color. B. Manhattan plots for pubescence color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pubescence color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of flower color using MLM. A. Density distribution of flower color. B. Manhattan plots for flower color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for flower color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leaf color using MLM. A. Density distribution of leaf color. B. Manhattan plots for leaf color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leaf color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of mature pod color using MLM. A. Density distribution of mature pod color. B. Manhattan plots for mature pod color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for mature pod color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed coat luster using MLM. A. Density distribution of seed coat luster. B. Manhattan plots for seed coat luster. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed coat luster. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of isoflavone content using MLM. A. Density distribution of isoflavone content. B. Manhattan plots for isoflavone content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for isoflavone content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of linoleic acid content using MLM. A. Density distribution of linoleic acid content. B. Manhattan plots for linoleic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for linoleic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of linolenic acid content using MLM. A. Density distribution of linolenic acid content. B. Manhattan plots for linolenic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for linolenic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of oleic acid content using MLM. A. Density distribution of oleic acid content. B. Manhattan plots for oleic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for oleic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of palmitic acid content using MLM. A. Density distribution of palmitic acid content. B. Manhattan plots for palmitic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for palmitic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of crude protein content using MLM. A. Density distribution of crude protein content. B. Manhattan plots for crude protein content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for crude protein content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of alanine content using MLM. A. Density distribution of alanine content. B. Manhattan plots for alanine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for alanine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of arginine content using MLM. A. Density distribution of arginine content. B. Manhattan plots for arginine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for arginine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of aspartic acid content using MLM. A. Density distribution of aspartic acid content. B. Manhattan plots for aspartic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for aspartic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of glutamate content using MLM. A. Density distribution of glutamate content. B. Manhattan plots for glutamate content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for glutamate content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of glycine content using MLM. A. Density distribution of glycine content. B. Manhattan plots for glycine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for glycine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of histidine content using MLM. A. Density distribution of histidine content. B. Manhattan plots for histidine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for histidine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of isoleucine content using MLM. A. Density distribution of isoleucine content. B. Manhattan plots for isoleucine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for isoleucine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leucine content using MLM. A. Density distribution of leucine content. B. Manhattan plots for leucine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leucine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of lysine content using MLM. A. Density distribution of lysine content. B. Manhattan plots for lysine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for lysine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of methionine content using MLM. A. Density distribution of methionine content. B. Manhattan plots for methionine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for methionine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of phenylalanine content using MLM. A. Density distribution of phenylalanine content. B. Manhattan plots for phenylalanine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for phenylalanine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of proline content using MLM. A. Density distribution of proline content. B. Manhattan plots for proline content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for proline content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of serine content using MLM. A. Density distribution of serine content. B. Manhattan plots for serine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for serine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of threonine content using MLM. A. Density distribution of threonine content. B. Manhattan plots for threonine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for threonine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of tyrosine content using MLM. A. Density distribution of tyrosine content. B. Manhattan plots for tyrosine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for tyrosine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of valine content using MLM. A. Density distribution of valine content. B. Manhattan plots for valine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for valine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of total amino acids content using MLM. A. Density distribution of total amino acids content. B. Manhattan plots for total amino acids content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for total amino acids content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of hundred-grain weight using MLM. A. Density distribution of one hundred seed weight. B. Manhattan plots for hundred-grain weight. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for hundred-grain weight. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pod number per plant using MLM. A. Density distribution of pod number per plant. B. Manhattan plots for pod number per plant. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pod number per plant. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pod size using MLM. A. Density distribution of pod size. B. Manhattan plots for pod size. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pod size. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed number per plant using MLM. A. Density distribution of seed number per plant. B. Manhattan plots for seed number per plant. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed number per plant. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed number per pod using MLM. A. Density distribution of seed number per pod. B. Manhattan plots for seed number per pod. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed number per pod. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed size using MLM. A. Density distribution of seed size. B. Manhattan plots for seed size. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed size. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed weight per plant using MLM. A. Density distribution of seed weight per plant. B. Manhattan plots for seed weight per plant. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed weight per plant. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
Gene expression validation of different haplotypes/phenotypes for five candidate genes. A. Phenotype distribution (left) and expression level (right) of GL3 for different haplotypes. B. Phenotype distribution (left) and expression level (right) of GSTL3 for different haplotypes. C. Phenotype distribution (left) and expression level (right) of GSTT1b for different haplotypes. D. Phenotype distribution (left) and expression level (right) of CKX3 for different haplotypes. E. Phenotype distribution (left) and expression level (right) of CYP85A2 for different haplotypes.
References
- 1.Sedivy E.J., Wu F., Hanzawa Y. Soybean domestication: the origin, genetic architecture and molecular bases. New Phytol. 2017;214:539–553. doi: 10.1111/nph.14418. [DOI] [PubMed] [Google Scholar]
- 2.Lam H.M., Xu X., Liu X., Chen W., Yang G., Wong F.L., et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Z., Jiang Y., Wang Z., Gou Z., Lyu J., Li W., et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol. 2015;33:408–414. doi: 10.1038/nbt.3096. [DOI] [PubMed] [Google Scholar]
- 4.Fang C., Ma Y., Wu S., Liu Z., Wang Z., Yang R., et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017;18:161. doi: 10.1186/s13059-017-1289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan J., Lv S., Hu M., Gao Z., He H., Ma Q., et al. Single-molecule sequencing assists genome assembly improvement and structural variation inference. Mol Plant. 2016;9:1085–1087. doi: 10.1016/j.molp.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Wang W., Mauleon R., Hu Z., Chebotarov D., Tai S., Wu Z., et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang N., Liu J., Gao Q., Gui S., Chen L., Yang L., et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat Genet. 2019;51:1052–1059. doi: 10.1038/s41588-019-0427-6. [DOI] [PubMed] [Google Scholar]
- 8.Schmutz J., Cannon S.B., Schlueter J., Ma J., Mitros T., Nelson W., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 10.Purfield D.C., McParland S., Wall E., Berry D.P. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS One. 2017;12:e0176780. doi: 10.1371/journal.pone.0176780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowell S., Korniliev P., Falcão A., Ismail A., Gregorio G., Mezey J., et al. Genome-wide association and high-resolution phenotyping link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nat Commun. 2016;7:10527. doi: 10.1038/ncomms10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yano K., Yamamoto E., Aya K., Takeuchi H., Lo P.C., Hu L., et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet. 2016;48:927–934. doi: 10.1038/ng.3596. [DOI] [PubMed] [Google Scholar]
- 13.Meng S., He J., Zhao T., Xing G., Li Y., Yang S., et al. Detecting the QTL-allele system of seed isoflavone content in Chinese soybean landrace population for optimal cross design and gene system exploration. Theor Appl Genet. 2016;129:1557–1576. doi: 10.1007/s00122-016-2724-0. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Wang X., Lu Y., Bhusal S.J., Song Q., Cregan P.B., et al. Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding. Mol Plant. 2018;11:460–472. doi: 10.1016/j.molp.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Messina M. A brief historical overview of the past two decades of soy and isoflavone research. J Nutr 2010;140:1350S–4S. [DOI] [PubMed]
- 16.Braidot E., Zancani M., Petrussa E., Peresson C., Bertolini A., Patui S., et al. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.) Plant Signal Behav. 2008;3:626–632. doi: 10.4161/psb.3.9.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W., Dubos C., Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Nie X., Tan J.L.H., Berger F. Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc Natl Acad Sci U S A. 2013;110:15479–15484. doi: 10.1073/pnas.1305175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 20.Kim T.W., Hwang J.Y., Kim Y.S., Joo S.H., Chang S.C., Lee J.S., et al. Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell. 2005;17:2397–2412. doi: 10.1105/tpc.105.033738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Bruyne L., Höfte M., De Vleesschauwer D. Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol Plant. 2014;7:943–959. doi: 10.1093/mp/ssu050. [DOI] [PubMed] [Google Scholar]
- 22.Kwon M., Fujioka S., Jeon J.H., Kim H.B., Takatsuto S., Yoshida S., et al. A double mutant for the CYP85A1 and CYP85A2 genes of Arabidopsis exhibits a brassinosteroid dwarf phenotype. J Plant Biol. 2005;48:237–244. [Google Scholar]
- 23.Northey J.G., Liang S., Jamshed M., Deb S., Foo E., Reid J.B., et al. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat Plants. 2016;2:16114. doi: 10.1038/nplants.2016.114. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Perez J.M., Serralbo O., Vanstraelen M., Gonzalez C., Criqui M.C., Genschik P., et al. Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C) Plant J. 2008;53:78–89. doi: 10.1111/j.1365-313X.2007.03312.x. [DOI] [PubMed] [Google Scholar]
- 25.Dar A.A., Choudhury A.R., Kancharla P.K., Arumugam N. The FAD2 gene in plants: occurrence, regulation, and role. Front Plant Sci. 2017;8:1789. doi: 10.3389/fpls.2017.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D., Sun L., Li S., Wang W., Ding Y., Swarm S.A., et al. Elevation of soybean seed oil content through selection for seed coat shininess. Nat Plants. 2018;4:30–35. doi: 10.1038/s41477-017-0084-7. [DOI] [PubMed] [Google Scholar]
- 27.Liu X.Q., Rong J.Y., Liu X.Y. Best linear unbiased prediction for linear combinations in general mixed linear models. J Multivariate Anal. 2008;99:1503–1517. [Google Scholar]
- 28.Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang C., Layer R.M., Faust G.G., Lindberg M.R., Rose D.B., Garrison E.P., et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods. 2015;12:966–968. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., Stecher G., Peterson D., Tamura K. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics. 2012;28:2685–2686. doi: 10.1093/bioinformatics/bts507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham G., Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014;9:e93766. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raj A., Stephens M., Pritchard J.K. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan J., Zou D., Li C., Zhang Z., Song S., Wang X. SR4R: an integrative SNP resource for genomic breeding and population research in rice. Genomics Proteomics Bioinformatics. 2020;18:173–185. doi: 10.1016/j.gpb.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin L., Zhang H., Tang Z., Xu J., Yin D., Zhang Z., et al. rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics Proteomics Bioinformatics. 2021;19:619–628. doi: 10.1016/j.gpb.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grant D., Nelson R.T., Cannon S.B., Shoemaker R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010;38:D843–D846. doi: 10.1093/nar/gkp798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., et al. The Genome Sequence Archive Family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021;19:578–583. doi: 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song S., Tian D., Li C., Tang B., Dong L., Xiao J., et al. Genome Variation Map: a data repository of genome variations in BIG Data Center. Nucleic Acids Res. 2018;46:D944–D949. doi: 10.1093/nar/gkx986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The density distribution of SNPs on chromosomes.
Allele frequency distribution between soybean subpopulations.
Selective sweep analysis for 250 soybean accessions. A. Selective sweep analysis by Tajima’ D combined with θπ. B. Selective sweep analysis by Fst combined with θπ ratios. Red dots present the top 5% of selected windows. C. Venn diagram of genes screened by two selective sweep analysis methods. D. Venn diagram of genes screened by two selective sweep analysis methods and ROH analysis.
Phenotype correlations between 50 soybean traits.
GWAS of pod height at bottom using MLM. A. Density distribution of pod height at bottom. B. Manhattan plots for pod height at bottom. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pod height at bottom. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of effective branch number using MLM. A. Density distribution of effective branch number. B. Manhattan plots for effective branch number. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for effective branch number. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pubescence density using MLM. A. Density distribution of pubescence density. B. Manhattan plots for pubescence density. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pubescence density. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of defollation using MLM. A. Density distribution of defollation. B. Manhattan plots for defollation. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for defollation. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of inflorenscence length using MLM. A. Density distribution of inflorenscence length. B. Manhattan plots for inflorenscence length. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for inflorenscence length. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leaf shape using MLM. A. Density distribution of leaf shape. B. Manhattan plots for leaf shape. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leaf shape. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leaflet size using MLM. A. Density distribution of leaflet size. B. Manhattan plots for leaflet size. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leaflet size. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of lodging using MLM. A. Density distribution of lodging. B. Manhattan plots for lodging. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for lodging. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of the number of nodes on the main stem using MLM. A. Density distribution of the number of nodes on the main stem. B. Manhattan plots for the number of nodes on the main stem. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for the number of nodes on the main stem. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of plant height using MLM. A. Density distribution of plant height. B. Manhattan plots for plant height. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for plant height. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of plant type using MLM. A. Density distribution of plant type. B. Manhattan plots for plant type. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for plant type. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of stem termination using MLM. A. Density distribution of podding habit. B. Manhattan plots for stem termination. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for stem termination. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed crack using MLM. A. Density distribution of seed crack. B. Manhattan plots for seed crack. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed crack. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of stem diameter using MLM. A. Density distribution of stem diameter. B. Manhattan plots for stem diameter. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for stem diameter. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of stem intension using MLM. A. Density distribution of stem intension. B. Manhattan plots for stem intension. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for stem intension. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pubescence color using MLM. A. Density distribution of pubescence color. B. Manhattan plots for pubescence color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pubescence color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of flower color using MLM. A. Density distribution of flower color. B. Manhattan plots for flower color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for flower color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leaf color using MLM. A. Density distribution of leaf color. B. Manhattan plots for leaf color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leaf color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of mature pod color using MLM. A. Density distribution of mature pod color. B. Manhattan plots for mature pod color. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for mature pod color. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed coat luster using MLM. A. Density distribution of seed coat luster. B. Manhattan plots for seed coat luster. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed coat luster. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of isoflavone content using MLM. A. Density distribution of isoflavone content. B. Manhattan plots for isoflavone content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for isoflavone content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of linoleic acid content using MLM. A. Density distribution of linoleic acid content. B. Manhattan plots for linoleic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for linoleic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of linolenic acid content using MLM. A. Density distribution of linolenic acid content. B. Manhattan plots for linolenic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for linolenic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of oleic acid content using MLM. A. Density distribution of oleic acid content. B. Manhattan plots for oleic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for oleic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of palmitic acid content using MLM. A. Density distribution of palmitic acid content. B. Manhattan plots for palmitic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for palmitic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of crude protein content using MLM. A. Density distribution of crude protein content. B. Manhattan plots for crude protein content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for crude protein content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of alanine content using MLM. A. Density distribution of alanine content. B. Manhattan plots for alanine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for alanine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of arginine content using MLM. A. Density distribution of arginine content. B. Manhattan plots for arginine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for arginine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of aspartic acid content using MLM. A. Density distribution of aspartic acid content. B. Manhattan plots for aspartic acid content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for aspartic acid content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of glutamate content using MLM. A. Density distribution of glutamate content. B. Manhattan plots for glutamate content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for glutamate content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of glycine content using MLM. A. Density distribution of glycine content. B. Manhattan plots for glycine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for glycine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of histidine content using MLM. A. Density distribution of histidine content. B. Manhattan plots for histidine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for histidine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of isoleucine content using MLM. A. Density distribution of isoleucine content. B. Manhattan plots for isoleucine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for isoleucine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of leucine content using MLM. A. Density distribution of leucine content. B. Manhattan plots for leucine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for leucine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of lysine content using MLM. A. Density distribution of lysine content. B. Manhattan plots for lysine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for lysine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of methionine content using MLM. A. Density distribution of methionine content. B. Manhattan plots for methionine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for methionine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of phenylalanine content using MLM. A. Density distribution of phenylalanine content. B. Manhattan plots for phenylalanine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for phenylalanine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of proline content using MLM. A. Density distribution of proline content. B. Manhattan plots for proline content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for proline content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of serine content using MLM. A. Density distribution of serine content. B. Manhattan plots for serine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for serine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of threonine content using MLM. A. Density distribution of threonine content. B. Manhattan plots for threonine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for threonine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of tyrosine content using MLM. A. Density distribution of tyrosine content. B. Manhattan plots for tyrosine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for tyrosine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of valine content using MLM. A. Density distribution of valine content. B. Manhattan plots for valine content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for valine content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of total amino acids content using MLM. A. Density distribution of total amino acids content. B. Manhattan plots for total amino acids content. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for total amino acids content. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of hundred-grain weight using MLM. A. Density distribution of one hundred seed weight. B. Manhattan plots for hundred-grain weight. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for hundred-grain weight. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pod number per plant using MLM. A. Density distribution of pod number per plant. B. Manhattan plots for pod number per plant. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pod number per plant. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of pod size using MLM. A. Density distribution of pod size. B. Manhattan plots for pod size. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for pod size. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed number per plant using MLM. A. Density distribution of seed number per plant. B. Manhattan plots for seed number per plant. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed number per plant. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed number per pod using MLM. A. Density distribution of seed number per pod. B. Manhattan plots for seed number per pod. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed number per pod. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed size using MLM. A. Density distribution of seed size. B. Manhattan plots for seed size. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed size. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
GWAS of seed weight per plant using MLM. A. Density distribution of seed weight per plant. B. Manhattan plots for seed weight per plant. Negative log10 P values from a genome-wide scan are plotted against SNP positions of 20 chromosomes. C. Quantile-quantile plot for seed weight per plant. The horizontal red line indicates the significant threshold (10−5). Trait-associated SNPs above the significant threshold are colored in red.
Gene expression validation of different haplotypes/phenotypes for five candidate genes. A. Phenotype distribution (left) and expression level (right) of GL3 for different haplotypes. B. Phenotype distribution (left) and expression level (right) of GSTL3 for different haplotypes. C. Phenotype distribution (left) and expression level (right) of GSTT1b for different haplotypes. D. Phenotype distribution (left) and expression level (right) of CKX3 for different haplotypes. E. Phenotype distribution (left) and expression level (right) of CYP85A2 for different haplotypes.
Data Availability Statement
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive [47] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: CRA002552), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa. The variation data are publicly accessible at the Genome Variation Map database (GVM: GVM000076), and are publicly accessible at https://ngdc.cncb.ac.cn/gvm [48].