Abstract
Glyphosate-based herbicides (GBHs) are massively used in agriculture. However, few studies have investigated the effects of glyphosate-based herbicides on avian species although they are largely exposed via their food. Here, we investigated the potential reversibility of the effects of chronic dietary exposure to glyphosate-based herbicides in broiler hens. For 42 days, we exposed 32-week-old hens to glyphosate-based herbicides via their food (47 mg/kg/day glyphosate equivalent, glyphosate-based herbicides, n = 75) corresponding to half glyphosate’s no-observed-adverse-effect-level in birds. We compared their performance to that of 75 control animals (CT). Both groups (glyphosate-based herbicides and control animals) were then fed for 28 additional days without glyphosate-based herbicides exposure (Ex-glyphosate-based herbicides and Ex-control animals). Glyphosate-based herbicides temporarily increased the plasma glyphosate and AMPA (aminomethylphosphonic acid) concentrations. Glyphosate and aminomethylphosphonic acid mostly accumulated in the liver and to a lesser extent in the leg muscle and abdominal adipose tissue. Glyphosate-based herbicides also temporarily increased the gizzard weight and plasma oxidative stress monitored by TBARS (thiobarbituric acid reactive substances). Glyphosate-based herbicides temporarily decreased the cecal concentrations of propionate, isobutyrate and propionate but acetate and valerate were durably reduced. The cecal microbiome was also durably affected since glyphosate-based herbicides inhibited Barnesiella and favored Alloprevotella. Body weight, fattening, food intake and feeding behavior as well as plasma lipid and uric acid were unaffected by glyphosate-based herbicides. Taken together, our results show possible disturbances of the cecal microbiota associated with plasma oxidative stress and accumulation of glyphosate in metabolic tissues in response to dietary glyphosate-based herbicides exposure in broiler hens. Luckily, glyphosate-based herbicides at this concentration does not hamper growth and most of the effects on the phenotypes are reversible.
Keywords: birds, glyphosate, metabolism, cecal microbiome, oxidative stress
Introduction
Glyphosate (Gly) is the most widely used herbicide in agriculture worldwide. It is a broad-spectrum herbicide with a generalized effect on all types of crops. Since it is a non-selective product, its commercialization is often coupled with that of genetically modified crops designed to resist the action of the herbicide, enabling farmers to spread increasingly large amounts on their fields without destroying their own crops (Martins-Gomes et al., 2022), while wild plants start to develop natural resistances to it. In animals and plants, Gly is metabolized into CO2 and aminomethylphosphonic acid (AMPA) by the enzyme glyphosate oxidoreductase (Mesnage et al., 2015). Gly’s herbicidal effect is due to disruption of the shikimate pathway, which produces aromatic amino acids in plants and in some microorganisms (Schönbrunn et al., 2001). Since humans and animals do not use the shikimate pathway to produce amino acids, Gly is not supposed to have any adverse effect on their health. However, several studies have shown that glyphosate-based herbicide (GBH) formulations can induce tissue damage (Jasper et al., 2012; Larsen et al., 2014), act as endocrine disruptors in various models (Walsh et al., 2000; Romano et al., 2010; Gill et al., 2018), and induce developmental issues in rats brain (Cattani et al., 2021). The use of GBHs is therefore very controversial, since scientific organizations have drawn contrasting conclusions about its dangerousness and recent studies have shown that populations are widely exposed to it (Grau et al., 2022).
Beside their controversial hypothetic effects on human health, GBHs are suspected to have ecotoxicological effects and to be part of the numerous factors leading to the current biodiversity crash. The fate of bird populations is of particular concern. European wild birds are going through a massive decline (Inger et al., 2015) and pesticides (with Gly as the prime representative) are suspected to be major actors of this loss. Gly residues are detectable in soil, water and food (Gill et al., 2018; Fogliatto et al., 2020) which allows them to threaten non-target species. Moreover, it is now established that, like for many pesticides, GBH’s toxicity is enhanced by (if not conditioned to) the presence of other components such as coformulants in herbicide formulations (Bradberry et al., 2004; Kim et al., 2013; Mesnage et al., 2019). Some of them are designed to reduce leaf surface tension and thus to allow penetration of the water-soluble Gly into the plant system first, and then into its cell’s membranes (Mesnage et al., 2019). It is therefore more relevant to study the toxicity of GBHs, rather than that of Gly alone. Few studies of this type have been conducted on poultry, while these animals are frequently exposed to GBHs through their diet. Most studies indeed show that GBHs administered in good farming practice conditions do not have any adverse effect on birds themselves, but rather on their foods and habitats (Gill et al., 2018). However, a recent study shows that GBHs can decrease liver catalase activity and reduce testosterone levels in Japanese quail. The gut microbiome is also disrupted, with a possible suppression of beneficial microorganisms (Ruuskanen et al., 2020). The exposure doses used in this study (12–20 mg Gly/kg body weight/day) being at least five times lower than the NOAEL (no-observed-adverse-effect level; 100 mg Gly/kg body weight/day) reported by the European Food Safety Authority (EFSA), more investigations are needed to characterize the impact of Gly and GBHs on avian models. The host gut microbiome is involved in many processes other than digestion such as xenobiotic detoxication, immune system homeostasis and vitamin synthesis (Samsel and Seneff, 2013). It has been shown that diet composition can induce changes in the gut microbiome bacterial community, and that these changes could have drastic impacts on the host’s biology (metabolism, immunity, behavior etc.) (Sommer and Bäckhed, 2013). Regarding GBHs, a 2013 study demonstrated a reduction of beneficial bacteria in the gastrointestinal tract microbiome after oral administration of GBHs in poultry, while highly pathogenic bacteria were found to be resistant (Shehata et al., 2013). They could therefore induce pathogenic dysbiosis in hens’ gut microbiome.
Thus, the aim of our study was to investigate the effects of a GBH-enriched diet on adult hens’ metabolism and on different biomolecular stress markers, using a dose equivalent to 47 mg Gly/kg body weight/day, half EFSA’s NOAEL, for 6 weeks. We also assessed the reversibility of the potential effects detected, by following the animals for 4 weeks after withdrawal of GBH from their diet. Meanwhile, Gly and AMPA were assayed in plasma and metabolic tissues (leg muscles, liver and abdominal adipose tissue). Detoxication processes were assayed by measuring mRNA expression of the enzymes cytochrome P450 (CYP) and GST (glutathione S-transferase) in animals’ liver, which is the main tissue of biotransformation. We also studied the impact of this diet on gut microbiome composition and diversity. To our knowledge, it is the first study evaluating the potential reversibility of changes in metabolism-related parameters after chronic dietary exposure to a GBH in broiler hens.
Materials and methods
Ethical issues
All experimental procedures were performed in accordance with the French National Guidelines for the care and use of animals for research purposes (certificate of authorization to experiment on living animals APAFIS number 21549–2019071809504554v3, approval date: 6 November 2021, Ministry of Agriculture and Fish Products, and a notice of the ethics committee of Val de Loire No. 19).
Animals
All animals (150 female chicks of the commercial breed ROSS 308) were obtained at 1 day of age from a local hatchery (Boye Accouvage La Villonniere 79,310 La Boissière en Gatine, France) and reared at “Pôle Expérimental Avicole de Tours” (INRAE, Nouzilly, France) according to traditional breeding conditions. In our experiment, all 150 hens (32 weeks old) were used. They were divided into groups of five birds in 30 pens, each pen with an area of 3 m2. The design of the experiment is summarized in Figure 1. The timeline is represented in days. Seventy-five hens (15 pens) were exposed for 42 days to GBH via their food (GBH hens, a GBH dose equivalent to 47 mg Gly/kg body weight/day), and 75 hens (15 pens) were fed with a regular diet without GBH (control animals, CT) (day 0 to day 42 of the protocol). After that, all animals were fed with a regular diet (day 43 to day 70 of the protocol, Ex-GBH and Ex-CT hens). During this protocol, blood samples were collected from hens to quantify Gly and its metabolite AMPA within their blood plasma. At day 42, exposure to GBH was stopped, and nine CT hens and nine GBH hens were slaughtered to recover biological samples. At day 70, 12 Ex-CT hens and 12 Ex-GBH hens were slaughtered to recover biological samples. All animals were killed by electrical stunning and bled out, as recommended by the ethical committee.
FIGURE 1.
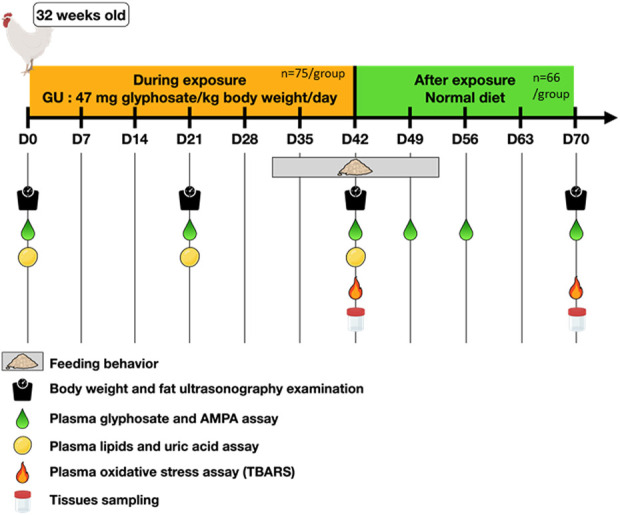
Experimental design applied to hens. Hens (32 weeks old; n = 75) were dietary exposed to GBH (Glyphosate-Based Herbicide, corresponding to a dose of 47 mg glyphosate/kg body weight/day, group named GBH) for 42 days straight, then they received a normal diet (not exposed to GBH, same diet, group named ex GBH) for the 28 following days. Their respective controls (CT, n = 75 and Ex-CT, n = 66) received a normal diet for the whole time. During these 70 days, hens were regularly weighed and ultrascanned to determine fat thickness, their feeding behavior was assayed by counting pecks at the diet and their blood was sampled for glyphosate, AMPA, lipids, uric acid and oxidative stress assays. Nine and twelve animals of both groups were euthanized at D42 and D70, respectively, for tissue sampling.
Diet composition
Hens (32 weeks old) received a restricted laying diet according to Hendrix Genetics recommendation. The composition of the diet is shown in Supplemental Table 1. For the GBH animals, this diet was mixed with Gallup super 360 in our laboratory in accordance with the directives of the Directions Départementales de la Protection des Populations (Departmental Directorate for the Protection of Populations). Mixing was carried out by a technician with “Certiphyto” certification for the handling of phytosanitary products, as recommended by French law. Gallup super 360, named GBH within the text, was obtained from Axereal (Monnaie, France); it contained 360 g/L Gly (485.8 g/L isopropylamine salt). Animals were fed with either feed containing GBH (n = 75) or control feed (n = 75) from the first week of the protocol to week 6 (Figure 1). The control feed contained low measurable Gly and AMPA concentrations (0.21 mg/kg feed for Gly and undetectable levels for AMPA as determined by Phytocontrol, Nimes, France). The GBH feed contained 1,250 mg/kg feed of Gly and 0.30 mg/kg feed of AMPA, as determined by Phytocontrol. Hens were food-restricted as recommended by the provider, and their food consumption was 200 g/day. Thus, the concentration in the feed corresponded to a dose of 47 mg Gly equivalent/kg body weight/day. From day 43 to day 70, all hens were fed with control feed (Ex-GBH n = 63 and Ex-CT n = 63).
TABLE 1.
Oligonucleotide primer sequences used for RT-qPCR.
| Tissue | Gene | Primer F | Primer R | References |
|---|---|---|---|---|
| GAPDH | ACGGATTTGGTCGTATTGGG | TGATTTTGGAGGATCTCGC | Grandhaye et al. (2021) | |
| EEF1α | AGCAGACTTTGTGACCTTGCC | TCACATGAGACAGACGGTTGC | ||
| β-actin | ACGGAACCACAGTTTATCATC | GTCCCAGTCTTCAACTATACC | ||
| Liver | CYP1A1 | AATGCTCGTTTCAGTGCCTTC | CCTCCCCTGTCCTTTTCTCC | Cong et al. (2019) |
| CYP1A2 | AACCCAGAGCGTTTCCTCAA | CTCCCACTTGCCTATGTTTTCC | ||
| CYP2C | CAAAATGGAACAGGAGAAAGAGAAC | CCCGCAAGGAACAAGTCAA | ||
| CYP3A | CCAAGCTATGCTCTTCACCG | TCAGGCTCCACTTACGGTCT | ||
| CYP2A6 | CTGCAGAGAATGGCATGAAG | CCTGCAAGACTGCAAGGAA | Wang et al. (2018) | |
| EPHX1 | GAAGATGTCAGGCGGATGTT | CAGGAGAGTCATTCAAACCACA | ||
| GSTA3 | AGACCAGAGCCATCCTCAAC | TGCCAGTCCTTCCACATACA | ||
| GSTA4 | GCTACATCGCAGGGAAATACA | TGGAGAGAAAGGAAACACCAA | ||
| FXR | AAAGCCGTTCTGTGCGTT | GGATTGGTGGGGTTCCTG | Dai et al. (2020) | |
| CYP3A37 | AAATCAGACAGCAATGGGAGC | GGTAAGCCAGGTAACCAAGTGT | ||
| BSEP | TGCAAAGCAAAGGAGACT | GCAATGGATAATGGAGGG | ||
| CYP1A5 | CTCTGCTCTGTTCDCAAAGCGTCTC | GCTCGCCTGCDCACCDCATCDCACT | ||
| CYP1A4 | CDCAGGACGGAGGCTGACDCAAGG | GCDCAGGATGGTGGTGAGGAAGA | ||
| Ces1 | TGACCATTCAATATCGCC | ACACTTTCTCCTCCCGCT | ||
| SLC O 1B3 | CAGGACTCTCGCTGGGTGG | TGGCTTTCAGGGGCTTTTT | ||
| CYP2H1 | TCATCCACGAAATCCAAAG | GATGGGAGACAGCAAAGG | ||
| CYP2H2 | GGCCCGGATGGAGCTATT | TTGCCGCCGAGGTGACTA | ||
| Gizzard | Hsp27 | ACACGAGGAGAAACAGGATGAG | ACTGGATGGCTGGCTTGG | Zhao et al. (2016) |
| Hsp40 | GGGCATTCAACAGCATAGA | TTCACATCCCCAAGTTTAGG | ||
| Hsp60 | AGCCAAAGGGCAGAAATG | TACAGCAACAACCTGAAGACC | ||
| Hsp70 | CGGGCAAGTTTGACCTAA | TTGGCTCCCACCCTATCTCT | ||
| Hsp90 | TCCTGTCCTGGCTTTAGTTT | AGGTGGCATCTCCTCGGT | ||
| NF-κB | TCAACGCAGGACCTAAAGACAT | GCAGATAGCCAAGTTCAGGATG | Xing et al. (2015) | |
| iNOS | CCTGGAGGTCCTGGAAGAGT | CCTGGGTTTCAGAAGTGGC | ||
| COX-2 | TGTCCTTTCACTGCTTTCCAT | TTCCATTGCTGTGTTTGAGGT | ||
| PTGES | GTTCCTGTCATTCGCCTTCTAC | CGCATCCTCTGGGTTAGCA | ||
| TNF-α | GCCCTTCCTGTAACCAGATG | ACACGACAGCCAAGTCAACG | ||
| Proventriculus | PGA5 | TCCGTCTACCTGAGCAAGGAT | AAGCAGGCGACGTACTTGTT | Al-Khalaifah et al. (2020) |
| PGC | ATCGGGATTGAGGACTTCGC | TGAAGACCTGGTTGGGAACG | ||
| Caecum | Chemerin | CGCGTGGTGAAGGATGTG | CGACTGCTCCCTAAAGAGGAACT | Estienne et al. (2020) |
| CMKLR1 | CGGTCAACGCCATTTGGT | GGGTAGGAAGATGTTGAAGGAA | ||
| IgA | GTCACCGTCACCTGGACTACA | ACCGATGGTCTCCTTCACATC | Grandhaye et al. (2020) | |
| IFNα | CAACGACACCATCCTGGACA | GGGCTGCTGAGGATTTTGAA | Garrido et al. (2018) | |
| IFNβ | TCCTGCAACCATCTTCGTCA | CACGTCTTGTTGTGGGCAAG | ||
| IL-1β | AGGCTCAACATTGCGCTGTA | CTTGTAGCCCTTGATGCCCA | Grandhaye et al. (2020) | |
| IL-6 | GCTTCGACGAGGAGAAATGC | GCCAGGTGCTTTGTGCTGTA | Garrido et al. (2018) | |
| IL-8 | CTGCGGTGCCAGTGCATTAG | AGCACACCTCTCTTCCATCC | ||
| Spleen | IFNα | CAACGACACCATCCTGGACA | GGGCTGCTGAGGATTTTGAA | Garrido et al. (2018) |
| IFNβ | TCCTGCAACCATCTTCGTCA | CACGTCTTGTTGTGGGCAAG | ||
| IL-1β | AGGCTCAACATTGCGCTGTA | CTTGTAGCCCTTGATGCCCA | Grandhaye et al. (2020) | |
| IL-8 | CTGCGGTGCCAGTGCATTAG | AGCACACCTCTCTTCCATCC | Garrido et al. (2018) | |
| IL-22 | TGTTGTTGCTGTTTCCCTCTTC | CACCCCTGTCCCTTTTGGA |
Count of pecks at the diet
Feeding behavior was quantified by counting pecks at the diet over the daily diet distribution. This count was carried out in three hens per pen during the 8 secondes on D31, D32, D35, D36 (during GBH exposure) and D49, D50, D51 and D52 (after exposure).
Biological samples
Blood samples from 10 hens were collected from the occipital sinus into heparin tubes on different days during the experiment (days 0, 21 and 42 during GBH exposure and days 49, 56 and 70 after exposure). Blood samples were centrifuged (5,000 × g for 10 min at 4°C) and stored at −20°C before use for Gly and AMPA assays (Phytocontrol, Nimes, France). Tissue samples were obtained at different points in the experiment [day 42 of the protocol during GBH exposure (n = 9 GBH and n = 9 CT) and day 70 after GBH exposure (n = 12 Ex-GBH and n = 12 Ex-CT)] by dissection after slaughtering.
Gly and aminomethylphosphonic acid assays in hen plasma
Gly and AMPA concentrations were measured in the blood plasma of hens after a derivatization reaction using FMOC-Cl (9-fluorenylmethyl chloroformate), in collaboration with Dr S El Balkhi (Service de Pharmacologie, Toxicologie et Pharmacovigilance, Limoges, France) as previously described (Serra et al., 2021).
Lipid and uric acid assays in hen plasma
Plasma concentrations of triglycerides, uric acid, phospholipids and cholesterol were determined by enzymatic assay using specific kits from Biolabo SAS (Maizy, France): triglycerides (reference: LP80519), uric acid (reference: 80,351), phospholipids (reference: 99,105) and cholesterol (reference: 80,106, Biolabo SAS, Maizy, France). The measurements were performed according to the manufacturer’s protocol. For all these assays, the inter- and intra-assay coefficient variations were <15%.
Body weight gain and measurement of tissue index
Chickens were individually weighed on days 0, 21, 42 and 70. Body weight was recorded and, based on the differences, the body weight gain per day was calculated [(final body weight − initial body weight)/number of days)] for each period D0–D21, D21–D42 and D42–D70 (Figure 1). The weight of organs (liver, spleen, heart, kidney, brain, gizzard) and abdominal adipose tissue (AAT) collected on the 42nd and 70th days of the protocol was determined and the weight of the organs or AAT as a percentage of the body weight was calculated and an organ/tissues index was shown as described by Pandey et al. (2019).
Plasma thiobarbituric acid reactive substances (TBARS) assay
Lipid peroxidation, as determined through measuring the amount of MDA (malondialdehyde) that reacts with 2-thiobarbituric acid, was used to estimate oxidative stress (Armstrong and Browne, 1994). Blood samples were collected into EDTA-treated tubes, then gently shaken and kept and handled on wet ice. The plasma was separated by centrifuging the blood samples at 1,000 × g for 10 min at 4°C, then transferred to 1.5 ml microcentrifuge tubes and stored at −80°C. The TBARS values of the EDTA-treated plasma were measured using the modified method of Grotto et al. (2007). A standard curve for 1,1,3,3-tetramethoxypropane was used, and the concentration was expressed as nmol MDA/mL solution.
Measurement of liver ATP concentration
Liver total proteins from 9 CT, 9 GBH, 12 Ex-CT and Ex-12 GBH hens were extracted using lysis buffer (1 M Tris (pH 7.4), 0.15 M NaCl, 1.3 mM EDTA, 1 mM EGTA, 43–23 mM VO, 0.1 M NaF, 1% NH2PO4, 0.5% Triton) and an Ultra-Turrax instrument for grinding, according to the manufacturer’s recommendations (Invitrogen by Life Technologies, Villebon-sur-Yvette, France). Lysates were centrifuged for 20 min at 16,000 × g and 4°C, and the supernatants collected. Lysate protein concentrations were then measured using the bicinchoninic acid (BCA) protein assay (Interchim, Montluçon, France). The ATP assay was performed using a Promega CellTiter® Luminescent Cell Viability Assay. Briefly, the assay buffer and the substrate were equilibrated to room temperature, then the buffer was transferred to the substrate and gently mixed with it to obtain a homogeneous solution. After a 30 min equilibration of the cell plate to room temperature, protein lysates (100 µL) were put into a 96-well plate and CellTiter-Glo reagent (100 µL) was added to each well. The plate was orbitally mixed for 2 min and incubated at room temperature for 10 min. The ATP concentration was then measured using a luminometer. Luminescence at the integration time 1,000 (ms) was read using an Ascent Luminoskan Luminometer (Thermo Scientific, Illkirch, France). Lysates’ ATP concentration was normalized with the previously determined total protein concentration.
Measurement of the gene expression in tissues
Total RNA from 9 CT, 9 GBH, 12 Ex-CT and 12 Ex-GBH hens was extracted from hens’ liver, gizzard, proventriculus, cecum and spleen using TRIzol RNA Isolation Reagents and an Ultra-Turrax instrument for grinding, according to the manufacturer’s recommendations (Invitrogen by Life Technologies, Villebon-sur-Yvette, France). The purity and concentrations of the obtained RNA were checked via their A260/A280 ratios using a Nanodrop machine. cDNA was obtained by reverse transcription of 2 µg of the total RNA in 20 µL of a mix containing each deoxyribonucleotide triphosphate (dATP, dTTP, dGTP, dCTP; 0.5 mM), 2 M RT Buffer, 15 μg/μL oligodT, 0.125 U of ribonuclease inhibitor and 0.05 U of Moloney murine leukemia virus reverse transcriptase (MMLV); the mixture was kept for 1 h at 37°C. Quantitative PCR was performed using a mix of 3 µL of cDNA and 8 µL of SYBR Green Supermix 1X Reagent (Bio-Rad, Marnes-la-Coquette, France) with 250 nM of specific primers (Invitrogen by Life Technologies, Villebon-sur-Yvette, France) given in Table 1. Samples were set up in duplicate in a 384-well plate and a MyiQ Cycle Device (Bio-Rad, Marnes-la-Coquette, France) was used to apply the following procedure: incubation (2 min at 50°C), denaturation (10 min at 95°C) and 40 PCR cycles (30 s at 95°C, 30 s at 60°C, 30 s at 72°C). Relative expression of genes was related to the geometric mean of the expression of three reference genes (GAPDH (glyceraldehyde-3-phosphate dehydrogenase), ACTB (actin B) and EEF1α (eukaryotic elongation factor 1 alpha)). For each target gene, expression was calculated according to primer efficiency (E) and quantification cycle (Cq), where expression = E − Cq. Then, relative expression of the target gene to the three reference genes was analyzed.
Nuclear magnetic resonance (NMR) metabolomics
1H NMR spectra for the metabolic fingerprinting of plasma samples were obtained at 300 K on a Bruker Avance III HD 600 MHz NMR spectrometer (Bruker Biospin, Rheinstetten, Germany) operating at 600.13 MHz for 1H resonance frequency using an inverse detection 5 mm 1H-13C-15N-31P cryoprobe attached to a cryoplatform (the pre-amplifier cooling unit). Plasma samples were prepared, and the analyses performed as previously described (Chamorro-García et al., 2021), with slight modifications. For all spectra, a total of 128 transients were collected into 65,536 data points using a spectral width of 20 ppm, a relaxation delay of 5 s and an acquisition time of 2.72 s. All free induction decays were then multiplied by an exponential function with a line broadening factor of 0.3 Hz prior to Fourier transform. All spectra were manually phase- and baseline-corrected, and referenced to the chemical shift of glucose (δ 5.24 ppm).
Analysis of short-chain fatty acids (SCFAs) in cecal samples
Approximately 35 mg of cecal content was weighed, snap-frozen and stored at −80°C until analysis. The samples were extracted with water and proteins precipitated with phosphotungstic acid. 2-Ethylbutyrate was added to supernatants at a ratio of 1: 4 as an internal standard. The SCFA content was determined from a 0.3 µL volume of supernatant by gas chromatography (Agilent 7890B gas chromatograph, Agilent Technologies, Les Ulis, France) equipped with a split-splitless injector, a flame-ionization detector and a fused silica capillary column (15 m × 0.53 mm × 0.5 µm; Supelco, Saint-Quentin-Fallavier, France). The carrier gas (H2) flow rate was 10 ml/min. The oven temperature was initially set at 100°C for 10 min, then increased from 100 to 180°C at a rate of 20 C/min and held for 2 min. The detector temperature was 240°C. Samples were analyzed in duplicate. The peaks obtained were integrated using OpenLAB Chemstation software (Agilent Technologies, Les Ulis, France).
Microbiome analysis
DNA from bacteria was extracted from cecal content [CT (n = 5), GBH (n = 9), Ex-GBH (n = 10), Ex-CT (n = 5)] using a G’NOME DNA isolation kit (MP Biomedicals, Strasbourg, France) (Furet et al., 2009). The V3–V4 region of the 16S rRNA genes was amplified using MolTaq (Molzym, Plaisir, France), 50 ng DNA and the primers V3F: TACGGRAGGCAGCAG and V4R: ATCTTACCAGGGTATCTAATCCT (Kozich et al., 2013). Purified amplicons were sequenced using MiSeq sequencing technology (Illumina) on the GeT-PLaGe platform (Toulouse, France). The sequences were submitted to the Short-Read Archive with accession number PRJNA741111. Paired-end reads obtained from MiSeq sequencing were analyzed using the Galaxy-supported pipeline named FROGS (Find, Rapidly, OTUs (Operational Taxonomic Units) with Galaxy Solution) (Escudié et al., 2018). For the preprocessing, reads with a length ≥380 bp were kept. The clustering and chimera removal tools followed the guidelines of FROGS (Escudié et al., 2018). Assignation was performed using SILVA138 16S pintail100. OTUs with abundance lower than 0.005% were removed from the analysis (Bokulich et al., 2013).
Statistical analysis
GraphPad Prism® software (version 6) was used for all analyses (except for microbiome and NMR metabolomic analysis). All data are reported as means ± standard error of mean (SEM). Outliers were identified by the Rout method and removed. We performed t-tests to compare means at D42 and D70, or two-way ANOVA followed by Tukey’s HSD tests. Regarding gut microbiome analysis, all statistical analyses were performed using R software (version 4.2.0; R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.). The microbiota composition analysis was performed using the mia package (Ernst et al., 2022). For the variability within bacterial communities ($\alpha$ diversity), Chao, Shannon and Faith indices were computed. The effect of exposure to GBH on $\alpha$ diversity was investigated using Kruskal–Wallis tests followed by Dunn’s post-hoc tests and was considered significant if p < 0.05. For the differences between bacterial communities ($\beta$ diversity), the Bray–Curtis matrix and Unifrac distance were computed and then visualized with non-metric multidimensional scaling (NMDS) plots.
For the differential analysis, OTUs with a prevalence below 0.05% were filtered out as well as those with a low number of reads (OTUs with sum counts of less than 0.05% of the sum of all counts). Then, OTU counts were agglomerated at the genus level and relative abundances were computed at each taxonomic level. As recommended by Nearing et al. (2022), three differential analysis methods were used (ALDEx2 (ANOVA-like differential expression 2) (Fernandes et al., 2014), ANCOM-BC (analysis of compositions of microbiomes with bias correction (Lin and Peddada, 2020) and DESeq2 (differential gene expression analysis based on the negative binomial distribution (Love et al., 2014)) and we focused on the common results. All p-values were adjusted with the Benjamini–Hochberg correction. A difference is significant if the adjusted p-value is below 0.05 and if the size effect is above 1 for ALDEx2 or if the logFC (log fold change) is above 2 for DESeq2. Finally, sparse partial least-squares discriminant analysis (sPLS-DA) was computed on the centered log ratio (CLR)-transformed data using the mixOmics package (Lê Cao et al., 2016; Rohart et al., 2017). The choice of optimal values for the sparsity parameters and the evaluation of classification was performed using a 5-fold cross-validation and 100 repeats.
All analyses were first performed considering all four groups (CT, GBH, Ex-CT and Ex-GBH) separately. Because no significant difference was detected between CT and Ex-CT groups, they were combined and renamed CT + Ex-CT for the microbiome analysis.
Results
Chronic dietary glyphosate-based herbicide exposure did not change chickens’ feeding behavior
One parameter of the hens’ feeding behavior was estimated by counting the number of times hens pecked at the diet during and after GBH exposure. As shown in Figure 2, no significant difference was detected between GBH and CT hens and between Ex-GBH and Ex-CT hens. All hens made significantly fewer pecks at the diet after exposure than during GBH exposure (p < 0.001).
FIGURE 2.
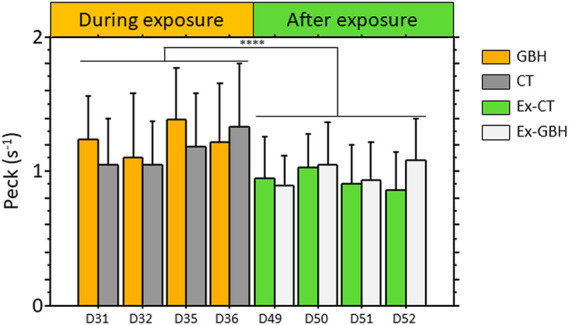
Frequency of pecks for feeding made by control hens (CT, n = 45) and those exposed to GBH (GBH, n = 45) at the manger during (D31, D32, D35 and D36) and after (D49, D50, D51 and D52; Ex-CT, n = 45; Ex-GBH, n = 45) GBH exposure. Results are presented as means ± SEM. ****p ≤ 0.0001 CT: Control, GBH: Glyphosate-Based Herbicide, Ex-CT: Ex-Control, Ex-GBH.
Chronic dietary glyphosate-based herbicide exposure did not affect chickens’ weight gain but increased gizzard weight
Weight gain determined at D0–D21, D21–D42 and D42–D70 is presented in Figure 3A. No significant difference was detected, either during (GBH vs. CT hens) or after GBH exposure (Ex-GBH vs. Ex-CT animals). In addition, no significant effect of GBH on chickens’ fat thickness was detected, either during or after GBH exposure (Figure 3B). Liver, spleen, heart, abdominal adipose tissue (AAT), kidney, brain and gizzard indices are presented in Table 2. During GBH exposure (D42), no significant effect was detected except for gizzards, which were significantly heavier in GBH as compared to CT hens (p < 0.05). Furthermore, after exposure (D70), spleens and kidneys were heavier and lighter (p < 0.05), respectively, in Ex-GBH hens compared to Ex-CT hens.
FIGURE 3.
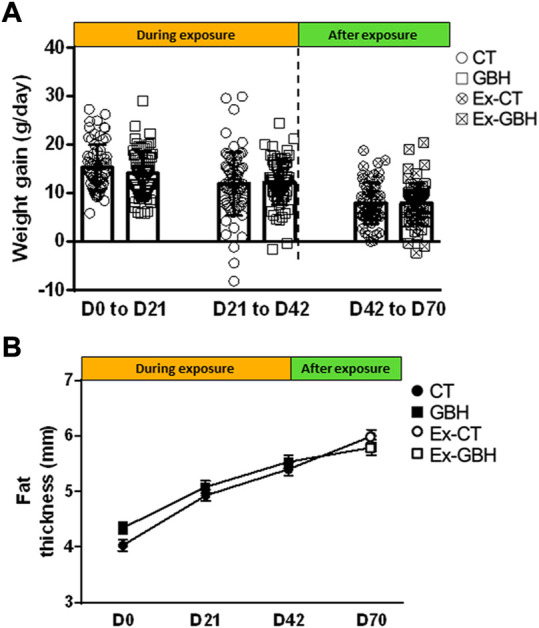
Body weight gained (A) and fat thickness (B) during dietary GBH exposure (D0 to D42; CT, n = 75; GBH, n = 75) and after (D42 to D70; Ex-CT, n = 66; Ex-GBH, n = 66) dietary GBH exposure. Results are presented as means ± SEM (A) and as means (B). CT: Control, GBH: Glyphosate-Based Herbicide, Ex-CT: Ex-Control, Ex-GBH.
TABLE 2.
Hens organ index [Tissue weight (g)×100)/(Body weight (g)] in GBH exposed and control animals during the period of exposure (42nd day of protocol, D42, CT n = 9, GBH n = 9) and after GBH exposure (70th day of protocol, D70, Ex-CT, n = 12, Ex-GBH, n = 12).
| D42 | p Value | D70 | p Value | |||
|---|---|---|---|---|---|---|
| CT | GBH | Ex-CT | Ex-GBH | |||
| Liver | 1.59 | 1.68 | 0.502 | 1.65 | 1.64 | 0.930 |
| Spleen | 0.069 | 0.062 | 0.318 | 0.059 | 0.075* | 0.015 |
| Heart | 0.360 | 0.336 | 0.367 | 0.359 | 0.375 | 0.427 |
| AAT | 2.04 | 2.12 | 0.734 | 2.30 | 2.42 | 0.148 |
| Kidney | 0.154 | 0.151 | 0.902 | 0.377 | 0.324 | 0.064 |
| Brain | 0.092 | 0.097 | 0.097 | 0.091 | 0.088 | 0.667 |
| Gizzard | 0.737 | 0.888*** | 0.0003 | 0.812 | 0.842 | 0.408 |
CT, control; GBH, Ex-CT, Ex-Control; Ex-GBH, animals that have been GBH, exposed for 42 days then non-exposed until D70, Ex-AAT, abdominal adipose tissue, Index = (Tissue weight (g)×100)/(Body weight (g)). Results are presented as means. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. CT, control; GBH, Ex-CT, Ex-Control, Ex-GBH.
Gly and aminomethylphosphonic acid accumulated in plasma and tissues after chronic dietary glyphosate-based herbicide exposure in chickens
Plasma Gly and AMPA concentrations were assayed during GBH exposure and normal diet. The results are presented in Table 3. Plasma Gly and AMPA concentrations reached a peak between D0 and D21 before progressively decreasing to values approximatively twice as low at D42 (1.7- and 1.9-fold decrease, respectively) in GBH animals. Concentrations continued to decrease after GBH exposure. Gly and AMPA accumulations in tissues are presented in Figure 4. Both molecules were found in liver (Gly: 8.10 mg/kg and AMPA: 2.40 mg/kg), AAT (Gly: 2.00 mg/kg and AMPA: 0.13 mg/kg) and leg muscle (Gly: 0.86 mg/kg and AMPA: 0.09 mg/kg) in GBH hens at D42 and were observed in smaller amounts in Ex-GBH at D70 (after GBH exposure): 3.11 and 0.74 mg/kg, 0.80 and 0.04 mg/kg and 0.09 and 0.07 mg/kg in liver, AAT and leg muscle, respectively. In CT and Ex-CT animals, Gly and AMPA were almost undetectable in the three tissues at D42 and D70.
TABLE 3.
Glyphosate and AMPA concentrations in hen’s plasma during (n = 10) and after (n = 10) the dietary GBH exposure period.
| Time (day) | (Glyphosate] (ng/ml) | (AMPA] (ng/ml) | |
|---|---|---|---|
| During exposure | D0 | 5.37 ± 0.60a | 0 ± 0a |
| D21 | 1,549.02 ± 89.37d | 19.82 ± 1.75d | |
| D42 | 910.59 ± 117.29c | 10.15 ± 0.61c | |
| After exposure | D49 | 297.54 ± 7.96b | 6.24 ± 0.26b |
| D56 | 160.99 ± 9.12ab | 4.45 ± 1.03ab | |
| D70 | 67.21 ± 6.87ab | 1.59 ± 0.52a |
Results are presented as means ± SEM; letters indicate significant differences detected by Two-way ANOVA, and Tukey HSD’s test for pair-wise comparisons (p < 0.05).
FIGURE 4.
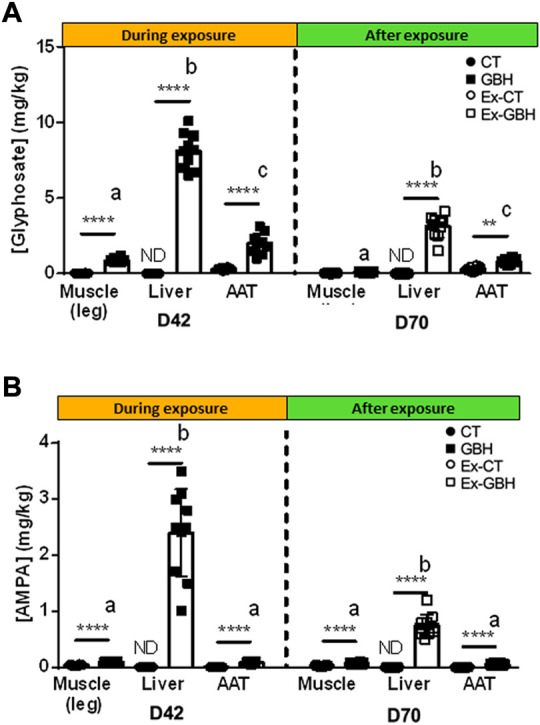
Glyphosate (A) and AMPA (B) concentrations in hens’ leg muscles, liver and abdominal adipose tissue (AAT) during (D42) and after (D70) GBH exposure. Results are presented as means ± SEM. p-values express mean differences between CT (n = 10) and GBH (n = 10) groups, and Ex-CT (n = 10) and Ex-GBH (n = 10); **p ≤ 0.01, ****p ≤ 0.0001; letters indicate significant differences detected by two-way ANOVA (p < 0.05) followed by Tukey’s HSD test for comparing mean tissue concentrations at D42 and D70 separately. CT: control, GBH: Glyphosate-Based Herbicide, Ex-CT: Ex-Control, Ex-GBH.
Chronic dietary glyphosate-based herbicide exposure reversibly increased plasma oxidative stress in chickens
Plasma oxidative stress was measured using the TBARS index, which quantifies the MDA concentration in plasma. The results are presented in Figure 5. At D42, the TBARS index was significantly (p < 0.05) higher in GBH hens than in CT hens. No significant difference was observed between Ex-GBH and Ex-CT hens after GBH exposure at D70.
FIGURE 5.
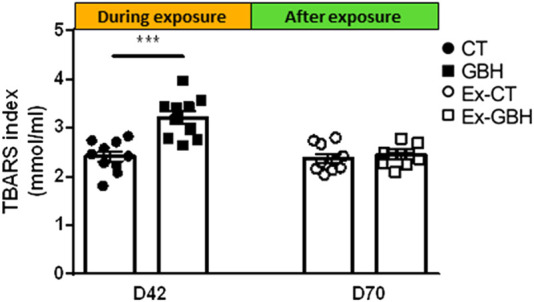
TBARS index in hens’ plasma during (D42; CT, n = 10 and GBH, n = 10) and after (D70; Ex-CT, n = 10 and Ex-GBH, n = 10) GBH exposure. Results are presented as means ± SEM. ***p ≤ 0.001. CT: Control, GBH: Glyphosate-Based Herbicide, Ex-CT: Ex-Control, Ex-GBH.
Chronic dietary glyphosate-based herbicide exposure did not modulate plasma lipids and uric acid concentrations
Plasma triglyceride, cholesterol, phospholipid and uric acid concentrations were assayed at D0, D21 and D44 (Supplemental Figure S1A–D). No significant effect of GBH exposure on any of these parameters was detected.
Chronic dietary glyphosate-based herbicide exposure did not trigger biotransformation enzyme transcription in liver but enhanced IgA transcript levels in chickens
The mRNA expression of liver biotransformation enzymes was measured by RT-qPCR. The results are presented in Table 4. No significant effect was detected on any of the transcripts of which we measured the level, either during exposure (CT vs. GBH hens) or after exposure (Ex-CT vs. Ex-GBH). In the cecum, expression of the immunoglobulin A gene (IgA) was significantly (p < 0.05) enhanced by GBH treatment after exposure in Ex-GBH hens as compared to Ex-CT hens whereas no significant effect was detected during exposure (CT vs. GBH hens). We next determined the expression levels of genes involved in the inflammatory and immune responses. No significant effect was detected for chemerin, CMKLR1, IFNα, IFNβ, IL-1β, IL-6 and IL-8 (Table 4). mRNA expression of stress-related genes in the gizzard was also measured and is shown in Table 4 iNOS expression was significantly (p < 0.05) higher in the Ex-GBH group than in the Ex-CT group after exposure and that of HSP70 was significantly (p < 0.05) higher in the Ex-CT group than in the Ex-GBH group after exposure. No significant effect on any of these genes was detected during exposure (CT vs. GBH hens). No significant effect on the mRNA expression of some digestive genes (PGA5 and PGC) was detected in the proventriculus (Table 4). The TNF-γ protein concentration in hens’ plasma was assayed and the results are presented in Supplemental Figure 2A. No significant effect of GBH exposure was detected, either at D42 or D70.
TABLE 4.
Relative mRNA expression of various genes in hen’s organs at the end of GBH exposure (D42, CT, n = 9 and GBH, n = 9) hens) and after dietary GBH exposure (D70, Ex-CT, n = 12 and Ex-GBH, n = 12 hens).
| Organ | D42 | D70 | |||||
|---|---|---|---|---|---|---|---|
| CT | GBH | p.value | Ex-CT | Ex-GBH | p.value | ||
| Liver | CYP1A2 | 10.47 | 8.74 | 0.578 | 16.42 | 12.31 | 0.210 |
| CYP1A4 | 0.110 | 0.130 | 0.598 | 0.170 | 0.190 | 0.835 | |
| CYP2A6 | 20.0 | 18.4 | 0.696 | 15.1 | 16.28 | 0.741 | |
| CYP2C | 11.4 | 9.86 | 0.498 | 12.4 | 11.40 | 0.793 | |
| CYP3A37 | 6.87 | 6.78 | 0.956 | 5.11 | 5.16 | 0.966 | |
| CYP3A4 | 0.300 | 0.35 | 0.401 | 0.35 | 0.310 | 0.425 | |
| CYP3A80 | 0.460 | 0.42 | 0.681 | 1.98 | 0.740* | 0.027 | |
| CYP2H1 | 22.8 | 22.0 | 0.854 | 16.7 | 18.4 | 0.661 | |
| CYP2H2 | 13.0 | 13.44 | 0.905 | 10.2 | 12.4 | 0.432 | |
| GSTA3 | 3.10 | 2.92 | 0.700 | 5.65 | 3.86 | 0.200 | |
| GSTA4 | 4.71 | 3.50 | 0.054 | 4.43 | 4.32 | 0.861 | |
| EPHX1 | 0.500 | 0.45 | 0.541 | 0.40 | 0.44 | 0.763 | |
| FXR | 0.760 | 0.78 | 0.881 | 1.49 | 1.24 | 0.461 | |
| SL O 1B3 | 0.710 | 0.71 | 0.972 | 0.71 | 0.77 | 0.759 | |
| Gizzard | COX2 | 0.201 | 0.132 | 0.188 | 0.188 | 0.142 | 0.194 |
| HSP27 | 39.9 | 33.8 | 0.637 | 64.1 | 53.4 | 0.360 | |
| HSP40 | 94.1 | 93.1 | 0.835 | 91.3 | 93.3 | 0.582 | |
| HSP60 | 0.533 | 0.361 | 0.113 | 0.707 | 0.628 | 0.310 | |
| HSP70 | 3.18 | 2.48 | 0.443 | 4.31 | 2.29* | 0.015 | |
| HSP90 | 2.24 | 1.49 | 0.249 | 1.87 | 1.55 | 0.544 | |
| iNOS | 0.028 | 0.022 | 0.412 | 0.023 | 0.042** | 0.003 | |
| NFκB | 0.079 | 0.059 | 0.417 | 0.104 | 0.099 | 0.760 | |
| PTGES | 0.087 | 0.054 | 0.168 | 0.074 | 0.055 | 0.245 | |
| TNFa | 0.036 | 0.029 | 0.154 | 0.023 | 0.033 | 0.052 | |
| Proventriculus | PGA5 | 32,266 | 38,158 | 0.330 | 26,653 | 27,223 | 0.899 |
| PGC | 5,916 | 7,495 | 0.114 | 6,874 | 6,838 | 0.971 | |
| Caecum | Chemerin | 0.336 | 0.337 | 0.986 | 0.272 | 0.257 | 0.731 |
| CMKLR1 | 0.017 | 0.017 | 0.984 | 0.065 | 0.048 | 0.301 | |
| IgA | 12.3 | 8.49 | 0.364 | 4.46 | 6.70* | 0.041 | |
| IFNα | 0.071 | 0.086 | 0.620 | 0.434 | 0.290 | 0.230 | |
| IFNβ | 0.002 | 0.005 | 0.179 | 0.024 | 0.011 | 0.100 | |
| IL-1β | 0.013 | 0.007 | 0.095 | 0.020 | 0.016 | 0.338 | |
| IL-6 | 0.001 | 0.001 | 0.895 | 0.004 | 0.002 | 0.077 | |
| IL-8 | 0.040 | 0.025 | 0.185 | 0.049 | 0.032 | 0.242 | |
| Spleen | IFNα | 0.031 | 0.025 | 0.607 | 0.112 | 0.061 | 0.108 |
| IFNβ | 3.50 | 2.18 | 0.301 | 7.90 | 6.00 | 0.414 | |
| IL-1β | 0.006 | 0.004 | 0.182 | 0.010 | 0.014 | 0.279 | |
| IL-8 | 0.076 | 0.075 | 0.964 | 0.065 | 0.038* | 0.036 | |
| IL-22 | 0.035 | 0.021 | 0.273 | 0.093 | 0.024** | 0.003 | |
Results are presented as mean of the pltarget gene mRNA, expression relative to the geometric mean of three housekeeping genes expression (GAPDH, EEF1α and β-actin); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. CT, control; GBH, Ex-CT, Ex-Control, Ex-GBH .
Chronic dietary glyphosate-based herbicide modified the plasma concentration of glycerol and six amino acids
Metabolites whose concentrations in hens’ plasma were significantly affected by GBH exposure are mentioned in Table 5. Plasma glycerol, isoleucine, lysine, methionine and tyrosine concentrations were significantly increased (p < 0.05), while plasma glycine and leucine concentrations were significantly reduced in GBH as compared to CT animals (p < 0.05).
TABLE 5.
Significant plasma metabolites identified in GBH dietary exposed (GBH, n = 10) as compared to control hens (CT, n = 10) at day 42 (D42).
| Metabolites | FC | Pvalue |
|---|---|---|
| Glycerol | 1.05 | 0.024 |
| Glycine | 0.941 | 0.045 |
| Isoleucine | 1.11 | 0.025 |
| Leucine | 0.926 | 0.024 |
| Lysine | 1.53 | 0.028 |
| Methionine | 1.18 | 0.028 |
| Tyrosine | 1.42 | 0.049 |
FC (Fold Change) = GBH/CT (CT, control, GBH). Results are presented as means.
Chronic dietary glyphosate-based herbicide exposure changed SCFA concentrations in chickens’ cecum
Cecal contents were subjected to gas chromatography to measure the cecal SCFA concentration. SCFAs in hens’ cecum were assayed and the results are shown in Table 6. Cecal acetate, propionate, isobutyrate, isovalerate and valerate concentrations were significantly (p < 0.05) reduced during exposure in GBH as compared to control animals (CT). All of them were restored in Ex-GBH chickens’ cecum after GBH exposure, as compared to their respective controls (Ex-CT), except for acetate and valerate, which were still significantly (p < 0.05) reduced.
TABLE 6.
Short-chain fatty acid concentration in cecal content (µmol/g) during dietary GBH (GBH, D42, n = 8) exposure in control (CT, n = 8) and after dietary GBH exposure (Ex-CT, D70, n = 12 and Ex-GBH, n = 11 hens).
| D42 | D70 | |||||
|---|---|---|---|---|---|---|
| CT | GBH | p.value | Ex-CT | Ex-GBH | p.value | |
| Acetate | 28.0 | 18.0** | 0.009 | 25.8 | 16.6* | 0.022 |
| Propionate | 8.36 | 5.43* | 0.022 | 8.02 | 5.63 | 0.126 |
| Isobutyrate | 0.770 | 0.510** | 0.004 | 0.730 | 0.59 | 0.194 |
| Butyrate | 2.08 | 1.64 | 0.293 | 1.85 | 1.30 | 0.166 |
| Isovalerate | 0.530 | 0.389* | 0.041 | 0.510 | 0.450 | 0.377 |
| Valerate | 0.710 | 0.516* | 0.044 | 0.620 | 0.390* | 0.033 |
| Caproate | 0.000 | 0.000 | 0.020 | 0.000 | 0.068 | |
| Total isoAGCC | 1.30 | 0.900** | 0.006 | 1.24 | 1.04 | 0.233 |
| Total AGCC | 41.5 | 26.4* | 0.015 | 38.0 | 25.2* | 0.045 |
Results are presented as means; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. CT, control; GBH, Ex-CT, Ex-Control, Ex-GBH.
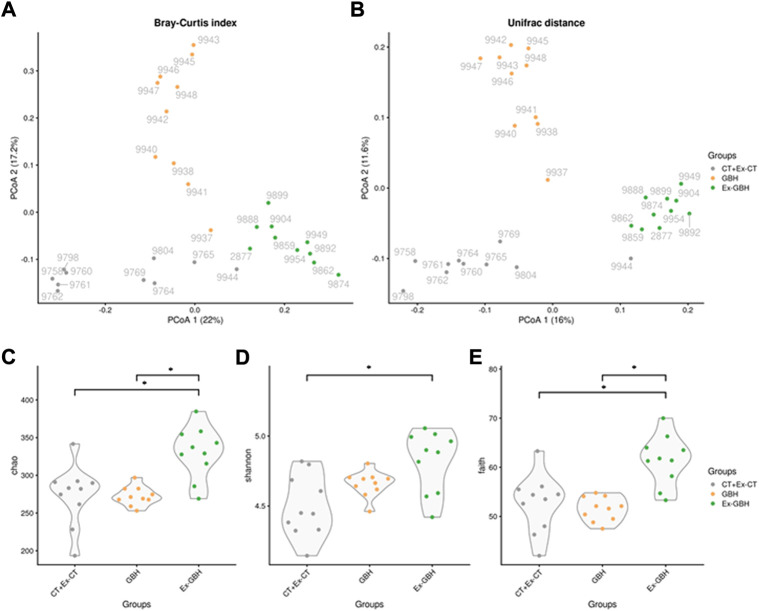
Chronic dietary glyphosate-based herbicide exposure increased gut microbiome diversity
GBH durably changed the composition of the microbiota, which was not completely resilient (Figures 6A,B). Surprisingly, GBH increased the diversity of the microbiota in a delayed manner. Indeed, the diversity indices (Chao, Shannon and Faith indices) became different at the final time point even though they were similar at the end of the GBH exposure (Figures 6C–E). The delayed impact of GBH was reflected in eight individual taxa that were different from the control at the end of the experiment but not just after the GBH exposure (Figure 7). For example, the Bacteroidales F082 family was not immediately affected by GBH administration but was significantly enhanced in the Ex-GBH group. Similarly, the Synergistes (family Synergistaceae) were favored by GBH at the final time point only. The other groups that showed a delayed impact were: Akkermansia (family Akkermansiaceae), DTU089 (family Clostridiaceae), Paraprevotella (family Paraprevotella), S50 (family Rikenellaceae) and Treponema (family Treponemataceae). Perhaps unsurprisingly, nine taxa were immediately impacted by GBH: the amounts of Muribaculaceae family, Alloprevotella (family Prevotellaceae), Porphyromonadaceae and Candidatus Vestibaculum were significantly higher after exposure in GBH and Ex-GBH groups than in the CT + Ex-CT group. The abundance of Barnesiella was significantly decreased whereas that of Colidextribacter was significantly higher in the GBH group than in the CT + Ex-CT group. The abundance of Coprobacter was different in all three groups: it was significantly lower in the GBH group than in the CT + Ex-CT group and was significantly higher in Ex-GBH than in the other groups. DTU089 abundance was significantly lower in the Ex-GBH group than in the CT + Ex-CT group. GCA-900066575 abundance was significantly higher in the Ex-GBH group than in GBH hens. Ruminococcus abundance was significantly higher in Ex-GBH and CT + Ex-GBH groups than in the GBH group. Abundance of the S50 wastewater-sludge group was significantly higher in Ex-GBH than in the GBH group. Synergistes abundance was significantly higher in Ex-GBH than in the CT + Ex-GBH group. Clostridia vadin BB60 abundance was significantly higher in the CT + Ex-CT group than in the GBH group. sPLS-DA analysis was performed to explain group variations. Two components have been identified. Most of the differential taxa identified by sPLS-DA analysis was already identified by the previous analysis. The first component (X-variate 1) explains 16% of the variations and allows to separate GBH and CT + Ex-CT hens from Ex-GBH hens. Second component (X-variate 2) explains 13% of the variations and allows to separate GBH hens from CT + Ex-CT hens. For each component, most contributing taxa were identified based on their abundance. The first component has 5 most contributing taxa. Its most contributing taxon is Treponema, more abundant in Ex-GBH group, followed by Prevotellaceae, more abundant in GBH group, Paraprevotella, a non-identified F082 family bacterium, more abundant in Ex-GBH group and Butyricicoccus which is more abundant in CT + Ex-CT group. The second component has 5 most contributing taxa. Its most contributing taxon is Muribaculaceae, more abundant in GBH group, followed by Bacteroidales, more abundant in GBH group too, itself followed by Flavobacteriales, more abundant in CT + Ex-CT group and Candidatus Vestibculum and Synergistes, more abundant in GBH and CT + Ex-CT group.
FIGURE 6.
Microbiome β-diversity in cecal content of hens exposed to GBH for 42 days (GBH, n = 9), then non-exposed until D70 (Ex-GBH, n = 12) and controls at D42 and D70 taken together (CT + Ex-CT, n = 10), collected during (D42) and after GBH exposure (D70) measured by principal coordinates analysis (PCoA) on Bray–Curtis index (A) and Unifrac distance (B). Results are presented as individual values. Microbiome α-diversity in the very same groups measured by Chao (C), Shannon (D) and Faith indices (E). *p ≤ 0.05. CT + Ex-CT: Control and ex-Control taken together, GBH: Glyphosate-Based Herbicide, Ex-GBH.
FIGURE 7.
Relative abundance of differential taxa in cecal content of hens exposed to GBH for 42 days (GBH, n = 9), exposed for 42 days then non-exposed until D70 (Ex-GBH, n = 12) and controls at D42 and D70 taken together (CT + Ex-CT, n = 10), commonly identified by ALDEx2, DESeq2 and ANCOM-BC methods. A bar between two groups indicates that the relative abundance is significant between these two groups. Taxa are identified at order, family and genus levels Results are presented as means ± SEM. *p ≤ 0.05. CT + Ex-CT: Control and ex-Control taken together, GBH: Glyphosate-Based Herbicide, Ex-GBH.
Discussion
Our results show for the first time possible disturbances of the cecal microbiota associated with plasma oxidative stress and the accumulation of Gly in metabolic tissues in response to chronic dietary GBH exposure in breeder broiler hens (Figure 8). Furthermore, some of these alterations are potentially reversible and were observed without any variation of growth performance (Figure 8).
FIGURE 8.
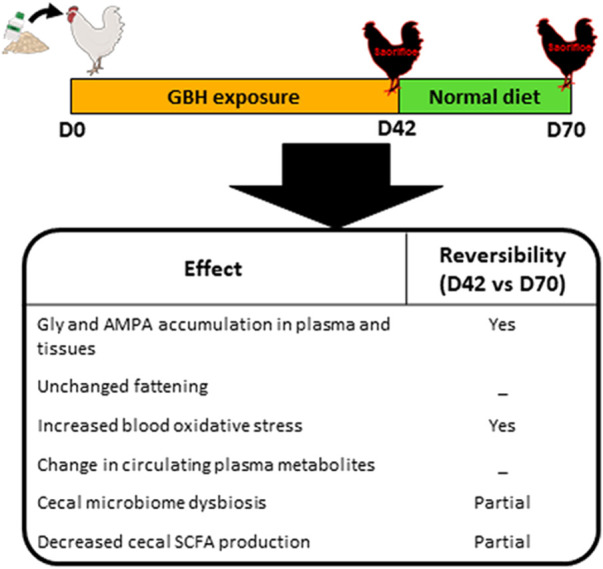
Schematic representation of effects of dietary GBH exposure in hens and their potential reversibility on glyphosate and AMPA accumulation in plasma and tissues, fattening, plasma oxidative stress, plasma metabolites, cecal microbiome dysbiosis and cecal SCFA production.
Indeed, when we monitored hens’ body weight and fat thickness during and after chronic dietary GBH exposure, we did not observe any differences in either of these two parameters between the hens fed with GBH and those fed without GBH. These results are in accordance with those obtained after a 13-weeks study performed on rats (Panzacchi et al., 2018). Moreover, triglyceride, cholesterol and phospholipid concentrations in plasma were not affected and the uric acid assay did not show any difference with the control either, suggesting that fatty acid and purine metabolism is not affected by the GBH diet. However, metabolomic analysis revealed that the concentrations of some serum metabolites were altered. A recent study shows that in Gly exposed humans, several metabolites levels in serum can be altered (Zhang et al., 2022). These alterations could be linked to metabolism dysfunctions, including fatty acids metabolism and purine biosynthesis. The authors also found that TCA cycle intermediates were altered following to the exposition. In our study, all altered serum metabolites were amino acids (except for glycerol). More specifically, five out of eight of the significantly affected metabolites were ketogenic amino acids, including two glucoformers. Also, all the metabolites we identified were reported several times as potential biomarkers for human colorectal cancer detection (Ni et al., 2014). Among them, leucine, lysine and tyrosine can be oxidized to generate acetyl-CoA for ketone body synthesis; methionine and isoleucine are also convertible into propionyl-CoA, then into succinyl-CoA to enter the TCA cycle (Newsholme et al., 2011; Chiang, 2014; Kumari, 2018). This suggests a potential disruption of mitochondrial activity. However, when we determined the ATP concentration in hens’ livers, we observed no significant effect of the treatment. The effect of such variations in amino acid circulation therefore remains unclear.
Still, it is worth noticing that Gly and AMPA mainly accumulated in the liver and, to a lesser extent, in leg muscles and AAT. However, no significant effect on liver and AAT weight was detected. Gly and AMPA were also detectable in plasma at both D42 (during exposure) and D70 (after exposure), where they were accompanied by an increase in oxidative stress only at D42. Oxidative stress induction has widely been demonstrated in mammals exposed to Gly and AMPA (Kwiatkowska et al., 2014; Owagboriaye et al., 2019; Turkmen et al., 2019). This effect is reversible since plasma TBARS levels drop back to CT values after exposure. The maximum Gly and AMPA concentrations were measured on the 21st day of the diet, and the values measured later on the 42nd day were lower. This suggests that their degradation/excretion rate is lower than their intake rate before D21 but becomes greater during the following days. In order to clarify this observation, we quantified transcript levels of biotransformation enzymes. A recent study has shown that in ovo injections of Gly and GBH trigger modulations in the mRNA expression of cytochromes and other biotransformation-related genes in chick embryos (Fathi et al., 2020). Yet, in adult hens, we did not detect any significant effect on the expression of biotransformation-related genes, whether they encode for Phase I (cytochromes) or Phase II enzymes (GSTs). CYP enzymes enable detoxication of xenobiotics by catalyzing redox and hydrolysis reactions, modifying their chemical properties and thus their toxicity. GSTs catalyze the conjugation reaction of GSH to xenobiotics, which enhances their hydrosolubility and thus their elimination in urine and feces. It has been shown that in rats, 98% of administered Gly is excreted as the unchanged parent compound (Panzacchi et al., 2018), which could explain the lack of action of Phase I and Phase II enzymes on it, which would have turned it into modified compounds. Moreover, a 2000 risk assessment reported that orally administered Gly and AMPA are weakly biotransformed in animals (Williams et al., 2000). However, Gly concentrations in the liver do not quickly drop to negligible values after exposure. Yet, as previously mentioned, GBH’s suspected toxicity is not due to Gly alone but also to the surfactants contained in GBH formulations (Bradberry et al., 2004; Kim et al., 2013; Mesnage et al., 2019). Studies have shown that some surfactants are even more toxic than the active ingredient. One example is lipid-based POEAs (polyethoxylated tallow amines) (Martins-Gomes et al., 2022) which are now banned in the EU but still allowed in the United States The presence of these coformulants could explain the high and sustainable accumulation of Gly in the liver, but we would have expected them to trigger the expression of biotransformation enzymes. The exact composition of most GBHs is, however, strictly confidential. We are therefore unable to determine which coformulants could be responsible for this phenomenon.
Considering that hens’ spleens were heavier in the Ex-GBH group than in the Ex-CT group, we assumed a disruption at the immune system level. We therefore investigated immune system gene transcript levels in several immune tissues and observed an increase in IgA mRNA expression in the cecum of Ex-GBH animals compared to that in controls. Since IgA is known to be involved in gut microbiome dynamics (Shi et al., 2017), we suspected potential disruption to hens’ gut microbiome. The main effects of GBH dietary exposure observed in the present study are actually at the gut microbiome level. It is now well established that GBH and Gly are able to induce changes in microbial communities, especially in the gut of exposed animals (Lozano et al., 2018; Mao et al., 2018). A recent study has shown that orally administered Gly and GBH trigger inhibition of the shikimate pathway in the gut microbiome of Sprague Dawley rats. This inhibition is coupled with increased levels of Akkermansia muciniphila (Mesnage et al., 2021). Interestingly, our results rather show a decrease in Akkermansia abundance, which is not restored after exposure. In humans, the shikimate pathway is mainly achieved by A. muciniphila (Mesnage and Antoniou, 2020), which, extrapolated to poultry, could make the crushing of its abundance expectable and consistent since it is the pathway targeted by Gly. Our results also show that Barnesiella and Clostridia vadin BB60 abundance follows the same scheme, as does that of Ruminococcus whose abundance follows a trend toward restoration after Gallup exposure, implicating various potential neuropsychiatric disorders (Barnett et al., 2022). On the other hand, Muribaculaceae, Porphyromonadaceae, Alloprevotella, Candidatus Vestibaculum and Colidextribacter abundances are significantly increased by GBH exposure and follow a trend toward restoration after GBH exposure. Interestingly, Colidextribacter species are known to be positively correlated with oxidative stress (Wang et al., 2021). The impact of GBH may be similar between rats and poultry, since GBH exposure also increases Alloprevotella in rats’ microbiome (Dechartres et al., 2019). However, to our knowledge, no evident link between variations in the populations of other above-mentioned bacteria and GBH exposure has been established. It has nevertheless previously been shown that a decrease in one bacterial population can induce an increase in other populations (Ruuskanen et al., 2020). We therefore assume that some of the community variations observed here are rather due to inner ecological competition triggered by the reduction in abundance of some taxa than to a direct antibiotic effect of the herbicide. Moreover, Gly is not deleterious to all bacteria: some are able to degrade the molecule (Mesnage and Antoniou, 2020) for phosphorus supply or energy production (Hove-Jensen et al., 2014; Strilbyska et al., 2021). Several recent studies suggest an increase in α-diversity in various models when individuals are exposed to glyphosate or GBHs (Tang et al., 2020; Castelli et al., 2021; Mesnage et al., 2021, 2022). Our results are similar to these findings, except that α-diversity is not increased in exposed animals, but in formerly exposed animals. It is surprising that adding a perturbation does not have any effect on a system but the removal of this perturbation does produce some significant effects on it. To our knowledge, no previous study has reported this kind of observation following to the administration of dietary GBH. However, since we observe some effects on the gut microbiome at the family and genus levels, we can hypothesize that cecal bacteria quickly accustom to the GBH presence, probably by gaining genetic resistances to it. They could however not accustom to its withdrawal as quickly. Since a genetic resistance, for instance to an antibiotic, is often a disadvantage in absence of it, GBH withdrawal could free up an ecological niche for other bacteria, resulting in an increase in α-diversity.
SFCA concentrations in animal feces are also good indicators of potential disruptions in the gut microbiome. Dietary carbohydrates that reach the large intestine without having previously been lysed into smaller molecules in the small intestine are metabolized by bacteria into SCFAs. These molecules are critical in maintaining animals’ health, as they are the primary energy source for colonocytes and are involved in the production of hormones which act on their metabolism (blood glucose regulation, fat and protein digestion, satiation promotion) (Barnett et al., 2022). Ruminococcus (Ruminococcaceae) is part of the gut bacteria able to produce SCFAs from dietary carbohydrates (Barnett et al., 2022) that we identified as reversibly diminished by GBH exposure. Our data are also in good agreement with those observed in rats (Dechartres et al., 2019) where Ruminococcaceae abundance was lowered by exposure to GBH. Akkermansia has also been reported to be an SCFA (propionate) producer in humans (Morrison and Preston, 2016). Our results show a global diminution of all cecal SCFAs during GBH administration, which are restored after exposure for acetate and valerate only. We can therefore hypothesize a link between the fall of these bacteria and that of cecal SFCA levels. Since SFCAs are also suspected to be involved in neuroendocrine regulation and gut microbiome–brain communication, disturbance in their dynamics suggests potential physiological and mental disorders (Silva et al., 2020). A 2017 study revealed depressive-like behaviors in young adult rats when exposed to a GBH (Cattani et al., 2017). However, we did not detect any effect on hens’ feeding behavior. At this point, it therefore seems important to remember the correlational nature of such studies. Since correlation does not imply causation, more studies are needed to confirm direct guilt of GBH/Gly/surfactants in disruption of the gut microbiome and cecal SFCA production and on the disorders they trigger. Other effects of GBH have been demonstrated in other cells. For instance, it has been shown that low GBH levels could induce oxidative stress and impair Ca2+-mediated functions in rat Sertoli cells, resulting in a reduced male fertility (de Liz Oliveira Cavalli et al., 2013). Future works could therefore focus on the potential effects of GBH administration on male and female reproductive functions.
Conclusion
Here, we show that half the current NOAEL of dietary Gly does not induce a direct impact on hens’ metabolism (at least on the various parameters determined in our 150 animals), though it reversibly triggers oxidative stress in blood plasma and impacts several gut microbes. Investigating the link between GBH and fertility is a direction that future works should probably take, in order to constitute a robust database allowing characterization of the actual impact that the molecule and its formulations have on animal and human health.
Acknowledgments
The authors are grateful to all the persons from the experimental unit (INRAE, PEAT, Centre Val de Loire DOI: 10.15454/1.5572326250887292E12) who take care of animals. We also thank to Christine Leterrier for her help in the feeding behavior analysis. All NMR experiments were performed on the instruments of the MetaToul-AXIOM platform, partner of the national infrastructure of metabolomics and fluxomics: MetaboHUB (MetaboHUB-ANR-11-INBS-0010). We also thank to Laurent Cauquil for the European Nucleotide Archive submission.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJNA741111.
Ethics statement
The animal study was reviewed and approved by ethics committee of Val de Loire No. 19.
Author contributions
MF, AE, CR, MC, PF, and JD. contributed to the overall approach and design of experiments. GL, MF. and JD performed statistical data analysis. CS and EV determined the fattening of animals by using ultrasound. CL and JL realized the analyses of food intake behavior and RG participated to the RTqPCR analysis. EM performed the analyses of SCFA concentrations in hen’s caecum and AH and OZ realized the caecal microbiome analysis. MF and JD. wrote the manuscript. All authors critically revised the manuscript and approved the final version.
Funding
The authors thank the Région Centre Val de Loire for the financial support (Project number 32000858, HAPOFERTI).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.974688/full#supplementary-material
Glossary
- Actin B:
Actin Beta ALDEx2: ANOVA-Like Differential Expression 2
- AMPA:
Amino-Methyl-Phosphonic Acid
- ANCOM-BC:
Analysis of Compositions of Microbiomes with Bias Correction
- BCA:
bicinchoninic acid
- BSEP:
ATP binding cassette subfamily B member 11
- Ces1:
Carboxylesterase 1
- CMKLR1:
Chemerin Chemokine-Like Receptor 1
- COX-2:
Cyclooxygenase 2
- Cq:
quantification cycle
- CT:
Control
- CYP1A1:
Cytochrome P450 Family 1 Subfamily A Member 1
- CYP1A2:
Cytochrome P450 Family 1 Subfamily A Member 2
- CYP1A4:
Cytochrome P450 Family 1 Subfamily A Member 4
- CYP1A5:
Cytochrome P450 Family 1 Subfamily A Member 5
- CYP2A6:
Cytochrome P450 family 2 subfamily A member 6
- CYP2C:
Cytochrome P450 Family 2 Subfamily C
- CYP2H1:
Cytochrome P4502H1
- CYP2H2:
Cytochrome P4502H2
- CYP3A:
Cytochrome P450, family 3, subfamily A
- CYP3A37:
Cytochrome P450, family 3, subfamily A member 37
- EPHX1:
Epoxide Hydrolase 1
- FMOC-Cl:
9-fluorenylmethyl chloroformate
- FROGS:
Find, Rapidly, OTUs, Operational Taxonomic Units
- FXR:
Farnesoid X receptor
- GAPDH:
Glyceraldehyde-3-phosphate dehydrogenase
- GBH:
Glyphosate-based Herbicides
- Gly:
Glyphosate
- GSH:
Glutathione
- GSTA3:
Glutathione S-Transferase Alpha 3
- GSTA4:
Glutathione S-Transferase Alpha 4
- GSTs:
Glutathione S-Transferase
- Hsp27:
Heat shock protein 27
- Hsp40:
Heat shock protein 40
- Hsp60:
Heat shock protein 60
- Hsp70:
Heat shock protein 70
- Hsp90:
Heat shock protein 90
- IFNα:
Interferon alpha
- IFNβ:
Interferon beta
- IgA:
Immunoglobulin A
- IL-1β:
Interleukin-1 beta
- IL-22:
Interleukin-22
- IL-6:
Interleukin-6
- IL-8:
Interleukin-8
- iNOS:
Inducible nitric oxide synthase
- MDA:
MalonDiAldehyde
- MMLV:
Moloney murine leukemia virus reverse transcriptase
- NF-κB:
Nuclear factor-kappa B
- NMDS:
non-metric multidimensional scaling
- NMR:
Nuclear magnetic resonance metabolomics
- NOAEL:
Non observed adverse effect level
- OTUs:
Operational Taxonomic Units
- PGA5:
Pepsinogen A5
- PGC:
Progastricsin
- POEA:
Polyethoxylated tallow amines
- PTGES:
Prostaglandin E Synthase
- SCFA:
Short-Chain Fatty Acid
- TCA cycle:
tricarboxylic acid cycle
- TNF-α:
Tumor Necrosis Factor-alpha
References
- Al-Khalaifah H. S., Shahin Sara. E., Omar A. E., Mohammed H. A., Mahmoud Hala. I., Ibrahim D. (2020). Effects of graded levels of microbial fermented or enzymatically treated dried brewer’s grains on growth, digestive and nutrient transporter genes expression and cost effectiveness in broiler chickens. BMC Vet. Res. 16, 424. 10.1186/s12917-020-02603-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D., Browne R. (1994). “The analysis of free radicals, lipid peroxides, antioxidant enzymes and compounds related to oxidative stress as applied to the clinical chemistry laboratory,” in Free Radicals in diagnostic medicine advances in experimental medicine and biology. Editor Armstrong D. (Boston, MA: Springer US; ), 43–58. 10.1007/978-1-4615-1833-4_4 [DOI] [PubMed] [Google Scholar]
- Barnett J. A., Bandy M. L., Gibson D. L. (2022). Is the use of glyphosate in modern agriculture resulting in increased neuropsychiatric conditions through modulation of the gut-brain-microbiome Axis? Front. Nutr. 9, 827384. 10.3389/fnut.2022.827384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradberry S. M., Proudfoot A. T., Vale J. A. (2004). Glyphosate poisoning. Glyphosate Poisoning Toxicol. Rev. 23, 159–167. 10.2165/00139709-200423030-00003 [DOI] [PubMed] [Google Scholar]
- Castelli L., Balbuena S., Branchiccela B., Zunino P., Liberti J., Engel P., et al. (2021). Impact of chronic exposure to sublethal doses of glyphosate on honey bee immunity, gut microbiota and infection by pathogens. Microorganisms 9, 845. 10.3390/microorganisms9040845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattani D., Cesconetto P. A., Tavares M. K., Parisotto E. B., De Oliveira P. A., Rieg C. E. H., et al. (2017). Developmental exposure to glyphosate-based herbicide and depressive-like behavior in adult offspring: Implication of glutamate excitotoxicity and oxidative stress. Toxicology 387, 67–80. 10.1016/j.tox.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Cattani D., Struyf N., Steffensen V., Bergquist J., Zamoner A., Brittebo E., et al. (2021). Perinatal exposure to a glyphosate-based herbicide causes dysregulation of dynorphins and an increase of neural precursor cells in the brain of adult male rats. Toxicology 461, 152922. 10.1016/j.tox.2021.152922 [DOI] [PubMed] [Google Scholar]
- Chamorro-García R., Poupin N., Tremblay-Franco M., Canlet C., Egusquiza R., Gautier R., et al. (2021). Transgenerational metabolomic fingerprints in mice ancestrally exposed to the obesogen TBT. Environ. Int. 157, 106822. 10.1016/j.envint.2021.106822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. (2014). “Liver physiology: Metabolism and detoxification,” in Pathobiology of human disease (Elsevier; ), 1770–1782. 10.1016/B978-0-12-386456-7.04202-7 [DOI] [Google Scholar]
- Cong Y., Chi Q., Teng X., Li S. (2019). The protection of selenium against cadmium-induced mitochondrial damage via the cytochrome P450 in the livers of chicken. Biol. Trace Elem. Res. 190, 484–492. 10.1007/s12011-018-1557-x [DOI] [PubMed] [Google Scholar]
- Dai D., Pan Y., Zeng C., Liu S., Yan Y., Wu X., et al. (2020). Activated FXR promotes xenobiotic metabolism of T-2 toxin and attenuates oxidative stress in broiler chicken liver. Chem. Biol. Interact. 316, 108912. 10.1016/j.cbi.2019.108912 [DOI] [PubMed] [Google Scholar]
- de Liz Oliveira Cavalli V. L., Cattani D., Heinz Rieg C. E., Pierozan P., Zanatta L., Benedetti Parisotto E., et al. (2013). Roundup disrupts male reproductive functions by triggering calcium-mediated cell death in rat testis and Sertoli cells. Free Radic. Biol. Med. 65, 335–346. 10.1016/j.freeradbiomed.2013.06.043 [DOI] [PubMed] [Google Scholar]
- Dechartres J., Pawluski J. L., Gueguen M., Jablaoui A., Maguin E., Rhimi M., et al. (2019). Glyphosate and glyphosate‐based herbicide exposure during the peripartum period affects maternal brain plasticity, maternal behaviour and microbiome. J. Neuroendocrinol. 31, e12731. 10.1111/jne.12731 [DOI] [PubMed] [Google Scholar]
- Escudié F., Auer L., Bernard M., Mariadassou M., Cauquil L., Vidal K., et al. (2018). Frogs: Find, rapidly, OTUs with Galaxy solution. Bioinformatics 34, 1287–1294. 10.1093/bioinformatics/btx791 [DOI] [PubMed] [Google Scholar]
- Estienne A., Reverchon M., Partyka A., Bourdon G., Grandhaye J., Barbe A., et al. (2020). Chemerin impairs in vitro testosterone production, sperm motility, and fertility in chicken: Possible involvement of its receptor CMKLR1. Cells 9, 1599. 10.3390/cells9071599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi M. A., Han G., Kang R., Shen D., Shen J., Li C. (2020). Disruption of cytochrome P450 enzymes in the liver and small intestine in chicken embryos in ovo exposed to glyphosate. Environ. Sci. Pollut. Res. Int. 27, 16865–16875. 10.1007/s11356-020-08269-3 [DOI] [PubMed] [Google Scholar]
- Fernandes A. D., Reid J. N., Macklaim J. M., McMurrough T. A., Edgell D. R., Gloor G. B. (2014). Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2, 15. 10.1186/2049-2618-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogliatto S., Ferrero A., Vidotto F. (2020). “Current and future scenarios of glyphosate use in Europe: Are there alternatives?,” in Advances in agronomy (Elsevier; ), 219–278. 10.1016/bs.agron.2020.05.005 [DOI] [Google Scholar]
- Furet J.-P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., et al. (2009). Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR: Human and farm animal faecal microbiota. FEMS Microbiol. Ecol. 68, 351–362. 10.1111/j.1574-6941.2009.00671.x [DOI] [PubMed] [Google Scholar]
- Garrido D., Alber A., Kut E., Chanteloup N. K., Lion A., Trotereau A., et al. (2018). The role of type I interferons (IFNs) in the regulation of chicken macrophage inflammatory response to bacterial challenge. Dev. Comp. Immunol. 86, 156–170. 10.1016/j.dci.2018.04.025 [DOI] [PubMed] [Google Scholar]
- Gill J. P. K., Sethi N., Mohan A., Datta S., Girdhar M. (2018). Glyphosate toxicity for animals. Environ. Chem. Lett. 16, 401–426. 10.1007/s10311-017-0689-0 [DOI] [Google Scholar]
- Grandhaye J., Douard V., Rodriguez-Mateos A., Xu Y., Cheok A., Riva A., et al. (2020). Microbiota changes due to grape seed extract diet improved intestinal homeostasis and decreased fatness in parental broiler hens. Microorganisms 8, 1141. 10.3390/microorganisms8081141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandhaye J., Hmadeh S., Plotton I., Levasseur F., Estienne A., LeGuevel R., et al. (2021). The adiponectin agonist, AdipoRon, inhibits steroidogenesis and cell proliferation in human luteinized granulosa cells. Mol. Cell. Endocrinol. 520, 111080. 10.1016/j.mce.2020.111080 [DOI] [PubMed] [Google Scholar]
- Grau D., Grau N., Gascuel Q., Paroissin C., Stratonovitch C., Lairon D., et al. (2022). Quantifiable urine glyphosate levels detected in 99% of the French population, with higher values in men, in younger people, and in farmers. Environ. Sci. Pollut. Res. Int. 29, 32882–32893. 10.1007/s11356-021-18110-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotto D., Santa Maria L. D., Boeira S., Valentini J., Charão M. F., Moro A. M., et al. (2007). Rapid quantification of malondialdehyde in plasma by high performance liquid chromatography–visible detection. J. Pharm. Biomed. Anal. 43, 619–624. 10.1016/j.jpba.2006.07.030 [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B., Zechel D. L., Jochimsen B. (2014). Utilization of glyphosate as phosphate source: Biochemistry and genetics of bacterial carbon-phosphorus lyase. Microbiol. Mol. Biol. Rev. 78, 176–197. 10.1128/MMBR.00040-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inger R., Gregory R., Duffy J. P., Stott I., Voříšek P., Gaston K. J. (2015). Common European birds are declining rapidly while less abundant species’ numbers are rising. Ecol. Lett. 18, 28–36. 10.1111/ele.12387 [DOI] [PubMed] [Google Scholar]
- Jasper R., Locatelli G. O., Pilati C., Locatelli C. (2012). Evaluation of biochemical, hematological and oxidative parameters in mice exposed to the herbicide glyphosate-Roundup®. Interdiscip. Toxicol. 5, 133–140. 10.2478/v10102-012-0022-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Hong J., Gil H., Song H., Hong S. (2013). Mixtures of glyphosate and surfactant TN20 accelerate cell death via mitochondrial damage-induced apoptosis and necrosis. Toxicol. Vitro 27, 191–197. 10.1016/j.tiv.2012.09.021 [DOI] [PubMed] [Google Scholar]
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A. (2018). “Citric acid cycle,” in Sweet biochemistry (Elsevier; ), 7–11. 10.1016/B978-0-12-814453-4.00002-9 [DOI] [Google Scholar]
- Kwiatkowska M., Huras B., Bukowska B. (2014). The effect of metabolites and impurities of glyphosate on human erythrocytes (in vitro). Pestic. Biochem. Physiol. 109, 34–43. 10.1016/j.pestbp.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Larsen K., Najle R., Lifschitz A., Maté M. L., Lanusse C., Virkel G. L. (2014). Effects of sublethal exposure to a glyphosate-based herbicide formulation on metabolic activities of different xenobiotic-metabolizing enzymes in rats. Int. J. Toxicol. 33, 307–318. 10.1177/1091581814540481 [DOI] [PubMed] [Google Scholar]
- Lê Cao K.-A., Costello M.-E., Lakis V. A., Bartolo F., Chua X.-Y., Brazeilles R., et al. (2016). MixMC: A multivariate statistical framework to gain insight into microbial communities. PLoS ONE 11, e0160169. 10.1371/journal.pone.0160169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Peddada S. D. (2020). Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514. 10.1038/s41467-020-17041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano V. L., Defarge N., Rocque L.-M., Mesnage R., Hennequin D., Cassier R., et al. (2018). Sex-dependent impact of Roundup on the rat gut microbiome. Toxicol. Rep. 5, 96–107. 10.1016/j.toxrep.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q., Manservisi F., Panzacchi S., Mandrioli D., Menghetti I., Vornoli A., et al. (2018). The ramazzini institute 13-week pilot study on glyphosate and roundup administered at human-equivalent dose to Sprague Dawley rats: Effects on the microbiome. Environ. Health 17, 50. 10.1186/s12940-018-0394-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Gomes C., Silva T. L., Andreani T., Silva A. M. (2022). Glyphosate vs. Glyphosate-based herbicides exposure: A review on their toxicity. J. Xenobiot. 12, 21–40. 10.3390/jox12010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R., Antoniou M. N. (2020). Computational modelling provides insight into the effects of glyphosate on the shikimate pathway in the human gut microbiome. Curr. Res. Toxicol. 1, 25–33. 10.1016/j.crtox.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R., Benbrook C., Antoniou M. N. (2019). Insight into the confusion over surfactant co-formulants in glyphosate-based herbicides. Food Chem. Toxicol. 128, 137–145. 10.1016/j.fct.2019.03.053 [DOI] [PubMed] [Google Scholar]
- Mesnage R., Bowyer R. C. E., El Balkhi S., Saint-Marcoux F., Gardere A., Ducarmon Q. R., et al. (2022). Impacts of dietary exposure to pesticides on faecal microbiome metabolism in adult twins. Environ. Health. 21, 46. 10.1186/s12940-022-00860-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R., Defarge N., Spiroux de Vendômois J., Séralini G. E. (2015). Potential toxic effects of glyphosate and its commercial formulations below regulatory limits. Food Chem. Toxicol. 84, 133–153. 10.1016/j.fct.2015.08.012 [DOI] [PubMed] [Google Scholar]
- Mesnage R., Teixeira M., Mandrioli D., Falcioni L., Ducarmon Q. R., Zwittink R. D., et al. (2021). Use of shotgun metagenomics and metabolomics to evaluate the impact of glyphosate or roundup mon 52276 on the gut microbiota and serum metabolome of sprague-dawley rats. Environ. Health Perspect. 129, 017005. 10.1289/EHP6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. J., Preston T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200. 10.1080/19490976.2015.1134082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nearing J. T., Douglas G. M., Hayes M. G., MacDonald J., Desai D. K., Allward N., et al. (2022). Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 13, 342. 10.1038/s41467-022-28034-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme P., Stenson L., Sulvucci M., Sumayao R., Krause M. (2011). “Amino acid metabolism,” in Comprehensive biotechnology (Elsevier; ), 3–14. 10.1016/B978-0-08-088504-9.00002-7 [DOI] [Google Scholar]
- Ni Y., Xie G., Jia W. (2014). Metabonomics of human colorectal cancer: New approaches for early diagnosis and biomarker discovery. J. Proteome Res. 13, 3857–3870. 10.1021/pr500443c [DOI] [PubMed] [Google Scholar]
- Okamoto A. S., Filho R. L. A., Milbradt E. L., Moraes A. C. I., Vellano I. H. B., Guimarães-Okamoto P. T. C. (2018). Bacterial communication between lactobacillus spp. isolated from poultry in the inhibition of Salmonella heidelberg—Proof of concept. Poult. Sci. 97, 2708–2712. 10.3382/ps/pey141 [DOI] [PubMed] [Google Scholar]
- Owagboriaye F., Dedeke G., Ademolu K., Olujimi O., Aladesida A., Adeleke M. (2019). Comparative studies on endogenic stress hormones, antioxidant, biochemical and hematological status of metabolic disturbance in albino rat exposed to roundup herbicide and its active ingredient glyphosate. Environ. Sci. Pollut. Res. Int. 26, 14502–14512. 10.1007/s11356-019-04759-1 [DOI] [PubMed] [Google Scholar]
- Pandey A., Dabhade P., Kumarasamy A. (2019). Inflammatory effects of subacute exposure of roundup in rat liver and adipose tissue. Dose. Response. 17, 1559325819843380. 10.1177/1559325819843380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzacchi S., Mandrioli D., Manservisi F., Bua L., Falcioni L., Spinaci M., et al. (2018). The ramazzini institute 13-week study on glyphosate-based herbicides at human-equivalent dose in Sprague Dawley rats: Study design and first in-life endpoints evaluation. Environ. Health 17, 52. 10.1186/s12940-018-0393-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F., Gautier B., Singh A., Lê Cao K.-A. (2017). mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752. 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano R. M., Romano M. A., Bernardi M. M., Furtado P. V., Oliveira C. A. (2010). Prepubertal exposure to commercial formulation of the herbicide glyphosate alters testosterone levels and testicular morphology. Arch. Toxicol. 84, 309–317. 10.1007/s00204-009-0494-z [DOI] [PubMed] [Google Scholar]
- Ruuskanen S., Rainio M. J., Gómez-Gallego C., Selenius O., Salminen S., Collado M. C., et al. (2020). Glyphosate-based herbicides influence antioxidants, reproductive hormones and gut microbiome but not reproduction: A long-term experiment in an avian model. Environ. Pollut. 266, 115108. 10.1016/j.envpol.2020.115108 [DOI] [PubMed] [Google Scholar]
- Samsel A., Seneff S. (2013). Glyphosate’s suppression of cytochrome P450 enzymes and amino acid biosynthesis by the gut microbiome: Pathways to modern diseases. Entropy 15, 1416–1463. 10.3390/e15041416 [DOI] [Google Scholar]
- Serra L., Estienne A., Bourdon G., Ramé C., Chevaleyre C., Didier P., et al. (2021). Chronic dietary exposure of roosters to a glyphosate-based herbicide increases seminal plasma glyphosate and AMPA concentrations, alters sperm parameters, and induces metabolic disorders in the progeny. Toxics 9, 318. 10.3390/toxics9120318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata A. A., Schrödl W., Aldin Alaa. A., Hafez H. M., Krüger M. (2013). The effect of glyphosate on potential pathogens and beneficial members of poultry microbiota in vitro . Curr. Microbiol. 66, 350–358. 10.1007/s00284-012-0277-2 [DOI] [PubMed] [Google Scholar]
- Shi N., Li N., Duan X., Niu H. (2017). Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 4, 14. 10.1186/s40779-017-0122-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva Y. P., Bernardi A., Frozza R. L. (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 11, 25. 10.3389/fendo.2020.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F., Bäckhed F. (2013). The gut microbiota — Masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Strilbyska O. M., Tsiumpala S. A., Kozachyshyn I. I., Strutynska T., Burdyliuk N., Lushchak V. I., et al. (2021). The effects of low-toxic herbicide roundup and glyphosate on mitochondria. EXCLI J. 14, 183–196. 10.17179/excli2021-4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q., Tang J., Ren X., Li C. (2020). Glyphosate exposure induces inflammatory responses in the small intestine and alters gut microbial composition in rats. Environ. Pollut. 261, 114129. 10.1016/j.envpol.2020.114129 [DOI] [PubMed] [Google Scholar]
- Turkmen R., Birdane Y. O., Demirel H. H., Kabu M., Ince S. (2019). Protective effects of resveratrol on biomarkers of oxidative stress, biochemical and histopathological changes induced by sub-chronic oral glyphosate-based herbicide in rats. Toxicol. Res. 8, 238–245. 10.1039/C8TX00287H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh L. P., McCormick C., Martin C., Stocco D. M. (2000). Roundup inhibits steroidogenesis by disrupting steroidogenic acute regulatory (StAR) protein expression. Environ. Health Perspect. 108, 769–776. 10.1289/ehp.00108769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li W., Muhammad I., Sun X., Cui X., Cheng P., et al. (2018). Biochemical basis for the age-related sensitivity of broilers to aflatoxin B1. Toxicol. Mech. Methods 28, 361–368. 10.1080/15376516.2018.1428258 [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang S., Zhong R., Wan F., Chen L., Liu L., et al. (2021). Olive fruit extracts supplement improve antioxidant capacity via altering colonic microbiota composition in mice. Front. Nutr. 8, 645099. 10.3389/fnut.2021.645099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. M., Kroes R., Munro I. C. (2000). Safety evaluation and risk assessment of the herbicide roundup and its active ingredient, glyphosate, for humans. Regul. Toxicol. Pharmacol. 31, 117–165. 10.1006/rtph.1999.1371 [DOI] [PubMed] [Google Scholar]
- Xing M., Zhao P., Guo G., Guo Y., Zhang K., Tian L., et al. (2015). Inflammatory factor Alterations in the gastrointestinal tract of cocks overexposed to arsenic trioxide. Biol. Trace Elem. Res. 167, 288–299. 10.1007/s12011-015-0305-8 [DOI] [PubMed] [Google Scholar]
- Zhang L., Gui S., Wang J., Chen Q., Zeng J., Liu A., et al. (2020). Oral administration of green tea polyphenols (TP) improves ileal injury and intestinal flora disorder in mice with Salmonella typhimurium infection via resisting inflammation, enhancing antioxidant action and preserving tight junction. J. Funct. Foods 64, 103654. 10.1016/j.jff.2019.103654 [DOI] [Google Scholar]
- Zhang Q., Liu X., Gao M., Li X., Wang Y., Chang Y., et al. (2022). The study of human serum metabolome on the health effects of glyphosate and early warning of potential damage. Chemosphere 298, 134308. 10.1016/j.chemosphere.2022.134308 [DOI] [PubMed] [Google Scholar]
- Zhao P., Zhang K., Guo G., Sun X., Chai H., Zhang W., et al. (2016). Heat shock protein alteration in the gastrointestinal tract tissues of chickens exposed to arsenic trioxide. Biol. Trace Elem. Res. 170, 224–236. 10.1007/s12011-015-0462-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJNA741111.