Abstract
Pseudomonas aeruginosa is able to use nitrate for both assimilation and anaerobic respiration. One set of genes, designated snr (for “shared nitrate reduction”), have been recently cloned and partially characterized. In this study, we demonstrate that the snr-1 gene encodes a predicted 52.5-kDa protein that is 82% similar to a unique cytochrome c of Desulfomonile tiedjei DCB-1. Importantly, the Snr-1 protein sequence of P. aeruginosa differed from that of the cytochrome c of D. tiedjei primarily in the first 25 amino acids, which are required for membrane attachment in D. tiedjei. In P. aeruginosa, the Snr-1 protein hydropathy profile indicates that it is a soluble protein. An isogenic snr-1::Gm insertional mutant was unable to grow aerobically with nitrate as a sole nitrogen source or anaerobically with nitrate as an electron acceptor. Complementation of the snr-1::Gm mutant with the snr-1 gene restored the wild-type phenotypes. Interestingly, anaerobic growth rates were significantly higher in the snr-1 mutant harboring a multicopy plasmid containing snr-1. In contrast, aerobic growth rates of the restored mutant using nitrate as the sole nitrogen source were similar to those of the wild type. Transcriptional lacZ fusions demonstrated that snr-1 was not regulated by molybdate, oxygen, or nitrate.
A few organisms possess the ability to both assimilate and dissimilate nitrate. One example of a bacterium capable of both processes is Pseudomonas aeruginosa. Pseudomonas aeruginosa possesses a metabolism entirely different from that of enteric bacteria. It does not ferment and is an obligate respirer. In contrast to the enterics, P. aeruginosa is a denitrifier. Both nas and nar gene mutants, molybdenum cofactor (MoCo)-processing mutants, and mutants defective in an FNR-like protein designated ANR, have been isolated in P. aeruginosa (4, 20, 24, 31). Thus, P. aeruginosa is a good organism with which to study the role of both pathways and is an excellent biological system for purposes of unveiling molecular differences between systems with different evolutionary backgrounds.
Since the first step of each nitrate reduction pathway is the identical reduction of nitrate to nitrite, it has been suggested for some time now that common or shared gene products may exist in P. aeruginosa (6, 7). One set of genes has been clearly demonstrated to be shared and is concerned with the transport of molybdate and the synthesis of the MoCo. The generic genetic nomenclature for these genes is mol. MoCo is found in the catalytic subunit of both assimilatory and dissimilatory nitrate reductase enzyme complexes of P. aeruginosa (6). Single-gene mutations in MoCo synthesis in this organism result in the inability to utilize nitrate by either pathway. MoCo mutants also will not grow on hypoxanthine (Hx) as a sole nitrogen source because the metabolism of Hx requires the MoCo-containing xanthine dehydrogenase enzyme. The inability to utilize Hx is an indicator of a defect in the synthesis of MoCo or transport of molybdate across the membrane. MoCo is a highly conserved cofactor that is utilized by a wide variety of different enzymes, and there is good evidence that various MoCo-requiring enzyme systems have a common MoCo (17).
There is a second set of shared genes in P. aeruginosa that do not involve MoCo synthesis. Mutations in this set of genes also result in the inability to assimilate or dissimilate nitrate to nitrite (6, 7, 9). The designation for these genes is snr (for “shared nitrate reduction”) (9). The snr mutants of P. aeruginosa PAO1 are deficient in both assimilation and dissimilation of nitrate but are still able to grow on Hx. Some of the snr and mol genes of P. aeruginosa have been recently isolated and cloned (9).
An examination of 12 snr mutants of P. aeruginosa PAO1, along with complementation studies, indicated the existence of four snr genetic loci. The snr-1, snr-2, and snr-3 genes are designated PA3032 (bp 3,396,727 to 3,395,324), PA2613 (bp 2,954,817 to 2,955,142), and PA3256 (bp 3,642,833 to 3,641,871), respectively, based on the location found in the Pseudomonas genome project (www.pseudomonas.com), while the location of snr-4 is unknown (9). Here we report that the snr-1 gene is required for assimilation or respiration of nitrate to nitrite. The snr-1 gene product is characterized with respect to the transcriptional regulation and predicted physiological function of its gene product.
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa PAO1 MJ mutants (class I and II) were previously isolated in this laboratory by Goldflam and Rowe (6). The genes representing the class I and class II loci were subcloned and characterized in a previous study (9). The vectors pUCP18/19, pUCGM, pZ1918G, and pEX100T were kindly provided by H. P. Schweizer (Department of Microbiology, Colorado State University) (21 to 23). All bacteria were grown from single-colony isolates or overnight cultures in Luria broth (10.0 g of tryptone, 5.0 g of yeast extract, and 5.0 g of NaCl per liter [pH 7.2]). For nitrate assimilation, the basal salts medium of Vogel and Bonner (VB) was used (10.0 g of K2HPO4, 3.5 g of NaH2PO4, 0.4 g of MgSO4, 4.0 g of citrate, 5.0 g of glucose, and 10.0 g of KNO3 per liter) (29). For nitrate dissimilation, nutrient broth (20.0 g/liter) was supplemented with 0.5% (wt/vol) glucose and 1.0% (wt/vol) KNO3. Anaerobic conditions were created by adding 10.0% (wt/vol) Oxyrase enzyme to solid media or broth. Aerobic liquid cultures were grown at 37°C with shaking at 300 rpm unless otherwise indicated. Culture volumes were 1/10 of the total Erlenmeyer flask volume to ensure proper aeration. Solid media were made by the addition of 1.5% Bacto Agar (Difco) to the appropriate broth medium. Antibiotics were used for Escherichia coli at the following concentrations (micrograms per milliliter): ampicillin, 100; kanamycin, 50; and gentamicin, 15. For P. aeruginosa, gentamicin and carbenicillin were used at 300 and 500 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, or relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17supE44 relA1 ΔlacU169 (φ80lacZΔM15) | Bethesda Research Laboratories |
| SM10 | Mobilizer strain, Kmr | 25 |
| P. aeruginosa | ||
| PAO1 | Wild type | |
| MJ2 | Defect in nitrate assimilation, nitrate dissimilation (snr mutant) | 6 |
| MJ4 | Defect in nitrate assimilation, nitrate dissimilation (mol mutant) | 6 |
| Plasmids | ||
| pCP13 | Tcr Kmr Mob IncP (23 kb); cosmid cloning vector derived from pLAFR1 | 9 |
| pAD1695 | Tcr Mob IncP (ca. 49 kb); ca. 26-kb chromosomal DNA from P. aeruginosa (partially digested with HindIII cloned into pCP13) | 9 |
| pAD1696 | Tcr Mob IncP (ca. 47.3 kb); ca. 24.3-kb chromosomal DNA from P. aeruginosa (partially digested with HindIII cloned into pCP13) | 9 |
| pRK2013 | Kmr ColE1-Tra (RK2)+ (ca. 50 kb) | 3 |
| pUCP18/19 | Apr, broad-host-range pUCP18/19 derivatives | 21 |
| pKS- | ColE1, Apr, lacZ cloning vector | Stratagene |
| pUCGM | ColE1, Apr GmraacCl | 22 |
| pZ1918G | ColE1, Apr GmrlacZ-aacC1 cassette plasmid | 23 |
| pSNR1 | Apr (ca. 7.3 kb); ca. 2.8-kb chromosomal DNA from P. aeruginosa (digested with SalI cloned into pUCP18) | This study |
| pEX100T | ColE1 AproriT mob sacB for construction of mutants | 23 |
| pEX100T-snr-1 | Apr Gmr, pEX100T + 2.8-kb HincII snr-1-containing fragment of pAD1696 (pSNR50) | This study |
| pEX100T-snr-1::Gm | Apr Gmr, pEX100T::snr-1 + 850-bp fragment of pUCGM (pSNR50G) | This study |
| pEX100T-snr-1::lacZ | Apr Gmr, pEX100T::snr-1 + 4.05-kb fragment of pZ1918G (pSNR50L) | This study |
Abbreviations: Apr, ampicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; IncP, incompatibility group P; Tra+, self-transmissible.
Molecular methods.
Plasmid purification, restriction endonuclease analysis, dephosphorylation, ligation, and transformation were all carried out using techniques described by Sambrook et al. (19). DNA transformation was performed in E. coli DH5α and SM10 as plasmid recipients (25). 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml) was routinely added to agar medium to detect the presence of insert DNA. Transformation of plasmids into P. aeruginosa PAO1 was performed as previously described (8). Triparental matings, as described previously by Goldberg and Ohman (5), were used to mobilize plasmids into P. aeruginosa. Chromosomal DNA was isolated from P. aeruginosa as previously described for gram-negative bacteria (1). Southern blot analysis was performed as previously described (27). Restriction fragments were recovered from agarose gels by using SeaPlaque low-melting-point agarose (FMC BioProducts, Rockland, Maine) or a GeneClean II kit (Bio 101, Inc., La Jolla, Calif.).
Amino acid sequence homology and hydropathy profiles of Snr-1.
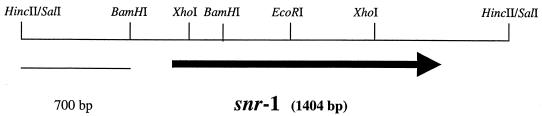
The location of the snr-1 gene has been identified as PA3032 (bp 3,396,727–3,395,324 bp) in the P. aeruginosa genome (www.pseudomonas.com). Plasmid pAD1696 was digested with HincII, and fragments were subsequently subcloned into the broad-host-range vector pUCP18 (21). These recombinant plasmids were transformed into the nitrate reduction-deficient snr-1 (class II) MJ2 mutant as previously described (9). After transformation, the bacteria were plated onto selective VBA medium supplemented with 1% KNO3 and 500 μg of carbenicillin per ml. The agar cultures were incubated aerobically or anaerobically at 37°C for 48 to 72 h. After incubation, strains harboring the insert that restored both assimilatory and dissimilatory nitrate reductase activities of the class II snr-1 MJ2 mutant were selected for further analysis. The HincII insert was sequenced, and the snr-1 gene was identified. The partial restriction map of snr-1 on the 2.8-kb HincII fragment is shown in Fig. 1. The snr-1 gene encodes a predicted protein of 52.5 kDa that is 82% similar to a unique cytochrome c of Desulfomonile tiedjei DCB-1. In contrast to P. aeruginosa, D. tiedjei is a sulfate-reducing obligate anaerobe that was originally identified in an enrichment culture for methanogens (16, 17). However, when sulfate is not present as an electron acceptor, D. tiedjei is able to perform anaerobic respiration in a process called dehalogenation (16, 17).
FIG. 1.
Restriction map of the snr-1 gene. A partial restriction map of the 2.8-kb HincII fragment containing the snr-1 gene is shown. The thick arrow indicates the location and direction of the snr-1 gene on this fragment.
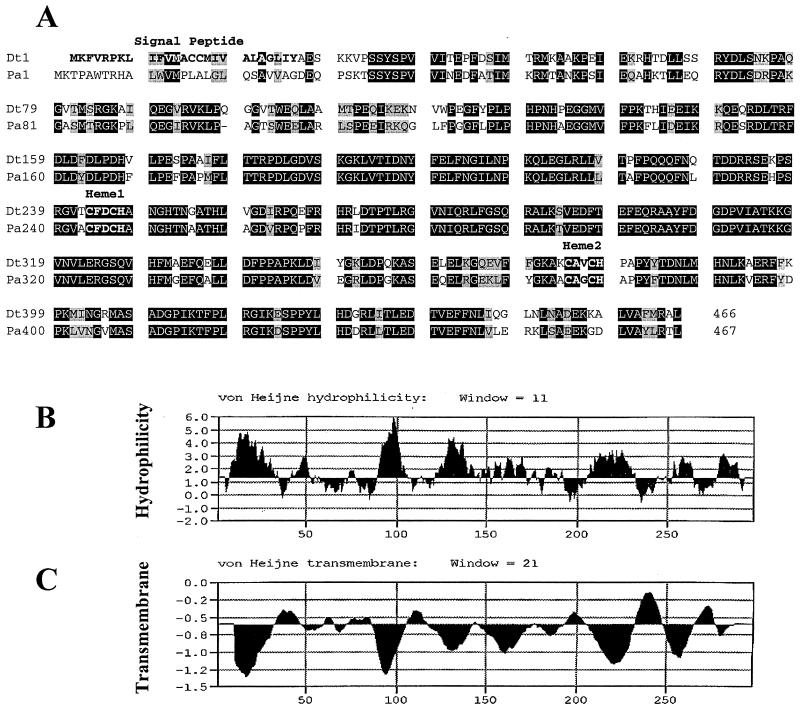
Cytochrome c of D. tiedjei was identified as a 52.6-kDa membrane-bound, diheme protein with an extremely low midpoint potential of −342 mV (14). It is believed to act as an electron donor for the dehalogenation respiratory system (14). Two heme motifs, which we believe are important as electron donors, are found in both sequences (Fig. 2A). The heme motifs are class I c-type motifs commonly found in mitochondrial cytochrome c (14). The CXXCH motif (Fig. 2A) represents the location where the heme groups covalently bind to the protein structure. A closer examination of the two sequences shows a high level of conserved regions located in and around both heme motifs. This leads us to believe that these areas are necessary for overall protein folding and functionality. One significant difference between the D. tiedjei cytochrome c and that of Snr-1 is that the former harbors a signal sequence (Fig. 2A). The N-terminal signal sequence of the D. tiedjei cytochrome c predicts that it is localized to the inner membrane (14). The hydropathy profile of Snr-1 shown in Fig. 2B suggests that it is a soluble protein. Figure 2C indicates that there are no transmembrane sequences within this protein. This is important since the assimilatory nitrate reduction system of P. aeruginosa is located in the cytoplasm.
FIG. 2.
Sequence homology comparisons and hydropathy profiles of Snr-1. (A) Comparison of the predicted amino acid sequence of the Snr-1 protein of P. aeruginosa PAO1 (Pa) with the predicted amino acid sequence of cytochrome c of D. tiedjei DCB-1 (Dt). The signal peptide of D. tiedjei is labeled and in boldface type. The two c-type heme motifs are labeled and in boldface type. (B) Hydropathy profile of Snr-1, generated using the Mac Vector (6.1.1) computer program. Any measurement greater than 0.00 represents a hydrophilic amino acid. (C) Profile of Snr-1 transmembrane segments. Any measurement above 0.00 is considered a transmembrane segment.
Construction of an snr-1::Gm mutant.
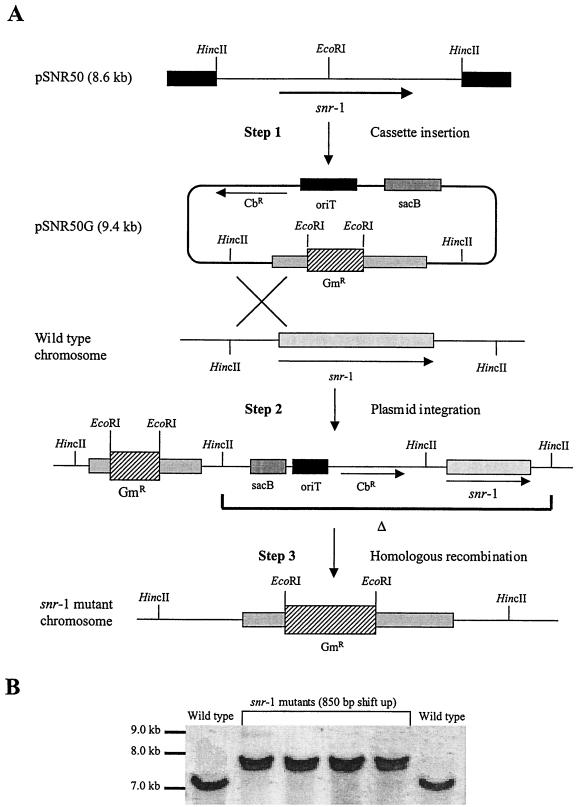
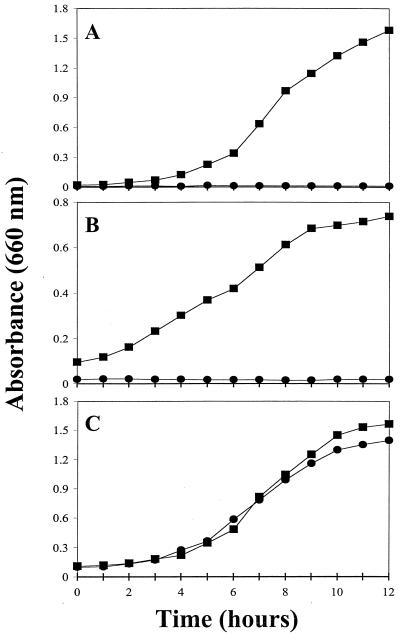
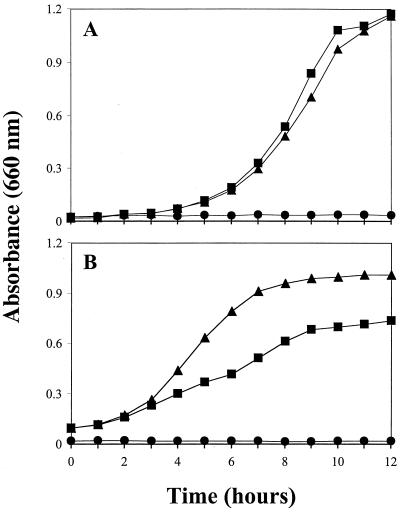
To confirm the overall importance of snr-1 to both nitrate reduction systems, we constructed an isogenic snr-1 mutant. An snr-1::Gm mutant was created by gene replacement as previously described, using the sucrose counterselection technique (23). The location and methods of introduction of the gentamicin cassette are shown in Fig. 3A. The integration and mutagenesis of snr-1 into the P. aeruginosa chromosome was confirmed by Southern blot analysis (Fig. 3B). Screened mutants were selected and grown both aerobically on VB medium with 1.0% nitrate and anaerobically on nutrient agar supplemented with 1.0% nitrate. The experimental cultures for aerobic growth studies of 1.0% nitrate and 0.1% Hx assimilation were inoculated with overnight cultures and subsequently grown at 37°C with shaking at 300 rpm. Experimental cultures for anaerobic growth studies of nitrate dissimilation were inoculated with overnight shaker-grown starter cultures and incubated at 37°C in Wheaton bottles flushed with argon followed by continuous magnetic stirring to prevent clumping of cells. Samples (3 ml) for both aerobic and anaerobic growth analysis were obtained from flask or bottle cultures at 1-h intervals, and growth was measured spectrophotometrically at an absorbance of 660 nm. Studies were done in triplicate, and the data were analyzed statistically using a nonparametric Kruskal-Wallis test (26). As shown in Fig. 4A and B, little or no growth was observed in the snr-1::Gm mutant. Even after a 72-h incubation, the mutant failed to display any organized CFU under aerobic or anaerobic conditions (data not shown). The aerobic growth rate during assimilation for wild-type bacteria was 1.67 h−1 (Fig. 4A). The growth rate during anaerobic respiration was 0.62 h−1 (Fig. 4B). There was essentially no growth of the mutant under either assimilation or denitrification conditions. To confirm that Snr-1 does not affect the MoCo of P. aeruginosa, we grew the snr-1::Gm mutant and wild-type organisms on VB medium supplemented with 0.1% Hx as the sole nitrogen source (Fig. 4C). The growth rates on Hx were 1.66 h−1 for wild-type bacteria and 1.54 h−1 for the snr-1::Gm mutant, thus demonstrating that disruption of the snr-1 gene does not affect the activity of the molybdoenzyme xanthine dehydrogenase.
FIG. 3.
Construction of an snr-1::Gm mutant and subsequent Southern blot analysis. (A) Mutagenesis of snr-1 in P. aeruginosa PAO1 by homologous recombination. Briefly, a ∼2.8-kb HincII snr-1-containing fragment of pAD1696 was subcloned into the SmaI site of pEX100T, forming pSNR50. As shown with step 1, an 850-bp gentamicin resistance (GmR) cassette derived from pUCGM was ligated into the unique EcoRI site within snr-1, forming pSNR50G. The subsequent plasmid was mobilized into P. aeruginosa PAO1 by triparental mating and colonies were selected for gentamicin resistance and finally for sucrose (counterselectable marker). In step 2, the transferred pSNR50G is integrated into the homologous chromosomal location of the wild-type strain. In step 3, homologous recombination takes place by selecting for sucrose-resistance. Sucrose-resistant and gentamicin-resistant bacteria were screened for the loss of carbenicillin resistance (CbR) and then screened for the loss of nitrate metabolism both aerobically on VBA supplemented with 1.0% nitrate and anaerobically on nutrient agar supplemented with 1.0% nitrate. (B) Southern blot analysis of genomic DNA from P. aeruginosa PAO1 and four putative snr-1 mutants cut with SmaI. The gentamicin cassette inserted into the EcoRI site within snr-1 would correspond with a ∼850-bp increase of the 7-kb SmaI fragment.
FIG. 4.
Growth during assimilatory nitrate utilization, anaerobic nitrate respiration, and Hx utilization. Growth was compared between wild-type PAO1 (■) and the snr-1::Gm mutant (●). (A) Assimilatory nitrate utilization on VB-medium with 1.0% KNO3 as the sole nitrogen source. (B) Anaerobic nitrate respiratory growth on nutrient broth containing 1.0% KNO3 as the terminal electron acceptor. (C) Utilization of Hx on VB medium using 0.1% Hx as the sole nitrogen source.
Complementation of the snr-1::Gm mutant.
The snr-1::Gm mutant of P. aeruginosa was complemented with pSNR1, containing a 2.8-kb SalI fragment with the intact snr-1 gene. The snr-1 mutant carrying pSNR1 was initially derived from single-colony isolates and grown in overnight cultures in VB broth at 37°C. Aerobic and anaerobic growth studies were performed as described above. The aerobic growth rate of the complemented mutant was comparable to that of the wild-type (Fig. 5A). The aerobic assimilatory growth rate of the wild-type organism was determined to be 1.67 h−1, while that of the complemented snr-1 mutant was calculated to be 1.64 h−1 (Fig. 5A). In contrast, we observed enhanced anaerobic growth of the complemented mutant containing multiple copies of the snr-1 gene compared to the wild-type organism (Fig. 5B). The anaerobic growth rate for wild-type bacteria was 0.61 h−1, while the growth rate for the complemented mutant containing multiple copies of snr-1 was 1.22 h−1 (Fig. 5B). When we applied an analysis of variance (ANOVA) test was applied to the calculated growth rates, we found them to be significantly different (K-W test, one-way ANOVA, P < 0.03, n = 3). This suggests that in the wild-type bacteria the level of Snr-1 limits anaerobic growth. Aerobically, multiple copies of Snr-1 did not have such an effect on the growth rate of this bacterium.
FIG. 5.
Aerobic assimilatory and anaerobic respiratory growth of a genetically complemented snr-1 mutant. (A) Aerobic growth curves of PAO1 (■), the snr-1::Gm mutant (●), and the genetically complemented snr-1 mutant (▴) with the 2.8-kb SalI fragment containing the snr-1 insert in a multiple-copy plasmid (10 to 20/cell), (pSNR1) in VB broth with 1.0% KNO3 as the sole nitrogen source. (B) Anaerobic growth curve of PAO1 (■), the snr-1::Gm mutant (●), and the genetically complemented snr-1 mutant (▴) in nutrient broth with 1.0% KNO3 as the terminal electron acceptor.
Transcriptional regulation studies using chromosomal snr-1::lacZ fusions.
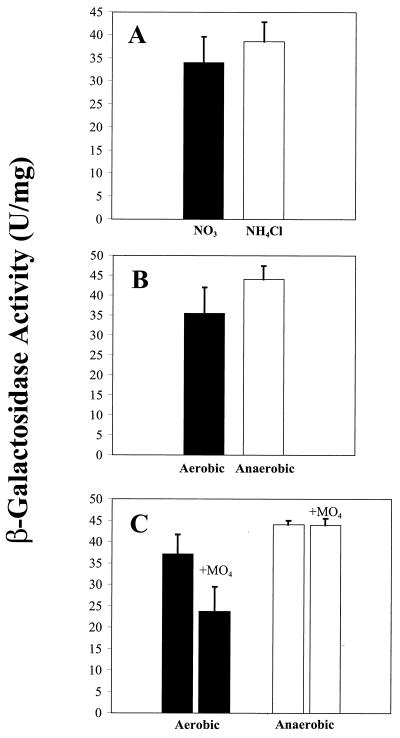
To examine the transcriptional regulation of snr-1, we performed β-galactosidase assays on cell extracts of P. aeruginosa containing a single-copy chromosomal snr-1::lacZ fusion. A 4.05-bp lacZ::Gm cassette was cloned into the unique XhoI site within the snr-1 gene contained in the pSNR50 plasmid. Merodiploid (single-crossover) colonies were selected and tested on medium containing X-Gal and gentamicin and confirmed by carbenicillin resistance. Single blue, Gmr Cbr colonies were selected and grown under appropriate growth conditions. The addition of 10 μM molybdate to overnight cultures was used to examine the effect of molybdate on expression. Cells were grown overnight at 37°C and collected by centrifugation (12,000 × g) for 10 min. They were subsequently washed in Z buffer (16.1 g of Na2HPO4, 5.5 g of NaH2PO4, 0.75 g of KCl, 0.25 g of MgSO4, and 2.7 ml of β-mercapthoethanol per liter) and assayed for β-galactosidase activity. The final activity was expressed in international units with a millimolar extinction coefficient of 3.1 using o-nitrophenyl-β-d-glucopyranoside (ONPG) (14). These studies were designed to examine the effects of snr-1 expression using oxygen, molybdate, ammonium, and nitrate, which are known to affect the regulation of nar and nas loci. Figure 6A and B demonstrate that the levels of snr-1 expression with the addition of nitrate or ammonium chloride and with or without oxygen are similar. We also examined the effect of molybdate on snr-1 expression both aerobically and anaerobically. Although there was a slight decrease in snr-1 expression under aerobic assimilatory conditions, molybdate had no effect on anaerobic snr-1 expression (Fig. 6C). An ANOVA statistical test was applied to all three sets of data (K-W, one-way ANOVA, P < 0.05, n = 3) to confirm that there was no significant difference. Thus, the snr-1 gene does not appear to be transcriptionally regulated by oxygen, molybdate, or nitrate.
FIG. 6.
β-Galactosidase production by P. aeruginosa harboring a single-copy snr-1::lacZ fusions under various growth conditions. (A) Expression of snr-1 under aerobic assimilatory growth in VB broth using 1.0% KNO3 as the sole nitrogen source and 0.1% ammonium chloride as the sole nitrogen source. (B) Expression of the snr-1 gene during aerobic growth in VB broth with 1.0% KNO3 as the sole nitrogen source and under conditions of anaerobic respiration using nutrient broth with 1.0% KNO3 as the terminal electron acceptor. (C) Expression of the snr-1 gene during aerobic growth in VB broth using 1.0% KNO3 as the sole nitrogen source with the addition of 10 μM molybdate and during anaerobic growth in nutrient broth with 1.0% KNO3 as the terminal electron acceptor with the addition of 10 μM molybdate. These experiments were performed in triplicate, and the data are expressed as the mean and standard error.
Conclusions.
The snr-1 gene codes for a protein which is 82% similar to the cytochrome c of D. tiedje, which has been implicated in the process of reductive dehalogenation (13, 15, 16). The Snr-1 protein is required for both the assimilatory and respiratory nitrate-to-nitrite reduction in P. aeruginosa. By analogy to the D. tiedjei system, the Snr-1 protein is likely to be involved in a necessary oxidation-reduction reaction in both assimilatory and respiratory nitrate reduction systems. Interestingly, Fewson and Nicholas (2) presented biochemical evidence in 1961 that a cytochrome c might be part of the respiratory nitrate reductase complex of P. aeruginosa. Nitrate reduction in P. aeruginosa involves the traditional nar genes originally identified in Escherichia coli (28) (www.pseudomonas.com), but in addition the Pseudomonas genome contains two open reading frames similar to narK of E. coli, which codes for a nitrite exporter (18). However, the nas system of P. aeruginosa appears to be quite different from that of the well-defined system of Klebsiella pneumoniae (10–12, 30) based on the homology found in the Pseudomonas genome (www.pseudomonas.com). The fact that the Snr-1 protein is soluble suggests that it may play an intermediate role in the reduction of the membrane respiratory nitrate reductase complex but that it may play a direct role in the reduction of the soluble assimilatory nitrate reductase. Thus, this modification of the traditional nitrate reduction models of enteric nitrate reduction may be significant to specific denitrifiers, which also possess the ability to assimilate nitrate.
Nucleotide sequence accession number.
GenBank accession number AF053982 has been assigned to the nucleotide sequence of the snr-1 genetic locus.
Acknowledgments
This work was supported in part through Public Health Service grant A1-40541 to D. J. Hassett and through the University of Dayton Council Summer Fellowships to Edward Kerschen, who is a doctoral candidate at the Department of Biology, University of Dayton.
We thank Al Darzins for providing the P. aeruginosa plasmid libraries as well as extensive consultation early on in the project, Wei Ping Shi for his preliminary work on the snr genes, and Ju-Fang Ma for help with the construction of the snr-1::Gm mutant.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates and John Wiley-Interscience; 1987. [Google Scholar]
- 2.Fewson C A, Nicholas J D. Nitrate reductase from Pseudomonas aeruginosa. Biochim Biophys Acta. 1961;49:335–349. doi: 10.1016/0006-3002(61)90133-0. [DOI] [PubMed] [Google Scholar]
- 3.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galmand M, Gamper M, Zimmermann A, Hass D. Positive FNR-line control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991;173:1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldflam M, Rowe J J. Evidence of gene sharing in the nitrate reduction systems of Pseudomonas aeruginosa. J Bacteriol. 1983;155:1446–1449. doi: 10.1128/jb.155.3.1446-1449.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez D, Rowe J J. Gas chromatographic assay for in vitro complementation of Pseudomonas aeruginosa mutants deficient in nitrate reduction. Appl Environ Microbiol. 1985;49:24–27. doi: 10.1128/aem.49.1.24-27.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irani V R, Rowe J J. Enhancement of transformation in Pseudomonas aeruginosa PAO1 by Mg2+ and heat. BioTechniques. 1997;22:54–56. doi: 10.2144/97221bm09. [DOI] [PubMed] [Google Scholar]
- 9.Irani V R, Darzins A, Rowe J J. snr, new genetic loci common to the nitrate reduction systems of Pseudomonas aeruginosa PAO1. Curr Microbiol. 1997;35:9–13. doi: 10.1007/s002849900202. [DOI] [PubMed] [Google Scholar]
- 10.Lin J T, Goldman B S, Stewart V. Structures of the genes nasA and nasB, encoding assimilatory nitrate and nitrite reductase in Klebsiella pneumoniae M5a1. J Bacteriol. 1993;175:2370–2378. doi: 10.1128/jb.175.8.2370-2378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J T, Goldman B S, Stewart V. The nasFEDCBA operon for nitrate and nitrite assimilation in Kelbsiella pneumoniae M5a1. J Bacteriol. 1994;176:2551–2559. doi: 10.1128/jb.176.9.2551-2559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J T, Stewart V. Nitrate assimilation by bacteria. Adv Microb Physiol. 1998;38:1–30. doi: 10.1016/s0065-2911(08)60014-4. [DOI] [PubMed] [Google Scholar]
- 13.Louie T M, Ni S, Xun L, Mohn W W. Purification, characterization, and gene sequence analysis of a novel cytochrome c co-induced with reductive dechlorination activity in Desulfomonile tiedjei DCB-1. Arch Microbiol. 1997;168:520–527. doi: 10.1007/s002030050530. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 72–74. [Google Scholar]
- 15.Mohn W W, Tiedje J M. Catabolic thiosulfate disproportionation and carbon dioxide reduction in strain DCB-1, a reductively dechlorinating anaerobe. J Bacteriol. 1990;172:2065–2070. doi: 10.1128/jb.172.4.2065-2070.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohn W W, Tiedje J M. Microbial reductive dehalogenation. Microbiol Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopalan K V, Johnson J L. The pterin molybdenum cofactor. J Biol Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- 18.Rowe J J, Ubbink-Kok T, Molenaar D, Konings W N, Driessen A J M. NarK is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol Microbiol. 1994;12:579–586. doi: 10.1111/j.1365-2958.1994.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sawers R G. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol Microbiol. 1991;122:263–270. doi: 10.1111/j.1365-2958.1991.tb00793.x. [DOI] [PubMed] [Google Scholar]
- 21.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 22.Schweizer H P. Small broad-host range gentamicin resistance cassette for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 23.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 24.Sias S R, Ingraham J H. Isolation and analysis of mutants of Pseudomonas aeruginosa unable to assimilate nitrate. Arch Microbiol. 1979;122:263–270. doi: 10.1007/BF00411289. [DOI] [PubMed] [Google Scholar]
- 25.Simon R, Priefer V, Puehller A. A broad-host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 26.Sokal R R, Rohlf F J. The principles and practice of statistics in biological research. 3rd ed. New York, N.Y: W. H. Freeman & Co.; 1994. [Google Scholar]
- 27.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 28.Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988;52:190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogel H, Bonner D M. Acetylornithinase of Escherichia coli, partial purification and properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 30.Wu Q, Stewart V. NasFED proteins mediate assimilatory nitrate reductase and nitrite transport in Klebsiella oxytoca (pneumoniae) M5a1. J Bacteriol. 1998;180:1311–1322. doi: 10.1128/jb.180.5.1311-1322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmerman A, Reimmann C, Galimand M, Hass D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991;5:1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]