Abstract
Microcin B17 is a peptide antibiotic that inhibits DNA replication in Escherichia coli by targeting DNA gyrase. Previously, two independently isolated microcin B17-resistant mutants were shown to harbor the same gyrB point mutation that results in the replacement of tryptophan 751 by arginine in the GyrB polypeptide. We used site-directed mutagenesis to construct mutants in which tryptophan 751 was deleted or replaced by other amino acids. These mutants exhibit altered DNA gyrase activity and different levels of resistance to microcin B17.
Type II DNA topoisomerases are enzymes that alter the topological state of DNA molecules by catalyzing strand transfer through transient, double-stranded breaks (24). Among them, bacterial DNA gyrase is the only topoisomerase known to be able to add negative supercoils to covalently closed circular DNA (11). Thanks to this unique property, DNA gyrase contributes to maintaining the supercoiling level required for bacterial DNA replication, transcription, and recombination (8, 24). The reaction involves the formation of a covalent protein-DNA intermediate and is coupled to the hydrolysis of ATP. Escherichia coli DNA gyrase is an A2B2 tetramer. The site for DNA breakage and rejoining is located in the A subunit, encoded by the gyrA gene, whereas the site for ATP binding and hydrolysis lies in the B subunit, encoded by the gyrB gene (19).
DNA gyrase is essential for cell viability, and therefore it is a prime target for antibacterial therapy. There are two classes of DNA gyrase inhibitors, those that compete with ATP for binding to the enzyme (e.g., coumarins), and those that act by trapping the gyrase-DNA reaction intermediate, the so-called cleavable complex (e.g., quinolones) (9, 17).
Microcin B17 (MccB17) is a hydrophobic peptide antibiotic that is active against most enterobacteria (2) and inhibits DNA gyrase by trapping the gyrase-DNA cleavable complex (23). As a result, DNA replication is blocked, the SOS response is induced, and massive degradation of DNA ensues (14). MccB17 is produced by Escherichia coli cells that harbor the plasmid-borne mcb operon (13, 22). The antibiotic is synthesized in the ribosome as a 69-amino-acid precursor (5) that undergoes two steps of posttranslational modification, the formation of eight heterocyclic rings (four oxazole and four thiazole rings), and the cleavage of a 26-amino-acid-long leader peptide (27).
In a previous work, two MccB17-resistant mutants were isolated independently by unrelated in vivo methods. Interestingly, both mutants carried the same AT→GC transition at position 2251 of the gyrB gene, which results in the replacement of tryptophan 751 by arginine (W751R) in the C terminus of the GyrB polypeptide (23). To investigate the role of this residue in the resistance to Mcc B17 and in the activity of DNA gyrase, we used site-directed mutagenesis to generate an in-frame deletion of this codon and to replace tryptophan 751 (W751) with other amino acids.
Site-directed mutagenesis of codon 751 of E. coli gyrB.
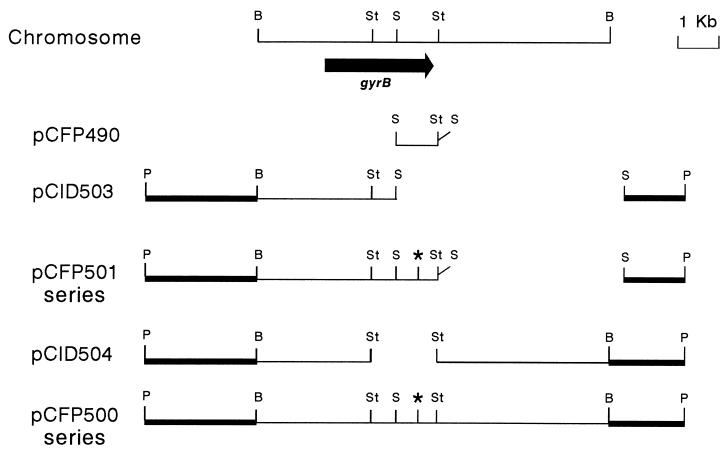
To obtain a template for site-directed mutagenesis, we amplified by PCR a 988-bp DNA fragment containing the 3′ end of the E. coli K-12 gyrB gene. This fragment stretches from the unique intragenic SalI site to the extragenic StuI site located downstream of gyrB (Fig. 1). For subsequent manipulation, we added a SalI site to the sequence of the lower primer that we used. The PCR fragment was cloned into the pMOSBlue phagemid T-vector (recombinant plasmid pCFP490) and sequenced to confirm that no artifactual mutations were introduced during PCR amplification.
FIG. 1.
Plasmids used for construction of gyrB mutants. Thick lines indicate sequences from pBR322, and thin lines indicate chromosomal sequences. Stars represent the engineered point mutations. B, BamHI; P, PstI; S, SalI; St, StuI.
Site-directed mutagenesis was performed by the Kunkel method (16), using plasmid pCFP490 as the template. We replaced the W751 residue with lysine (W751K), glutamic acid (W751E), phenylalanine (W751F), glycine (W751G), and histidine (W751H). In addition, we also constructed an in-frame deletion of the W751 residue (ΔW751). All plasmid inserts were sequenced after mutagenesis to confirm that the intended mutation had indeed taken place and that undesired mutations were absent.
The DNA fragments containing the desired mutations were recovered by SalI digestion of pCFP490 derivatives and cloned in the unique SalI site of plasmid pCID503 (7) (Fig. 1), which contains the 5′ end of gyrB, to assemble a complete, mutated gyrB gene (recombinant plasmids pCFP501K, pCFP501E, pCFP501F, pCFP501G, pCFP501H, and pCFP501ΔW).
Mutation ΔW751 abolishes GyrB function.
We tested whether the mutant gyrB genes encoded functional GyrB polypeptides. Strain LE316 (18), which carries a gyrB(Ts) mutant gene on its chromosome, was transformed with the pCFP501 series of plasmids. Transformants harboring plasmids that carried gyrB W751 substitutions E, F, G, H, and K grew at the restrictive temperature (42°C), so we concluded that these mutations do not abolish GyrB function. In addition, all transformants carrying plasmids with gyrB W751 substitutions displayed higher resistance to MccB17 than strain LE316, harboring a wild-type gyrB plasmid, when tested by the cross-streaking method (22) at 42°C.
On the other hand, transformants harboring plasmid pCFP501ΔW did not grow at 42°C. To discern whether the effect of the ΔW751 mutation on GyrB function was temperature dependent [i.e., whether the mutation generated a GyrB(Ts) polypeptide unable to complement the gyrB(Ts) mutation of strain LE316], we performed the following experiment. All derivatives of plasmid pCID503 (including pCFP501ΔW) carry the gyrB320 mutation (7), which confers resistance to coumermycin A1, a coumarin antibiotic. In contrast, strain LE316 is sensitive to coumermycin A1 at 30°C (7). LE316 harboring pCFP501ΔW did not grow at 30°C in the presence of coumermycin A1 (16 μg ml−1), while LE316 harboring other plasmids from the pCFP501 series (also carrying gyrB320) did grow. This result indicated that the in-frame deletion of codon 751 abolishes GyrB function irrespective of temperature.
Construction of chromosomal gyrB mutants.
We intended to replace the mutant alleles for the chromosomal wild-type allele by homologous recombination. To obtain a longer homology region downstream of the mutation point, we subcloned the 1.5-kb StuI fragment from every pCFP501 series plasmid into the StuI site of plasmid pCID504, creating recombinant plasmids pCFP500E, pCFP500F, pCFP500G, pCFP500H, and pCFP500K. Plasmid pCID504 (7) carries the same gyrB320 (Cour) mutation harbored by pCID503 (Fig. 1).
Allelic replacement was carried out by transforming a recBC sbcBC strain with the linearized plasmids of the pCFP500 series (26) and selecting for resistance to coumermycin A1. To determine which Cour clones harbored the desired gyrB W751 mutations, chromosomal DNA from each Cour clone was used as a template for allele-specific PCR amplifications of a 472-bp intragenic gyrB fragment. Finally, all gyrB W751 mutations were P1 transduced to strain RYC1010 (RYC1000 λp recA cIind) (23) for tests of MccB17 sensitivity and DNA gyrase function. The selection procedure that we used was identical to the one outlined above (resistance to coumermycin A1 plus allele-specific PCR). The resultant strains carried the desired substitutions: W751E (strain CFP1020E), W751F (CFP1020F), W751G (CFP1020G), W751H (CFP1020H), and W751K (CFP1020K).
Phenotypic characterization of gyrB mutants.
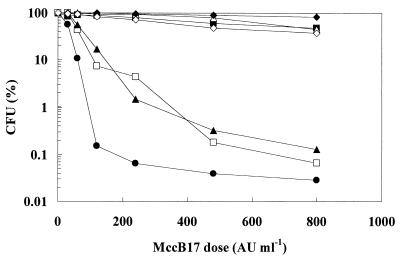
We assayed the effect of MccB17 on the viability of the gyrB mutants (Fig. 2). As controls, we used the isogenic wild-type strain RYC1020 (RYC1010 gyrB320) and the MccB17r mutant isolated by in vivo methods [RYC1030; relevant genotype, RYC1020 gyrB301 (W751R)]. Strains were grown in 20 ml of M63 glucose minimal medium to an optical density at 600 nm of 1.5. Cells were then collected by centrifugation and resuspended in 4 ml of fresh M63 medium. Aliquots of cells were treated with different doses of MccB17 at 37°C for 1 h, plated out on Luria-Bertani (LB) medium, and incubated at 37°C to determine the number of CFU. MccB17 stocks were obtained and titrated as described before (14).
FIG. 2.
Effect of MccB17 on the viability of E. coli gyrB mutant strains. Aliquots of cells were treated with different doses of MccB17 at 37°C for 1 h, plated out on LB medium, and incubated at 37°C to determine the number of CFU. Values are the means of three independent experiments. Symbols: ●, wild-type RYC1020; ■, CFP1020E W751E mutant; □, CFP1020F W751F mutant; ▵, CFP1020G W751G mutant; ▴, CFP1020H W751H mutant; ⧫, CFP1020K W751K mutant; ◊, RYC1030 W751R mutant. AU, antibiotic units; CFU (%), percentage of CFU surviving after MccB17 treatment.
While all mutant strains were less sensitive to MccB17 than the wild-type strain, the higher levels of resistance were due to mutations which replaced large hydrophilic polar side chains (lysine, glutamic acid, and arginine) or very small side chains (glycine) with the hydrophobic side chain of tryptophan (Fig. 2). Interestingly, mutants that harbored aromatic-for-aromatic substitutions of residue 751, such as phenylalanine (nonpolar) or histidine (polar) for tryptophan, retained a significant level of MccB17 sensitivity (Fig. 2). Taken together, these results suggest that removal of the aromatic side chain of tryptophan plays a major role in bringing about MccB17 resistance, while increasing the polarity of the side chain of residue 751 is not sufficient by itself to prevent the inhibition of DNA gyrase by MccB17.
We next tested the effect of gyrB mutations on growth rate. No differences in growth rate were observed in LB rich medium. However, strains CFP1020E and CFP1020K grew more slowly in M63 glucose minimal medium than the wild-type strain (data not shown). This result suggested that DNA gyrase activity might be altered in these mutants.
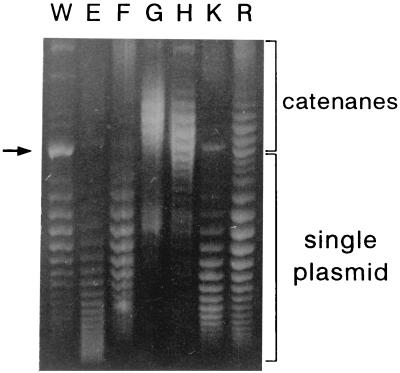
To explore this possibility, we assayed the effect of gyrB W751 mutations on the supercoiling of a reporter plasmid (Fig. 3). Wild-type strain RYC1020 (W) and its gyrB mutant derivatives (E, F, G, H, K, and R) were transformed with multicopy plasmid pPH16 (6). Strains were grown in LB medium supplemented with 40 μg of ampicillin per ml, and plasmid DNA was isolated by using Plasmid DNA Midi kits (Qiagen). Plasmid topoisomers were separated in 0.8% agarose–chloroquine gels as described (15). Plasmids extracted from strains CFP1020E and CFP1020K showed higher levels of negative supercoiling than the plasmid extracted from the control strain (Fig. 3, lanes W, E, and K), which might account for the slower growth rate of these two mutant strains, since it is known that excess negative supercoiling impairs many cellular processes (24). Most of the plasmids isolated from strains CFP1020G and CFP1020H appeared as catenanes (Fig. 3, lanes G and H). This result indicated that separation of daughter molecules after plasmid replication, a process dependent on the decatenating activity of DNA gyrase, was taking place in those mutants at an unusually slow rate. Finally, plasmids extracted from strains CFP1020F and RYC1030 (W751R mutant) showed a supercoiling level roughly similar to that of the plasmid extracted from the wild-type strain (Fig. 3, lanes W, F, and R).
FIG. 3.
Effect of gyrB mutations on the supercoiling of a reporter plasmid. Plasmid pPH16 topoisomers isolated from wild-type strain RYC1020 (W) and its gyrB mutant derivatives (E, F, G, H, K, and R) were separated in 0.8% agarose–chloroquine gels. Electrophoresis was performed in 90 mM Tris-borate (pH 8.3)–10 mM EDTA–10 μg of chloroquine per ml at 3 V cm−1 for 18 h. Under these conditions, the more negatively supercoiled topoisomers migrate faster. The arrow indicates the position of pPH16 DNA after in vitro treatment with DNA topoisomerase I to achieve complete relaxation.
Taken together, our results highlight the role of the tryptophan 751 residue of the GyrB polypeptide in the biological activity of DNA gyrase. In fact, the alignment of sequences of type II DNA topoisomerases from different organisms reveals extensive similarities in the sequence stretch that includes this residue (Fig. 4). In bacteria and archaea, residues at the position equivalent to E. coli GyrB W751 are tryptophan or arginine. Remarkably, the MccB17-resistant mutants isolated by in vivo methods carry the W751R substitution, which in our experiments showed a normal growth rate with a supercoiling level similar to that of the wild type. Other substitutions generated by in vitro mutagenesis resulted in either altered DNA gyrase activity (high supercoiling levels concomitant with a diminished growth rate [W751E and W751K] or a deficiency in decatenating activity [W751G and W751H]) or low-level resistance to MccB17 (W751F).
FIG. 4.
Alignment of amino acid sequences of type II DNA topoisomerases from different organisms. Prokaryotic type II topoisomerases include GyrB polypeptides from DNA gyrase and ParE polypeptides from DNA topoisomerase IV. Stars indicate identical amino acids. The residues equivalent to E. coli GyrB tryptophan 751 appear in bold.
Although the crystal structure of the E. coli GyrB N-terminal domain has already been solved (25), no data are available for the C-terminal domain. However, the crystal structure of a large fragment of yeast type II topoisomerase containing the domain homologous to the GyrB C terminus (the so-called B′ domain) has been solved at a resolution of 2.7 Å (3). The position equivalent to E. coli GyrB W751 is occupied by arginine 615 in the yeast enzyme (Fig. 4), a residue that is located in the B′ fragment, as part of the α8 helix. It should be noted that in most prokaryotic type II topoisomerases, α8 helix would be either kinked or truncated six residues downstream of W751 due to the existence of a proline residue in the position equivalent to Saccharomyces cerevisiae leucine 621 (Fig. 4). The results of recent studies on the mechanisms of type II DNA topoisomerases support a model in which the enzyme acts as an ATP-dependent molecular clamp with two gates for the strand passage (20, 21). In this model, the DNA segment to be transported (T-segment) enters through the gate on one side of the enzyme, crosses the cleaved, enzyme-bound DNA segment (G-segment), and, moving between the two monomers of the enzyme, exits through a second gate on the opposite side of the complex (3, 10, 20, 21). Interestingly, the B′ domain is part of the entry gate, and so it is tempting to speculate on a putative direct interaction of the hydrophobic MccB17 with GyrB W751 and residues in its spatial vicinity, an interaction that may block strand passage.
MccB17 is the only peptide inhibitor of DNA gyrase whose resistance mutations map within the B′ domain of GyrB. Mutations conferring resistance to MccB17 have been shown to exert very diverse effects on in vivo DNA gyrase activity. Therefore, MccB17 should be a valuable tool for probing the role of the B′ domain in the DNA gyrase protein complex.
Acknowledgments
F. J. del Castillo was the recipient of a fellowship from Fundación Ramón Areces.
REFERENCES
- 1.Adachi T, Mizuuchi M, Robinson E A, Appella E, O'Dea M H, Gellert M, Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987;15:771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baquero F, Moreno F. The microcins. FEMS Microbiol Lett. 1984;23:117–124. [Google Scholar]
- 3.Berger J M, Gamblin S J, Harrison S C, Wang J C. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Peebles C L, Cozzarelli N R. A topoisomerase from Escherichia coli related to DNA gyrase. Proc Natl Acad Sci USA. 1979;76:6110–6114. doi: 10.1073/pnas.76.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davagnino J, Herrero M, Furlong D, Moreno F, Kolter R. The DNA replication inhibitor microcin B17 is a forty-three-amino-acid protein containing sixty percent glycine. Proteins. 1986;1:230–238. doi: 10.1002/prot.340010305. [DOI] [PubMed] [Google Scholar]
- 6.del Castillo F J, Leal S C, Moreno F, del Castillo I. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol Microbiol. 1997;25:107–115. doi: 10.1046/j.1365-2958.1997.4391813.x. [DOI] [PubMed] [Google Scholar]
- 7.del Castillo I, Vizán J L, Rodríguez-Sáinz M C, Moreno F. An unusual mechanism for resistance to the antibiotic coumermycin A1. Proc Natl Acad Sci USA. 1991;88:8860–8864. doi: 10.1073/pnas.88.19.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984;48:273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drlica K, Franco R J. Inhibitors of DNA topoisomerases. Biochemistry. 1988;27:2253–2259. doi: 10.1021/bi00407a001. [DOI] [PubMed] [Google Scholar]
- 10.Fass D, Bogden C E, Berger J M. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat Struct Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 11.Gellert M, Mizuuchi K, O'Dea M H, Nash H A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA. 1976;73:3872–3875. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellert M, Fisher L M, O'Dea M H. DNA gyrase: purification and properties of a fragment of the gyrase B protein. Proc Natl Acad Sci USA. 1979;76:6289–6293. doi: 10.1073/pnas.76.12.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genilloud O, Moreno F, Kolter R. DNA sequence, products, and transcriptional pattern of the genes involved in production of the DNA replication inhibitor microcin B17. J Bacteriol. 1989;171:1126–1135. doi: 10.1128/jb.171.2.1126-1135.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrero M, Moreno F. Microcin B17 blocks DNA replication and induces the SOS system in Escherichia coli. J Gen Microbiol. 1986;132:393–402. doi: 10.1099/00221287-132-2-393. [DOI] [PubMed] [Google Scholar]
- 15.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 17.Maxwell A. DNA gyrase as a drug target. Trends Microbiol. 1997;5:102–109. doi: 10.1016/S0966-842X(96)10085-8. [DOI] [PubMed] [Google Scholar]
- 18.Orr E, Fairweather N F, Holland I B, Pritchard R H. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177:103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- 19.Reece R J, Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 20.Roca J, Wang J C. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 21.Roca J, Berger J M, Harrison S C, Wang J C. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc Natl Acad Sci USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.San Millán J L, Hernández-Chico C, Pereda P, Moreno F. Cloning and mapping of the genetic determinants for microcin B17 production and immunity. J Bacteriol. 1985;163:275–281. doi: 10.1128/jb.163.1.275-281.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vizán J L, Hernández-Chico C, del Castillo I, Moreno F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 1991;10:467–476. doi: 10.1002/j.1460-2075.1991.tb07969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 25.Wigley D B, Davies G J, Dodson E J, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 26.Winans S C, Elledge S J, Krueger J H, Walker G C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yorgey P, Lee J, Kördel J, Vivas E, Warner P, Jebaratnam D, Kolter R. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc Natl Acad Sci USA. 1994;91:4519–4523. doi: 10.1073/pnas.91.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]