To the Editor: Multiple sublineages of the omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have dominated the coronavirus disease 2019 pandemic since December 2021. BA.5 is currently the dominant omicron sublineage, representing more than 50% of new cases since early July 2022 and exhibiting the greatest ability to escape neutralizing antibodies among all the SARS-CoV-2 variants to date. Another omicron sublineage, BA.2.75, recently emerged with a slow but alarmingly steady increase in prevalence. As of August 19, 2022, BA.2.75 has been detected in at least 35 countries and in 20 U.S. states1 and is being monitored as the next potentially predominant globally circulating variant. The ability of BA.2.75 to escape vaccine-induced neutralizing antibodies is of high interest.
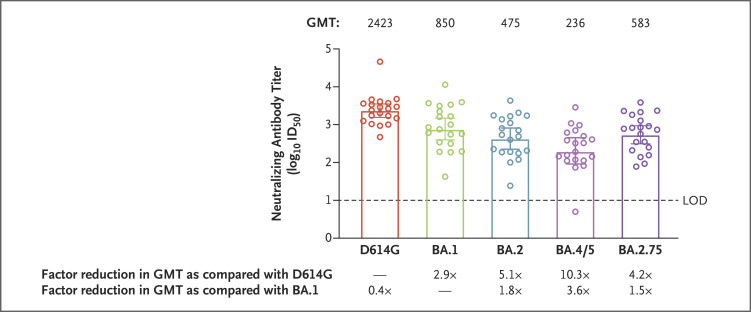
In a phase 2 clinical trial, we characterized the neutralization susceptibility of BA.2.75 through the use of serum samples obtained 29 days after a 50-μg booster dose of the mRNA-1273 vaccine (Moderna) from 20 adults who had received primary vaccination with two 100-μg doses of mRNA-1273 (Table S1 in the Supplementary Appendix, available with the full text of this letter at NEJM.org).2 The neutralization assay used lentivirus-based pseudoviruses and was performed in 293T cells that were stably transduced to overexpress angiotensin-converting enzyme 2.3 The 50% inhibitory dilution (ID50) geometric mean titers (GMTs) for the 20 serum samples obtained 1 month after the mRNA-1273 booster dose were 2.5 (95% confidence interval [CI], 1.8 to 3.4) times higher against BA.2.75 (GMT, 583) than they were against BA.5 (GMT, 236) and more closely resembled the titers against the BA.1 (GMT, 850) and BA.2 (GMT, 475) sublineages that drove the initial omicron wave (Figure 1). Thus, unlike BA.5, the BA.2.75 omicron sublineage has not evolved toward greater escape from mRNA-1273–induced neutralizing antibodies than the original BA.1 and BA.2 sublineages but nonetheless remains 4.2 (95% CI, 2.7 to 6.3) times less sensitive to neutralization than the prototypic D614G variant (Figure 1 and Table S2).
Figure 1. Neutralization of SARS-CoV-2 D614G and Omicron Variants after mRNA-1273 Booster Dose.
In a phase 2 trial, 20 serum samples were obtained from participants 18 years of age or older (mean age, 55.3) who had received a two-dose primary vaccination series and a single 50-μg booster dose of mRNA-1273 (Table S1 in the Supplementary Appendix).2 The 50% inhibitory dilution (ID50) geometric mean titers (GMTs) of the samples against ancestral severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) D614G and omicron sublineages (BA.1, BA.2, BA.4 and BA.5 [BA.4/5], and BA.2.75) were assessed at day 29 after administration of the booster dose. The median time from the second dose of primary vaccination to the booster dose was 221 days.4 Data regarding neutralization of D614G and BA.1 by these samples have been reported previously.4 The spike mutations of Wuhan-1 (D614G) and the four sublineages of the omicron variant are provided in Figure S1.5 Mutations in the spike of the BA.2.75 pseudovirus relative to Wuhan-1 are T19I, L24S, P25del, P26del, A27del, G142D, K147E, W152R, F157L, I210V, V213G, G257S, G339H, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, G446S, N460K, S477N, T478K, E484A, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, and N969K. Differences in receptor-binding domain spike mutations of BA.2.75 and BA.4/5 include N460K (present in BA.2.75 and absent in BA.4/5), L452R (present in BA.4/5 and absent in BA.2.75), F486V (present in BA.4/5 and absent in BA.2.75), and G339H in BA.2.75, instead of G339D in BA.4/5.5 The GMT values relative to D614G and BA.1 for each variant were obtained by first calculating the mean difference in log10 scale; the mean was then back-transformed to the original scale for presentation. The 95% confidence intervals for the GMT reductions were calculated on the basis of the distribution of differences in the log-transformed values and were then back-transformed to the original scale for presentation (Table S2); these 95% confidence intervals are shown as 𝙸 bars in the figure. The circles represent individual samples. The limit of detection (LOD [dashed line]) of the assays was 10. Values below the LOD were assigned a value of 5 for the calculation of summary statistics.
The spike mutations in BA.2.75, including the mutations in the receptor-binding domain (a major target for neutralizing antibodies), most closely resemble those in BA.2 (Fig. S1). Indeed, two receptor-binding domain mutations that emerged in BA.5 (L452R and F486V) are absent in BA.2.75, a finding that possibly explains why BA.2.75 most closely resembled BA.2 with respect to its neutralization by serum samples from persons who had received mRNA-1273 booster doses. In addition, the N460K mutation that arose in the receptor-binding domain of BA.2.75 does not appear to substantially contribute to escape. The increasing spread and possible competitive advantage of BA.2.75 over BA.5 is most likely due to factors other than heightened neutralization escape. Overall, vaccine efficacy against BA.2.75 is expected to be similar to that against BA.1 and BA.2. Whether and by how much another vaccine booster, particularly one containing an omicron spike, would elicit a more potent response against BA.2.75 and future SARS-CoV-2 variants remains of high interest for vaccine development.
Supplementary Appendix
Disclosure Forms
Access to patient-level data and supporting clinical documents by qualified external researchers may be available on request and subject to review.
This letter was published on September 9, 2022, at NEJM.org.
Footnotes
Supported by the Office of the Administration for Strategic Preparedness and Response, Biomedical Advanced Research and Development Authority (contract number, 75A50120C00034) and Moderna. The development of the microneutralization assay was funded by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services (NIAID Preclinical Services contract number, HHSN272201800013I/75N93020F00002).
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Outbreak.info. SARS-CoV-2 (hCoV-19) mutation reports location tracker. 2022. (https://outbreak.info/location-reports).
- 2.Chu L, McPhee R, Huang W, et al. A preliminary report of a randomized controlled phase 2 trial of the safety and immunogenicity of mRNA-1273 SARS-CoV-2 vaccine. Vaccine 2021;39:2791-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen X, Tang H, McDanal C, et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 2021;29(4):529-539.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pajon R, Doria-Rose NA, Shen X, et al. SARS-CoV-2 omicron variant neutralization after mRNA-1273 booster vaccination. N Engl J Med 2022;386:1088-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyke KE, Atmar RL, Islas CD, et al. Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 omicron variant. Cell Rep Med 2022;3:100679-100679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.