Abstract
Background
T cell-mediated antitumor immunity has a vital role in cancer prevention and treatment; however, the immune-suppressive tumor microenvironment (TME) constitutes a significant contributor to immune evasion that weakens antitumor immunity. Here, we explore the relationship between nucleus accumbens-associated protein-1 (NAC1), a nuclear factor of the BTB (broad-complex, Tramtrack, bric a brac)/POZ (Poxvirus, and Zinc finger) gene family, and the TME.
Methods
Adoptive cell transfer (ACT) of mouse or human tumor antigen (Ag)-specific CD8+ cytotoxic T lymphocytes (CTLs) was tested in an immunocompetent or immunodeficient mouse model of melanoma with or without expression of NAC1. The effects of NAC1 expression on immune evasion in tumor cells were assessed in vitro and in vivo. CRISPR/Cas9, glycolysis analysis, retroviral transduction, quantitative real-time PCR, flow cytometric analysis, immunoblotting, database analyses were used to screen the downstream target and underlying mechanism of NAC1 in tumor cells.
Results
Tumorous expression of NAC1 negatively impacts the CTL-mediated antitumor immunity via lactate dehydrogenase A (LDHA)-mediated suppressive TME. NAC1 positively regulated the expression of LDHA at the transcriptional level, which led to higher accumulation of lactic acid in the TME. This inhibited the cytokine production and induced exhaustion and apoptosis of CTLs, impairing their cell-killing ability. In the immunocompetent and immunodeficient mice, NAC1 depleted melanoma tumors grew significantly slower and had an elevated infiltration of tumor Ag-specific CTLs following ACT, compared with the control groups.
Conclusions
Tumor expression of NAC1 contributes substantially to immune evasion through its regulatory role in LDHA expression and lactic acid production. Thus, therapeutic targeting of NAC1 warrants further exploration as a potential strategy to reinforce cancer immunotherapy, such as the ACT of CTLs.
Keywords: Immunotherapy, Immunomodulation, Tumor Escape, Tumor Microenvironment
WHAT IS ALREADY KNOWN ON THIS TOPIC
Expression of nucleus accumbens-associated protein-1 (NAC1) is closely associated with tumor development and poor prognosis in various types of cancers.
WHAT THIS STUDY ADDS
The crucial relationship between NAC1 and the immune-suppressive tumor microenvironment (TME) is identified. Tumorous NAC1 is classified as a critical determinant of antitumor immunity. In addition, NAC1 promotes immune evasion through lactate dehydrogenase A-mediated production of lactic acid, contributing to an acidic and immune-suppressive TME.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Targeting NAC1 in tumor cells may represent a novel strategy that significantly strengthens the adoptive T cell transfer-based cancer immunotherapy, thus warranting further investigation.
Introduction
Immunotherapy has shown significant potential as a powerful approach to treat cancers by harnessing the body’s immune system and numerous studies have demonstrated promising results of cancer immunotherapy. For instance, immunotherapy based on adoptive cell transfer (ACT) of ex vivo activated and expanded tumor-infiltrating T lymphocytes (TILs), has shown favorable clinical outcomes in patients with metastatic melanoma,1–3 one of the most aggressive and fatal neoplasms responsible for over 80% of skin cancer-related deaths.4 5 Yet, despite enormous advances in cancer immunotherapy, its clinical efficacy and benefits remain less satisfactory due to a variety of factors that limit antitumor immunity, and among those factors, the immune-suppressive tumor microenvironment (TME) is a major hindrance to successful treatment of cancer including melanoma. To circumvent the immune-evasive TME and enhance the efficacy of immunotherapeutic intervention, discovering novel and effective TME targets would be a pressing need.
Nucleus accumbens-associated protein-1 (NAC1), encoded by the NACC1 gene, is a transcriptional co-factor belonging to the BTB (broad-Complex, Tramtrack and bric a brac)/POZ (Poxvirus and Zinc finger) gene family.6 7 Expression of NAC1 is closely associated with tumor development and poor prognosis in various types of cancers such as melanoma,8 urethral,9 ovarian,10 and lung cancer.11 We previously reported that NAC1 promotes autophagic response,12 disables cellular senescence,13 and facilitates oxidative stress resistance during cancer progression.14 More recently, we showed that NAC1 is a positive regulator of glycolysis in ovarian cancer through its stabilization of HIF-1α.15 The upregulation of glycolysis by tumor cells may contribute substantially to an acidic TME that suppresses the antitumor immune response and promotes cancer development.
Better understandings of the molecular mechanism behind tumor resistance to the ACT may help develop novel approaches to enhance the efficacy and benefits of cancer immunotherapy. Here, we report that NAC1 expression in melanoma cells contributes substantially to immune evasion through its positive regulation of lactate dehydrogenase A (LDHA) expression, leading to increased lactic acid (LA) production. Depleting tumor NAC1 can effectively suppress LDHA transcription, enhance the functional status of cytotoxic CD8+ T cells and reinforce the efficacy of ACT against melanoma.
Materials and methods
Cell lines and mice
B16-F10 (CRL-6475) and A2058 (CRL-11147) cell lines were obtained from ATCC (Manassas, VA). Platinum-E (Plat-E) cell lines were purchased from Cell Biolabs (San Diego, California, USA). B16-F10 cells transfected to express chicken ovalbumin (OVA) (B16-OVA) have been previously described.16 OT-I TCR Tg mice, B6. Thy1.1 Tg mice and NSG (NOD-scid IL2Rgnull) mice, 6–8 weeks, were purchased from The Jackson Laboratory (Bar Harbor, Maine, USA).
Cell culture
A2058, and Plat-E cell lines were cultivated in DMEM medium supplemented with 10% heat-inactivated fetal calf serum, 0.5% penicillin/streptomycin. B16-OVA were cultivated in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 0.5% penicillin/streptomycin. All reagents were from Sigma-Aldrich (St Louis, MI).
PBMC isolation
PBMCs were isolated from healthy donor blood samples from the Gulf Coast Regional Blood Center (Houston, Texas, USA). Mononuclear cells were isolated by the density gradient centrifugation using Ficoll-Paque PLUS from Sigma-Aldrich (GE17-1440-02, St Louis, Michigan, USA).
CD8+ T cell purification, expansion, and transduction
Murine CD8+ T cells were isolated from the pooled lymph nodes and spleen of OT-I TCR Tg mice by the magnetic bead separation using the MojoSort Mouse CD8 T Cell Isolation Kit from BioLegend (#480008, San Diego, California, USA). Purified CD8+ T cells were activated with anti-CD3 (5 µg/mL, plate-coated) and anti-CD28 (5 µg/mL, soluble) and cultured in RPMI containing 10% FBS, 100 µg/mL penicillin/streptomycin, 2 mM L-glutamine, 20 mg/mL NEAA, and 5 µl/mL β-mercaptoethanol from Sigma-Aldrich (#M6250, St Louis, Michigan, USA). Human CD8+ T cells were isolated from PBMCs by magnetic bead separation using the MojoSort human CD8 T Cell Isolation Kit from BioLegend (#480012, San Diego, California, USA). Purified CD8+ T cells were activated and expanded with anti-CD3 (5 µg/mL, plate-coated) and anti-CD28 (5 µg/mL, soluble), human rIL-2 (100 U/mL) and cultured in RPMI containing 10% FBS, 100 µg/mL penicillin/streptomycin, 2 mM L-glutamine, 20 mg/mL NEAA, and 5 µl/mL β-mercaptoethanol from Sigma-Aldrich (St Louis, Michigan, USA).
Two days after activation, human CD8+ T cells were transduced with anti-tyrosinase TCR construct. The retroviral plasmid pMSGV1 backbone integrated with anti-tyrosinase TCR was a gift from Dr. Richard Morgan as previously described.17 The construct was transfected into the packing cell line Plat-E. After 48 hours, the retrovirus-enriched supernatant was harvested and purified with 0.45 µM filter from Sigma-Aldrich (#SLHV004SL, St Louis, Michigan, USA). Retrovirus was enriched, and CD8+ T cells were transduced with a RetroNectin (#T202)-coated plate according to the manufacturer’s instructions from Takara Bio (San Jose, California, USA)
Plasmid transfection and retroviral transduction
B16-OVA mouse melanoma cells were transfected with CRISPR plasmids (BTBD14B CRISPR/Cas9 KO Plasmid (m) from Santa Cruz Biotech (#sc-426213, Dallas, Texas, USA) to specifically knockout the expression of NACC1. Transfected GFP+ cells were sorted using a high-speed cell sorter. In parallel, we used B16-OVA cell line as a non-transfected control (WT).
In the human melanoma cell line A2058, NACC1 was knocked out with CRISPR plasmids (BTBD14B CRISPR/Cas9 KO Plasmid (h)) from Santa Cruz Biotech (#sc-410250, Dallas, Texas, USA). Transfected GFP+ cells were sorted using a high-speed cell sorter. Non-transfected cells (WT) served as control. cDNA of LDHA was obtained from Dr. Sunmin Kang (Emory University, Georgia, USA). We subcloned the cDNA of LDHA into the retroviral vector backbone pMIG (#9044, Addgene) as previously described.18 Cloning was confirmed by PCR amplification and gene sequencing. The pMIG-LDHA plasmid was transfected into the packaging cell line of Plat-E. After 48 hours, the retrovirus-enriched supernatant was harvested and purified with 0.45 µM filter from Sigma-Aldrich (St Louis, MI). NAC1 KO B16-OVA and NAC1 KO A2058 tumor cells were cultured with this viral supernatant overnight with 10 µg/mL polybrene from Sigma-Aldrich (St Louis, MI). The transduced GFP+ cells were sorted using a high-speed cell sorter. In parallel, we used the pMIG transduced NAC1 KO B16-OVA and NAC1 KO A2058 tumor cells as mock controls.
In vitro analysis of CD8+ T cells
For cytotoxicity analysis, after 24 hours of activation 5×105 CD8+ T cells were cultured with WT B16-OVA and NAC1 KO B16-OVA tumor cells in the ratio of 1:5 and 1:10 respectively. Cytotoxicity was measured by the CytoTox 96 Non-Radioactive Cytotoxicity Assay from Promega (#G1780, Madison, Wisconsin, USA). For cytokine expression analysis, the CD8+ T cells were cultured with the conditional medium (CM) that was collected from WT B16-OVA and NAC1 KO B16-OVA melanoma cells for 12 hours. Before harvesting the cells, the cells were blocked with the Monensin Solution from BioLegend (#420701, San Diego, California, USA) for 4 hours. For apoptosis analysis, after 12 hours of activation the CD8+ T cells were incubated with the CM that was collected from WT B16-OVA and NAC1 KO B16-OVA cells for 12 hours, in the absence or presence of L-LA from Sigma-Aldrich (St Louis, MI). The apoptosis analysis was performed using APC Annexin V Apoptosis Detection Kit (#640920) with the Aqua Live/Dead (#423101) from BioLegend (San Diego, California, USA).
Glycolysis analysis
For lactate production analysis, the LA concentration in the supernatants of cells cultured for 24 hours was measured enzymatically using a Lactate-Glo Assay from Promega (#J5021, Madison, Wisconsin, USA). For glucose-uptake production analysis, the cells were cultured for 24 hours with an associated fresh medium before harvesting these cells. Intracellular glucose concentration was quantified enzymatically using a Glucose Uptake-Glo Assay from Promega (#J1341, Madison, Wisconsin, USA, USA). For the Seahorse glycolysis analyzer: 10 000 cells/well were seeded in a Seahorse XF96 cell culture plate and were allowed to adhere overnight. The next day, plates were further incubated at 37°C in a non-CO2 incubator for 30 min, followed by testing with Glycolytic Rate Assay Kit of Agilent Seahorse XF (#103344-100). One hour before measurement, cell culture media was replaced with Seahorse XF DMEM with pH 7.4, containing 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose. The following concentrations of each drug were used for extracellular acidification rate (ECAR) acquisitions: 2-DG 50 µM, Rotenone and Antimycin A, 0.5 µM. All reagents were from Agilent (Santa Clara, California, USA).
Quantitative real-time PCR
Total RNA of WT B16-OVA, NAC1 KO B16-OVA, WT A2058 and NAC1 KO A2058 tumor cells was obtained using the RNeasy Mini Kits from QIAGEN (#74104, Germantown, Maryland, USA). Complementary DNA was synthesized with a Maxima H Minus First Strand cDNA Synthesis Kit and amplified by qPCR with PowerUp SYBR Green Master Mix (#A25742, Thermo Scientific, Massachusetts, USA) using the CFX96 Touch Real-Time PCR Detection System from Bio-Rad (Hercules, California, USA). The primer sequences are included in online supplemental table 1.
jitc-2022-004856supp009.pdf (37.2KB, pdf)
Flow cytometric analysis
For mouse tissue, each tumor was minced using the mouse Tumor Dissociation Kit from Miltenyi Biotec (#130-096-730, Auburn, California, USA). All samples were then washed with flow cytometry buffer, and the cells were further passed through a 100 µm cell strainer. The samples were incubated for 30 min at 4°C in the dark with the following antibodies: APC-PD-1 (#135210), BV711-TIM3 (#134021), PE-IL-2 (#503808), FITC-IFN-γ (#505806), APC-Thy1.2 (#140331), Pacific Blue-Granzyme B (#515408), PE-TNFα (#506306), PE-CD4 (#100408), APC-perforin (#154304), APC-Annexin V (#640941) and FITC-CD8 (#100706) from BioLegend (San Diego, California, USA). Intracellular staining was performed after incubation of single-cell suspensions with BD GolgiStop from BD Biosciences (#AB_2869012, San Diego, California, USA) in medium for 4 hours using Intracellular Staining Permeabilization Wash Buffer and Fixation Buffer from BioLegend (#421002, San Diego, California, USA). Stained cell populations were acquired by LSRFortessa from BD Biosciences (Franklin, New Jersey, USA), and the results were analyzed by using FlowJo software from Tree Star (Ashland, Oregon, USA).
Immunoblotting
Cells were lysed in ice-cold RIPA Lysis Buffer (#89900) for 30 min. Pierce protease inhibitors and Halt phosphatase inhibitors from Thermo Fisher Scientific (Waltham, Massachusetts, USA). Protein concentration was measured using the Bio-Rad protein assay kit (#5000002, Hercules, California, USA). Equal protein concentrations were loaded per condition. Proteins were separated with Nu-PAGE 4–12% Bis-Tris gels (#NP0321BOX) using MES x 1 running buffer (#NP0002) at 150V constant. Protein was transferred using the wet transfer Xcell II Blot Module. Membranes were incubated in blocking buffer (5% milk in TBST) for 1 hour before incubation with primary antibodies at 4℃ overnight. After washing 3 times for 10 min with TBST, membranes were incubated with HRP-conjugated antibodies for 1 hour at room temperature. Primary antibodies used were NAC1 (#ab29047, Abcam), β-actin (#664802, BioLegend), and LDHA (#AF7304, R&D system). Other reagents were from Thermo Fisher Scientific (Waltham, Massachusetts, USA).
Murine melanoma models
For the B16 melanoma model, 1×106 WT B16-OVA or NAC1 KO B16-OVA melanoma cells were s.c. inoculated into the right flank of B6. Thy1.1 mice (N=5). In vitro activated OT-I CD8+ T cells (5×106) were washed and resuspended in cold PBS before i.v. injection into B6. Thy1.1 mice through the tail vein when the tumor sizes reached around 50 mm3. Tumor sizes were calculated as V=long diameter × (short diameter/2).2 When mice-bearing tumors reach a maximum size of 2000 mm3, tumors were prepared for analysis.
In the mouse model of human melanoma, 1×106 human WT A2058 or NAC1 KO A2058 melanoma cells were s.c. inoculated into the right flank of NSG mice. Human CD8+ cells were activated and transduced with an anti-tyrosinase TCR, and the transduced GFP+ cells were resuspended in cold PBS before intravenous injection in NSG mouse (5×106/mouse) through the tail vein when the tumor sizes reached around 50 mm3.
Histology and immunohistochemistry
Tumor tissues were fixed with 10% neutral formalin solution (VWR, West Chester, Pennsylvania, USA), and the fixed samples were prepared and stained with H&E as described.18The tissues were frozen in cryovials on dry ice immediately following resection for immunofluorescent microscopy. Cryosectioning and immunofluorescent staining were performed as described.18 FITC-CD8 and Alexa Fluor 647 CD90.2 from BioLegend (San Diego, California, USA) were used to detect the tumor-infiltrating OT-I CD8+ T cells.
Database analyses
Kaplan-Meier estimation curves for overall survival (OS) of individual metastatic melanoma patients were generated with the microarray analysis and visualization platform R2 (http://r2.amc.nl) by using the ‘R2: Tumor Skin Cutaneous Melanoma – The Cancer Genome Atlas (TCGA) – 470 – rsem – tcgars’. Correlation of LDHA expression with NACC1 was determined with the ‘R2: Tumor Skin Cutaneous Melanoma – TCGA – 470 – rsem – tcgars’ dataset containing data from 470 melanoma patients (http://cancergenome.nih.gov). Pearson’s correlation was calculated with the transform 2log setting. The NAC1 expression of skin cancers was determined with The Cancer Cell Line Encyclopedia (https://depmap.Org/portal/ccle/), including 70 types of human skin cancer cell line. The infiltration level of CD8+ T cells in SKCM was determined with Tumor Immune Estimation Resource (TIMER2.0) with official instruction.19
Statistical analysis
Paired or unpaired Student’s t-test or one-way analysis of variance was performed to analyze the differences between the groups, using GraphPad Prism (GraphPad Software, San Diego, Clifornia, USA); significance was set at 5%.
Results
Expression of NAC1 is associated with the prognosis of melanoma patients and proliferation of tumor cells
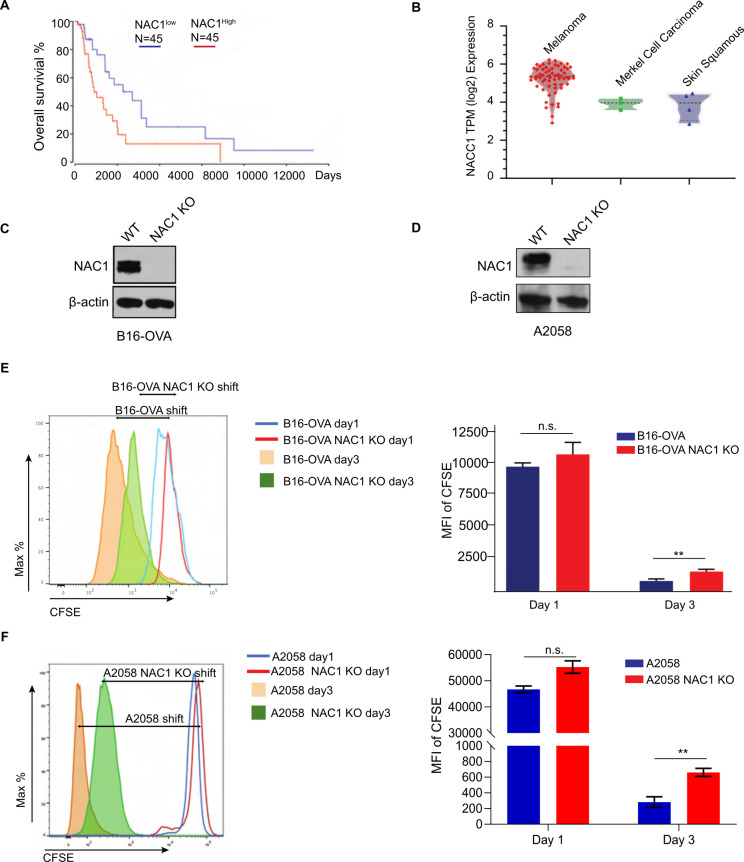
Our analysis of TCGA dataset showed that patients with high NAC1 expression had a greatly reduced OS time as compared with the patients with low NAC1 expression (figure 1A). NAC1 expression was higher in melanomas than in Merkel cell and skin squamous carcinomas (figure 1B). We first confirmed that NAC1 is knocked out in mouse B16 melanoma cells (figure 1C) and human A2058 melanoma cells (figure 1D) by Western blots. We then observed that NAC1 KO in tumor cells remarkably inhibited their proliferation, as assayed by cell counting (online supplemental figure 1C and online supplemental figure 1D) and cell division (figure 1E, F). These results suggest a role of NAC1 in tumor development and progression.
Figure 1.
Effects of NAC1 on proliferation of melanoma cells and development of melanoma. (A) Survival of melanoma patients with high expression (top 30%; red) and low expression of NAC1 (bottom 30%; blue), calculated with the ‘R2 R2: Tumor Skin Cutaneous Melanoma-TCGA-470-rsem-tcgars’ dataset (http://r2.amc.nl) from the TCGA database. (B) The distribution of NAC1 expression in human skin cancer cell lines (melanoma, Merkel cell carcinoma, and skin squamous carcinoma), determined by the CCLE database (https://portals.broadinstitute.org/ccle). (C) Protein expression of NAC1 and β-actin in WT and NAC1-deficient (NAC1 KO) mouse B16-OVA melanoma cells by Western blots. (D) Protein expression of NAC1 and β-actin in WT and NAC1 KO human A2058 melanoma cells by Western blots. (E) Cell division rates of WT and NAC1 KO mouse B16-OVA melanoma cells were analyzed by CFSE between day 1 and day 3. (F) Cell division rates of WT and NAC1 KO human A2058 melanoma cells were analyzed by CFSE between day 1 and day 3. In both (E, F), the tumor cells were labeled with CFSE dye (5 µM) prior to seeding onto the 48-well tissue culture plates. The CFSE intensities were analyzed by flow cytometer (left panel) and quantified (right panel) by the mean fluorescence intensity (MFI) of CFSE. One representative of three identical experiments is shown. **p ≤0.01. CCLE, cancer cell line encyclopedia; CFSE, carboxyfluorescein succinimidyl ester; NAC1, nucleus accumbens-associated protein-1; n.s., not significant; OVA, ovalbumin; TCGA, The Cancer Genome Atlas.
jitc-2022-004856supp001.pdf (360.7KB, pdf)
NAC1 supports glycolysis of melanoma cells
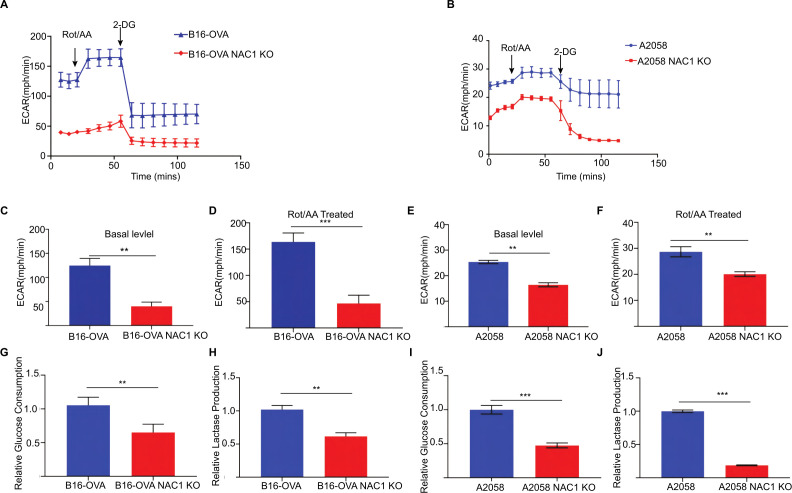
We previously reported that NAC1 plays a critical role in promoting glycolysis in hypoxic ovarian cancer cells.15 Consistently, we observed that melanoma cells with depletion of NAC1 showed a decreased ECAR when compared with the control cells (figure 2A,B). Furthermore, the basal level (figure 2C, E) and the maximum level (figure 2D, F) of ECAR were significantly lower in NAC1 KO B16-OVA cells and NAC1 KO A2058 cells when compared with B16-OVA and A2058 WT control cells. Treatment of the melanoma cells with rotenone and antimycin A (Rot/AA), inhibitors of the oxidative phosphorylation pathway, caused an elevated ECAR level in the WT control cells. However, the NAC1 KO tumor cells could not raise the ECAR level due to insufficient glycolysis support. Moreover, the lactate production and glucose uptake in the cellular supernatants of these melanoma cells was reduced (figure 2G–J). As expected, mouse NAC1 KO B16-OVA cells and human NAC1 KO A2058 cells secreted significantly less lactate compared with WT cells and altered glucose consumption compared with WT cells in vitro. Although OVA is not a natural Ag of mouse tumors, here we used B16-OVA cells as a model for cancer immunotherapy because expression of OVA can facilitate strong immune responses to tumor Ags. These observations demonstrated that NAC1 plays a critical role in supporting metabolic reprogramming in melanoma cells.
Figure 2.
Depleting NAC1 decreases the glycolytic rate of melanoma cells. (A) Time series of ECAR measurements in WT and NAC1 KO B16-OVA cells by the Seahorse Metabolic Analyzer. (B) Time series of ECAR measurements in WT and NAC1 KO A2058 cells by the Seahorse Metabolic Analyzer. Data are represented as mean±SEM; n=6 wells per condition from two independent experiments. (C) Basal glycolysis rates of WT and NAC1 KO B16-OVA cells. (D) Maximum glycolysis rates of WT and NAC1 KO B16-OVA cells. (E) Basal glycolysis rates of WT and NAC1 KO A2058 cells. (F) Maximum glycolysis rates of WT and NAC1 KO A2058 cells. In (C–F), six wells per condition from two independent experiments; One-way ANOVA with multiple comparisons correction. (G) Quantification of relative glucose consumption in WT and NAC1 KO B16-OVA cells. (H) Quantification of lactase production in the supernatant of WT and NAC1 KO B16-OVA cells. (I) Quantification of relative glucose consumption in WT and NAC1 KO A2058 cells. (J) Quantification of lactase production in the supernatant of WT and NAC1 KO A2058 cells. **p≤0.01, ***p≤0.001. ANOVA, analysis of variance; ECAR, extracellular acidification rate; NAC1, nucleus accumbens-associated protein-1; OVA, ovalbumin.
Depletion of tumorous NAC1 invigorates cytotoxic CD8+ T cells
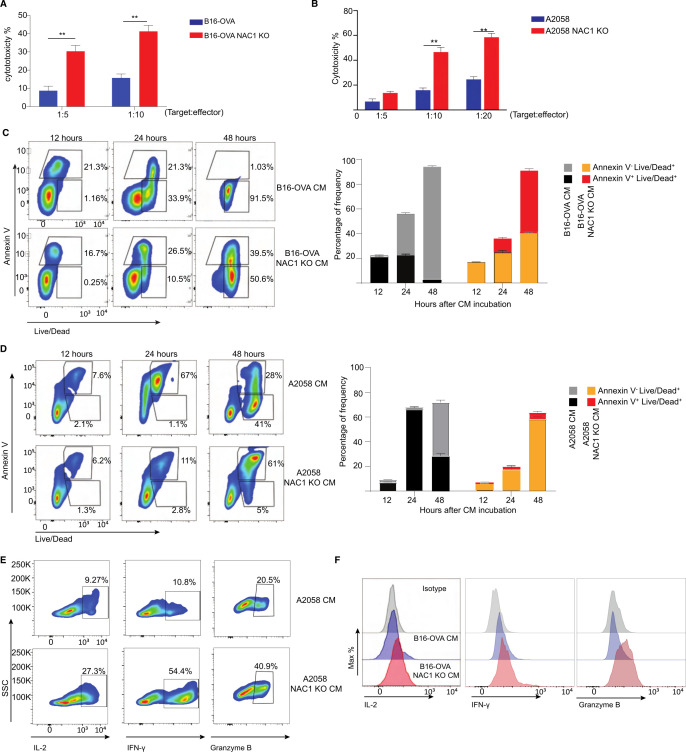
The activity of immune cells can be impacted by the metabolic alteration of tumor cells.20 21 Tumor cells have high glycolytic activity, leading to their secretion and accumulation of lactate and subsequent development of an acidic TME. The TME affects the development and function of immune cells through numerous avenues. For instance, massive LA production from tumor cells was reported to inhibit T cell cytotoxicity and effector functions.22–25 We recently reported that expression of NAC1 negatively regulates the suppressive activity of regulatory T cells (Tregs).26 Therefore, we next determined whether the tumorous expression of NAC1 affects the cytocidal effect/activity of CD8+ T cells. We co-cultured WT or NAC1 KO B16-OVA cells with CD8+ T cells prepared from the OT-I T cell receptor (TCR) transgenic mice, which specifically recognizes ovalbumin (OVA) present on B16-OVA cells. The cytotoxicity of CD8+ T cells against tumor cells was significantly increased in NAC1 KO B16-OVA cells compared with that in WT cells (figure 3A). To further validate the effects of NAC1 on the cytotoxicity of human CD8+ T cells, we used human A2058 melanoma cells, which expressed tyrosinase Ag, and constructed tyrosinase-specific human CD8+ T cells by transduction with a retroviral vector of anti-tyrosinase TCR (online supplemental figure 2). Similar results were obtained from the co-culture of human WT or NAC1 KO A2058 melanoma cells with the tyrosinase-specific human CD8+ T cells (figure 3B). To explore how expression of NAC1 in tumor cells alters the cytocidal effect of CD8+ T cells, we determined the effect of the CM from the cultures of WT B16-OVA, NAC1 KO B16-OVA, WT A2058, or NAC1 KO A2058 cells on CD8+ T cells. In these experiments, activated CD8+ T cells were incubated in the CM for 12, 24, or 48 hours. The incubation period was followed by assessments of CD8+ T cell apoptosis and function. Apoptosis of CD8+ T cells cultured in NAC1 KO B16-OVA or A2058 CM was significantly reduced compared with those cultured in the WT CM (figure 3C, D). Moreover, CD8+ T cells cultured in the NAC1 KO tumor CM produced more significant amounts of cytokines IFN-γ, granzyme Β, and IL-2, than those cultured in the media of control tumor cells (figure 3E, F). This indicates that the expression of NAC1 in tumors can impair the activity of CD8+ T cells. In addition, CD8+ T cells cultured in the media of WT tumor cells had a higher percentage of PD-1+ TIM-3+ than those cultured in the NAC1 KO tumor CM (online supplemental figure 3), suggesting that tumorous NAC1 may induce exhaustion of CD8+ T cells.
Figure 3.
Depleting tumorous NAC1 strengthens the cytotoxicity of CD8+ T cells. (A) WT or NAC1 KO B16-OVA cells (targets) were co-cultured with OT-I CD8+ T cells (effectors) (ratio: 1:5 and 1:10) for 6 hours. Per cent cytotoxicity was calculated as (1- (R5/R0)) x 100, where R5 = (target cells as % of total of 6 hours) / (effector cells as % of total of 6 hours), R0 = (target cells as % of total of 0 h) / (effector cells as % of total at 0 h). (B) WT or NAC1 KO A2058 cells were co-cultured with anti-tyrosinase TCR transduced human CD8+ T cells (effectors) (ratio: 1:5, 1:10, 1:20) for 6 hours. Per cent cytotoxicity was calculated as (1- (R5/R0)) x 100, where R5 = (target cells as % of total at 6 hours) / (effector cells as % of total of 6 hours), R0 = (target cells as % of total of 0 hour) / (effector cells as % of total of 0 hour). (C) Representative contour plots of Annexin V and Live-Dead expression on the CM-treated OT-I CD8+ T cells (left panel). Frequencies of the indicated Annexin V+ and Live-Dead+ expressing populations (right panel). After incubation with WT conditional medium (CM) or NAC1 KO B16-OVA CM for 12, 24, and 48 hours, respectively. Apoptotic rates were assessed by flow cytometry. One representative experiment out of three is shown. (D) Representative contour plots of Annexin V and Live-Dead expression on the CM-treated human CD8+ T cells (left panel). Frequencies of the indicated Annexin V+ and Live-Dead+ expressing populations (right panel). After incubation with WT CM or NAC1 KO A2058 CM for 12, 24, and 48 hours, respectively. Apoptotic rates were assessed by flow cytometry. One representative experiment out of three is shown. (E) Histograms showing expression of the indicated cytokines after incubation with WT CM or NAC1 KO B16-OVA CM for 6 hours (E); WT CM and NAC1 KO A2058 CM (F). One representative of three identical experiments is shown. IFN-γ, Granzyme B, and IL-2 expression were determined by flow cytometry. **p≤ 0.01; one way analysis of variance with multiple comparision correction. NAC1, nucleus accumbens-associated protein-1; OVA, ovalbumin.
jitc-2022-004856supp002.pdf (339.9KB, pdf)
jitc-2022-004856supp003.pdf (589.2KB, pdf)
NAC1-mediated LDHA expression contributes to LA production
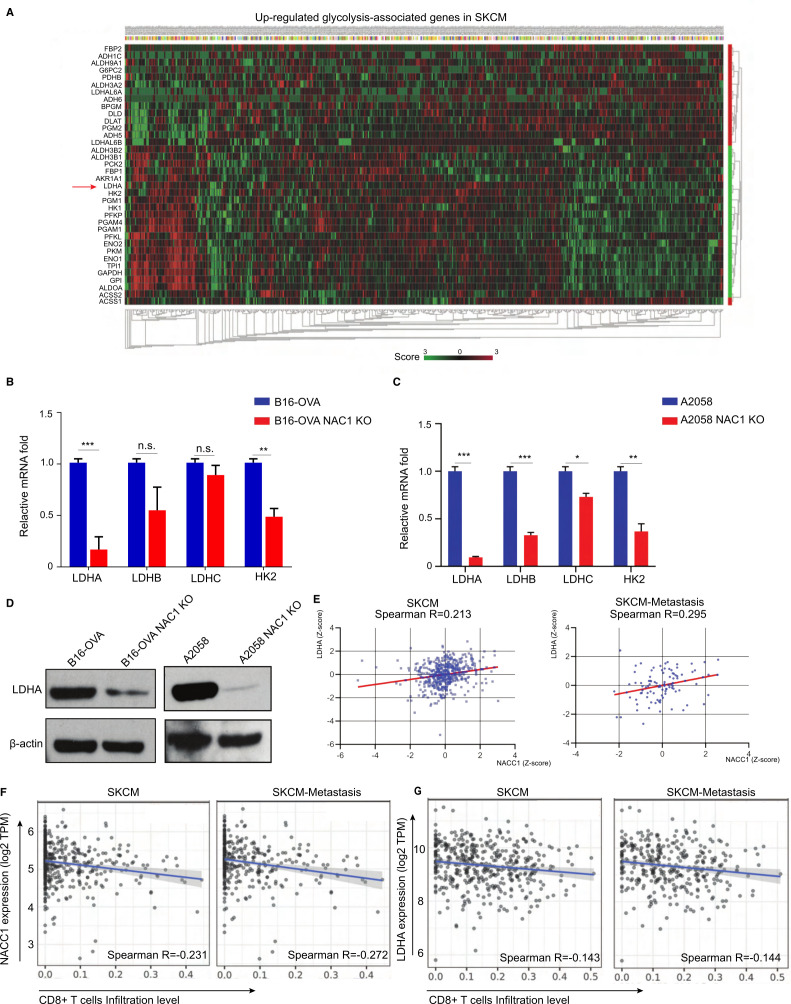
Because NAC1 has a critical role promoting glycolysis in melanoma cells (figure 2), we determined the impact of tumorous NAC1 on the glycolytic activity of CD8+ T cells. Using the TCGA-melanoma database, we analyzed the possible association between NACC1 and glycolysis-related genes (figure 4A). Among the positively correlated genes, we found that expression of LDHA demonstrated a strong correlation with NAC1 expression, consistent with ovarian cancer models.15 LDH family genes are the primary metabolic enzymes that convert pyruvate to lactate and vice versa. In addition, the enzymes also play a significant role in regulating nutrient exchange between tumor and stroma.27 28 To determine how NAC1 regulates the expression of LDHA, we tested if NAC1 affects mRNA expression of LDHA and other LDH family genes in the tumor cells. Using qPCR, we show that the LDHA and hexokinase 2 (HK2) mRNA levels were significantly lower in NAC1 KO B16-OVA and NAC1 KO A2058 tumor cells than in B16-OVA or A2058 cells (figure 4B, C). This indicates NAC1 plays a role in the regulation of LDHA transcription. To confirm this observation in melanoma cells, we compared the protein expression of LDHA in NAC1-expressing and NAC1 KO tumor cells through Western blots. LDHA protein was remarkably downregulated in NAC1 KO B16-OVA and A2058 cells, compared with that in WT cells (figure 4D). In addition, our analyses of the TCGA database found that LDHA expression is significantly correlated with NAC1 expression, with a higher correlation rate in patients with poor prognosis (Stage>III) (figure 4E). Using the TIMER, an algorithm developed to analyze the abundance of tumor-infiltrating immune cells comprehensively and in a flexible manner, we evaluated whether the NAC1-LDHA axis was involved in the tumor–immune interactions in melanoma patients. The expressions of NACC1 (encoding NAC1) and LDHA (encoding LDHA) are inversely correlated with cytotoxic T lymphocyte (CTL) infiltration in SKCM and SKCM-Metastasis (figure 4F, G).
Figure 4.
NAC1 promotes the expression of LDHA in tumor cells. (A) Heatmap of normalized RNA-seq reads (Z score), top upregulated glycolysis-associated genes in the TCGA SKCM database. (B, C) qRT-PCR analysis of mRNA of LDHA, LDHB, LDHC, and HK2 in WT and NAC1 KO B16-OVA cells (B) or WT and NAC1 KO A2058 cells (C). The results are presented relative to the level of GAPDH. n=5 per condition from two independent experiments. *p≤0.05, **p≤0.01, ***p≤0.001. (D) LDHA expression in whole-cell extracts by Western blots. β-actin was used as a loading control. One representative experiment out of three is shown. (E) Correlations of NACC1 with LDHA gene expression (Z-score) were determined in a dataset including 470 melanoma tumors and 368 melanoma-metastatic tumors (‘R2: Tumor Skin Cutaneous Melanoma-TCGA-470-rsem-tcgars’). Pearson’s correlation was calculated. (F) Correlations of CD8+ T cell infiltration with NACC1 expression in melanoma (SKCM, n=470) and metastasis melanoma (SKCM-metastasis, n=368), (G) Correlations of CD8+ T cell infiltration level with LDHA expression in melanoma (SKCM, n=470) and metastasis melanoma (SKCM-metastasis, n=368), were estimated using TIMER algorithm. Pearson’s correlation was calculated. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LDHA, lactate dehydrogenase A; NAC1, nucleus accumbens-associated protein-1; SKCM, skin cutaneous melanoma; TCGA, The Cancer Genome Atlas; TIMER, Tumor Immune Estimation Resource.
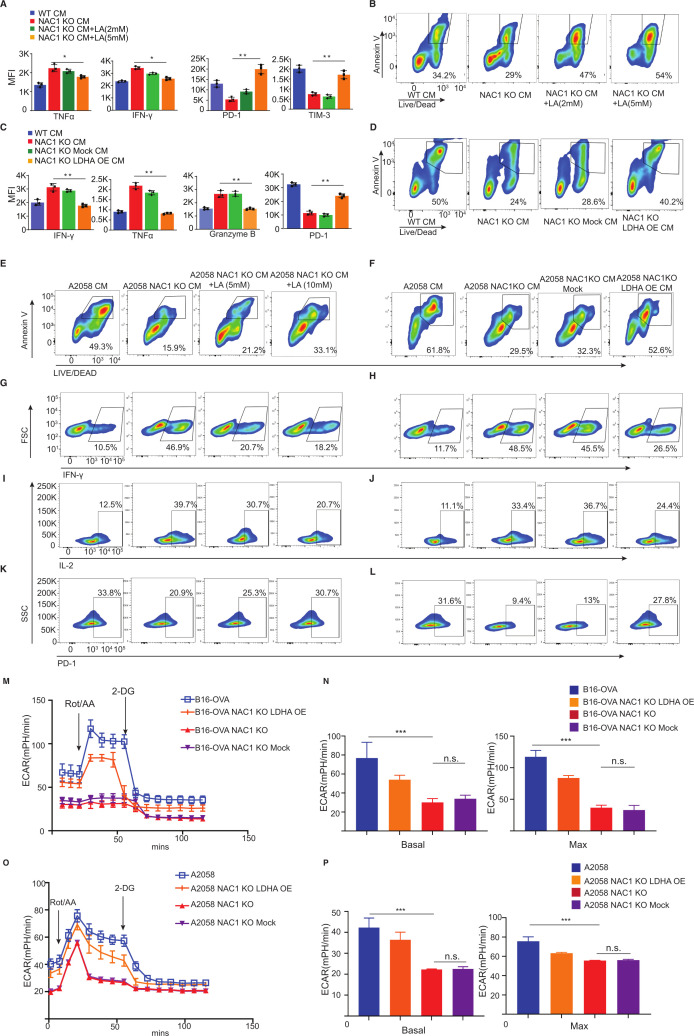
Next, we cultured mouse CD8+ T cells in the CM from NAC1 KO mouse B16-OVA cells and supplemented the medium with or without 2 mM or 5 mM LA. The addition of LA reduced the production of cytokines including TNF-alpha and IFN-gamma and increased the expression of PD-1 and TIM-3 of CD8+ T cells (figure 5A and online supplemental figure 5). The addition of LA also caused a higher percentage of apoptosis determined by Annexin V staining (figure 5B). In parallel, we transfected NAC1 KO tumor cells with an LDHA expression retroviral vector or a retroviral backbone to overexpress LDHA in NAC1 KO B16-OVA (online supplemental figure 6). We cultured mouse CD8+ T cells in the CM from NAC1 KO B16-OVA cells overexpressing LDHA (NAC1 KO LDHA OE) or an empty control (NAC1 KO Mock). We demonstrated that overexpression of LDHA conferred NAC1 KO mouse tumor cells with the ability to suppress the production of cytokines and increased expression of PD-1 (figure 5C and online supplemental figure 5) and apoptosis (figure 5D) of mouse cytotoxic CD8+ T cells. Similar results were obtained from human CD8+ T cells in the CM from NAC1 KO human A2058 cells, NAC1 KO human A2058 cells supplemented with the medium, with or without LA (5 mM or 10 mM), or NAC1 KO human A2058 cells overexpressing LDHA (NAC1 KO LDHA OE) (figure 5E–L).
Figure 5.
Effects of LDHA expression and lactic acid level on cytokine production, apoptosis, and exhaustion of CD8+ T cells. Mouse or human CD8+ T cells were incubated with CM from mouse WT or NAC1 KO B16-OVA cells, or human WT or NAC1 KO A2058 tumor cells for 24 hours. In parallel, some T cells were incubated with CM from NAC1 KO tumor cells overexpressing LDHA (NAC1 KO LDHA OE) or an empty control (NAC1 KO Mock), or CM supplemented with LA (2–5 mM for mouse T cells; 5–10 mM for human T cells). (A) The production of cytokines TNF-α or IFN-γ and expression of PD-1 and TIM-3 of mouse CD8+ T cells with CM from mouse WT or NAC1 KO B16-OVA cells, or CM supplemented with LA by flow cytometry. One-way ANOVA with multiple comparisons correction. *p≤0.05, **p≤0.01. (B) Apoptosis of mouse CD8+ T cells with CM from mouse WT or NAC1 KO B16-OVA cells, or CM supplemented with LA. (C) The production of cytokines and expression of PD-1 of mouse CD8+ T cells with CM from mouse WT or NAC1 KO B16-OVA cells, or CM from NAC1 KO tumor cells overexpressing LDHA or an empty control by flow cytometry. One-way ANOVA with multiple comparisons correction. **p≤0.01. (D) Apoptosis of mouse CD8+ T cells with CM from mouse WT or NAC1 KO B16-OVA cells, or CM from NAC1 KO tumor cells overexpressing LDHA or an empty control by Annexin V staining. (E, F) Apoptosis of human CD8+ T cells with CM from human WT or NAC1 KO A2058 cells, or CM supplemented with LA or CM from human NAC1 KO tumor cells overexpressing LDHA or an empty control by Annexin V staining. (G, H) The production of IFN-γ of human CD8+ T cells with CM from human WT or NAC1 KO A2058 cells, CM supplemented with LA, or CM from human NAC1 KO tumor cells overexpressing LDHA or an empty control. (I, J) The production of IL-2 of human CD8+ T cells with CM from human WT or NAC1 KO A2058 cells, CM supplemented with LA, or CM from human NAC1 KO tumor cells overexpressing LDHA or an empty control. (K, L) Expression of PD-1 of human CD8+ T cells with CM supplemented with LA, or CM from human NAC1 KO tumor cells overexpressing LDHA or an empty control. (M) ECAR of mouse tumor cells measured by the Seahorse Metabolic Analyzer. Data are represented as mean±SEM; n=6 per condition from two independent experiments. (N) Basal glycolysis rates (left panel) and maximum glycolysis rate (right panel) of mouse tumor cells. One-way ANOVA with multiple comparisons correction. ***p≤0.001. (O) ECAR of human tumor cells measured by the Seahorse Metabolic Analyzer. Data are represented as mean±SEM; n=6 per condition from two independent experiments. (P) Basal glycolysis rates (left panel) and maximum glycolysis rate (right panel) of human tumor cells. One-way ANOVA with multiple comparisons correction. ***p≤0.001. ANOVA, analysis of variance; CM, conditional medium; ECAR, extracellular acidification rate; LDHA, lactate dehydrogenase A; NAC1, nucleus accumbens-associated protein-1; OVA, ovalbumin.
jitc-2022-004856supp005.pdf (82KB, pdf)
jitc-2022-004856supp006.pdf (348.6KB, pdf)
To further prove that LDHA plays a role in metabolic reprogramming in melanoma cells expressing NAC1, we analyzed and compared their glycolysis profile of these mouse or human tumor cells (WT, NAC1 KO, NAC1 KO Mock, NAC1 KO LDHA OE). Overexpression of LDHA (LDHA-OE) could partially restore glycolytic activity in the NAC1 KO mouse B16-OVA cells (figure 5M, N) and human A2058 tumor cells (figure 5O, P). Taken together, these results suggest that NAC1-mediated LDHA expression plays a critical role in supporting an immune-suppressive TME through its promotion of LA production.
Depletion of tumorous NAC1 enhances the efficacy of ACT of CTLs against melanoma
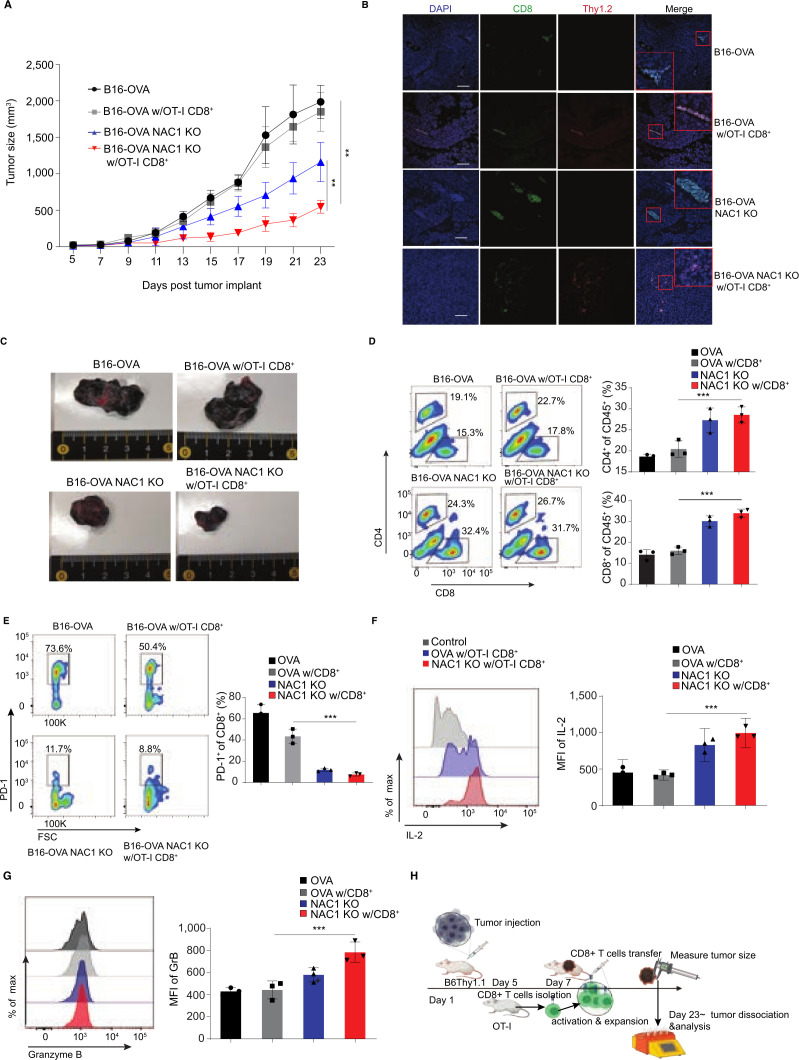
To evaluate the impact of tumorous expression of NAC1 on melanoma immunotherapy, we inoculated C57BL/6 congenic (B6. Thy1.1+) mice s.c. with either WT B16-OVA or NAC1 KO B16-OVA tumor cells (1×106 cells/mouse). Inoculation was followed by providing the mice with or without an ACT of OT-I CTLs (5×106 cells/mouse; Thy1.2+). We observed that the tumor sizes in the mice bearing WT B16-OVA tumors were significantly larger than the animals bearing NAC1 KO B16-OVA tumors (figure 6A). Transfer of OT-I CTLs had little effect on tumor growth in the mice inoculated with WT B16-OVA cells; however, the CTL treatment markedly inhibited tumor growth in the mice bearing NAC1 KO B16-OVA tumors (figure 6A, C). Moreover, we also observed that all (100%) NAC1 KO B16-OVA tumor bearing mice receiving OT-I CTLs survived to Day 23, compared with WT B16-OVA untreated control (40%) or WT B16-OVA group of mice treated with OT-I CTLs (60%) (data not shown). The potent inhibitory effect of CTLs on the growth of NAC1 KO tumors may be attributed to increased infiltration of Thy1.2+CD8+ cells into tumor tissues (figure 6B, D), less exhaustion of CTLs (as evidenced by low expression of PD-1) (figure 6E), and increased production of proinflammatory cytokines IL-2 and Granzyme B by CTLs (figure 6F, G and online supplemental figure 7).
Figure 6.
NAC1 deficiency increases CTL infiltration in tumors following an ACT in immune-competent mice. Mouse WT B16-OVA or NAC1 KO B16-OVA tumor cells (1×106) were injected s.c. in the flank of B6. Thy1.1+ mice (n=5), followed by treatment with or without the ACT of OT-I CD8+ T cells. (A) Graphical representation of the progression of tumor size. (B) OT-I CD8+ T cell infiltration (Thy1.2+CD8+) in tumors was determined by immunofluorescence staining. (C) Indicated tumors were harvested and photographed on day 23. (D) Flow cytometric analysis of tumor-infiltrating CD4+ T cells and CD8+ T cells. (E) Histograms show expression of PD-1+ (left panel) and quantitation of PD-1+ cells of total infiltrated Thy1.2+ CD8+ cells (right panel). (F) Histograms showing production of IL-2 (left panel) and quantitation of MFI of IL-2 of total infiltrated CD8+ cells (right panel). (G) Histograms showing production of Granzyme B (left panel) and quantitation of MFI of Granzyme B of total infiltrated CD8+ cells (right panel). (H) Schematic representation of experimental paradigm. B6 Thy 1.1 mice were injected with mouse or human tumor cells on Day 1 followed by antigen specific CD8+ T cell transfer on day 7. The tumor size was monitored until day 23 post injection of the melanoma cells. All the results shown are representative of three identical experiments. *p≤0.05, ***p≤0.001. ACT, adoptive cell transfer; CTL, cytotoxic T lymphocytes; MFI, mean fluorescence intensity; NAC1, nucleus accumbens-associated protein-1; OVA, ovalbumin.
jitc-2022-004856supp007.pdf (161KB, pdf)
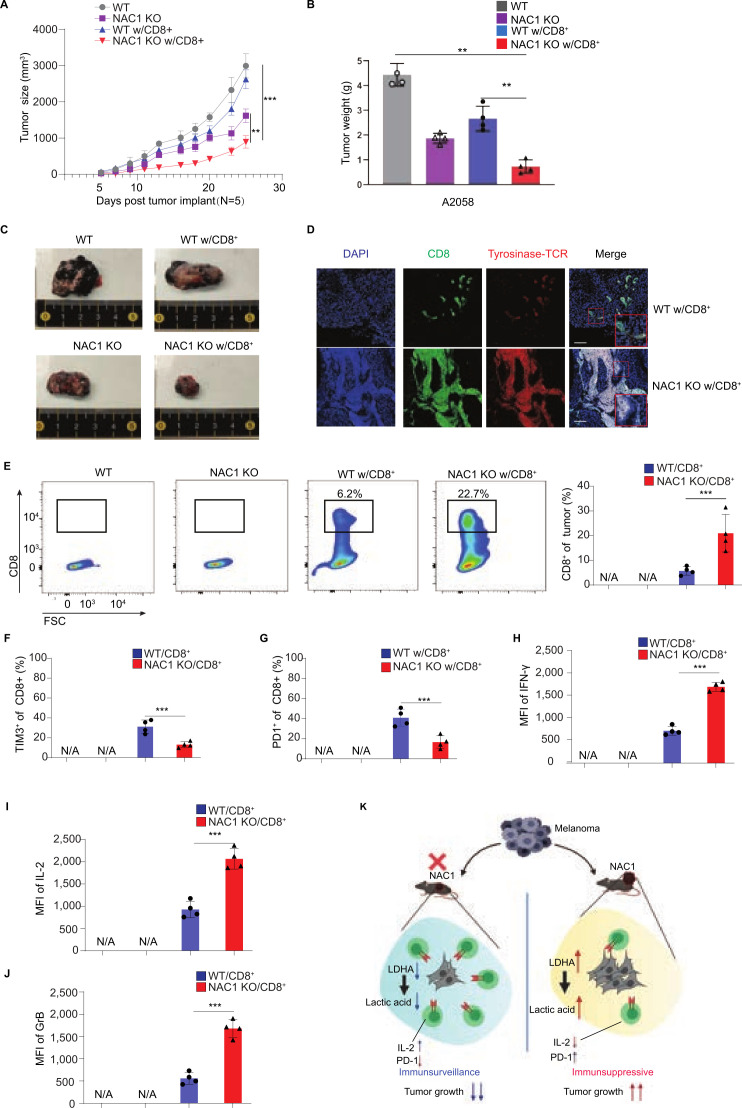
To further demonstrate the importance of the tumorous expression of NAC1 in antitumor immunity, we tested the humanized (NOD-scid IL2rgnull, NSG) mouse melanoma model. In these experiments, NSG immune-compromised mice were inoculated s.c. with either human WT A2058 or NAC1 KO A2058 melanoma cells into their flanks. When tumor sizes reached 100 mm3, the mice were injected with the human tyrosinase-specific CTLs via the tail vein. Tumor growth was significantly suppressed in NAC KO tumor-bearing mice receiving an ACT of CTLs than those without the CTL treatment (figure 7A–C). Analysis of tyrosinase-specific CTL infiltration in tumor tissues was conducted using immunofluorescence microscopy (figure 7D) and flow cytometry (figure 7E). These analyses revealed more significant amounts of CTLs in NAC1 KO tumors than in NAC1-expressing tumors. In addition, the CTLs in NAC1 KO tumors showed substantially higher levels of the cytokines, including IFN-γ, IL-2, and granzyme B (figure 7H–J and online supplemental figure 8). Consistently, the CTL exhaustion observed in NAC1-expressing tumor cells was significantly improved when tumor cells lost NAC1, as suggested by the reduced levels of TIM-3 and PD-1 (figure 7F–7G and online supplemental figure 8). Together, these results imply that the tumorous expression of NAC1 may contribute to immune evasion through an acidic TME caused by upregulation of the LDHA-mediated production of LA. Targeting NAC1 in tumor cells may restrict LA production and improve the TME, thereby reinforcing immunotherapy such as ACT-based immunotherapy (figure 7K).
Figure 7.
NAC1 deficiency increases tumor CTL infiltration following an ACT in immune-deficient mice. Human WT A2058 or NAC1 KO A2058 tumor cells (1×106) were s.c. injected into the flank of NSG mice, followed by treatment with or without the ACT of Tyrosinase-specific CD8+ T cells. (A) Graphical representation of the progression of tumor size. (B) The tumor weight was evaluated by scales and represented graphically. Values are mean±SEM of three independent experiments (n=5). (C) Indicated tumors were harvested and photographed at day 25. (D) CD8+ T cell infiltration (CD8+; green) in tumors (tyrosinase; red) was determined by immunofluorescence staining. Images are representative images taken from the tumors of three different mice. (E) Flow cytometric analysis of infiltrating CD8+ T cells (left panel) and quantitation of tumor infiltrating CD8+ T cells (right panel). Values are the mean±SEM of 3 independent experiments (n=5). (F) Flow cytometric analysis of infiltrating TIM-3+CD8+ T cells. (G) Flow cytometric analysis of infiltrating PD-1+CD8+ T cells. Values are the mean±SEM of 3 independent experiments (n=5). (H) Flow cytometric analysis of IFN-γ production of infiltrating CD8+ T cells. (I) Flow cytometric analysis of the IL-2 production of infiltrating CD8+ T cells. (J) Flow cytometric analysis of Granzyme B production of infiltrating CD8+ T cells. Values are the mean±SEM of four independent experiments (n=4). (K) Schematic representation of the hypothesis.**p≤0.01, ***p≤0.001. ACT, adoptive cell transfer; CTL, cytotoxic T lymphocytes; LDHA, lactate dehydrogenase A; MFI, mean fluorescence intensity; NAC1, nucleus accumbens-associated protein-1.
jitc-2022-004856supp008.pdf (193.2KB, pdf)
Discussion
The efficacy of antitumor immunity is often hindered by a multitude of factors that contribute to immune evasion. Identifying key molecular determinants involved in immune evasion may help develop novel and effective strategies to circumvent tumor cell resistance to immunotherapy. We recently reported that expression of NAC1 negatively regulates the suppressive activity of CD4+ Tregs26 and the formation of CD8+ memory T cells.29 In this study, we identified tumorous NAC1 as a critical determinant of antitumor immunity, and NAC1 promotes immune evasion through LDHA-mediated production of LA, contributing to an acidic and immune-suppressive TME.
Metabolic reprogramming of immune cells in the TME can impact antitumor therapeutic outcomes. A few connections between glycolytic metabolism and T cell regulation have been revealed.30–33 As the central players in the ACT, T cells develop rapid immune response through several stages, including initial cell growth followed by massive clonal expansion and differentiation, a contraction or death phase, and establishment and maintenance of immune memory. Metabolic reprogramming has important roles in these processes.34 35 During the initial growth phase, T cells undergo an activation-induced metabolic reprogramming, switching from the β-oxidation of fatty acids in T cells to glycolysis, pentose-phosphate and glutaminolytic pathways in activated T cells. This phase represents the engagement of biosynthesis to produce proteins, nucleic acids, lipids, carbohydrates, and other macromolecules for generation of new cells. Activated T cells upregulate glycolysis for their growth, proliferation, and function,36 but inhibiting glycolysis was reported to enhance CD8+ T cell memory and antitumor function.37 Whereas CD8+ T cells differentiate into CTLs, CD4+ T cells differentiate into either induced Tregs (iTregs) that suppress uncontrolled immune responses or cells of the TH1, TH2 or TH17 subset of T cells (effector T cells, Teffs) that mediate appropriate immune responses. Glycolysis promotes CD4+ T cell differentiation into various Teffs, and inhibition of glycolytic activity blocks this process but promotes Tregs.38–40 Glycolysis is also critical for function of Teffs. Of these various T cell subsets, the iTregs and memory T cells, mainly rely on lipid oxidation as a major source of energy, whereas CTLs and Teffs sustain high glycolytic activity and glutaminolytic activity. Lipid metabolism is believed to be the key metabolic pathway in Treg development and differentiation.41 42 Dendritic cells and macrophages also switch to glycolysis on activation even though they do not proliferate. Thus, metabolic reprogramming in various immune cells is intimately associated with their differentiation, survival, and function; yet the molecular mechanisms and pathways involved remain to be fully explored. Based on the functional association between metabolic reprogramming and T cell activation, expansion, and effector function, targeting T cell metabolism may provide new directions to modulate therapeutic immunity.
Metabolic reprogramming in tumor cells also impacts antitumor immunity. One of the hallmarks of cancer is a metabolic reprogramming, which supports macromolecule synthesis, bioenergetics demand, and cellular survival.43 TME may be severely affected by the metabolic status of tumor cells, and can be such that even the most potent immune cells can accomplish their cytocidal function.44 The potency of ACT is impacted by metabolic alteration of tumor cells,20 21 which have the capability to cope by enhancing alternative energy production mechanisms. While normal cells rely on respiration, malignant cells depend on glycolysis even in the presence of sufficient oxygen. Warburg metabolism consumes glucose and increases the production of LA, and this altered cellular metabolism causes the changes in the nutrient compositions in the TME. As both tumor and tumor stromal cells have high glycolytic activity, through secretion of lactate they can build an acidic TME that affects the development and function of immune cells. For instance, massive production of LA from tumor cells was reported to inhibit T cell cytotoxicity and effector functions.22–25 Tumor cells consume glucose more competently than T cells, and this deprives TILs of this key nutrient and thus weakens the cytotoxic functions of CD8+ T cells. Alterations in tumor cell metabolism can deprive TILs of essential nutrients that are required for effective response to the tumor cells, leading to immune evasion. The glucose-deficient TME may diminish CTLs activity through an immunosuppressive TME,45–47 and tumor cells with higher glycolytic activity have a strong capacity to evade immunosurveillance. On the other hand, cancer cells themselves can become impervious to cytocidal effects of ACT via reprogramming energy metabolism. Also, decreased expressions of tumor Ags and major histocompatibility complexes and increased expressions of inhibitory checkpoint molecules (eg, PD-L1) may provide a growth advantage to tumor cells through evading T cell-mediated immune destruction. It was reported that Warburg glycolysis was associated with tumor cell resistance to TNF-related apoptosis-inducing ligand (TRAIL)-induced cell death48 and chemotherapeutic agents such as paclitaxel and doxorubicin.49 We have reported that targeting the eukaryotic elongation factor-2 kinase-mediated glycolysis can sensitize cancer cells to paclitaxel and doxorubicin,50 and depletion of this kinase increases tumor cell sensitivity to TRAIL, curcumin, velcade, temozolomide, and AKT inhibitors via activating apoptosis.51–54 We had previously shown that NAC1 plays an important role in regulating glycolysis and hypoxia response.15 We recently reported that expression of NAC1 negatively regulates the suppressive activity of CD4+ Tregs26 and the formation of CD8+ memory T cells.29 Although several pathways, including NAC1, are known to promote metabolic reprogramming in tumor cells, whether and how they affect antitumor immunity remains largely unclear.
The antitumor immune response induced by CTLs can be weakened by acidification of the TME through the metabolic reprogramming of tumor cells.23 55 56 Practical approaches to restrain tumorous production of lactate may improve immune permissive-TME and strengthen immunotherapy. A recent study showed that bicarbonate administration to neutralize the acidic TME is a promising strategy to improve the efficacy of adoptively T cell transfer-based immunotherapy.57 Here, we show that high expression of NAC1 in melanoma cells is associated with poor prognosis of melanoma patients and reduced cytotoxicity of CTLs (figures 1 and 2). We also demonstrate that the effect of NAC1 on antitumor immunity is mediated through LDHA-regulated LA production (figures 3 and 4). Although studies from our group and others demonstrated that NAC1 promotes oncogenesis,9 12 14 58 59 the current study reveals a previously unappreciated role of this oncoprotein in modulating immune evasion.
It has been reported that LDHA-mediated LA production restrained CTL activity and function,55 and inhibitors of LDHA could strengthen the antitumor activity of CTLs both in vitro and in vivo.60 Our finding that NAC1 regulates LDHA expression, as well as LA production in melanoma cells, may provide NAC1 a new target for modulating the TME, suppressing immune evasion, and enhancing the efficacy of ACT. Notably, both immune-competent B6. Thy1.1 mouse model and immune-deficient NSG mice bearing human melanoma show that depletion of tumor NAC1 significantly enhances the therapeutic efficacy of ACT (figure 6 and figure 7) by improving the TME and invigorating CTLs. Whether depleting tumorous NAC1 affects the activity and function of other types of immune cells, such as CD4+ Teffs, dendritic cells, macrophages, and NK cells, remains to be determined. Because the status of these immune cells can also be affected by the glycolytic activity of tumor cells,55 61–63 in addition to improving the TME and invigorating CTLs, targeting NAC1 may modulate other immune cell-mediated antitumor immunity.
Successful cancer immunotherapy could be hindered by the barriers such as low amount of tumor Ag-specific T cells due to clonal erasure, poor activation of T cells, accumulation of tolerigenic Ag-presenting cells in the TME, and formation of a hypoxic and immunosuppressive TME. Notably, studies have indicated that the metabolic status of both immune cells and tumor cells can have a great impact on antitumor immunity.64 Immune cells and tumor cells can up-regulate glycolysis, the anaerobic metabolism of glucose into ATP, when they turn into a highly proliferative state, to meet their needs for large amounts of energy as building materials. Immune activation, acquisition of effector functions, and generation of immune memory are closely coupled with alterations in energy metabolism. In particular, the transition from quiescence to activation is associated with a significant and prolonged escalation of aerobic glycolysis (Warburg effect), which can supply ATP, glycolytic intermediates for biosynthesis of DNA and cellular structural materials. A better understanding of the metabolic reprogramming in both immune cells and tumor cells shall facilitate the development of more effective approaches to improving cancer immunotherapy. This study identified NAC1 as a key modulator of tumor immune evasion and demonstrates that its role is mediated through the LDHA-regulated production of LA. TME consists of tumor cells, tumor stromal cells and various immune cells. In addition to impact CTLs, NAC1 may play roles in other immune cells, which remains unknown. Nevertheless, our results suggest that targeting NAC1 in tumor cells may represent a novel strategy that significantly strengthens the adoptive T cell transfer-based cancer immunotherapy, thus warranting further investigation.
jitc-2022-004856supp004.pdf (1.4MB, pdf)
jitc-2022-004856supp010.pdf (56.2KB, pdf)
Acknowledgments
The authors acknowledge the assistance of the Integrated Microscopy and Imaging Laboratory at Texas A&M School of Medicine (RRID:SCR_021637).
Footnotes
YR, AK and JKD contributed equally.
Contributors: JS and J-MY designed the experiments, analyzed the data, and wrote this paper. YR, AK and JKD performed the experiments. H-YP, LW, DB, XX, XR and YZ analyzed the data. JS is the gurantor for the overall content of this work.
Funding: This work was supported by the National Institutes of Health Grant R01CA221867 to J-MY and JS, and R01AI121180 and R21AI167793, and Department of Defense Grant LC210150 to JS.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.Dudley ME, Wunderlich JR, Yang JC, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol 2005;23:2346–57. 10.1200/JCO.2005.00.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015;348:62–8. 10.1126/science.aaa4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 4.Donald Morton L L, Kirkwood JM, Wollman RC. Malignant Melanoma. In: Kuff RCB, Hait WN, Hong WK, et al., eds. Cancer medicine. 7 edn. Hamilton, London, New York: BC Decker Inc, 2006: 1644–62. [Google Scholar]
- 5.Goff SL, Robbins PF, El-Gamil M, et al. No correlation between clinical response to CTLA-4 blockade and presence of NY-ESO-1 antibody in patients with metastatic melanoma. J Immunother 2009;32:884–5. 10.1097/CJI.0b013e3181affbf0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama K, Nakayama N, Davidson B, et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proc Natl Acad Sci U S A 2006;103:18739–44. 10.1073/pnas.0604083103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Torrado R, Yamada D, Defossez P-A. Born to bind: the BTB protein-protein interaction domain. Bioessays 2006;28:1194–202. 10.1002/bies.20500 [DOI] [PubMed] [Google Scholar]
- 8.Jiao H, Jiang S, Wang H, et al. Upregulation of LINC00963 facilitates melanoma progression through miR-608/NACC1 pathway and predicts poor prognosis. Biochem Biophys Res Commun 2018;504:34–9. 10.1016/j.bbrc.2018.08.115 [DOI] [PubMed] [Google Scholar]
- 9.Morita K, Fujii T, Itami H, et al. NACC1, as a target of MicroRNA-331-3p, regulates cell proliferation in urothelial carcinoma cells. Cancers 2018;10:347. 10.3390/cancers10100347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shih I-M, Nakayama K, Wu G, et al. Amplification of the ch19p13.2 NACC1 locus in ovarian high-grade serous carcinoma. Mod Pathol 2011;24:638–45. 10.1038/modpathol.2010.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z. Circular RNA hsa_circ_0001588 promotes the malignant progression of lung adenocarcinoma by modulating miR-524-3p/NACC1 signaling. Life Sci 2020;259:118157. 10.1016/j.lfs.2020.118157 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Yang JW, Ren X, et al. NAC1 and HMGB1 enter a partnership for manipulating autophagy. Autophagy 2011;7:1557–8. 10.4161/auto.7.12.17910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Cheng Y, Ren X, et al. Dysfunction of nucleus accumbens-1 activates cellular senescence and inhibits tumor cell proliferation and oncogenesis. Cancer Res 2012;72:4262–75. 10.1158/0008-5472.CAN-12-0139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren Y-J, Wang X-H, Ji C, et al. Silencing of Nac1 expression induces cancer cells oxidative stress in hypoxia and potentiates the therapeutic activity of Elesclomol. Front Pharmacol 2017;8:804. 10.3389/fphar.2017.00804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Ren Y-J, Guo L-C, et al. Nucleus accumbens-associated protein-1 promotes glycolysis and survival of hypoxic tumor cells via the HDAC4-HIF-1α axis. Oncogene 2017;36:4171–81. 10.1038/onc.2017.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellone M, Cantarella D, Castiglioni P, et al. Relevance of the tumor antigen in the validation of three vaccination strategies for melanoma. J Immunol 2000;165:2651–6. 10.4049/jimmunol.165.5.2651 [DOI] [PubMed] [Google Scholar]
- 17.Frankel TL, Burns WR, Peng PD, et al. Both CD4 and CD8 T cells mediate equally effective in vivo tumor treatment when engineered with a highly avid TCR targeting tyrosinase. J Immunol 2010;184:5988–98. 10.4049/jimmunol.1000189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei F, Zhao B, Haque R, et al. In vivo programming of tumor antigen-specific T lymphocytes from pluripotent stem cells to promote cancer immunosurveillance. Cancer Res 2011;71:4742–7. 10.1158/0008-5472.CAN-11-0359 [DOI] [PubMed] [Google Scholar]
- 19.Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–14. 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho P-C, Liu P-S. Metabolic communication in tumors: a new layer of immunoregulation for immune evasion. J Immunother Cancer 2016;4:4:4. 10.1186/s40425-016-0109-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beavis PA, Slaney CY, Kershaw MH, et al. Reprogramming the tumor microenvironment to enhance adoptive cellular therapy. Semin Immunol 2016;28:64–72. 10.1016/j.smim.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 22.Mendler AN, Hu B, Prinz PU, et al. Tumor lactic acidosis suppresses CTL function by inhibition of p38 and JNK/c-Jun activation. Int J Cancer 2012;131:633–40. 10.1002/ijc.26410 [DOI] [PubMed] [Google Scholar]
- 23.Calcinotto A, Filipazzi P, Grioni M, et al. Modulation of microenvironment acidity reverses anergy in human and murine tumor-infiltrating T lymphocytes. Cancer Res 2012;72:2746–56. 10.1158/0008-5472.CAN-11-1272 [DOI] [PubMed] [Google Scholar]
- 24.Ohashi T, Akazawa T, Aoki M, et al. Dichloroacetate improves immune dysfunction caused by tumor-secreted lactic acid and increases antitumor immunoreactivity. Int J Cancer 2013;133:1107–18. 10.1002/ijc.28114 [DOI] [PubMed] [Google Scholar]
- 25.Fischer K, Hoffmann P, Voelkl S, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007;109:3812–9. 10.1182/blood-2006-07-035972 [DOI] [PubMed] [Google Scholar]
- 26.Yang J-M, Ren Y, Kumar A, et al. NAC1 modulates autoimmunity by suppressing regulatory T cell-mediated tolerance. Sci Adv 2022;8:eabo0183. 10.1126/sciadv.abo0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta VK, Sharma NS, Durden B, et al. Hypoxia-Driven oncometabolite L-2HG maintains stemness-differentiation balance and facilitates immune evasion in pancreatic cancer. Cancer Res 2021;81:4001–13. 10.1158/0008-5472.CAN-20-2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo G, Wang Y, Zhou Y, et al. Immune cell concentrations among the primary tumor microenvironment in colorectal cancer patients predicted by clinicopathologic characteristics and blood indexes. J Immunother Cancer 2019;7:179. 10.1186/s40425-019-0656-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Kumar A, Das JK, et al. Expression of NAC1 Restrains the Memory Formation of CD8+ T Cells during Viral Infection. Viruses 2022;14:1713. 10.3390/v14081713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck MD, O'Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med 2015;212:1345–60. 10.1084/jem.20151159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerriets VA, Rathmell JC. Metabolic pathways in T cell fate and function. Trends Immunol 2012;33:168–73. 10.1016/j.it.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity 2013;38:633–43. 10.1016/j.immuni.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol 2012;13:907–15. 10.1038/ni.2386 [DOI] [PubMed] [Google Scholar]
- 34.Pearce EL. Metabolism in T cell activation and differentiation. Curr Opin Immunol 2010;22:314–20. 10.1016/j.coi.2010.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Priyadharshini B, Turka LA. T-cell energy metabolism as a controller of cell fate in transplantation. Curr Opin Organ Transplant 2015;20:21–8. 10.1097/MOT.0000000000000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gubser PM, Bantug GR, Razik L, et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat Immunol 2013;14:1064–72. 10.1038/ni.2687 [DOI] [PubMed] [Google Scholar]
- 37.Sukumar M, Liu J, Ji Y, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest 2013;123:4479–88. 10.1172/JCI69589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michalek RD, Gerriets VA, Jacobs SR, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 2011;186:3299–303. 10.4049/jimmunol.1003613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerriets VA, Kishton RJ, Nichols AG, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest 2015;125:194–207. 10.1172/JCI76012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macintyre AN, Gerriets VA, Nichols AG, et al. The glucose transporter GLUT1 is selectively essential for CD4 T cell activation and effector function. Cell Metab 2014;20:61–72. 10.1016/j.cmet.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su W, Chapman NM, Wei J, et al. Protein Prenylation Drives Discrete Signaling Programs for the Differentiation and Maintenance of Effector Treg Cells. Cell Metab 2020;32:996–1011. 10.1016/j.cmet.2020.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Franco F, Tsui Y-C, et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol 2020;21:298–308. 10.1038/s41590-019-0589-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 44.McCarthy SA, Mufson RA, Pearce EJ, et al. Metabolic reprogramming of the immune response in the tumor microenvironment. Cancer Biol Ther 2013;14:315–8. 10.4161/cbt.23616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim S-O, Li C-W, Xia W, et al. EGFR signaling enhances aerobic glycolysis in triple-negative breast cancer cells to promote tumor growth and immune escape. Cancer Res 2016;76:1284–96. 10.1158/0008-5472.CAN-15-2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho P-C, Bihuniak JD, Macintyre AN, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 2015;162:1217–28. 10.1016/j.cell.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang C-H, Qiu J, O'Sullivan D, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 2015;162:1229–41. 10.1016/j.cell.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumazaki M, Shinohara H, Taniguchi K, et al. Perturbation of the Warburg effect increases the sensitivity of cancer cells to TRAIL-induced cell death. Exp Cell Res 2016;347:133–42. 10.1016/j.yexcr.2016.07.022 [DOI] [PubMed] [Google Scholar]
- 49.Tavares-Valente D, Baltazar F, Moreira R, et al. Cancer cell bioenergetics and pH regulation influence breast cancer cell resistance to paclitaxel and doxorubicin. J Bioenerg Biomembr 2013;45:467–75. 10.1007/s10863-013-9519-7 [DOI] [PubMed] [Google Scholar]
- 50.Cheng Y, Ren X, Yuan Y, et al. eEF-2 kinase is a critical regulator of Warburg effect through controlling PP2A-A synthesis. Oncogene 2016;35:6293–308. 10.1038/onc.2016.166 [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Cheng Y, Zhang L, et al. Inhibition of eEF-2 kinase sensitizes human glioma cells to TRAIL and down-regulates Bcl-xL expression. Biochem Biophys Res Commun 2011;414:129–34. 10.1016/j.bbrc.2011.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Y, Ren X, Zhang Y, et al. Integrated regulation of autophagy and apoptosis by EEF2K controls cellular fate and modulates the efficacy of curcumin and velcade against tumor cells. Autophagy 2013;9:208–19. 10.4161/auto.22801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng Y, Zhang Y, Zhang L, et al. MK-2206, a novel allosteric inhibitor of Akt, synergizes with gefitinib against malignant glioma via modulating both autophagy and apoptosis. Mol Cancer Ther 2012;11:154–64. 10.1158/1535-7163.MCT-11-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng Y, Ren X, Zhang Y, et al. eEF-2 kinase dictates cross-talk between autophagy and apoptosis induced by Akt inhibition, thereby modulating cytotoxicity of novel Akt inhibitor MK-2206. Cancer Res 2011;71:2654–63. 10.1158/0008-5472.CAN-10-2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brand A, Singer K, Koehl GE, et al. LDHA-Associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab 2016;24:657–71. 10.1016/j.cmet.2016.08.011 [DOI] [PubMed] [Google Scholar]
- 56.Müller B, Fischer B, Kreutz W. An acidic microenvironment impairs the generation of non-major histocompatibility complex-restricted killer cells. Immunology 2000;99:375–84. 10.1046/j.1365-2567.2000.00975.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, et al. Neutralization of tumor acidity improves antitumor responses to immunotherapy. Cancer Res 2016;76:1381–90. 10.1158/0008-5472.CAN-15-1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ueda SM, Yap KL, Davidson B, et al. Expression of fatty acid synthase depends on Nac1 and is associated with recurrent ovarian serous carcinomas. J Oncol 2010;2010:285191. 10.1155/2010/285191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jinawath N, Vasoontara C, Yap K-L, et al. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene 2009;28:1941–8. 10.1038/onc.2009.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cascone T, McKenzie JA, Mbofung RM, et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab 2018;27:977–87. 10.1016/j.cmet.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pucino V, Certo M, Bulusu V, et al. Lactate Buildup at the Site of Chronic Inflammation Promotes Disease by Inducing CD4+ T Cell Metabolic Rewiring. Cell Metab 2019;30:1055–74. 10.1016/j.cmet.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anagnostou V, Smith KN, Forde PM, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 2017;7:264. 10.1158/2159-8290.CD-16-0828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cong J, Wang X, Zheng X, et al. Dysfunction of natural killer cells by FBP1-Induced inhibition of glycolysis during lung cancer progression. Cell Metab 2018;28:243–55. 10.1016/j.cmet.2018.06.021 [DOI] [PubMed] [Google Scholar]
- 64.Herbel C, Patsoukis N, Bardhan K, et al. Clinical significance of T cell metabolic reprogramming in cancer. Clin Transl Med 2016;5:29. 10.1186/s40169-016-0110-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004856supp009.pdf (37.2KB, pdf)
jitc-2022-004856supp001.pdf (360.7KB, pdf)
jitc-2022-004856supp002.pdf (339.9KB, pdf)
jitc-2022-004856supp003.pdf (589.2KB, pdf)
jitc-2022-004856supp005.pdf (82KB, pdf)
jitc-2022-004856supp006.pdf (348.6KB, pdf)
jitc-2022-004856supp007.pdf (161KB, pdf)
jitc-2022-004856supp008.pdf (193.2KB, pdf)
jitc-2022-004856supp004.pdf (1.4MB, pdf)
jitc-2022-004856supp010.pdf (56.2KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.