Abstract
The yeast two-hybrid assay revealed that Bacillus subtilis DnaD, a possible component of the primosome and required for replication initiation, interacted with DnaA and DnaD itself. The mutant DnaD23 was incapable of interacting with DnaA but retained interaction with the wild-type DnaD. These results suggest that interaction between DnaD and DnaA is important for replication initiation.
Entry of the replicative DNA helicase at the replication origin is a very important step in replication initiation (1). In Escherichia coli, loading of the DnaB helicase into the origin (oriC) is carried out with the aid of two other proteins, DnaA and DnaC. DnaA, the initiator of replication, binds to specific sequences (DnaA boxes) within oriC and opens double-stranded DNA at AT-rich sequences adjacent to these boxes. This protein also interacts with DnaB helicase (12, 18) and is required for prepriming complex formation on DNA (20). DnaC forms a stable hexameric complex with DnaB, which then elicits single-stranded-DNA binding activity of DnaC (11). These results indicate that the DnaB-DnaC complex is loaded onto the unwound region at oriC with the aid of an interaction between DnaA and DnaB. After loading, DnaC is released from the complex by its own ATPase activity (1).
In Bacillus subtilis, more proteins seem to be engaged in entry of the DNA helicase at oriC. In addition to DnaA (13), four other proteins (DnaB, DnaC, DnaD, and DnaI) have been genetically proven to be required for replication initiation (2–3, 16–17, 21). DnaC is the counterpart of E. coli DnaB and, thus, probably acts as the replicative DNA helicase in B. subtilis (17). The three remaining proteins may be components of a primosome, based on studies of plasmid replication (4). In fact, DnaI showed a strong interaction with the DnaC helicase as examined by the yeast two-hybrid system (8). Here, we found using the same technique that DnaD interacts with DnaA and DnaD itself. When a mutant protein, DnaD23, was tested for these interactions, it was found to be active for interaction with the wild-type DnaD but inactive for interaction with DnaA. If the DnaD protein is a primosome component, interaction between DnaD and DnaA also appears to play an important role in loading the DnaC helicase onto DNA at oriC in B. subtilis.
DnaD interacts with DnaA and DnaD itself.
To examine interactions among Dna initiation proteins by the yeast two-hybrid system (6), they were fused to Gal4 binding domain (BD) and activation domain (AD) in plasmids pGBT9 and pGAD424 (Clontech), respectively. The AD fusions of all five Dna initiation proteins, including DnaD, and the BD fusions of DnaC and DnaI were already described (8). The remaining three BD fusions were constructed as follows: coding regions of dnaA, dnaB, and dnaD lacking the first five, five, and four codons, respectively, were amplified by PCR and fused to the Gal4 BD in frame by cloning between BamHI and PstI sites of pGBT9. The yeast two-hybrid assay using these plasmids revealed the following results (Table 1). (i) DnaD interacted with itself. (ii) DnaD also interacted with DnaA. However, in this case, only one combination (BD-DnaD to AD-DnaA) showed the interaction; the opposite combination (AD-DnaD to BD-DnaA) did not. AD-DnaD was active because it interacted with BD-DnaD. As BD-DnaA was detected in cell extracts from yeast cells bearing pGBT9 dnaA by immunoblotting with anti-DnaA antibody (data not shown), the failure of interaction with AD-DnaD may be due to interference with proper folding or a conformational change caused by fusion with the binding domain. (iii) DnaB interacted with itself, although the interaction was weak as shown by β-galactosidase activity. The yeast two-hybrid assay was carried out using the matchmaker two-hybrid system (Clontech) according to the supplier's manual, and β-galactosidase activities were measured as described previously (8).
TABLE 1.
Interaction of B. subtilis Dna initiation protein analyzed by the yeast two-hybrid system
| Protein fused to Gal4 BD | X-Gal assay (β-galactosidase activity)a
|
|||||
|---|---|---|---|---|---|---|
| Protein fused to Gal4 AD
| ||||||
| None | DnaA | DnaB | DnaC | DnaD | DnaI | |
| No | − (0) | − (71 ± 48) | − (0) | − (0) | − (0) | − (0) |
| DnaA | − (0) | − (0) | − (80 ± 64) | − (0) | − (23 ± 1) | − (0) |
| DnaB | − (25 ± 18) | − (0) | + (720 ± 30) | − (0) | − (0) | − (0) |
| DnaD | − (180 ± 125) | + (2,467 ± 80) | − (0) | − (0) | + (5,555 ± 391) | − (0) |
| DnaD23 | NDb | − (151 ± 19) | ND | ND | + (4,219 ± 305) | ND |
+, yeast colonies showed a blue color within 4 h under the assay conditions; −, still white even after 8 h of incubation. In each combination, five independent clones were used for this assay and all showed the same result (data not shown). X-Gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. The enzyme activity is represented as units in 1 ml of cell suspension with an optical density of 1.0 at 600 nm. One unit defines the amount of enzyme that hydrolyzes 1 pmol of MUG (4-methylumbelliferyl-β-d-galactoside) per min. Average units from three independent experiments and their standard errors are shown. As shown previously (8), BD-DnaC and BD-DnaI interacted with AD-DnaI and AD-DnaC, respectively, indicating that the latter two fusion proteins used in this study are active in yeast cells.
ND, not done.
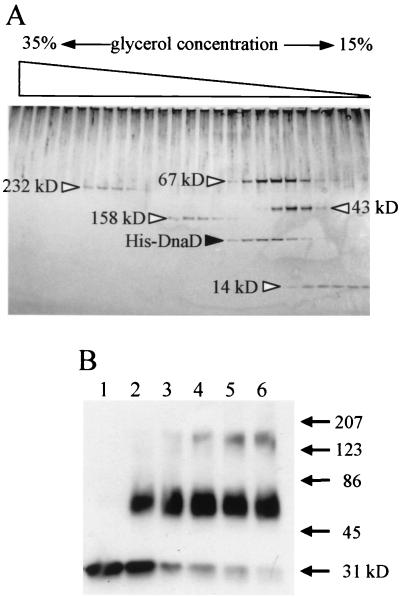
The DnaD-DnaD interaction was further examined biochemically. The whole coding region of the dnaD gene was amplified by PCR using primers with artificial NdeI and XhoI sites and cloned between these sites of pET-15b (Novagen) to fuse with the His tag in frame at the amino terminus. The His-DnaD protein was purified according to the supplier's manual, and oligomer formation was analyzed by glycerol density-gradient centrifugation (Fig. 1A). His-DnaD behaved as a slightly larger protein than the 67-kDa protein, one of the molecular mass markers, in the gradient. As the molecular mass of the monomer His-DnaD is about 31 kDa (Fig. 1B, lane 1), the result suggested that His-DnaD was present as a dimer or trimer. To conclude its oligomerization, cross-linking analysis with glutaraldehyde was carried out (Fig. 1B). From this analysis, His-DnaD was found to be present as a dimer, not a trimer, because no bands were detected around 86 kDa. However, with longer incubation with glutaraldehyde, another oligomer was observed between 123 and 207 kDa, although it is a small population in the input His-DnaD. Thus, His-DnaD is present mainly as a stable dimer but also may form another oligomer (probably a hexamer from its size).
FIG. 1.
Analysis of DnaD-DnaD interaction in vitro by glycerol density-gradient centrifugation (A) and cross-linking (B). (A) The purified His-tagged DnaD protein (1.8 μg) was loaded with molecular mass (MM) markers on a glycerol density gradient (15 to 35%; 2.4 ml) containing buffer A (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.01% Nonidet P-40, 100 mM NaCl, 1 mM dithiothreitol) with a cushion of 50% glycerol (0.1 ml) at the bottom of the gradient. The proteins were separated by centrifugation for 12 h at 160,000 × g using a TLS55 rotor and Optima TL Ultracentrifuge (Beckman). After centrifugation, the density gradient was fractionated into 24 samples (100 μl in each). Proteins in the fractions were detected by silver staining after separation on a sodium dodecyl sulfate-polyacrylamide gradient gel (10 to 20%) by electrophoresis. MM markers shown by open triangles are follows: catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and RNase A (14 kDa). (B) His-DnaD (1 μl; 220 ng/μl) was added into 50 μl of buffer A containing 0.01% glutaraldehyde to start cross-linking. After incubation at room temperature for 1, 3, 5, 7, and 9 min (lanes 2, 3, 4, 5, and 6, respectively), each reaction mixture was mixed with 50 μl of 2× sample loading buffer and heated at 95°C for 5 min. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by immunoblotting using a rabbit anti-DnaD antiserum. Lane 1, without glutaraldehyde. MM markers are shown with arrows.
The mutant DnaD23 maintains interaction with the wild-type DnaD but loses interaction with DnaA.
The dnaD23 mutation affects replication initiation at the restrictive temperature (2). To examine effects of the mutation on DnaD-DnaD and DnaD-DnaA interactions, its BD fusion (BD-DnaD23) was made and used for the yeast two-hybrid assay. As shown in Table 1, it interacted with DnaD but not with DnaA. The loss of interaction between BD-DnaD and AD-DnaA by introduction of the dnaD23 mutation confirmed that the DnaD part of BD-DnaD was certainly engaged in the interaction. These results raise the possibility that interaction between DnaD and DnaA is important for replication initiation in B. subtilis.
The amino-terminal domains of DnaD are engaged in DnaD-DnaA and DnaD-DnaD interactions.
To elucidate which domains of DnaD are involved in the two kinds of interactions, a series of deletion mutants of DnaD fused to the Gal4 BD were constructed and examined for their interactions with AD-DnaA and AD-DnaD by the yeast two-hybrid system. As shown in Fig. 2, BD-DnaD1–140 (containing the amino-terminal 140 amino acids of DnaD) showed interactions with both DnaA and DnaD itself. However, BD-DnaD1–133 and BD-DnaD1–104 lost interaction with DnaA but retained DnaD-DnaD interaction. These results suggest that an internal region (at least the 105th to 140th amino acids) of DnaD is specifically required for interaction with DnaA and that the amino-terminal half of DnaD involves a region required for DnaD-DnaD interaction. As we did not test another deletion mutant from the amino terminus of DnaD, the extent of the region required for interaction with DnaA was not elucidated. The dnaD23 mutation is positioned at the 166th amino acid. This mutation may affect the tertiary structure of DnaD, and thereby its interaction with DnaA is lost. A leucine zipper-like motif is found near the amino terminus of DnaD (21st to 35th leucines). As the leucine zipper motif mediates homo- and heterodimerization of proteins (5), the similar motif in DnaD may serve for the dimerization.
FIG. 2.
Interactions of various DnaD deletion proteins with DnaA and wild-type DnaD. Thick bars indicate DnaD portions fused to the Gal4 BD. These dnaD parts were amplified by PCR and cloned in pGBT9 as described for construction of BD-DnaD in the text. The carboxyl termini of the BD-DnaD proteins are shown by the amino acid positions in the wild-type DnaD at the right of the bars. Interaction was judged by change of color (+, blue, −, white) after 4 h of incubation in 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The location of the dnaD23 mutation is shown by the amino acid position (166th) with an arrow.
Possible role of interaction between DnaD and DnaA in replication initiation.
In B. subtilis, two noncoding regions upstream and downstream of dnaA are required for replication initiation (14) and initiation begins within the downstream region (15). DnaA binds to 9-mer sequences (DnaA boxes) within the two regions (7), and the binding causes local unwinding of duplex DNA at an AT-rich sequence within the downstream region (10). As DnaB, DnaD, and DnaI are probably components of a primosome (4), interaction between DnaD and DnaA may play a key role in bringing the primosome to oriC bound by DnaA. DnaB is reported to possess single-stranded-DNA binding activity (19), and, thus, it may help in loading the DnaC helicase into the unwound AT-rich region within oriC. The DnaC helicase would be loaded as a complex with DnaI because they interact with each other strongly (8). As DnaI contains the ATP-binding motif (9), it may be released from the complex by ATP hydrolysis after loading, like E. coli DnaC (1). In our two-hybrid assays, interactions between DnaB and other components of the primosome were not detected (Table 1). Therefore, further analyses using different methods are needed to elucidate the structure of the primosome, the role of DnaB, and the mechanism for entry of the DNA helicase at B. subtilis oriC.
Acknowledgments
We are grateful to C. Bruand and W. Firshein for providing the dnaD23 mutant and for critical reading of our manuscript, respectively.
This work was supported by Grants-in-Aid for Scientific Research (B) and for Scientific Research on Priority Area (C) from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Baker T A, Bell S P. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 2.Bruand C, Sorokin A, Serror P, Ehrlich S D. Nucleotide sequence of the Bacillus subtilis dnaD gene. Microbiology. 1995;141:321–322. doi: 10.1099/13500872-141-2-321. [DOI] [PubMed] [Google Scholar]
- 3.Bruand C, Ehrlich S D. The Bacillus subtilis dnaI gene is part of the dnaB operon. Microbiology. 1995;141:1199–1200. doi: 10.1099/13500872-141-5-1199. [DOI] [PubMed] [Google Scholar]
- 4.Bruand C, Ehrlich S D, Janniere L. Primosome assembly site in Bacillus subtilis. EMBO J. 1995;14:2642–2650. doi: 10.1002/j.1460-2075.1995.tb07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busch S J, Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990;6:36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- 6.Fields S, Song O. A novel genetic system to detect protein-protein interaction. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 7.Fukuoka T, Moriya S, Yoshikawa H, Ogasawara N. Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J Biochem. 1990;107:732–739. doi: 10.1093/oxfordjournals.jbchem.a123117. [DOI] [PubMed] [Google Scholar]
- 8.Imai Y, Ogasawara N, Ishigo-oka D, Kadoya R, Daito T, Moriya S. Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of nucleoids. Mol Microbiol. 2000;36:1037–1048. doi: 10.1046/j.1365-2958.2000.01928.x. [DOI] [PubMed] [Google Scholar]
- 9.Koonin E V. DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res. 1992;20:1997. doi: 10.1093/nar/20.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause M, Rückert B, Lurz R, Messer W. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J Mol Biol. 1997;274:365–380. doi: 10.1006/jmbi.1997.1404. [DOI] [PubMed] [Google Scholar]
- 11.Learn B A, Um S-J, Huang L, McMacken R. Cryptic single-stranded-DNA binding activities of the phage λ P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc Natl Acad Sci USA. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marszalek J, Kaguni J M. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- 13.Moriya S, Kato K, Yoshikawa H, Ogasawara N. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: amount of DnaA protein determines cells' initiation potential. EMBO J. 1990;9:2905–2910. doi: 10.1002/j.1460-2075.1990.tb07481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriya S, Atlung T, Hansen F G, Yoshikawa H, Ogasawara N. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol Microbiol. 1992;6:309–315. doi: 10.1111/j.1365-2958.1992.tb01473.x. [DOI] [PubMed] [Google Scholar]
- 15.Moriya S, Ogasawara N. Mapping of the replication origin of the Bacillus subtilis chromosome by the two-dimensional gel method. Gene. 1996;176:81–84. doi: 10.1016/0378-1119(96)00223-5. [DOI] [PubMed] [Google Scholar]
- 16.Ogasawara N, Moriya S, Mazza P G, Yoshikawa H. Nucleotide sequence and organization of dnaB gene and neighbouring genes on the Bacillus subtilis chromosome. Nucleic Acids Res. 1986;14:9989–9999. doi: 10.1093/nar/14.24.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto Y, Nakai S, Moriya S, Yoshikawa H, Ogasawara N. The Bacillus subtilis dnaC gene encodes a protein homologous to the DnaB helicase of Escherichia coli. Microbiology. 1995;141:641–644. doi: 10.1099/13500872-141-3-641. [DOI] [PubMed] [Google Scholar]
- 18.Seitz H, Weigel C, Messer W. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol Microbiol. 2000;37:1270–1279. doi: 10.1046/j.1365-2958.2000.02096.x. [DOI] [PubMed] [Google Scholar]
- 19.Sueoka N. Cell membrane and chromosome replication in Bacillus subtilis. Prog Nucleic Acid Res Mol Biol. 1998;59:35–53. doi: 10.1016/s0079-6603(08)61028-4. [DOI] [PubMed] [Google Scholar]
- 20.Sutton D M, Carr K M, Vicente M, Kaguni J M. Escherichia coli DnaA protein. J Biol Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- 21.Yoshikawa H, Wake R G. Initiation and termination of chromosome replication. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 507–528. [Google Scholar]