Abstract
The alternative sigma factor PvdS is required by Pseudomonas aeruginosa for initiation of transcription from pyoverdine (pvd) promoters. Two divergent PvdS-dependent promoters (pvdE and pvdF) were characterized by deletion analysis, and the minimal promoter region for each included a sequence element, the iron starvation (IS) box, that is present in other pvd promoters. Site-directed mutagenesis showed that the IS box elements were essential for promoter activity in vivo. Band shift assays and in vitro transcription experiments showed that a complex of PvdS and core RNA polymerase required the presence of an IS box in order to bind to and initiate transcription from pvd promoters. These results indicate that IS box elements participate in sequence-specific recognition by PvdS to enable initiation of transcription from pvd promoters and are likely to represent a −35 sequence element for this sigma factor.
Fluorescent pseudomonads are characterized by the production of yellow-green siderophores termed pyoverdines or pseudobactins that enable iron uptake under conditions where little free iron is available. These molecules are thought to be associated with biocontrol of fungal pathogens in the biosphere (7), and pyoverdine is required for virulence of the opportunist mammalian pathogen Pseudomonas aeruginosa (14). Production of pyoverdine by P. aeruginosa strain PAO is dependent on a protein, PvdS, that has been shown to be an alternative sigma factor protein (9, 27). The presence of PvdS is essential for the expression of all other pyoverdine synthesis genes that have been examined, including pvdA, pvdD, pvdE, pvcA to D, and ptxR (4, 8, 23, 24). These genes encode a variety of proteins that all contribute to the biosynthesis of pyoverdine. PvdS is also required for the synthesis of exotoxin A (18) and an extracellular proteinase, PrpL (P. J. Wilderman and M. L. Vasil, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. B-331, 2000), two enzymes that are secreted by P. aeruginosa. Expression of pvdS is regulated by the iron-responsive Fur repressor protein such that there is no transcription of pvdS and no production of pyoverdine or exotoxin A by cells grown in iron-rich media (reviewed in reference 25).
The DNA sequences that are recognized by PvdS to enable transcription from cognate promoters have not yet been identified. However, a DNA sequence element named the iron starvation (IS) box has been identified in the promoters of iron-regulated genes from Pseudomonas and been shown to be required for promoter function (20). The purpose of the experiments described here was to test the hypothesis that this sequence element is a DNA recognition site for PvdS. This was done through a detailed characterization of two divergent PvdS-dependent pvd promoters, the promoters of the pvdE and pvdF genes. Both of these genes are essential for pyoverdine production by P. aeruginosa PAO, with pvdE encoding an ATP binding cassette-2 type transporter that is likely to be involved in secretion of pyoverdine or a precursor (11) and pvdF encoding an enzyme required for synthesis of formyl-hydroxyornithine residues that are present in the pyoverdine produced by this strain (B. McMorran et al., unpublished data).
Identification of the pvdF gene transcriptional start sites.
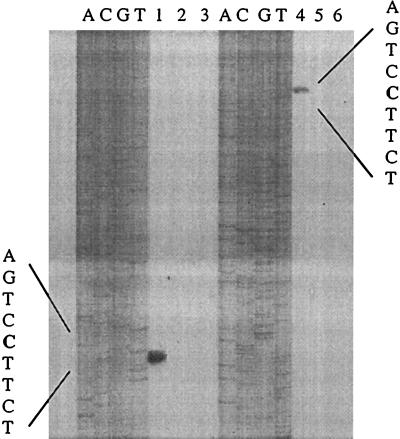
The strains and plasmids used in this study are described in Table 1, and the intergenic sequence between the pvdE and pvdF genes is shown in Fig. 1. As a prelude to promoter characterization, it was necessary to determine the transcriptional start site of the pvdF gene; the pvdE transcriptional start site has been mapped previously (20). The initiation site of the pvdF transcript was determined by primer extension analysis (Fig. 2) using an avian myeloblastosis virus reverse transcriptase primer extension kit (Promega). Primer extension analysis was carried out using 5′-end-labeled oligonucleotides, primer 1 (CCGCGGGGTTTCTCAGGGACCAGATATAGGCCAG), and primer 2 (CGCAGTCTGGTTCAGCGCCTCCACCAAGGACTCC). With both primers, the 3′ end of the major reaction product corresponded to a cytosine residue located 87 bp upstream from the pvdE transcript start site. No products were detected in experiments performed with RNA from bacteria grown in iron-rich medium (Fig. 2, lanes 2 and 5), consistent with expression of pvdF requiring the PvdS protein, which is absent from iron-rich cells (4).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| P. aeruginosa strains | ||
| OT11 | Pvd+ | 19 |
| OT11pvdS | pvdS::Kan Pvd− | 4 |
| Plasmids | ||
| pMP190 | Broad-host-range promoter probe vector; Cmr | 22 |
| pUC19 | Cloning vector: Apr | 28 |
| pUC::pvdE/F | pvdE-pvdF promoter region (−91 to +34 of pvdF) cloned into pUC19 | 27 |
| pUC::mut1 | pvdE-pvdF region with mutation in pvdE-proximal IS box | This study |
| pUC::mut2 | pvdE-pvdF region with mutation in pvdF-proximal IS box | This study |
| pUC::pvdD | pvdD promoter (−96 to +39) cloned into pUC19 | 27 |
| pUC::pvdDmut | pvdD promoter (−96 to +39) with mutation in IS box | This study |
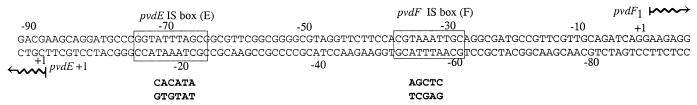
FIG. 1.
The pvdE-pvdF intergenic promoter region. The DNA sequence and the location of the pvdE IS box and the pvdE transcript start site have been described previously (20), and the sequence has been deposited with GenBank (accession no. U07359). The positions of the pvdF IS box and pvdF transcription start site (this work) are also shown. Numbering above the sequence is relative to the pvdF transcript start site, while numbering below the sequence is relative to the pvdE transcript start site. Mutations that were created at the IS box elements (see text) are shown in boldface underneath the wild-type sequences.
FIG. 2.
Identification of the 5′ end of pvdF transcript. RNA was prepared from P. aeruginosa grown in King's B medium (6) containing the iron chelator ethylenediamine-(o-hydroxy)phenylacetic acid (EDDA) (200 μg/ml) (lanes 1 and 4) or containing FeCl3 (60 μg/ml) (lanes 2 and 5) and used in primer extension analysis with primers 1 and 2. Lanes 3 and 6, reactions performed without RNA. Sequencing ladders of pvdF DNA obtained with the same oligonucleotides are shown and allowed the precise identification of the 5′ ends of the pvdF transcript. The DNA sequence corresponding to the transcription start site is shown, with the initiating nucleotide in boldface.
The DNA upstream from the 5′ end of the major pvdF transcript was examined for likely promoter sequences and regulatory elements. Recognition sequences have been suggested for P. aeruginosa RpoD and RpoN (21), but we were unable to identify similar sequences upstream from pvdF. However, a sequence matching 7 out of 10 nucleotides of the IS box motif [(G/C)CTAAATCCC] (20) was centered 33 bp upstream of the pvdF transcript start site (Fig. 1).
Mutational analysis of the pvdE and pvdF promoters.
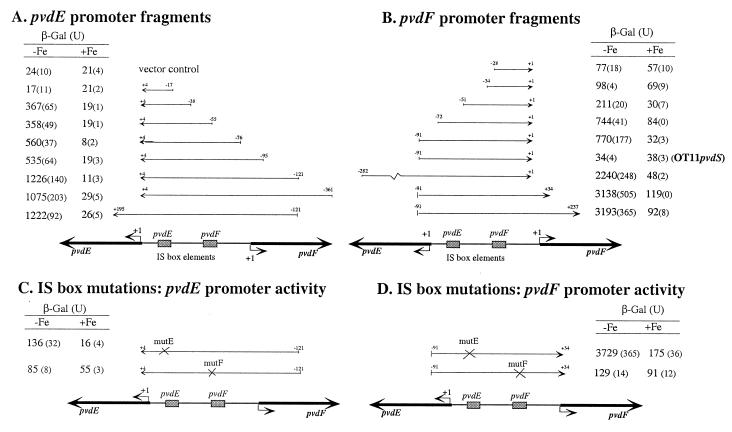
A series of 5′ deletions of each promoter was constructed in order to localize the DNA sequences necessary for promoter function. This was done by using PCR to amplify fragments with a shared 3′ end and different lengths of 5′ promoter sequence (Fig. 3A and B). The fragments were cloned into plasmid pMP190 (22) just upstream from a promoterless lacZ reporter gene, using XbaI and BglII restriction sites incorporated into the ends of each PCR product. Each construct was then introduced into P. aeruginosa OT11 (19), which is a derivative of strain PAO, and the amounts of β-galactosidase (β-Gal) produced by the bacteria were assayed (Fig. 3).
FIG. 3.
Promoter activities of the pvdE and pvdF promoter deletion fragments in P. aeruginosa. The positions of the fragments are shown relative to a map of the pvdE-pvdF promoter region, with the nucleotides present in each fragment given relative to the transcript start site of the relevant gene. The transcript start sites (+1) and positions of IS box elements are also shown. The amounts of β-Gal produced from each construct in P. aeruginosa during growth in iron-deficient (−Fe) and iron-replete (+Fe) media, with standard deviations from three independent assays in parentheses, were determined using the method of Miller (15) as described previously (4, 20). Promoter fragments cloned into pMP190 were oriented such that lacZ expression was dependent upon pvdE promoter activity (A and C) or pvdF promoter activity (B and D) as shown. Fragments with the wild-type promoter sequence (A and B) or with mutations in the IS box sequence elements (C and D) were used. Vector control, P. aeruginosa OT11 containing pMP190 vector DNA. OT11pvdS, the pMP190 promoter construct was transformed into OT11pvdS (3) and assays were carried out.
For the pvdE promoter the smallest fragment to have promoter activity extended from nucleotide −38 to +4. This fragment includes the IS box (nucleotides −14 to −23) shown previously to be required for promoter activity (20). The inclusion of DNA to nucleotide −76 resulted in higher levels of promoter activity, and this was further increased by inclusion of DNA to nucleotide −121. This suggests that DNA between nucleotides −76 and −55 and −121 to −95 may contain sequence elements that are important for complete pvdE promoter activity. The −76 to −55 region contains the second divergent IS box sequence (nucleotides −61 to −52), and it may be this element that contributes to increased promoter activity. Larger fragments extending further 5′ and/or 3′ than the −121/+4 fragment did not produce significantly different β-Gal levels (Fig. 3A), indicating that all of the sequences necessary for pvdE promoter activity were located within the −121/+4 region. None of the fragments had significant promoter activity when the bacteria were grown in iron-rich medium, consistent with previous studies on the iron-regulated nature of this promoter (11) and indicating that all promoter activity is PvdS dependent.
Expression from the pvdF promoter was abolished in a pvdS mutant, OT11pvdS, and was very greatly reduced in iron-rich cells in which transcription of pvdS is repressed (Fig. 3B). This showed that pvdF promoter activity is also PvdS dependent. The two smallest pvdF promoter fragments examined had low-level promoter activities in both iron-deficient and iron-starved cells (Fig. 3B). These levels of expression were not iron regulated and so did not reflect PvdS-dependent promoter activity, as pvdS is not transcribed in iron-rich cells (4). The smallest fragment to show iron-regulated promoter activity extended from nucleotide −51 to +1 (Fig. 3B). This DNA contains a sequence (nucleotides −36 to −27) similar to the IS box element (Fig. 1) (20). Further increases in promoter activity were observed with larger fragments, and the data indicate that sequences in the −72 to −51 and −282 to −91 regions contribute to promoter activity. The −72 to −51 region contains almost all of the IS box sequence (nucleotides −65 to −74) shown to be important for activity of the divergent pvdE promoter (Fig. 3) (20). Thus, this element may contribute to the activities of both promoters. Expression from the pvdF promoter was further increased by the presence of DNA downstream from the transcriptional start site (nucleotides +1 to +34), indicating that a sequence element in this region is required for maximal expression from this promoter. A study of the PvdS-dependent pvdA promoter also found that DNA downstream from the +1 site is required for promoter activity in P. aeruginosa (8). However, DNA downstream from the pvdE transcription start site, to nucleotide +195, did not increase pvdE promoter activity (Fig. 3A); it remains to be seen whether other pvd promoters need DNA elements downstream from the transcription start site for maximal activity. It should be noted that DNA downstream from nucleotide +1 is likely to be involved in promoter recognition by other extracytoplasmic-function (ECF) sigma factor proteins. DNA from nucleotide +50 to +120 of the crtl promoter (a CarQ-dependent promoter) is required for promoter activity (10). PCR mutagenesis of an ECF-dependent promoter from Escherichia coli, pfecA, revealed that residues clustered around the +13 site were required for promoter function (1).
Mutations were introduced into the IS box elements by PCR mutagenesis using a protocol developed by Datta (5) (Fig. 1) in order to assess their involvement in promoter activity more directly. The mutated fragments were cloned into pMP190 in both orientations, the resulting constructs were transformed into P. aeruginosa, and the production of β-Gal was assayed (Fig. 3C and D). Mutation of the IS box nearest to pvdE reduced pvdE promoter activity, consistent with a previous study (20), but had no effect on pvdF promoter activity (Fig. 3C and D, mutE). The mutation to the IS box nearest to pvdF abolished expression from the pvdF promoter and also greatly reduced expression from the pvdE promoter (Fig. 3C and D, mutF). This indicates that this IS box is important for expression from both promoters.
In vitro analysis of PvdS-IS box interactions.
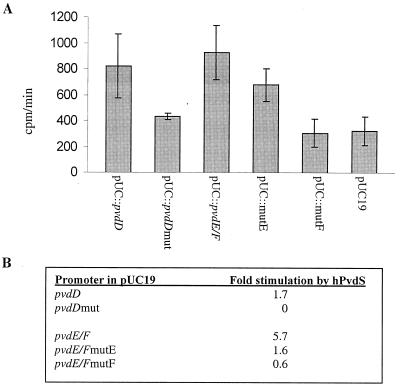
The above data, in conjunction with earlier studies (8, 20), indicate the critical role of IS box sequence elements in pvd promoter function in P. aeruginosa. These promoters are PvdS dependent and therefore must contain a PvdS recognition sequence. Furthermore, we (4) and others (8, 17) have previously shown that pvd promoters are active in E. coli, although only if the pvdS gene is present; promoter constructs carrying the site-directed IS box mutations were inactive in E. coli expressing pvdS (data not shown). Collectively, these data lead to the hypothesis that the IS box forms part of the DNA recognition sequence of the PvdS sigma factor. To test this hypothesis, promoter fragments containing wild-type and mutated IS box elements were used in in vitro transcription assays. PvdS was purified as a His-tagged protein (hPvdS) and incubated with core RNA polymerase and plasmid templates in an in vitro transcription assay as described previously (27). Experiments were carried out with the pvdD promoter, in which a mutation in the IS box abolishes promoter activity (20), as well as the pvdE and pvdF promoters. PvdS stimulated transcription from the pvdD promoter but failed to stimulate transcription from the pvdD promoter if the IS box had been mutated (Fig. 4). Mutation of the IS box nearest to pvdE (mutE) (Fig. 4) resulted in a reduction in PvdS-mediated transcription from the pvdE-pvdF promoter fragment, and mutating the other IS box in this fragment (mutF) (Fig. 4) virtually eliminated PvdS-mediated transcription (Fig. 4). These data are consisted with the hypothesis that the IS box forms part of the DNA recognition sequence of the PvdS sigma factor. They also suggest that the IS box nearest pvdF is required for PvdS-mediated expression from both of the divergent promoters, whereas the pvdE-proximal IS box is required only for transcription from one promoter. This is the same pattern as that found in vivo (Fig. 3C and D).
FIG. 4.
Activities of mutant promoter fragments in vitro with purified PvdS protein. (A) Purified hPvdS was incubated with core RNA polymerase and different plasmid templates as shown. Templates carried wild-type promoter fragments (pvdD [nucleotides −96 to +39] or pvdE-pvdF [−121 to +4 of pvdE and −92 to +34 of pvdF]) or mutations in the IS box of the pvdD promoter (pvdDmut), the pvdE-proximal IS box (mutE), or the pvdF-proximal IS box (mut F) (Fig. 1; Table 1); the mutation in the pvdD IS box is the same as that described previously (20). The rate of RNA production from each plasmid template was measured as described previously (27). Error bars indicate standard deviations. (B) Fold stimulation represents the ability of hPvdS to stimulate activity above that of core enzyme with a pvd promoter template minus the corresponding value obtained with a vector template, calculated using the following equation (2, 27): fold stimulation = [(hPvdS-core − hPvdS only)/core only]pUC::pvd template − [(hPvdS-core − hPvdS only)/core only]pUCvector template.
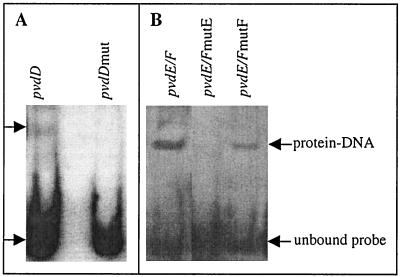
Interactions between promoter fragments and the core-PvdS complex were also analyzed using band shift assays. Core-PvdS bound the pvdD wild-type promoter fragment but failed to cause any shifting of the corresponding fragment containing a mutated IS box (Fig. 5). For the pvdE-pvdF promoter fragment, a mutation in the pvdE-proximal IS box (mutE) prevented binding of core-PvdS, indicating that PvdS requires this sequence to bind to the pvdE-pvdF promoter fragment. Core-PvdS still retarded a pvdE-pvdF promoter fragment containing the pvdF IS box mutation (mutF), although slightly less effectively than the wild-type promoter fragment, suggesting that this sequence is not essential for binding of the protein to the promoter fragments used in these experiments. Consistent with these results, a fragment containing only the pvdF IS box (nucleotides −72 to +34 of pvdF) failed to bind core-PvdS, whereas a fragment containing only the pvdE IS box (nucleotides −38 to +195 of pvdE) still showed binding by core-PvdS in gel shift assays (data not shown). Collectively these results indicate that PvdS requires the presence of an IS box to enable binding of RNA polymerase at both the pvdE-pvdF and pvdD promoter fragments.
FIG. 5.
Band shift assays with hPvdS and mutant promoters. (A) Band shift assays were carried out as described previously (27). Core enzyme (3.4 pmol) was incubated with hPvdS (13 pmol) and digoxigenin-labeled DNA fragments containing either the pvdD wild-type promoter fragment (nucleotides −96 to +39) or a pvdD promoter fragment containing a mutation in the IS box. Following electrophoresis and transfer to a nylon membrane, the DNA-protein complexes were detected by immunoblotting with antibodies against digoxigenin. (B) Core and hPvdS were incubated with the wild-type pvdE-pvdF promoter fragment (−121/+4 with respect to the pvdE transcription start site) or with promoter fragments containing a mutation in either the pvdE IS box (mutE) or the pvdF-proximal IS box (mutF), as shown. The positions of DNA-protein complexes are indicated by arrows.
It is surprising that the mutation in the pvdE-proximal IS box essentially abolished binding of core-PvdS to the promoter fragment, as this mutation had no effect on expression from the pvdF promoter (Fig. 3D). Similarly, the mutation in the pvdF-proximal IS box essentially abolished activity from both promoters (Fig. 3C and D and 4), whereas it reduced but did not prevent binding of core-PvdS to the promoter fragment (Fig. 5). These apparently conflicting data may reflect differences in the use of linearized DNA fragments in band shift assays, instead of the supercoiled plasmid templates of the in vitro transcription and in vivo studies, or may be a consequence of complex interactions between the two divergent promoters. It may also be possible that a complex of core-PvdS bound at the pvdE IS box is more stable in the gel shift assay than the complex formed at the pvdF IS box, reflecting a more stringent requirement of protein-DNA binding in this assay than in transcription assays.
Investigation of two other promoters, pvdY and ptxR, that have been shown to require PvdS for activity (24, 25) reveals that they also contain IS box-like sequences (Table 2). Thus, all promoters that have been shown to be PvdS dependent contain IS box sequences, further supporting the proposition that this sequence motif is recognized by PvdS during initiation of transcription from pvd genes. For those promoters whose transcription sites have been mapped, the IS box is at the −35 region of the promoter (with the exception of the pvdE promoter), consistent with other ECF family members, which tend to have a recognition motif centered at about −35 (10, 16). The position of the pvdE IS box, centered at −19, is likely to be atypical and a consequence of the divergent and interacting natures of the pvdE and pvdF promoters.
TABLE 2.
IS boxes in PvdS-dependent promotersa
| Promoter | IS box | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pvdA | −38 | C | T | T | A | A | A | T | T | C | −30 |
| pvdD | −39 | G | C | T | A | A | A | T | C | C | −29 |
| pvdE | −23 | G | C | T | A | A | A | T | A | C | −15 |
| pvdF | −38 | C | G | T | A | A | A | T | T | G | −30 |
| ptxRb | ? | A | G | T | A | A | A | T | A | G | ? |
| pvdY | ? | G | G | T | A | A | A | T | T | C | ? |
| Consensus | (G/C) | (G/C) | T | A | A | A | T | (T/A) | (C/G) | ||
The pvdA, -D, -E, and -F genes have been characterized and are required for pyoverdine synthesis (11, 12, 13, 26). The IS boxes of these genes have been mutated and shown to be required for activity (8, 20; this study). Expression of ptxR, which encodes a positive regulator of the pvc genes, and the pvdY gene, which encodes a 17-kDa protein of unknown function, has been shown to be PvdS dependent (24, 25). ?, the transcriptional start site is not known.
The transcriptional start site can be estimated on the basis of RNase protection data (24) as being between nucleotide −28 and −20 downstream from the IS box.
In summary, characterization of the divergently transcribed pvdE and pvdF promoters has allowed the minimal promoter regions to be determined and emphasizes the involvement of the IS box sequence. Gel shift and in vitro transcription data very strongly suggest that PvdS recognizes the IS box in pyoverdine promoters. Further studies will be required to unravel the complex protein-DNA interactions that take place at these promoters as well as to identify and characterize the other sequence elements that contribute to the activity of these promoters.
Acknowledgments
We are grateful to members of the Lamont laboratory for their comments on a preliminary version of the manuscript.
M.J.W. and B.J.M. were supported by postgraduate scholarships from the Health Research Council of New Zealand. This work was supported in part by a grant from the New Zealand Lotteries Health Research Board.
REFERENCES
- 1.Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. FecI belongs to a new subfamily of ς70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;18:163–174. doi: 10.1111/j.1365-2958.1995.mmi_18010163.x. [DOI] [PubMed] [Google Scholar]
- 2.Burgess R R, Travers A A. Purification of the RNA polymerase sigma factor. Methods Enzymol. 1971;21:500–506. [Google Scholar]
- 3.Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion cloning in Escherichia coli cells. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Cunliffe H E, Merriman T R, Lamont I L. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta A K. Efficient amplification using ‘megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res. 1995;23:4530–4531. doi: 10.1093/nar/23.21.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 7.Leong J. Siderophores: their biochemistry and possible role in the biochontrol of plant pathogens. Annu Rev Plant Phyt. 1986;26:187–209. [Google Scholar]
- 8.Leoni L, Ciervo A, Orsi N, Visca P. Iron-regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of Fur and PvdS on promoter activity. J Bacteriol. 1996;178:2299–2313. doi: 10.1128/jb.178.8.2299-2313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leoni L, Orsi N, de Lorenzo V, Visca P. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1481–1491. doi: 10.1128/jb.182.6.1481-1491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marinez-Argudo I, Ruiz-Vazquez R M, Murillo F J. The structure of an ECF-ς-dependent, light-inducible promoter from the bacterium Myxococcus xanthus. Mol Microbiol. 1998;30:883–893. doi: 10.1046/j.1365-2958.1998.01129.x. [DOI] [PubMed] [Google Scholar]
- 11.McMorran B J, Merriman M E, Rombel I T, Lamont I L. Characterisation of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene. 1996;176:55–59. doi: 10.1016/0378-1119(96)00209-0. [DOI] [PubMed] [Google Scholar]
- 12.McMorran B J. Pyoverdine promoters from P. aeruginosa. Ph.D. thesis. Dunedin, New Zealand: University of Otago; 1997. [Google Scholar]
- 13.Merriman T E, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer J M, Neely A, Stintzi A, Georges C, Holder I A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 16.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 17.Miyazaki H, Kato H, Nakazawa T, Tsuda M. A positive regulatory gene, pvdS, for expression of pyoverdin biosynthetic genes in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1995;248:17–24. doi: 10.1007/BF02456609. [DOI] [PubMed] [Google Scholar]
- 18.Ochsner U A, Johnson Z, Lamont I L, Cunliffe H E, Vasil M L. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol Microbiol. 1996;21:1019–1028. doi: 10.1046/j.1365-2958.1996.481425.x. [DOI] [PubMed] [Google Scholar]
- 19.Rombel I T, Lamont I L. DNA homology between siderophore genes from fluorescent Pseudomonads. J Gen Microbiol. 1992;138:181–187. doi: 10.1099/00221287-138-1-181. [DOI] [PubMed] [Google Scholar]
- 20.Rombel I T, McMorran B J, Lamont I L. Identification of a DNA sequence motif required for expression of iron-regulated genes in Pseudomonads. Mol Gen Genet. 1995;246:519–528. doi: 10.1007/BF00290456. [DOI] [PubMed] [Google Scholar]
- 21.Ronald S, Farinha M A, Allan B J, Kropinski A M. Cloning and physical mapping of transcriptional regulatory (sigma) factors from Pseudomonas aeruginosa. In: Gaili E, Silver S, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington D.C.: American Society for Microbiology; 1992. pp. 249–257. [Google Scholar]
- 22.Spaink H P, Okker R J H, Wiffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1J1. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 23.Stintzi A, Johnson Z, Stonehouse M, Ochsner U, Meyer J, Vasil M L, Poole K. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromphore and regulation of PtxR and PvdS. J Bacteriol. 1999;181:4118–4124. doi: 10.1128/jb.181.13.4118-4124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasil M L, Ochsner U A, Johnson Z, Colmer J A, Hamood A N. The Fur-regulated gene encoding the alternative sigma factor PvdS is required for iron-dependent expression of the LysR-type regulator PtxR in Pseudomonas aeruginosa. J Bacteriol. 1998;180:6784–6788. doi: 10.1128/jb.180.24.6784-6788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasil M L, Ochsner U A. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 26.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdine biosynthesis enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson M J, Lamont I L. Characterisation of a ECF sigma factor protein from Pseudomonas aeruginosa. Biochem Biophys Res Commun. 2000;273:578–583. doi: 10.1006/bbrc.2000.2996. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]