Abstract
Several hypotheses have been presented on the origin of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) from its identification as the agent causing the current coronavirus disease 19 (COVID-19) pandemic. So far, no solid evidence has been found to support any hypothesis on the origin of this virus, and the issue continue to resurface over and over again. Here we have unfolded a pattern of distribution of several mutations in the SARS-CoV-2 proteins in 24 geo-locations across different continents. The results showed an evenly uneven distribution of the unique protein variants, distinct mutations, unique frequency of common conserved residues, and mutational residues across these 24 geo-locations. Furthermore, ample mutations were identified in the evolutionarily conserved invariant regions in the SARS-CoV-2 proteins across almost all geo-locations studied. This pattern of mutations potentially breaches the law of evolutionary conserved functional units of the beta-coronavirus genus. These mutations may lead to several novel SARS-CoV-2 variants with a high degree of transmissibility and virulence. A thorough investigation on the origin and characteristics of SARS-CoV-2 needs to be conducted in the interest of science and for the preparation of meeting the challenges of potential future pandemics.
Keywords: SARS-CoV-2, Mutations, Furin cleavage site (FCS), Evenly-uneven, Invariant regions
1. Introduction
SARS-CoV-2 is the etiological agent causing the COVID-19 pandemic. Since its very onset, the understanding of the origin of the SARS-CoV-2 has been of utmost importance. In fact, this knowledge is crucial both for the successful fight against this virus, for better understanding of the mechanisms of the potential emergence of new pathogens, and for the meaningful analysis of the exposure risks [1], [2], [3], [4]. A great source for the unfolding of the roots of the COVID-19 pandemic is the access to the SARS-CoV-2 hub at the National Center for Biotechnology Information (NCBI) [5]. In this context, a careful time-based dynamic surveillance of mutations and associated functional changes in viral proteins are most productive due to the potential link to changes in general viral properties, such as transmissibility, immune-escape, pathogenesis, and virulence, among others [6]. The surveillance should focus on the analysis of the viral genome and identification of mutations [7], [8], [9]. At the beginning of the pandemic, the largely accepted consensus was that, compared to other RNA viruses (typically with smaller genomes), the SARS-CoV-2 mutation rate should be lower due to the presence of the proofreading protein ExoN-nsp14, whose function is to prevent excessive changes to the viral genome [10], [11]. In agreement with this hypothesis, the mutation rates of the coronaviruses are indeed low (10−6 per site per cycle) in comparison with those of other RNA viruses, such as the influenza A virus (FLUVA, which has a mutation rate of 2.3 × 10−5 per site per cycle) or Hepatitis C virus (HCV, with the mutation rate of 1.2 × 10−4 per site per cycle) [12], [13]. However, because the RNA genome of SARS-CoV-2 is long (between 29.8 kb and 29.9 kb, which is more than twice as long as the FLUVA genome of ~14 kb), the presence of the “proofreading” machinery is somehow “compensated” by the virus length [12], [14]. Because the SARS-CoV-2 multiplication rate is high (each infected person carries 109 to 1011 virions during peak infection and 1 mL of sputum might contain >107 viral RNA molecules, and since the SARS-CoV-2 mutation rate is 10−6 mutations/site/cycle, the chances of generating mutants is high [14], [15]. In fact, based on these numbers, it seems very likely that every site of the SARS-CoV-2 genome can be mutated more than once in the virions produced by each infected person. Therefore, SARS-CoV-2 is steadily mutating during continuous transmission among humans. In line with these considerations, a study based on the comparative analysis of then available 48,635 SARS-CoV-2 complete genomes with the reference SARS-CoV-2 Wuhan genome NC 045512.2 revealed an average of 7.23 mutations per sample [16]. Obviously, not all acquired mutations are retained, as mutations not leading to a viable progeny are eliminated. Therefore, a typical SARS-CoV-2 virus accumulates two single-letter mutations per month in its genome. This sums up to the retention rate of some 20–30 mutations per year, which is still significant [17]. The fact that the ex vivo multiplication of this virus in the relevant cells leads to shedding of a considerable number of mutants, including many mutants with defective genomes, represents an important constraint that makes impossible the formulation of any assumption from the landscape of mutations without RNA comparisons (see e.g., [18]).
SARS-CoV-2 sequences from COVID-19 patients showed that the receptor-binding domain (RBD) of the Spike (S) protein possessed eight mutations, which assist in initiating infection of the host cells [19], [20], [21]. Curiously, based on the analysis of the experimental evolution of two circulating SARS-CoV-2 lineages in Vero cells it was concluded that these lineages are characterized by different genome mutation rates, where a lineage of SARS-CoV-2 with the originally described S protein (D614) mutated at the rate of 3.7 × 10−6 nt−1 cycle−1, whereas the SARS-CoV-2 lineage carrying the D614G mutation in the S protein showed a mutation rate of 2.9 × 10−6 nt−1 cycle−1 [22]. Furthermore, it was also shown that the mutation accumulation was highly heterogeneous along the genome, with the spike gene accumulating mutations at a mean rate of 16 × 10−6 nt−1 per infection cycle, which is five times faster than the genome-average mutation rate [22].
Many of the mutations in SARS-CoV-2 are non-essential, and some are disadvantageous to the virus itself. Some mutations may allow the virus to propagate more easily from host to host, and these mutations make SARS-CoV-2 variants more transmissible [23]. The majority of the SARS-CoV-2 mutations do not appear to cause a more severe disease, but just make the virus more contagious [24]. The mutation rate is defined as the probability that a change in the genetic information is passed to the next generation [25], [26]. For viruses, a generation is simply defined as a cell infection cycle, which includes initiating attachment to the cell surface, entry, replication, encapsidation, and release of infectious particles [27]. It was previously reported that in RNA viruses, an inverse correlation exists between the mutation rates and genome size [28]. Coronaviruses have the largest genomes among RNA viruses (30–33 kb) and have acquired proofreading capacity in contrast to all other known RNA viruses [29], [30]. Though most mutations in the SARS-CoV-2 are expected to be either deleterious and swiftly purged or relatively neutral, a small proportion will affect functional properties of viral proteins and increase/decrease infectivity of the virus and disease severity or capability of a virus to interact with host immune system [31], [32]. In SARS-CoV-2, the average mutation rate remains low and steady, being much lower than for other RNA viruses, such as FLUVA, HIV, and HCV [33].
Such atypical characteristics have contributed to the resurfacing of the question of the origin of the SARS-CoV-2. So far, no clear animal progenitor or intermediary host has been confirmed. Therefore, in light of these observations, the hypothesis that SARS-CoV-2 originated as a leak from the Wuhan lab is taken seriously now. Primarily, a zoonotic source was thought to have spilled over to humans through the ‘wet market’ in Wuhan, China, where the virus was first detected in December 2019 [34], [35], [36], [37], [38]. But later, several other orthogonal hypotheses reverted to the old question about the SARS-CoV-2 origin [39], [40], [41], [42], [43]. It is clear that although it is very likely that SARS-CoV-2 has zoonotic roots and originated as a result of a transition between bats and humans, the available data also suggest that this transition is most likely to have necessitated an intermediate animal. Importantly, this view does not tell whether the spillover happened in an open environment setting or within a laboratory, as many virology laboratories use animal models. Furthermore, there is a second alternative, which should be taken seriously: transition from bats to humans has happened via ex vivo cultivation and adaptation of human cells. This is a daunting possibility, which, nevertheless, should be considered and discussed, as this type of experiment has been pursued in several laboratories world-wide. In this study, the apparent uneven distribution of the identified mutations in several proteins of SARS-CoV-2 across the 24 geo-locations questions the natural origin of the SARS-CoV-2, based on the prior knowledge from other beta-coronaviruses. Several other observations, such as mutations in invariant regions of the SARS-CoV-2 proteins, which are conserved across four other beta-coronaviruses, strengthen the case of the pseudo-natural origin of SARS-CoV-2.
2. Data acquisition and methods
2.1. Data and informatics
The amino acid sequences (complete) of SARS-CoV-2 spike (S), envelope (E), membrane (M), nucleocapsid (N), ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 from different geo-locations were exported in FASTA format from the NCBI database (http: // www.ncbi.nlm.nih.gov/) (as of May 29, 2021). To this end, the 24 geo-locations with relatively high frequency of SARS-CoV-2 proteins were chosen from six continents, individual SARS-CoV-2 proteins were searched and associated sequences were retrieved from the NCBI database. The Asian group comprises patients in India, Hong Kong, Bahrain, Bangladesh, and Pakistan. The Oceania group comprises Australian patients only, whereas the European group includes patients from Austria, France, Greece, Poland, Serbia, and Spain. The South American group contains patients from Peru and Chile. The African group contains patients from the Egypt, Ghana, and Tunisia. Finally, the North American group contains patients from California, Florida, Texas, Massachusetts, Minnesota, Michigan, and Pennsylvania. The retrieved FASTA files were processed in Matlab-2021a for extracting unique protein sequences from each geo-location. The frequencies of total and unique protein sequences are presented in Table 1 .
Table 1.
Frequencies and percentages of total and unique S, E, M, N, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 protein sequences in SARS-CoV-2 from 24 different geo-locations.
| Geo-locations | S |
E |
M |
N |
ORF3a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Unique | % | Total | Unique | % | Total | Unique | % | Total | Unique | % | Total | Unique | % | |
| Australia | 9919 | 1121 | 11.302 | 9919 | 38 | 0.3831 | 9919 | 38 | 0.3831 | 9919 | 213 | 2.147 | 9919 | 19 | 0.192 |
| Austria | 97 | 26 | 26.804 | 97 | 2 | 2.0619 | 97 | 2 | 2.0619 | 97 | 22 | 22.680 | 97 | 3 | 3.093 |
| Bahrain | 167 | 56 | 33.533 | 167 | 4 | 2.3952 | 167 | 4 | 2.3952 | 167 | 33 | 19.760 | 167 | 7 | 4.192 |
| Bangladesh | 402 | 98 | 24.378 | 402 | 11 | 2.7363 | 402 | 11 | 2.7363 | 402 | 53 | 13.184 | 402 | 9 | 2.239 |
| California | 15,616 | 3321 | 21.267 | 15,744 | 192 | 1.2195 | 15,744 | 192 | 1.2195 | 15,616 | 1345 | 8.613 | 15,615 | 104 | 0.666 |
| Chile | 290 | 25 | 8.621 | 290 | 2 | 0.6897 | 290 | 2 | 0.6897 | 290 | 16 | 5.517 | 290 | 3 | 1.034 |
| Egypt | 700 | 183 | 26.143 | 700 | 22 | 3.1429 | 700 | 22 | 3.1429 | 700 | 116 | 16.571 | 700 | 10 | 1.429 |
| Florida | 17,180 | 2527 | 14.709 | 17,324 | 131 | 0.7562 | 17,324 | 131 | 0.7562 | 17,180 | 973 | 5.664 | 17,178 | 65 | 0.378 |
| France | 90 | 19 | 21.111 | 90 | 4 | 4.4444 | 90 | 4 | 4.4444 | 90 | 6 | 6.667 | 90 | 3 | 3.333 |
| Ghana | 167 | 65 | 38.922 | 167 | 7 | 4.1916 | 167 | 7 | 4.1916 | 167 | 41 | 24.551 | 167 | 10 | 5.988 |
| Greece | 97 | 11 | 11.340 | 97 | 3 | 3.0928 | 97 | 3 | 3.0928 | 97 | 9 | 9.278 | 97 | 2 | 2.062 |
| Hong Kong | 228 | 48 | 21.053 | 230 | 5 | 2.1739 | 230 | 5 | 2.1739 | 228 | 28 | 12.281 | 228 | 3 | 1.316 |
| India | 813 | 178 | 21.894 | 830 | 20 | 2.4096 | 830 | 20 | 2.4096 | 813 | 86 | 10.578 | 813 | 7 | 0.861 |
| Massachusetts | 8856 | 1281 | 14.465 | 9045 | 92 | 1.0171 | 9045 | 92 | 1.0171 | 8856 | 625 | 7.057 | 8856 | 47 | 0.531 |
| Michigan | 9930 | 1297 | 13.061 | 9998 | 78 | 0.7802 | 9998 | 78 | 0.7802 | 9930 | 418 | 4.209 | 9930 | 38 | 0.383 |
| Minnesota | 13,046 | 2658 | 20.374 | 13,621 | 77 | 0.5653 | 13,621 | 77 | 0.5653 | 13,046 | 481 | 3.687 | 13,044 | 45 | 0.345 |
| Pakistan | 214 | 49 | 22.897 | 214 | 7 | 3.2710 | 214 | 7 | 3.2710 | 214 | 33 | 15.421 | 214 | 5 | 2.336 |
| Pennsylvania | 8779 | 1343 | 15.298 | 8913 | 105 | 1.1781 | 8913 | 105 | 1.1781 | 8779 | 643 | 7.324 | 8779 | 52 | 0.592 |
| Peru | 116 | 44 | 37.931 | 116 | 8 | 6.8966 | 116 | 8 | 6.8966 | 116 | 19 | 16.379 | 116 | 2 | 1.724 |
| Poland | 153 | 26 | 16.993 | 153 | 2 | 1.3072 | 153 | 2 | 1.3072 | 153 | 22 | 14.379 | 153 | 1 | 0.654 |
| Serbia | 146 | 23 | 15.753 | 146 | 3 | 2.0548 | 146 | 3 | 2.0548 | 146 | 22 | 15.068 | 145 | 1 | 0.690 |
| Spain | 134 | 36 | 26.866 | 134 | 4 | 2.9851 | 134 | 4 | 2.9851 | 134 | 21 | 15.672 | 134 | 3 | 2.239 |
| Texas | 9251 | 1546 | 16.712 | 9431 | 101 | 1.0709 | 9431 | 101 | 1.0709 | 9251 | 644 | 6.961 | 9251 | 61 | 0.659 |
| Tunisia | 58 | 30 | 51.724 | 58 | 3 | 5.1724 | 58 | 3 | 5.1724 | 58 | 22 | 37.931 | 57 | 1 | 1.754 |
| Geo-locations | ORF6 |
ORF7a |
OR7b |
ORF8 |
ORF10 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Unique | % | Total | Unique | % | Total | Unique | % | Total | Unique | % | Total | Unique | % | |
| Australia | 9919 | 19 | 0.192 | 9919 | 58 | 0.585 | 9919 | 14 | 0.141 | 9919 | 54 | 0.544 | 9919 | 16 | 0.161 |
| Austria | 97 | 3 | 3.093 | 97 | 5 | 5.155 | 95 | 2 | 2.105 | 26 | 3 | 11.538 | 97 | 2 | 2.062 |
| Bahrain | 167 | 7 | 4.192 | 167 | 18 | 10.778 | 167 | 4 | 2.395 | 145 | 17 | 11.724 | 167 | 3 | 1.796 |
| Bangladesh | 402 | 9 | 2.239 | 402 | 15 | 3.731 | 400 | 6 | 1.500 | 397 | 19 | 4.786 | 402 | 11 | 2.736 |
| California | 15,615 | 104 | 0.666 | 15,612 | 330 | 2.114 | 15,724 | 89 | 0.566 | 12,945 | 359 | 2.773 | 15,739 | 61 | 0.388 |
| Chile | 290 | 3 | 1.034 | 290 | 5 | 1.724 | 290 | 2 | 0.690 | 290 | 5 | 1.724 | 290 | 1 | 0.345 |
| Egypt | 700 | 10 | 1.429 | 700 | 20 | 2.857 | 700 | 11 | 1.571 | 697 | 34 | 4.878 | 700 | 8 | 1.143 |
| Florida | 17,178 | 65 | 0.378 | 17,161 | 314 | 1.830 | 17,305 | 63 | 0.364 | 7948 | 231 | 2.906 | 17,322 | 47 | 0.271 |
| France | 90 | 3 | 3.333 | 90 | 1 | 1.111 | 90 | 1 | 1.111 | 90 | 3 | 3.333 | 90 | 1 | 1.111 |
| Ghana | 167 | 10 | 5.988 | 167 | 10 | 5.988 | 167 | 7 | 4.192 | 69 | 12 | 17.391 | 167 | 3 | 1.796 |
| Greece | 97 | 2 | 2.062 | 96 | 2 | 2.083 | 97 | 1 | 1.031 | 97 | 4 | 4.124 | 97 | 1 | 1.031 |
| Hong Kong | 228 | 3 | 1.316 | 230 | 5 | 2.174 | 230 | 2 | 0.870 | 212 | 10 | 4.717 | 230 | 3 | 1.304 |
| India | 813 | 7 | 0.861 | 828 | 23 | 2.778 | 828 | 7 | 0.845 | 798 | 27 | 3.383 | 830 | 3 | 0.361 |
| Massachusetts | 8856 | 47 | 0.531 | 8853 | 184 | 2.078 | 9044 | 46 | 0.509 | 5264 | 137 | 2.603 | 9044 | 29 | 0.321 |
| Michigan | 9930 | 38 | 0.383 | 9927 | 199 | 2.005 | 9998 | 45 | 0.450 | 3061 | 77 | 2.516 | 9998 | 23 | 0.230 |
| Minnesota | 13,044 | 45 | 0.345 | 13,029 | 758 | 5.818 | 13,600 | 59 | 0.434 | 4619 | 118 | 2.555 | 13,608 | 29 | 0.213 |
| Pakistan | 214 | 5 | 2.336 | 212 | 6 | 2.830 | 206 | 2 | 0.971 | 208 | 10 | 4.808 | 212 | 3 | 1.415 |
| Pennsylvania | 8779 | 52 | 0.592 | 8779 | 202 | 2.301 | 8913 | 38 | 0.426 | 4564 | 135 | 2.958 | 8913 | 29 | 0.325 |
| Peru | 116 | 2 | 1.724 | 116 | 9 | 7.759 | 116 | 1 | 0.862 | 115 | 8 | 6.957 | 116 | 5 | 4.310 |
| Poland | 153 | 1 | 0.654 | 152 | 8 | 5.263 | 153 | 2 | 1.307 | 149 | 6 | 4.027 | 153 | 2 | 1.307 |
| Serbia | 145 | 1 | 0.690 | 146 | 3 | 2.055 | 146 | 1 | 0.685 | 146 | 6 | 4.110 | 146 | 2 | 1.370 |
| Spain | 134 | 3 | 2.239 | 134 | 2 | 1.493 | 130 | 2 | 1.538 | 62 | 3 | 4.839 | 134 | 3 | 2.239 |
| Texas | 9251 | 61 | 0.659 | 9251 | 190 | 2.054 | 9430 | 43 | 0.456 | 4626 | 154 | 3.329 | 9430 | 39 | 0.414 |
| Tunisia | 57 | 1 | 1.754 | 58 | 7 | 12.069 | 58 | 2 | 3.448 | 56 | 7 | 12.500 | 57 | 4 | 7.018 |
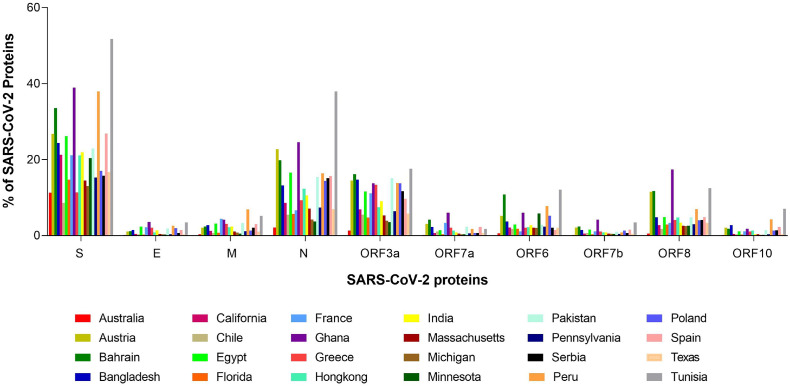
The percentages of each SARS-CoV-2 protein across the 24 geo-locations are presented in Fig. 1 , which indicates that the highest amounts of unique variations across the 24 geo-locations were observed for the S protein. Relatively less unique variations were distributed over the E and ORF3a proteins. Other proteins have a minimal number of unique variations. On the other hand, it was observed that most SARS-CoV-2 proteins possessed the highest unique variations in the viral isolates collected from Tunisia, Ghana, and Greece.
Fig. 1.
Percentage of each SARS-CoV-2 proteins across 24 geo-locations.
Furthermore, amino acid sequences of S, E, M, N, ORF3a, ORF6, ORF7a, ORF7b, and ORF8 proteins from four other coronaviruses Recombinant SARS-CoV (taxid-698,398), Bat SARS-CoV (taxid-442,736), SARS-CoV ExoN1 (taxid-627,440), and Bat SARS- like-CoV (taxid-1,508,227) were also downloaded from the NCBI database. In this study, all mutations in SARS-CoV-2 proteins were detected with reference to the SARS-CoV-2 reference sequence, which was deposited in January 2020 by Wu and co-workers formerly called “Wuhan seafood market pneumonia virus” (WSM, NC 045512) [44]. The frequencies of total and unique protein sequences analyzed in this study are presented in Table 2 .
Table 2.
Frequencies and percentages of S, E, M, N, ORF3a, ORF6, ORF7a, ORF7b, and ORF8 from four different types of CoVs.
| Protein | Total | Unique | Percentage | Protein | Total | Unique | Percentage |
|---|---|---|---|---|---|---|---|
| E-698398 | 80 | 6 | 7.5 | Spike-698,398 | 36 | 2 | 5.56 |
| E-442736 | 2 | 1 | 50 | Spike-627,440 | 18 | 2 | 11.11 |
| E-627440 | 15 | 5 | 33.3 | Spike-442,736 | 13 | 7 | 53.85 |
| E-1508227 | 2 | 1 | 50 | Spike-1,508,227 | 13 | 13 | 100 |
| M-698398 | 116 | 4 | 3.45 | ORF3a-442,736 | 2 | 1 | 50 |
| M-442736 | 2 | 1 | 50 | ORF3a-1,508,227 | 11 | 10 | 90.91 |
| M-627440 | 33 | 3 | 9.09 | ORF6-1508227 | 11 | 6 | 54.55 |
| M-1508227 | 2 | 1 | 50 | ORF6-442736 | 2 | 1 | 50 |
| N-698398 | 80 | 4 | 5 | ORF7a-442,736 | 2 | 1 | 50 |
| N-442736 | 2 | 1 | 50 | ORF7a-1,508,227 | 11 | 5 | 45.45 |
| N-627440 | 15 | 4 | 26.67 | ORF7b-1,508,227 | 11 | 2 | 18.18 |
| N-1508227 | 13 | 12 | 92.31 | ORF7b-442,736 | 2 | 1 | 50 |
| ORF8-1508227 | 10 | 7 | 70 | ||||
| ORF8-442736 | 2 | 1 | 50 |
The least unique variations of M proteins of four types of beta-coronaviruses were observed. Other proteins of four CoVs had several unique variations, unlike in the case of non-uniformity in unique variations in SARS-CoV-2 proteins.
3. Methods
CLUSTAL Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/) tools were used to conduct multiple sequence alignment and for mutation detection with reference to the reference sequence NC 045512 the web-server ViPR (https://www.viprbrc.org/brc/home.spg?decorator = corona) [45], [46], [47]. At each position of a given protein, the consensus residue is the allele with frequency >50 %, regardless of which coverage was considered. If no allele exceeds 50 %, Xaa (for an amino acid) indicates ambiguity [47]. The effect of mutation was predicted using a webserver, PredictSNP (https://loschmidt.chemi.muni.cz/predictsnp1/predictsnp.html) [48]. The statistical and mathematical computations were performed using Matlab software.
4. Results
4.1. Unique proteins variants and their mutations
Across the 24 geo-locations, the common amino acid residues which did not possess any mutations were named as invariant residues. These invariant residues of all unique protein variants from all 24 geo-locations in SARS-CoV-2, were extracted (Table 3) (Supplementary file-I). On the other hand, mutated residues common in all 24 geo-locations were also detected (Table 4) (Supplementary file-I).
Table 3.
Invariant-residues in SARS-CoV-2 proteins, which were common in all unique variants from all 24 geo-locations.
| S (0.39) |
E (4 %) |
M (9.46 %) |
N (1.91 %) |
ORF3a (0.73 %) |
ORF6 (1.64 %) |
ORF7a (0.83 %) |
ORF7b |
ORF8 (0.83 %) |
|
|---|---|---|---|---|---|---|---|---|---|
| 1-Met | 1-Met | 1-Met | 190-Asp | 1-Met | 1-Met | 1-Met | 1-Met | 1-Met | 1-Met |
| 953-Asn | 2-Tyr | 9-Thr | 192-Gly | 42-Pro | 8-Phe | ||||
| 1051-Ser | 3-Ser | 65-Phe | 193-Phe | 49-Thr | |||||
| 1054-Gln | 119-Leu | 195-Ala | 51-Ser | ||||||
| 1269-Lys | 121-Asn | 202-Gly | 52-Trp | ||||||
| 156-Leu | 203-Asn | 57-Thr | |||||||
| 174-Arg | 218-Ala | 58-Gln | |||||||
| 176-Leu | 219-Leu | 143-Lys | |||||||
| 177-Ser | 220-Leu | ||||||||
| 180-Lys | 222-Gln | ||||||||
| 181-Leu | |||||||||
Table 4.
Mutated residues in SARS-CoV-2 proteins that were common in all 24 geo-locations.
| S | E | M | N | ORF3a | ORF6 | ORF7a | ORF7b | ORF8 | ORF10 |
|---|---|---|---|---|---|---|---|---|---|
| D614G/C/N/A | NONE | NONE | R203E/K/M/S/T G204L/P/Q/R/T/V | 57H/E/L/N/R/Y | NONE | NONE | NONE | NONE | NONE |
Table 3 shows that the methionine residue (M) at the position 1 did not change in any of the SARS-CoV-2 proteins listed above, except in ORF10. In ORF10, all amino acid residues from position 1 to 38 were mutated. Even methionine at position 1 was changed to glycine in the only ORF10 sequence QKG88643 from Massachusetts, USA (collected on 18-03-2020). This mutation M1G was found to be a ‘neutral’ mutation as predicted through the webserver, PredictSNP. Note that there was no homologous sequence to QKG88643 with 100 % homology and 100 % query coverage (NCBI Blast). It is known that data is never without errors. The fact that an M1G mutation was found in ORF10 raises some concerns of the reliability of this observation. In fact, it is known that the N-terminal methionine is completely invariant in eukaryotic proteins because the AUG translation initiation codon of mRNAs is recognized by the anticodon of initiator methionine transfer RNA in eukaryotes (or the specialized formyl methionine transfer RNA in prokaryotes, mitochondria, and chloroplasts). Therefore, the protein synthesis is initiated universally with the amino acid methionine (or formyl methionine) that is invariantly present as the first residue of the newly synthesized polypeptide chain. The fact that we found that this is almost always the case, with only one M → G change suggests that this G can be due to a sequencing error. Although it also looks a bit strange as it would imply the presence of an AUG → GGG double mutation [49].
On the other hand, the number of common mutations in the SARS-CoV-2 proteins across 24 geo-locations was surprisingly low (Table 4). D614G was the only mutation possessed by each unique S protein variant from all 24 geo-locations. Similarly, each unique N protein variant from all 24 geo-locations possessed R203 and G204 with changes to multiple amino acids (Table 4). The unique ORF3a variants from all 24 geo-locations had the only common mutation at position 57 with changes to multiple amino acids H/E/L/N/R, and Y. It was noticed that not a single common mutation across 24 geo- locations was found in E, M, ORF6, ORF7a, ORF7b, ORF8, and ORF10. The fact that only very few mutations are spread everywhere and that the number of common mutations in the SARS-CoV-2 proteins across 24 geo-locations (e.g. D614G) were found to be surprisingly low is important, as it suggests that the virus was fairly well adapted to its human host from the early COVID-19 outset.
4.1.1. Spike protein variants and mutations
The total frequency of unique mutations possessed by the S protein of SARS-CoV-2 across the 24 geo-locations is presented in Table 5 . The outermost layer of the SARS-CoV-2 viral particle is made of a phospholipid membrane containing three proteins; the M protein in high abundance, the E coating proteins in relatively low abundance, and finally, the most importantly the S protein) [12], [50]. The S protein is a homotrimeric multifunctional glycoprotein, with its monomer being 1273-amino-acid-long polypeptide. It consists of the S1 and S2 subunits. The S1 subunit is further divided into the N-terminal domain (NTD) and C-terminal domain (CTD) and has a receptor-binding domain (RBD) that detects mammalian cellular receptors and is responsible for binding the viral particle to the host cell, whereas the S2 subunit is used for fusion to the cell membrane [12]. Angiotensin converting enzyme 2 (ACE-2) protein on the epithelial surface of the host cells is the primary entry receptor for SARS-CoV-2, and protein-protein interaction assays demonstrate high-affinity binding of the S protein to ACE2 [50], [25]. After binding to the host cell, the S protein is cleaved at the boundary between the S1 and S2 subunits, leading to the separation of the S1 and S2 domains and formation of the screw-like S2 fusion conformation composed of a spiral of trimeric protomers [51].
Table 5.
Number of unique S protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| Number (#) of mutations in S (MS) | 542 | 98 | 110 | 233 | 1107 | 63 |
| # of unique S sequences (US) | 1121 | 26 | 56 | 98 | 3321 | 25 |
| Avg. # of mutations per unit unique seqs. (MS/US) | 0.48 | 3.77 | 1.96 | 2.38 | 0.33 | 2.52 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in S (MS) | 213 | 995 | 28 | 179 | 11 | 115 |
| # of unique S sequences (US) | 183 | 2527 | 19 | 65 | 11 | 48 |
| Avg. # of mutations per unit unique seqs. (MS/US) | 1.16 | 0.39 | 1.47 | 2.75 | 1.00 | 2.40 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in S (MS) | 219 | 911 | 815 | 970 | 83 | 829 |
| # of unique S sequences (US) | 178 | 1281 | 1297 | 2658 | 49 | 1343 |
| Avg. # of mutations per unit unique seqs. (MS/US) | 1.23 | 0.71 | 0.63 | 0.36 | 1.69 | 0.62 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in S (MS) | 218 | 39 | 21 | 88 | 1122 | 55 |
| # of unique S sequences (US) | 44 | 26 | 23 | 36 | 1546 | 30 |
| Avg. # of mutations per unit unique seqs. (MS/US) | 4.95 | 1.50 | 0.91 | 2.44 | 0.73 | 1.83 |
Furthermore, trimers of the S protein are decorated with N-linked glycans that act as a glycan shield thwarting the host immune response [52]. Therefore, the surface-exposed S glycoprotein mediates entry into host cells, serves as the main target of neutralizing antibodies upon infection (in fact, it has immune recognition sites), and, being the most important protein for viral entry into cells, acts as the focal point of therapeutic and vaccine design [50], [53].
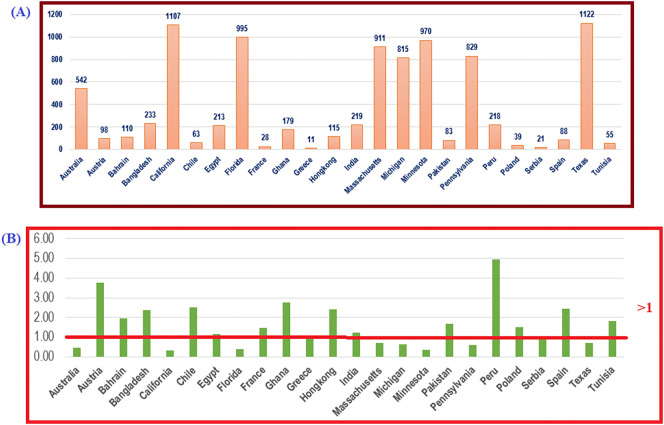
We observed that the highest number (495 %) of unique mutations possessed by unique S protein variants was from Peru, where 44 unique S sequences had 218 unique mutations. On the other side, the second-highest number of unique S protein variants from California possessed the lowest amount (33 %) of unique mutations. Fig. 2 shows the average numbers of mutations per unit unique S protein variants.
Fig. 2.
Geo-location-wise (A): total number of unique mutations and (B): average number of mutation(s) per unique S sequences.
Fig. 2 (B) shows that the probability of having triple mutants in any randomly chosen unique S protein variant from Austria is nearly 1, since the ratio (MS/US) is 3.77 > 3. Similarly, the probability of having more than quadruple mutants in a randomly chosen unique S protein variant from Peru is nearly 1, since the ratio (MS/US) is 4.95 > 4. Spectacularly, none of the unique S protein variants from the geo-locations in North America possessed more than one mutation, since the ratio in each case was <1, although the total number of unique S variants and mutations were relatively higher than those at other locations.
The total 23’Variants of Concern (VoC)’ and 25’Variants of Interest (VoI)’ mutations in the S protein were reported [54], [55], [56], [57]. Continent-wise, the frequency of common mutations were determined, as well as VoC, VoI among those common S protein mutations possessed by each continental geo-location (Table 6 ). It was interesting to note, since Australia was the only geo-location in Oceania considered in this study, that common mutations were not observed.
Table 6.
Continent-wise common mutations in the S protein and list of Variants of Concern (VoC), Variants of Interest (VoI) mutations in the S protein.
| Continent | Total # of common mutations in S | List of VoC on the continent | List of VoI on the continent |
|---|---|---|---|
| Asia | 4 | 614, 681 | 5, 142, 614, 681 |
| Europe | 1 | 614 | 614 |
| Africa | 22 | 80, 452, 484, 614, 681, 701 | 18, 26, 80, 484, 501, 570 |
| 614, 681, 716, 982, 1118 | |||
| 13, 18, 20, 26, 80, 138, 152, 190, | 5, 19, 67, 80, 95, 142, 154, 157, | ||
| North America | 487 | 215, 417, 452, 484, 501, 570, 614, 655, | 158, 253, 452, 477, 478, 484, 614, |
| 655, 681, 701, 716, 982, 1027, 1118, 1191 | 677, 681, 701, 950, 1071, 1176 | ||
| South America | 45 | 614 | 614 |
It was found that 487 common mutations in the S proteins were from patients from the seven geo-locations in North America, although the only common mutation across 24 geo-locations was D614G. Furthermore, it was noticed that all 23 VoC were presented in each geo-location from North America. On the other hand, the unique S proteins from the European geo-locations possessed only the D614G common mutation. In all African geo-locations, a moderate number of VoC and VoI were found, although the number of common mutations over the geo-locations was not relatively high compared to that of others (Table 7 ). Also, randomly chosen S protein variants from Ghana has a very high probability of acquiring double VoC/VoI mutants as the M S /U S ratio is 2.75.
Table 7.
Mutations in the unique furin-like cleavage site (FCS) of the S proteins.
| Accession | Lineage | Length | Geo Location | Collection Date | FCS (RRAR) |
|---|---|---|---|---|---|
| QVU70282 | B.1.1.7 | 1270 | USA: Massachusetts | 06-05-2021 | RRVR |
| QVU09331 | B.1.1.7 | 1270 | USA: California | 16-04-2021 | RRVR |
| QVI42615 | B.1.1.291 | 1273 | USA: California | 24-03-2021 | RRVR |
| QVI49490 | B.1.427 | 1273 | USA: California | 09-02-2021 | RRVR |
| QUD47347 | B.1.1.7 | 1270 | USA: Michigan | 05-04-2021 | RRSR |
| QUB14687 | B.1.2 | 1273 | USA: Michigan | 24-03-2021 | RRSR |
| QTU74764 | B.1.427 | 1273 | USA: California | 09-02-2021 | RRVR |
| QTS38722 | B.1.429 | 1271 | USA: Michigan | 15-03-2021 | RRSR |
| QTP22615 | B.1.243 | 1273 | USA: Massachusetts | 09-09-2020 | RRVR |
| QSS81313 | B.1.427 | 1273 | USA: California | 21-02-2021 | RRVR |
| QSL71584 | B.1.427 | 1273 | USA: California | 10-02-2021 | RRVR |
| QSL80009 | B.1.2 | 1273 | USA: Michigan | 11-02-2021 | RRSR |
| QRG20397 | B.1.243 | 1273 | USA: CA, Alameda County | 12-09-2020 | RRVR |
| QQX02259 | B.1.561 | 1273 | USA: California | 02-01-2021 | RRVR |
| QQN04304 | B.1.517 | 1273 | USA: Massachusetts | 27-11-2020 | RRVR |
Earlier, it was reported that ‘RRAR’ (amino acid positions: 682–685), a unique furin-like cleavage site (FCS) in the S protein, which was absent in other lineages beta-coronaviruses, such as SARS-CoV, caused high infectivity and transmissibility [58], [59], [60]. Even in this FCS, a single mutation at position 684 was noticed in some unique S protein variants from California, Massachusetts, and Michigan. Details of the protein accessions with associated information are presented in Table 7. The first such mutation, A684V, was reported in Massachusetts on September 9, 2020 (Accs. ID: QTP22615). Three days later, the same mutation was identified in California (QRG20397). The mutation A684V/S was ‘neutral’ (predicted using PredictSNP web-server), and hence it was expected that the ability to infect and transmit remains unchanged [48].
4.1.2. Envelope protein variants and mutations
The total frequency of unique mutations possessed by the E protein of SARS-CoV-2 across the 24 geo-locations is presented in Table 8 . Being the smallest of the major structural proteins of SARS-CoV-2, the E protein contains 75 residues [61]. Although this protein is highly conserved in different viral subtypes, its roles in viral invasion, replication and release are not fully elucidated. The E protein might cause membrane bending or scission at the budding site. Functions of the E protein in the viral particle envelope are determined by its interactions with other structural proteins. For example, the shape of the viral particle is maintained due to the interaction between the E and M proteins, which also promotes the viral release [62], [63]. Co-expression of the E and M proteins in host cells lead to the relocation of the S protein to the endoplasmic reticulum (ER)-Golgi intermediate region (ERGIC) or Golgi region [64]. Curiously, although the E protein is expressed at a high level in each infected cell, only a small fraction of this protein is inserted into the viral membrane, with most of the protein located at intracellular transport sites, which are related to the virus assembly and budding [65], [66], [67].
Table 8.
Number of unique E protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in E (ME) | 58 | 0 | 1 | 11 | 61 | 0 |
| # of unique E seqs. (UE) | 19 | 1 | 2 | 6 | 61 | 1 |
| Avg. # of mutations per unit unique seqs. (ME/UE) | 3.05 | 0.00 | 0.50 | 1.83 | 1.00 | 0.00 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in E (ME) | 12 | 43 | 1 | 5 | 1 | 1 |
| # of unique E seqs. (UE) | 16 | 49 | 2 | 6 | 2 | 2 |
| Avg. # of mutations per unit unique seqs. (ME/UE) | 0.75 | 0.88 | 0.50 | 0.83 | 0.50 | 0.50 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in E (ME) | 32 | 39 | 45 | 54 | 2 | 24 |
| # of unique E seqs. (UE) | 11 | 37 | 35 | 36 | 4 | 33 |
| Avg. # of mutations per unit unique seqs. (ME/UE) | 2.91 | 1.05 | 1.29 | 1.50 | 0.50 | 0.73 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in E (ME) | 11 | 5 | 0 | 1 | 31 | 1 |
| # of unique E seqs. (UE) | 3 | 3 | 1 | 2 | 33 | 2 |
| Avg. # of mutations per unit unique seqs. (ME/UE) | 3.67 | 1.67 | 0.00 | 0.50 | 0.94 | 0.50 |
Deletion of the E protein in vitro leads to a significant reduction in viral titer and maturity or production of incompetent offspring [68]. Several SARS-CoV-2 proteins, such as E, ORF3a, and ORF8, can act as viroporins, being able to self-assemble into oligomers that generate formation of ion channels [69], [70], [71]. This homo-oligomerization of the E protein depends on its transmembrane domain (TMD), with the homopentameric E protein acting as the viroporin involved in various functions, such as facilitation of the release of viral particles from host cells [72]. Mutation of the gene encoding the E protein is known to promote apoptosis [73]. Almost every unique E protein variant from Australia possessed triple mutations as the ratio M E /U E was 3.05 > 3. Likewise, in India, any E protein contains at least a double mutation (M E /U E = 2.91 > 1). Compared to this, a much higher number of unique mutations in the unique E proteins from Peru was observed, and any randomly chosen E protein from Peru contains quadruple mutations (M E /U E = 3.67 > 1). Based on the ratio M E /U E = 0 that each COVID-19 positive case in Austria, Chile, and Serbia was infected by the SARS-CoV-2 with the wild type E sequence (YP 009724392).
The 12 common mutations at positions 9, 21, 24, 41, 49, 55, 58, 62, 68, 71, 72, and 73 were detected in the unique E protein variants from geo-locations in North America. Among these 12 mutations, 8 mutations (at positions 49, 55, 58, 62, 68, 71, 72, and 73) were shared by the unique E variants from India. Among the 12 mutations, two mutations at positions 21 and 41 were shared with E variants from Bangladesh. No other common mutation was found in geo-locations in Asia, except for the single mutation at position 37 found in India and Bangladesh. E protein variants from the three African geo-locations shared only a single common mutation at position 71.
4.1.3. Membrane protein variants and mutations
The frequency of unique mutations possessed by the M protein of SARS-CoV-2 across the 24 geo-locations is presented in Table 9 . The SARS-CoV-2 M protein is a 222-residue-long transmembrane protein, which is the most abundant structural protein and which, together with the E protein plays a role in defining the shape of the viral envelope [74]. It was shown that M can adopt at least two different conformations, elongated and compact, with the elongated form being involved in the regulation of the membrane curvature and association with clusters of spikes [74]. Being three times large than the E protein, the M protein contains three transmembrane domains (TMD1-TMD3), whereas its N- and C-termini are exposed inside and outside the viral particle, respectively [75]. Different regions of the M protein serve diverse purposes, with the TMDs being able to bind to the S protein and engaged in the homotypic interaction of the M protein itself, and with the C-terminus being involved in the interaction with the N and E proteins [76], [77]. Furthermore, membrane bending and germination as well as the formation of the inner core of SARS-CoV-2 virus-like particles (VLPs) depend on the interaction of M with other structural proteins [75], [78]. In fact, VLP formation requires stable interaction between the M and N, M and E, and M and S proteins [78], [79].
Table 9.
Number of unique M protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in M (MM) | 34 | 1 | 3 | 16 | 139 | 1 |
| # of unique M seqs. (UM) | 38 | 2 | 4 | 11 | 192 | 2 |
| Avg. # of mutations per unit unique seqs. (MM/UM | 0.89 | 0.50 | 0.75 | 1.45 | 0.72 | 0.50 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in M (MM) | 19 | 92 | 3 | 6 | 17 | 4 |
| # of unique M seqs. (UM) | 22 | 131 | 4 | 7 | 3 | 5 |
| Avg. # of mutations per unit unique seqs. (MM/UM) | 0.86 | 0.70 | 0.75 | 0.86 | 5.67 | 0.80 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in M (MM) | 16 | 93 | 96 | 64 | 6 | 87 |
| # of unique M seqs. (UM) | 20 | 92 | 78 | 77 | 7 | 105 |
| Avg. # of mutations per unit unique seqs. (MM/UM) | 0.80 | 1.01 | 1.23 | 0.83 | 0.86 | 0.83 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in M (MM) | 12 | 1 | 2 | 3 | 94 | 2 |
| # of unique M seqs. (UM) | 8 | 2 | 3 | 4 | 101 | 3 |
| Avg. # of mutations per unit unique seqs. (MM/UM) | 1.50 | 0.50 | 0.67 | 0.75 | 0.93 | 0.67 |
A relatively large number of mutations were found in the M proteins from Greece. The ratio M M /U M = 5.67 > 5 for Greece implied that any randomly chosen M protein variants possessed five mutations (Table 9). In California, the highest number of unique M proteins possessed relatively very few mutations. Almost surely, no M protein from California contains more than one mutation (M M /U M = 0.72 < 1), whereas each M protein from Michigan and Massachusetts contains a single mutation (M M /U M > 1). Most of the unique M protein variants from Peru were likely to contain double mutations ((M M /U M = 1.5 > 1) (Table 9).
All North American geo-locations shared a sum of 24 mutations in the M protein variants at positions 2, 7, 17, 23, 28, 33, 34, 60, 69, 70, 81, 82, 85, 89, 98, 104, 109, 125, 142, 155, 173, 175, 208, and 209 (Supplementary file-I). On the other hand, not a single common mutation in the M protein was noticed in geo-locations from Asia and the same was observed in Africa and Europe. Each M protein from India shared 9 mutations with those of each North American geo-location, at positions 2, 17, 69, 70, 82, 104, 125, 142, and 209. Among the 24 common mutations from geo-locations in North America, only two mutations at positions 17 and 23 were shared with M proteins from Greece.
4.1.4. Nucleocapsid protein variants and mutations
The frequency of unique N protein mutations across the 24 geo-locations is presented in Table 10 . The N protein is an important 419-residue-long structural protein responsible for packaging of the viral RNA into helical ribonucleocapsids (RNPs), whereas interaction of this protein with the other structural SARS-CoV-2 proteins leads to the genome encapsidation during virion assembly [80], [81]. There are two highly conserved domains in the SARS-CoV-2 N protein, the N-terminal RNA binding domain (residues 46–174) and the C-terminal dimerization domain (residues 247–364), whereas the N- and C-terminal regions of this protein (residues 1–42 and 365–419) and the linker region (residues 176–246) are intrinsically disordered [82], [83], [84]. Importantly, disordered regions of the N protein can be phosphorylated and contain binding motifs for the regulatory host cell 14-3-3 proteins, with some of these motifs being mutated in natural SARS-CoV-2 variants [85], [86]. The N protein is abundantly produced during infection and is highly immunogenic [87].
Table 10.
Number of unique N protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in N (MN) | 200 | 21 | 38 | 86 | 362 | 16 |
| # of unique N seqs. (UN) | 213 | 22 | 33 | 53 | 1345 | 16 |
| Avg. # of mutations per unit unique seqs. (MN/UN) | 0.94 | 0.95 | 1.15 | 1.62 | 0.27 | 1.00 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in N (MN) | 83 | 356 | 7 | 34 | 9 | 32 |
| # of unique N seqs. (UN) | 116 | 973 | 6 | 41 | 9 | 28 |
| Avg. # of mutations per unit unique seqs. (MN/UN) | 0.72 | 0.37 | 1.17 | 0.83 | 1.00 | 1.14 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in N (MN) | 84 | 363 | 238 | 322 | 31 | 280 |
| # of unique N seqs. (UN) | 86 | 625 | 418 | 481 | 33 | 643 |
| Avg. # of mutations per unit unique seqs. (MN/UN) | 0.98 | 0.58 | 0.57 | 0.67 | 0.94 | 0.44 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in N (MN) | 20 | 20 | 24 | 17 | 286 | 24 |
| # of unique N seqs. (UN) | 19 | 22 | 22 | 21 | 644 | 22 |
| Avg. # of mutations per unit unique seqs. (MN/UN) | 1.05 | 0.91 | 1.09 | 0.81 | 0.44 | 1.09 |
It was observed that the least number of mutations was possessed by the unique N proteins from California (M N /U N = 0.27 < 1), whereas 53 unique N protein variants from Bangladesh had 86 mutations (M N /U N = 1.62 > 1) (Table 10). Every unique N protein-variant contain at least a single mutation, which is followed by the ratio (= 1.62 > 1). Likewise, each unique N variant from Bahrain, Peru, Chile, France, Greece, Hong Kong, India, Serbia, and Tunisia contain at least one mutation (for each geo-location (M N /U N = 1.62 ≥ 1). Furthermore, it was noticed that 153 mutations were shared among all unique N proteins from each geo-location in North America. Only 6 mutations at positions 3, 194, 202, 203, 204 and 377 were common across Asian geo-locations, whereas only two mutations at positions 203 and 204 were found in the N variants from the European geo-locations. There were 9 mutations at positions 9, 194, 202, 203, 204, 205, 220, 235, and 238 in the N proteins detected in the African geo-locations.
4.1.5. ORF3a protein variants and mutations
The frequency of unique ORF3a protein mutations across the 24 geo-locations is presented in Table 11 . The ORF3a is the largest SARS-CoV-2 accessory protein (275 amino acids long), which is a multifunctional protein involved in virulence, infectivity, ion channel activity, morphogenesis, and virus release [88]. Together with other SARS-CoV-2 ion-channel proteins (viroporins, ORF8a, and E) ORF3A plays a critical role in infection-induced tissue inflammation caused by the viroporin-mediated disruption of the lysosomes and redistribution of ions resulting in the expression of inflammatory cytokines, such as interleukin 1β (IL-1β), IL-6, and tumor necrosis factor (TNF) [89].
Table 11.
Number of unique ORF3a protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in ORF3a (M3a) | 151 | 16 | 28 | 51 | 264 | 15 |
| # of unique ORF3a seqs. (U3a) | 132 | 14 | 27 | 59 | 1073 | 16 |
| Avg. # of mutations per unit unique seqs. (M3a/U3a) | 1.14 | 1.14 | 1.04 | 0.86 | 0.25 | 0.94 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in ORF3a (M3a) | 56 | 264 | 9 | 27 | 25 | 13 |
| # of unique ORF3a seqs. (U3a) | 81 | 808 | 10 | 23 | 13 | 17 |
| Avg. # of mutations per unit unique seqs. (M3a/U3a) | 0.69 | 0.33 | 0.90 | 1.17 | 1.92 | 0.76 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in ORF3a (M3a) | 62 | 232 | 235 | 242 | 47 | 225 |
| # of unique ORF3a seqs. (U3a) | 73 | 468 | 389 | 456 | 32 | 561 |
| Avg. # of mutations per unit unique seqs. (M3a/U3a) | 0.85 | 0.50 | 0.60 | 0.53 | 1.47 | 0.40 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in ORF3a (M3a) | 16 | 23 | 19 | 14 | 247 | 12 |
| # of unique ORF3a seqs. (U3a) | 16 | 21 | 17 | 13 | 532 | 10 |
| Avg. # of mutations per unit unique seqs. (M3a/U3a) | 1.00 | 1.10 | 1.12 | 1.08 | 0.46 | 1.20 |
Furthermore, the ion channel activity of the SARS-CoV-2 ORF3a, E, and M proteins impedes with the apoptotic pathway [90]. ORF3a also plays a role in IL-1β maturation, activates the innate immune signaling receptor NLRP3 (NOD-, LRR-, and pyrin domain-containing 3) inflammasome, participates in the activation of the proinflammatory cytokine signaling transcription factors, such as STAT1, STAT2, IRF9, and NFKB1, and can affect type-I interferon (INT) activation, thereby acting as an IFN antagonist [89], [91], [92]. Via interaction with heme oxygenase-1 (HMOX1), ORF3a contributes to the heme catabolism and controls the anti-inflammatory system [89]. Finally, potent and durable antibody responses against SARS-CoV-2 ORF3a, ORF3b, ORF7a, and ORF8 proteins were found in children [93]. Therefore, mutations in this protein are expected to alter the host immune response to SARS-CoV-2 infection. From Table 11, it was observed that the least number of mutations was possessed by the ORF3a variants from California, where the highest number of unique ORF3a variants available was M 3a /U 3a = 0.25 < < 1. On the other hand, 13 ORF3a variants from Greece had 25 mutations altogether. Therefore, almost every ORF3a variant was likely to contain double mutations M 3a /U 3a = 1.92 ∼ = 2. Furthermore, each ORF3a variant from Australia, Austria, Bahrain, Chile, France, Ghana, Pakistan, Peru, Poland, Serbia, Spain, and Tunisia contains at least one mutation, that is Q57, but not more than two mutations, since the M 3a /U 3a ratio lies between 1 and 2.
A total of 167 common mutations in ORF3a variants across the North American geo-locations were detected, whereas the only common mutation, Q57 was detected in the European geo-locations. It was noted that unique ORF3a variants from Texas, Pennsylvania, Florida, Michigan, and Minnesota had common mutations at positions 243, 224, 255, 229, and 238, respectively, from California. ORF3a variants from African geo-locations share five common mutations at positions 57, 100, 155, 171, and 224. Also, three mutations at positions 57, 175, and 223 were possessed by the ORF3a variants from each Asian geo-location. It was noted that unique ORF3a variants shared 225 mutations among 264 in total in both California and Massachusetts.
4.1.6. ORF6 protein variants and mutations
The frequency of unique ORF6 protein mutations across the 24 geo-locations is presented in Table 13. SARS-CoV-2 ORF6 is a 61-amino-acid-long membrane-associated protein that acts as an interferon (IFN) antagonist. ORF6 contains a putative diacidic motif (DDEE) and lysosomal targeting motif (YSEL) and can increase viral replication by promoting appearance of virus-induced or virus associated vesicles due to the intracellular membrane rearrangements [94]. ORF6 and ORF8 can inhibit the type-I IFN signaling pathway [95]. For example, ORF6 interacts with the karyopherin import complex, thereby limiting the transcription factor STAT1 involved in down-regulation of the IFN pathway [84]. By analogy with SARS-CoV, in association with other SARS-CoV-2 proteins, such as M, NSP1 and NSP3, ORF6 and ORF3a can potentially impede IRF3 signaling, repress IFN expression, and promote degradation of IFNAR1 and STAT1 [89], [96]. ORF6 interacts with the NSP8 protein from the SARS-CoV-2 replicase complex, and during early infection, can increase infection titers at a low multiplicity of infection [95].
Table 13.
Number of unique ORF7a protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in ORF7a (M7a) | 59 | 5 | 15 | 21 | 120 | 5 |
| # of unique seqs. ORF7a (U7a) | 58 | 5 | 18 | 15 | 330 | 5 |
| Avg. # of mutations per unit unique seqs. (M7a) | 1.02 | 1.00 | 0.83 | 1.40 | 0.36 | 1.00 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in ORF7a (M7a) | 18 | 108 | 0 | 13 | 7 | 5 |
| # of unique ORF7a seqs. (U7a) | 20 | 314 | 1 | 10 | 2 | 5 |
| Avg. # of mutations per unit unique seqs. (M7a/M7a) | 0.90 | 0.34 | 0.00 | 1.30 | 3.50 | 1.00 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in ORF7a (M7a) | 25 | 114 | 110 | 103 | 5 | 105 |
| # of unique ORF7a seqs. (U7a) | 23 | 184 | 199 | 758 | 6 | 202 |
| Avg. # of mutations per unit unique seqs. (M7a/M7a) | 1.09 | 0.62 | 0.55 | 0.14 | 0.83 | 0.52 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in ORF7a (M7a) | 29 | 6 | 3 | 1 | 109 | 5 |
| # of unique ORF7a seqs. (U7a) | 9 | 8 | 3 | 2 | 190 | 7 |
| Avg. # of mutations per unit unique seqs. (M7a/M7a) | 3.22 | 0.75 | 1.00 | 0.50 | 0.57 | 0.71 |
The probability of having quadruple mutations in a chosen unique ORF6 variant from Bahrain was nearly 1 as the M 6 /U 6 ratio = 4.29 > 4 (Table 12 ). Almost certainly, each ORF6 variant from Hong Kong (M 6 /U 6 = 3.33 > 3) and Australia (M 6 /U 6 = 2.32 > 2) contains triple and double mutations, respectively. Also, it was noticed that no new ORF6 variant was detected in Poland, Serbia, and Tunisia.
Table 12.
Number of unique ORF6 protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in ORF6 (M6) | 44 | 2 | 30 | 8 | 59 | 2 |
| # of unique ORF6 seqs. (U6) | 19 | 3 | 7 | 9 | 104 | 3 |
| Avg. # of mutations per unit unique seqs. (M6/U6) | 2.32 | 0.67 | 4.29 | 0.89 | 0.57 | 0.67 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in ORF6 (M6) | 6 | 46 | 2 | 15 | 1 | 10 |
| # of unique ORF6 seqs. (U6) | 10 | 65 | 3 | 10 | 2 | 3 |
| Avg. # of mutations per unit unique seqs. (M6/U6) | 0.60 | 0.71 | 0.67 | 1.50 | 0.50 | 3.33 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in ORF6 (M6) | 5 | 45 | 45 | 57 | 4 | 38 |
| # of unique ORF6 seqs. (U6) | 7 | 47 | 38 | 45 | 5 | 52 |
| Avg. # of mutations per unit unique seqs. (M6/U6) | 0.71 | 0.96 | 1.18 | 1.27 | 0.80 | 0.73 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in ORF6 (M6) | 1 | 0 | 0 | 2 | 55 | 0 |
| # of unique ORF6 seqs. (U6) | 2 | 1 | 1 | 3 | 61 | 1 |
| Avg. # of mutations per unit unique seqs. (M6/U6) | 0.50 | 0.00 | 0.00 | 0.67 | 0.90 | 0.00 |
There were 25 common mutations in ORF6 variants in each geo-location of North America, whereas no common mutation in ORF6 was found in the European geo-locations. Likewise, in Asian and African geo-locations, no common mutation was detected for the ORF6 variants.
4.1.7. ORF7a protein variants and mutations
The frequency of unique ORF7a protein mutations across the 24 geo-locations is presented in Table 13 . ORF7a is a 121-residue-long type I transmembrane protein, which may function during early infection, interacts with the structural proteins M, E, and S, therefore being involved in viral replication and assembly, and, via interaction with the E protein, can promote apoptosis [97], [98], [99], [89]. Furthermore, ORF7a induces chemokines and pro-inflammatory cytokines including RANTES and IL-8 [84]. ORF7b is a putative viral accessory protein encoded from subgenomic (sg) RNA, where the ORF7b initiation codon overlaps with the ORF7a stop codon in a −1 shifted ORF [100]. This 43-residue-long protein can be found in association with intracellular viral particles, and also in purified virions in the Golgi compartment [100]. The overall roles of ORF7a and ORF7b in SARS-CoV-2 replication are poorly understood [97]. It was pointed out that SARS-CoV ORF7a and ORF8 genes are most similar to bat coronavirus sequences, their SARS-CoV-2 counterparts are closer to pangolin coronavirus homologs [101]. Furthermore, using supervised sequence space walking in database searches, it was shown that SARS-CoV-2 proteins ORF7a and ORF8 are characterized by the remote, non-trivial sequence similarities [101].
The ratio M 7a /U 7a > 3 in Greece and Peru implied that most unique variants must have at least three mutations (Table 13). Unique ORF7a variants from Australia, Austria, Bangladesh, Chile, Egypt, Ghana, Hong Kong, India, Pakistan, and Serbia must contain at least a single mutation as in each case, the ratio was found greater than or equal/near to 1. Furthermore, it was observed that no new ORF7a sequence was found among 90 infected patients in France, so far.
Ninety-two common mutations were detected in the unique ORF7a variants in the North American geo-locations, whereas no common mutation was observed in the European geo-locations. Only one common mutation at position 28 in Asian geo- locations, and another single common mutation at position 14 in ORF7a were found in African countries. ORF7a protein sequences from Austria had four mutations at positions 79, 99, 102, and 103, commonly found in each geo-location in North America. Likewise, all unique mutations in ORF7a variants detected in Greece, Poland, and Serbia were present in each North American geo-location.
4.1.8. ORF7b protein variants and mutations
The frequency of unique ORF7b protein mutations across the 24 geo-locations is presented in Table 14 . Compared to the wild type ORF7b (YP 009725318), no new ORF7b variants were found in France, Greece, Peru, and Serbia, whereas only one variant other than the wild ORF7b was found in Austria, Chile, Hong Kong, Pakistan, Poland, Spain, and Tunisia. Each ORF7b variant from Australia and India contained at least a single mutation. There were 17 common mutations at positions 2, 3, 4, 5, 6, 8, 10, 13, 14, 15, 18, 31, 32, 34, 40, 42, and 43 in all North American geo-locations. No ORF7b variants from North America possessed double mutations based on the ratio M 7b /U 7b < 1 for each North American geo-location (Table 14).
Table 14.
Number of unique ORF7b protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in ORF7b (M7b) | 19 | 1 | 3 | 5 | 40 | 1 |
| # of unique. ORF7b seqs (U7b) | 14 | 2 | 4 | 6 | 89 | 2 |
| Avg. # of mutations per unit unique seqs. (M7b/U7b) | 1.36 | 0.50 | 0.75 | 0.83 | 0.45 | 0.50 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in ORF7b (M7b) | 8 | 36 | 0 | 15 | 0 | 1 |
| # of unique. ORF7b seqs (U7b) | 11 | 63 | 1 | 7 | 1 | 2 |
| Avg. # of mutations per unit unique seqs. (M7b/U7b) | 0.73 | 0.57 | 0.00 | 2.14 | 0.00 | 0.50 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in ORF7b (M7b) | 10 | 35 | 34 | 30 | 1 | 26 |
| # of unique. ORF7b seqs (U7b) | 7 | 46 | 45 | 59 | 2 | 38 |
| Avg. # of mutations per unit unique seqs. (M7b/U7b) | 1.43 | 0.76 | 0.76 | 0.51 | 0.50 | 0.68 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in ORF7b (M7b) | 0 | 1 | 0 | 1 | 30 | 1 |
| # of unique. ORF7b seqs (U7b) | 1 | 2 | 1 | 2 | 43 | 2 |
| Avg. # of mutations per unit unique seqs. (M7b/U7b) | 0.00 | 0.50 | 0.00 | 0.50 | 0.70 | 0.50 |
4.1.9. ORF8 protein variants and mutations
The frequency of unique ORF8 protein mutations across the 24 geo-locations is presented in Table 15 . ORF8 in SARS-CoV-2 is a unique 121-residue-long accessory protein (neither ORF7a nor ORF8 genes are found in the gamma or delta coronavirus groups), which being characterized by prominent structural plasticity and high sequence diversity is suggested to have important roles in SARS-CoV-2 pathogenicity and the ability of virus to spread [102]. ORF8 interacts with the major histocompatibility complex (MHC) class-I molecules and down-regulates their surface expression in various cell types [29]. Inhibition of ORF8 function might represent a strategy to improve the special immune surveillance and accelerate the eradication of SARS-CoV-2 in vivo [103]. Therefore, the ORF7a/ORF8 superfamily of SARS-CoV-2 proteins from the immunoglobulin superfamily might serve as a key system for immune evasion, similar to those found in adenoviruses, herpesviruses, and poxviruses [101], [104]. Based on the presence of remote sequence similarities between the ORF7a and ORF8 proteins and the fact that although the ORF7a is more constrained, ORF8 is subjected to fast evolution, it was hypothesized that ORF7a serves as a conserved template, to generate fast evolving variants, such as ORF8, thereby distorting immune responses of the host [101].
Table 15.
Number of unique ORF8 protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in ORF8 (M8) | 33 | 2 | 14 | 23 | 117 | 4 |
| # of unique ORF8 seqs. (U8) | 54 | 3 | 17 | 19 | 359 | 5 |
| Avg. # of mutations per unit unique seqs. (M8/U8) | 0.61 | 0.67 | 0.82 | 1.21 | 0.33 | 0.80 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in ORF8 (M8) | 26 | 114 | 2 | 43 | 3 | 9 |
| # of unique ORF8 seqs. (U8) | 34 | 231 | 3 | 12 | 4 | 10 |
| Avg. # of mutations per unit unique seqs. (M8/U8) | 0.76 | 0.49 | 0.67 | 3.58 | 0.75 | 0.90 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in ORF8 (M8) | 30 | 89 | 69 | 65 | 9 | 69 |
| # of unique ORF8 seqs. (U8) | 27 | 137 | 77 | 118 | 10 | 135 |
| Avg. # of mutations per unit unique seqs. (M8/U8) | 1.11 | 0.65 | 0.90 | 0.55 | 0.90 | 0.51 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in ORF8 (M8) | 7 | 5 | 5 | 3 | 78 | 6 |
| # of unique ORF8 seqs. (U8) | 8 | 6 | 6 | 3 | 154 | 7 |
| Avg. # of mutations per unit unique seqs. (M8/U8) | 0.88 | 0.83 | 0.83 | 1.00 | 0.51 | 0.86 |
In each geo-location, wild type ORF8 protein mutated several times and emerged as a set of unique ORF8 variants in each geo-location. Every unique ORF8 variant from India and Bangladesh contains at least one mutation as the ratio in each case was >1 (Table 15). A total of 32 shared mutations were identified across geo-locations in North America. It was noticed that L84 was the only common mutation found in Asian and African geo-locations.
4.1.10. ORF10 protein variants and mutations
The frequency of unique ORF10 protein mutations across the 24 geo-locations is presented in Table 16 .
Table 16.
Number of unique ORF7b protein mutations possessed in each geo-location.
| Continent |
Oceania |
Europe |
Asia |
Asia |
N-America |
S-America |
|---|---|---|---|---|---|---|
| Geo-location | Australia | Austria | Bahrain | Bangladesh | California | Chile |
| # of mutations in ORF10 (M10) | 13 | 1 | 2 | 9 | 29 | 0 |
| # of unique ORF10 seqs. (U10) | 16 | 2 | 3 | 11 | 61 | 1 |
| Avg. # of mutations per unit unique seqs. (M10/U10) | 0.81 | 0.50 | 0.67 | 0.82 | 0.48 | 0.00 |
| Continent |
Africa |
N-America |
Europe |
Africa |
Europe |
Asia |
|---|---|---|---|---|---|---|
| Geo-location | Egypt | Florida | France | Ghana | Greece | Hong Kong |
| # of mutations in ORF10 (M10) | 6 | 29 | 0 | 2 | 0 | 2 |
| # of unique ORF10 seqs. (U10) | 8 | 47 | 1 | 3 | 1 | 3 |
| Avg. # of mutations per unit unique seqs. (M10/U10) | 0.75 | 0.62 | 0.00 | 0.67 | 0.00 | 0.67 |
| Continent |
Asia |
N-America |
N-America |
N-America |
Asia |
N-America |
|---|---|---|---|---|---|---|
| Geo-location | India | Massachusetts | Michigan | Minnesota | Pakistan | Pennsylvania |
| # of mutations in ORF10 (M10) | 2 | 23 | 16 | 20 | 2 | 22 |
| # of unique ORF10 seqs. (U10) | 3 | 29 | 23 | 29 | 3 | 29 |
| Avg. # of mutations per unit unique seqs. (M10/U10) | 0.67 | 0.79 | 0.70 | 0.69 | 0.67 | 0.76 |
| Continent |
S-America |
Europe |
Europe |
Europe |
N-America |
Africa |
|---|---|---|---|---|---|---|
| Geo-location | Peru | Poland | Serbia | Spain | Texas | Tunisia |
| # of mutations in ORF10 (M10) | 8 | 1 | 1 | 2 | 21 | 2 |
| # of unique ORF10 seqs. (U10) | 5 | 2 | 2 | 3 | 39 | 4 |
| Avg. # of mutations per unit unique seqs. (M10/U10) | 1.60 | 0.50 | 0.50 | 0.67 | 0.54 | 0.50 |
ORF10 is a 38-residue-long accessory protein, which is unique for SARS-CoV-2. This highly ordered, hydrophobic, and thermally stable protein contains at least one transmembrane region [105], [106]. The ORF10 interacts with an E3 ubiquitin ligase complex CRL2ZY G11B containing Cullin-2, RBX1, Elongin B, Elongin C, and ZYG11B [107], [108], [109]. This CRL2ZY G11B hijacking by ORF10 suggests a role of this protein in ubiquitylation and subsequent proteasomal degradation of the cellular antiviral proteins [108]. Although ORF10 may negatively affect the antiviral protein degradation process through interaction with the E3 ubiquitin ligase complex CRL2ZY G11B, no evidence of ORF10 regulating or being regulated by CRL2ZY G11B was detected [89], [108]. Earlier pandemic analysis of more than two million sequence data of SARS-CoV-2 infected patients from the open COVID-19 dashboard revealed that although most residues of this protein can be mutated, ORF10 contains the hot spots (A8, I13, and V30, which show high mutation rates) and cold spots (N5, N25, and N36, which are mostly conserved) [110]. However, the consequences of these ORF10 variants to the viral transmission, reinfection, as well as disease severity or patient death are not verified as of yet [110].
The ratio M 10 /U 10 = 0 implied that the wild type ORF10 (YP 009725255), no new ORF10 protein emerged in Chile, France, and Greece, although every amino acid contained mutations at each position starting from 1 to 38. In all 24 geo-locations, every unique ORF10 variant possessed only a single mutation (as in each case 0 < M 10 /U 10 < 2) (Table 16). In North American geo-locations, a set of common mutations in ORF10 variants at positions 4, 8, 10, 23, 24, 27, 28, 30, and 37 were identified. No other continental geo-locations have common mutations in ORF10. It was noted that an ORF10 variant (QKG88643.1) possessed the M1G mutation.
4.2. Mutations in the invariant residue regions of various proteins of SARS-CoV-2
The ORF10 is the unique SARS-CoV-2 protein present, which is not present in any other beta-coronavirus. So, except for the ORF10, other unique protein variants of four types of beta-coronaviruses were obtained from the NCBI database (Table 2) Further, sequence-based homology analysis using the Clustal-Omega webserver of each unique protein variant of four types with reference protein sequence (NC 045512-China) was conducted (Supplementary file-II). Based on the alignment, invariant residue regions of length greater than three amino acids were detected (Table 17). From the results of sequence alignment, it was observed that the SARS-CoV-2 reference protein sequences of NC 045512 with a set of invariant residues were shared by those proteins of four other different types of beta-coronaviruses. There are several invariant regions identified in all proteins as indicated in Table 17. Each of the S, E, M, N, ORF3a, ORF6, ORF7a, ORF7b, and ORF8 proteins of five different coronaviruses shared 29, 4, 9, 11, 6, 1, 3, 2, and 2 invariant residue regions. Furthermore, the largest invariant region with a length of 101 residues was identified in the S protein. These invariant regions possibly serve as sets of functional units in the respective proteins, indicating why they were conserved in the beta-coronavirus family.
Table 17.
Invariant regions and domain specifications in proteins of four type of CoVs.
| Protein | Invariant residues | Total # of residues | Protein | Invariant residues | Total # of residues | Protein | Invariant residues | Total # of residues |
|---|---|---|---|---|---|---|---|---|
| S | 34–38 | 5 | E | 3–24 | 22 | ORF3a | 31–36 | 4 |
| S | 102–104 | 3 | E | 26–36 | 11 | ORF3a | 53–58 | 4 |
| S | 165–167 | 3 | E | 43–54 | 12 | ORF3a | 135–142 | 8 |
| S | 189–191 | 3 | E | 57–67 | 11 | ORF3a | 154–162 | 9 |
| S | 281–284 | 4 | ORF3a | 244–255 | 12 | |||
| S | 310–320 | 11 | Protein | Invariant residues | Total # of residues | ORF3a | 262–275 | 14 |
| S | 374–383 | 10 | M | 5–11 | 7 | |||
| S | 418–429 | 12 | M | 16–26 | 11 | Protein | Invariant residues | Total # of residues |
| S | 509–518 | 10 | M | 41–51 | 11 | ORF6 | 1–15 | 15 |
| S | 520–528 | 9 | M | 53–75 | 23 | |||
| S | 538–546 | 9 | M | 98–124 | 27 | |||
| S | 591–603 | 13 | M | 135–144 | 10 | |||
| S | 608–618 | 11 | M | 156–167 | 12 | |||
| S | 659–674 | 16 | M | 170–187 | 18 | |||
| S | 751–767 | 17 | M | 198–210 | 13 | Protein | Invariant residues | Total # of residues |
| S | 797–809 | 13 | ORF7a | 15–31 | 17 | |||
| S | 814–833 | 18 | ORF7a | 37–58 | 22 | |||
| S | 846–867 | 22 | Protein | Invariant residues | Total # of residues | ORF7a | 75–93 | 19 |
| S | 885–921 | 37 | N | 38–62 | 25 | |||
| S | 944–1044 | 101 | N | 66–78 | 13 | |||
| S | 1074–1083 | 10 | N | 81–93 | 13 | |||
| S | 1090–1096 | 7 | N | 104–119 | 16 | |||
| S | 1115–1122 | 8 | N | 132–151 | 20 | Protein | Invariant residues | Total # of residues |
| S | 1134–1163 | 30 | N | 158–181 | 24 | ORF7b | 6–25 | 19 |
| S | 1165–1190 | 26 | N | 217–231 | 15 | ORF7b | 27–33 | 4 |
| S | 1192–1207 | 16 | N | 243–266 | 24 | |||
| S | 1209–1229 | 21 | N | 270–289 | 20 | Protein | Invariant residues | Total # of residues |
| S | 1234–1246 | 13 | N | 297–325 | 28 | ORF8 | 35–38 | 3 |
| S | 1262–1273 | 12 | N | 350–375 | 26 | ORF8 | 88–91 | 3 |
Over time and due to intraspecies evolution, SARS-CoV-2 proteins have acquired several mutations even in the invariant regions. The total frequency and respective percentage of mutations detected in each invariant residue window of all proteins are presented in Table 18.
Table 18.
Frequency and respective percentage of mutations detected in each invariant residue window of S proteins.
| S proteins invariant residues |
Number of mutations |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Invariant residues | Total # of residues | Domain | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts |
| 34–38 | 5 | S1 | 0 | 5 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 1 | 5 |
| 102–104 | 3 | S1 | 0 | 3 | 0 | 0 | 0 | 3 | 3 | 1 | 3 | 3 | 3 |
| 165–167 | 3 | S1 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 3 | 3 |
| 189–191 | 3 | S1 | 0 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 3 | 1 | 3 |
| 281–284 | 4 | S1 | 0 | 4 | 0 | 0 | 0 | 4 | 4 | 0 | 4 | 4 | 4 |
| 310–320 | 11 | S1 | 1 | 11 | 0 | 0 | 0 | 0 | 11 | 0 | 11 | 11 | 11 |
| 374–383 | 10 | S1 | 0 | 7 | 0 | 0 | 0 | 10 | 4 | 0 | 3 | 3 | 10 |
| 418–429 | 12 | S1 | 0 | 12 | 0 | 0 | 0 | 3 | 1 | 0 | 3 | 1 | 12 |
| 509–518 | 10 | S1 | 0 | 10 | 6 | 0 | 0 | 0 | 10 | 0 | 10 | 9 | 10 |
| 520–528 | 9 | S1 | 1 | 9 | 4 | 0 | 0 | 0 | 2 | 0 | 9 | 2 | 9 |
| 538–546 | 9 | S1 | 0 | 9 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 9 |
| 591–603 | 13 | S1 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 4 |
| 608–618 | 11 | S1 | 1 | 11 | 1 | 1 | 1 | 1 | 6 | 1 | 6 | 4 | 4 |
| 659–674 | 16 | S1 | 0 | 16 | 0 | 0 | 0 | 0 | 14 | 1 | 15 | 7 | 4 |
| 751–767 | 17 | S2 | 0 | 8 | 1 | 0 | 0 | 0 | 14 | 0 | 11 | 7 | 14 |
| 797–809 | 13 | S2 | 0 | 6 | 2 | 0 | 0 | 0 | 5 | 0 | 11 | 1 | 13 |
| 814–833 | 18 | S2 and S2’ | 0 | 11 | 0 | 0 | 0 | 0 | 9 | 14 | 18 | 8 | 19 |
| 846–867 | 22 | S2’ | 0 | 12 | 0 | 0 | 0 | 2 | 8 | 3 | 6 | 5 | 5 |
| 885–921 | 37 | S2’ | 2 | 16 | 0 | 0 | 0 | 3 | 5 | 1 | 36 | 31 | 8 |
| 944–1044 | 101 | S2’ | 2 | 88 | 1 | 1 | 1 | 3 | 14 | 1 | 72 | 64 | 28 |
| 1074–1083 | 10 | S2’ | 0 | 4 | 0 | 1 | 0 | 2 | 4 | 1 | 5 | 3 | 2 |
| 1090–1096 | 7 | S2’ | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 5 | 7 | 2 |
| 1115–1122 | 8 | S2’ | 1 | 4 | 1 | 0 | 1 | 1 | 5 | 1 | 5 | 3 | 5 |
| 1134–1163 | 30 | S2’ | 0 | 24 | 1 | 0 | 1 | 0 | 8 | 3 | 19 | 8 | 9 |
| 1165–1190 | 26 | S2’ | 0 | 25 | 0 | 0 | 0 | 3 | 12 | 1 | 24 | 7 | 12 |
| 1192–1207 | 16 | S2’ | 0 | 10 | 1 | 0 | 1 | 0 | 7 | 1 | 16 | 5 | 5 |
| 1209–1229 | 21 | S2’ (1214–1229-TMD) | 0 | 19 | 0 | 1 | 1 | 1 | 8 | 2 | 21 | 5 | 14 |
| 1234–1246 | 13 | S2’ (1234-TMD) | 2 | 13 | 0 | 0 | 0 | 0 | 9 | 0 | 8 | 4 | 7 |
| 1262–1273 | 12 | S2’ | 0 | 4 | 0 | 0 | 1 | 0 | 3 | 1 | 4 | 4 | 5 |
| S proteins invariant residues |
Number of mutations |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Invariant residues | Total # of residues | Domain | India | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| 34–38 | 5 | S1 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 5 | 2 | 0 | 0 | 2 |
| 102–104 | 3 | S1 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 3 | 1 | 1 | 0 | 3 |
| 165–167 | 3 | S1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 3 |
| 189–191 | 3 | S1 | 1 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 3 | 1 | 0 | 0 | 2 |
| 281–284 | 4 | S1 | 0 | 0 | 0 | 4 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 4 |
| 310–320 | 11 | S1 | 0 | 1 | 0 | 11 | 0 | 11 | 1 | 0 | 11 | 0 | 0 | 11 | 11 |
| 374–383 | 10 | S1 | 4 | 0 | 0 | 0 | 2 | 2 | 3 | 10 | 9 | 10 | 0 | 0 | 0 |
| 418–429 | 12 | S1 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 3 | 0 | 0 | 0 | 0 |
| 509–518 | 10 | S1 | 0 | 10 | 0 | 1 | 0 | 10 | 0 | 0 | 10 | 1 | 1 | 0 | 10 |
| 520–528 | 9 | S1 | 0 | 9 | 0 | 0 | 0 | 3 | 2 | 0 | 8 | 1 | 0 | 0 | 9 |
| 538–546 | 9 | S1 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 591–603 | 13 | S1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 5 | 1 | 0 | 0 | 1 |
| 608–618 | 11 | S1 | 1 | 1 | 1 | 2 | 2 | 11 | 2 | 1 | 6 | 1 | 1 | 1 | 2 |
| 659–674 | 16 | S1 | 0 | 0 | 0 | 1 | 0 | 16 | 0 | 0 | 11 | 1 | 0 | 0 | 6 |
| 751–767 | 17 | S2 | 0 | 0 | 0 | 0 | 0 | 7 | 1 | 1 | 14 | 0 | 0 | 1 | 9 |
| 797–809 | 13 | S2 | 5 | 0 | 0 | 2 | 0 | 5 | 3 | 0 | 10 | 13 | 0 | 0 | 1 |
| 814–833 | 18 | S2 and S2’ | 6 | 1 | 0 | 1 | 0 | 7 | 3 | 0 | 10 | 4 | 1 | 0 | 2 |
| 846–867 | 22 | S2’ | 0 | 0 | 0 | 0 | 0 | 15 | 1 | 0 | 10 | 0 | 0 | 0 | 2 |
| 885–921 | 37 | S2’ | 1 | 0 | 1 | 1 | 0 | 6 | 1 | 0 | 32 | 0 | 1 | 1 | 3 |
| 944–1044 | 101 | S2’ | 1 | 2 | 0 | 2 | 1 | 48 | 5 | 0 | 93 | 4 | 3 | 3 | 12 |
| 1074–1083 | 10 | S2’ | 2 | 0 | 0 | 1 | 0 | 10 | 8 | 1 | 4 | 1 | 1 | 0 | 3 |
| 1090–1096 | 7 | S2’ | 1 | 1 | 0 | 1 | 0 | 4 | 7 | 0 | 5 | 2 | 1 | 0 | 1 |
| 1115–1122 | 8 | S2’ | 3 | 1 | 1 | 2 | 0 | 8 | 1 | 0 | 6 | 2 | 1 | 2 | 2 |
| 1134–1163 | 30 | S2’ | 3 | 0 | 0 | 3 | 1 | 14 | 3 | 0 | 25 | 0 | 3 | 0 | 6 |
| 1165–1190 | 26 | S2’ | 5 | 2 | 0 | 0 | 0 | 16 | 2 | 0 | 24 | 3 | 0 | 0 | 2 |
| 1192–1207 | 16 | S2’ | 1 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 16 | 0 | 0 | 2 | 0 |
| 1209–1229 | 21 | S2’ (1214–1229-TMD) | 0 | 0 | 0 | 3 | 0 | 14 | 1 | 1 | 21 | 0 | 0 | 0 | 4 |
| 1234–1246 | 13 | S2’ (1234-TMD) | 1 | 0 | 0 | 1 | 0 | 7 | 4 | 1 | 13 | 2 | 1 | 0 | 7 |
| 1262–1273 | 12 | S2’ | 2 | 0 | 1 | 0 | 1 | 4 | 7 | 0 | 9 | 4 | 1 | 1 | 4 |
In all invariant regions of the S protein, unique variants from California, Florida, Texas, Minnesota, and Massachusetts possessed several mutations (Table 18). Notably, unique S protein variants from California, Texas, and Minnesota possessed correspondingly 93, 88, and 72 distinct mutations in the invariant region of 101 amino acid residues. Among 29 invariant regions, only seven of the S proteins from Tunisia had a minimal number of mutations, with a maximum of two in each region. Likewise, S protein variants from Spain, Poland, Serbia, Greece, and France showed a minimal number of mutations in nine, eight, five, four, and seven invariant regions, respectively. S protein variants from other geo-locations possessed a relatively (with regard to the North American geo-locations) smaller number of mutations in the invariant regions. In >50 % of the 29 invariant regions, S protein variants from India, Bangladesh, Austria, Egypt, and Pakistan possessed a small number of mutations (Table 18). It was noteworthy that in India, Bangladesh, Austria, Egypt, and Pakistan, only a maximum of five mutations were found in the largest invariant region of the S2 domain of the S proteins.
Several mutations were identified in the S1, S2, and S2’ domains of the S protein (Table 18). The S1 domain of the S protein attaches the virion to the cell membrane by interacting with the host ACE2 receptor, initiating the infection. Also, the S2 domain contributes to the fusion of the virion and cellular membranes by acting as a class-I viral fusion protein, and the S2’ domain acts as a viral fusion peptide which is unmasked following the S2 cleavage occurring after virus endocytosis [111]. These functions might be modified due to several mutations occurring in the invariant regions (postulated as important functional sites for the virus). Whether these mutations in the invariant regions in the S1, S2 and S2’ domains would increase the infectivity of the virus is not clear but definitely remains a matter of concern.
Invariant regions in the E, M, and N proteins of five CoVs which include SARS-CoV-2 too, are presented in Table 19 . There were 4, 9, and 11 invariant regions identified in the E, M, and N proteins, respectively.
Table 19.
Frequency and respective percentage of mutations detected in each invariant residue window of the E, M, and N proteins.
| Number of mutations | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| E | 3–24 | 0 | 11 | 0 | 0 | 0 | 7 | 10 | 1 | 7 | 12 | 8 | 2 |
| E | 26–36 | 0 | 3 | 0 | 0 | 0 | 2 | 1 | 1 | 11 | 11 | 1 | 0 |
| E | 43–54 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 9 | 4 | 9 | 9 |
| E | 57–67 | 0 | 5 | 1 | 0 | 0 | 0 | 4 | 0 | 11 | 5 | 11 | 11 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| E | 3–24 | 0 | 0 | 2 | 0 | 11 | 5 | 0 | 15 | 3 | 0 | 0 | 7 |
| E | 26–36 | 0 | 0 | 0 | 0 | 5 | 2 | 0 | 11 | 2 | 0 | 0 | 11 |
| E | 43–54 | 0 | 0 | 0 | 0 | 6 | 1 | 0 | 10 | 0 | 0 | 0 | 12 |
| E | 57–67 | 0 | 0 | 1 | 0 | 8 | 0 | 0 | 9 | 0 | 0 | 0 | 11 |
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| M | 5–11 | 1 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 2 | 3 | 0 |
| M | 16–26 | 0 | 4 | 0 | 0 | 0 | 1 | 4 | 0 | 3 | 11 | 3 | 1 |
| M | 41–51 | 0 | 2 | 0 | 0 | 0 | 8 | 4 | 1 | 1 | 7 | 11 | 0 |
| M | 53–75 | 0 | 9 | 1 | 1 | 0 | 1 | 10 | 0 | 8 | 7 | 18 | 4 |
| M | 98–124 | 0 | 15 | 0 | 0 | 0 | 0 | 6 | 1 | 6 | 16 | 7 | 2 |
| M | 135–144 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 3 | 6 | 1 |
| M | 156–167 | 0 | 10 | 0 | 0 | 0 | 0 | 8 | 0 | 2 | 0 | 4 | 0 |
| M | 170–187 | 0 | 10 | 0 | 0 | 0 | 0 | 5 | 1 | 4 | 2 | 2 | 0 |
| M | 198–210 | 0 | 3 | 0 | 0 | 0 | 0 | 4 | 0 | 2 | 4 | 2 | 1 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| M | 5–11 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 3 | 1 | 1 | 0 | 3 |
| M | 16–26 | 0 | 8 | 0 | 0 | 4 | 0 | 0 | 10 | 0 | 0 | 0 | 1 |
| M | 41–51 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 9 | 0 | 0 | 0 | 1 |
| M | 53–75 | 0 | 0 | 1 | 1 | 9 | 4 | 0 | 20 | 0 | 0 | 0 | 4 |
| M | 98–124 | 1 | 0 | 0 | 0 | 16 | 3 | 0 | 15 | 8 | 1 | 0 | 3 |
| M | 135–144 | 1 | 0 | 1 | 0 | 3 | 0 | 0 | 10 | 0 | 0 | 0 | 3 |
| M | 156–167 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| M | 170–187 | 0 | 0 | 1 | 1 | 2 | 1 | 0 | 5 | 0 | 0 | 0 | 4 |
| M | 198–210 | 1 | 4 | 1 | 0 | 5 | 0 | 0 | 5 | 1 | 1 | 0 | 2 |
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| N | 38–62 | 0 | 8 | 0 | 0 | 0 | 0 | 8 | 0 | 6 | 8 | 9 | 1 |
| N | 66–78 | 0 | 3 | 0 | 1 | 0 | 0 | 3 | 3 | 13 | 3 | 13 | 1 |
| N | 81–93 | 1 | 4 | 0 | 1 | 0 | 0 | 5 | 0 | 13 | 5 | 12 | 2 |
| N | 104–119 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 7 | 16 | 16 | 1 |
| N | 132–151 | 0 | 15 | 3 | 1 | 0 | 1 | 12 | 1 | 16 | 16 | 18 | 3 |
| N | 158–181 | 0 | 12 | 0 | 2 | 0 | 0 | 13 | 1 | 17 | 7 | 21 | 2 |
| N | 217–231 | 1 | 15 | 1 | 1 | 0 | 0 | 12 | 0 | 15 | 5 | 15 | 2 |
| N | 243–266 | 0 | 18 | 0 | 0 | 2 | 1 | 15 | 1 | 11 | 14 | 21 | 2 |
| N | 270–289 | 0 | 17 | 0 | 0 | 1 | 0 | 18 | 1 | 18 | 6 | 20 | 1 |
| N | 297–325 | 2 | 21 | 1 | 0 | 0 | 1 | 17 | 0 | 29 | 8 | 13 | 3 |
| N | 350–375 | 1 | 19 | 1 | 0 | 1 | 2 | 17 | 2 | 14 | 16 | 26 | 6 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| N | 38–62 | 0 | 0 | 1 | 0 | 14 | 0 | 0 | 11 | 1 | 1 | 0 | 4 |
| N | 66–78 | 0 | 0 | 2 | 0 | 13 | 2 | 0 | 6 | 6 | 0 | 0 | 4 |
| N | 81–93 | 0 | 0 | 1 | 0 | 13 | 1 | 0 | 7 | 3 | 1 | 0 | 4 |
| N | 104–119 | 0 | 0 | 0 | 0 | 11 | 1 | 0 | 15 | 0 | 0 | 0 | 1 |
| N | 132–151 | 3 | 0 | 1 | 0 | 15 | 6 | 0 | 16 | 2 | 2 | 0 | 7 |
| N | 158–181 | 0 | 0 | 1 | 0 | 19 | 4 | 1 | 22 | 3 | 0 | 0 | 5 |
| N | 217–231 | 3 | 0 | 1 | 0 | 12 | 2 | 0 | 15 | 7 | 2 | 1 | 4 |
| N | 243–266 | 1 | 0 | 0 | 1 | 16 | 2 | 1 | 21 | 1 | 1 | 1 | 16 |
| N | 270–289 | 0 | 0 | 0 | 0 | 20 | 2 | 0 | 17 | 3 | 0 | 0 | 20 |
| N | 297–325 | 2 | 0 | 1 | 0 | 29 | 3 | 2 | 19 | 4 | 5 | 0 | 29 |
| N | 350–375 | 0 | 1 | 0 | 0 | 19 | 7 | 1 | 20 | 1 | 2 | 1 | 8 |
No mutation was identified in the E protein variants from Tunisia, Serbia, Poland, Hong Kong, Greece, France (Table 19). On the other hand, the E protein variants from Chile, Bahrain, Austria, Australia, Texas, Pennsylvania, Minnesota, Michigan, Massachusetts, Florida, and California had a significant number of mutations in each invariant region. Very few mutations were identified in the E protein variants from India, Bangladesh, Spain, Peru, Egypt, Ghana, and Pakistan. M protein variants in the North American and Oceanian geo-locations contained various mutations in each identified invariant region. In contrast, few mutations in the M proteins in the rest of the geo-locations, were detected in some invariant regions (Table 19). N proteins from California, Texas, Minnesota, Michigan, Massachusetts, Pennsylvania, Florida, India, Bangladesh, Egypt, and Australia had many mutations in each invariant region. In some of the invariant regions, few mutations were detected in the N proteins from the rest of the geo-locations.
Mutations in the invariant regions of the SARS-CoV-2 ORF proteins are listed in Table 20 . There were 6, 1, 3, 2, and 2 invariant regions found in ORF3a, ORF6, ORF7a, ORF7b, and ORF8 variants, respectively.
Table 20.
Frequency and respective percentage of mutations detected in each invariant residue window of ORF3a, ORF6, ORF7a, ORF7b, and ORF8 proteins.
| Number of mutations | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| ORF3a | 31–36 | 0 | 5 | 0 | 1 | 0 | 0 | 6 | 0 | 6 | 5 | 5 | 1 |
| ORF3a | 53–58 | 1 | 5 | 2 | 2 | 2 | 1 | 6 | 1 | 4 | 5 | 6 | 4 |
| ORF3a | 135–142 | 0 | 4 | 1 | 0 | 0 | 0 | 4 | 1 | 5 | 2 | 8 | 0 |
| ORF3a | 154–162 | 1 | 9 | 0 | 1 | 1 | 0 | 4 | 0 | 9 | 9 | 9 | 1 |
| ORF3a | 244–255 | 0 | 12 | 1 | 0 | 1 | 3 | 7 | 2 | 12 | 12 | 6 | 3 |
| ORF3a | 262–275 | 0 | 13 | 0 | 0 | 0 | 0 | 14 | 0 | 12 | 13 | 12 | 1 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| ORF3a | 31–36 | 0 | 1 | 0 | 1 | 6 | 3 | 1 | 6 | 0 | 1 | 1 | 2 |
| ORF3a | 53–58 | 1 | 1 | 2 | 1 | 5 | 3 | 1 | 6 | 3 | 1 | 1 | 5 |
| ORF3a | 135–142 | 0 | 0 | 0 | 0 | 8 | 0 | 1 | 8 | 1 | 1 | 0 | 7 |
| ORF3a | 154–162 | 0 | 1 | 1 | 1 | 9 | 2 | 0 | 9 | 1 | 0 | 0 | 9 |
| ORF3a | 244–255 | 1 | 6 | 0 | 1 | 12 | 3 | 1 | 11 | 3 | 0 | 0 | 4 |
| ORF3a | 262–275 | 1 | 0 | 0 | 0 | 13 | 1 | 0 | 14 | 2 | 1 | 0 | 5 |
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| ORF6 | 1–15 | 0 | 13 | 0 | 0 | 0 | 0 | 7 | 1 | 13 | 11 | 12 | 3 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| ORF6 | 1–15 | 0 | 0 | 2 | 1 | 11 | 0 | 1 | 14 | 4 | 11 | 1 | 12 |
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| ORF7a | 15–31 | 0 | 13 | 0 | 0 | 0 | 2 | 9 | 1 | 8 | 11 | 15 | 1 |
| ORF7a | 37–58 | 0 | 22 | 1 | 1 | 3 | 8 | 19 | 1 | 20 | 22 | 21 | 2 |
| ORF7a | 75–93 | 0 | 19 | 0 | 1 | 0 | 0 | 19 | 1 | 19 | 19 | 19 | 3 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| ORF7a | 15–31 | 0 | 0 | 1 | 0 | 9 | 2 | 1 | 17 | 4 | 1 | 0 | 5 |
| ORF7a | 37–58 | 1 | 0 | 1 | 0 | 22 | 1 | 1 | 22 | 3 | 2 | 1 | 9 |
| ORF7a | 75–93 | 0 | 0 | 1 | 0 | 19 | 4 | 0 | 19 | 3 | 3 | 1 | 8 |
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| ORF7b | 6–25 | 0 | 14 | 0 | 0 | 0 | 0 | 12 | 1 | 13 | 18 | 17 | 5 |
| ORF7b | 27–33 | 0 | 5 | 0 | 0 | 0 | 0 | 4 | 0 | 3 | 4 | 5 | 1 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| ORF7b | 6–25 | 0 | 0 | 8 | 0 | 17 | 3 | 0 | 19 | 1 | 1 | 0 | 12 |
| ORF7b | 27–33 | 0 | 0 | 4 | 0 | 5 | 2 | 0 | 6 | 2 | 0 | 0 | 2 |
| Protein | Invariant residues | Tunisia | Texas | Spain | Serbia | Poland | Peru | Pennsylvania | Pakistan | Minnesota | Michigan | Massachusetts | India |
| ORF8 | 35–38 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 0 | 4 | 4 | 4 | 0 |
| ORF8 | 88–91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 2 | 0 |
| Protein | Invariant residues | Hong Kong | Greece | Ghana | France | Florida | Egypt | Chile | California | Bangladesh | Bahrain | Austria | Australia |
| ORF8 | 35–38 | 0 | 0 | 4 | 1 | 4 | 1 | 0 | 4 | 1 | 1 | 0 | 1 |
| ORF8 | 88–91 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
ORF3a variants in the North American and Oceanian geo-locations had several mutations in each invariant region, whereas very few mutations were detected in some invariant regions (not in all) of ORF3a in India, Bangladesh, Egypt, and Chile (Table 20).
No mutations at the invariant region in ORF6 variants were found in Tunisia, Spain, Serbia, Poland, Peru, Hong Kong, Greece, and Egypt. On the other hand, a handful of mutations in the invariant region were detected in the rest of the geo-locations. In the North American geo-locations, the number of mutations in ORF3a proteins was relatively big. In the North American geo-locations, in the invariant regions, a significant number of mutations in ORF3a proteins were found. A small number of mutations were found in the invariant regions of the ORF7a variant in the rest of the geo-locations with the exception of Tunisia, Hong Kong, Greece, and France (Table 20).
No mutations were found in the ORF7b invariant regions for the ORF7b proteins from Tunisia, Spain, Serbia, Poland, Peru, Hong Kong, Greece, France, Chile, and Austria. On the contrary, a significant number of mutations were detected in the two invariant regions of ORF7b from the rest of the geo-locations.
In two invariant regions, ORF8 variants from California possessed four mutations in each region, and in other North American geo-locations several mutations were also detected in the two invariant regions. However, in most geo-locations, such as India, Tunisia, Spain, France, Greece, and so on, no mutations were found in the two invariant regions (Table 20).
5. Discussion and remarks
We would like to emphasize here that the protein sequences analyzed in this study were collected from 24 geo-locations across all six continents (essentially worldwide), as per availability of the public data in NCBI at the time of the assembly of the datasets on May 29, 2021. Therefore, this work represents a historical snapshot of the SARS-CoV-2 evolution based on then available data. We recognize that the newer SARS-CoV-2 variants have shown higher transmissibility but lower fatality rates, and our prediction of the severity includes transmissibility as well. One should keep in mind that the high transmission rates increase the probability of the emergence of new SARS-CoV-2 variants, some of which might be fatal as well.
Variants of S, E, M, N, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 proteins of SARS-CoV-2 from 24 geo-locations in six continents were analyzed in this study. In each geo-location, a non-uniform frequency distribution of unique variants of all ten proteins was noticed despite the identical number of total proteins. Clearly, various mutations in a given protein gave rise to several unique variants. Therefore, it turned out that during the intraspecies evolution of a given SARS-CoV-2 RNA genome, this later expressed variable amounts/rates of mutations in different genomic segments, which yielded irregularity in the frequency of protein variants (Table 20). Therefore, it is clear that each SARS-CoV-2 genome from each geo-location is characterized by the non-uniform frequency of unique protein variants. Notably, it was not the case for the other beta-coronaviruses. Furthermore, it was noticed that the total number of common invariant residues and common mutations possessed by each unique set of protein variants from all 24 geo-locations were significantly small. In most of the proteins, neither common invariant residues nor mutated residues were found. Therefore, a significantly large percentage of mutations in each protein variant of SARS-CoV-2 is unevenly or non-uniformly distributed over each of the 24 geo-locations. Therefore, an equally evenly uneven pattern of distribution of unique variants of ten SARS-CoV-2 proteins over the 24 geo-locations was observed. It was anticipated that if sets of common invariant residues are markedly small, then common mutations must be significantly large. But this expected natural flow was not observed.
In spite of the factors behind this behavior, the S glycoprotein remains the main target for mutations reported so far, as it presents the main structure for the SARS-CoV-2 attachment to host cells. Recent articles have reported a mouse-adapted WBP-1 SARS-CoV-2 strain (through several in vivo passages of the Wuhan-Hu-1 (NC 045512) strain) characterized by two (Q493K and Q498H) mutations in its RBD [112], [113], [114], [115], [116], [117], [118], [119]. Both mutations seem responsible for converting resistant mice susceptible to SARS-CoV-2 because of the compatibility of the host ACE2 receptor with the mutated RBD in the SARS-CoV-2 S protein. Therefore, avoiding changes in the dynamics of the spread of this virus seems impossible due to the continuous appearance of new SARS-CoV-2 variants with novel mutations in viral proteins that affect efficiency of transmissibility. This is illustrated by the fact that two naturally emerging mutations in the S protein (Q493K and Q498H) of SARS-CoV-2 from the mouse-adapted strain WBP-1 showed increased infectivity in BALB/c mice caused by the enhanced affinity of the S protein RBD to the mouse ACE2 receptor. The severe lung infections in mice closely resemble lung pathologies and symptoms in COVID-19 patients. Furthermore, a number of SARS-CoV-2 strains found in several countries have naturally acquired the Q493K mutation in the S protein RBD, which may allow the virus to efficiently bind to mouse ACE2 and infect mice. Therefore, it was proposed that the Q493K and Q498H mutations in the RBD could serve as an indicator of SARS-CoV-2 variants that represent the potential risk to public health and that could emerge at the human-mouse interface [112]. Taken together, these results send an important message, indicating that the presence of the tight human-animal interactions would be expected to serve as a source of the appearance of novel infectious agents as a result of the zoonotic spillover and/or indicate that the highly virulent SARS-CoV-2 could be a man-made virus [120], [121], [122].
The frequency of distinct mutations possessed by the SARS-CoV-2 proteins in North American geo-locations, especially in California, was relatively minimal. In particular, no unique S protein variant contains more than one mutation in each sequence. It was also noticed that a significantly large number of common mutations in the S protein (487, 45, 22, 4, and 1 common mutations, respectively, were found in North America, South America, Africa, Asia, and Africa) in all of the SARS-CoV-2 proteins were found in North American geo-locations, unlike in other continental geo-locations. Therefore, it is possible that the uneven mutations across the geo-locations may be due to ethnicity of the population of these locations. Thus, such a non-uniform frequency of shared mutations on different continents led to the single mutation at position 614. A question arises in this regard: why do mutational factors vary in different geo-locations? Are they dependent on viral or host factors or both? The uneven distribution certainly demands a thorough investigation of demographic correlation with several factors of mutations of SARS-CoV-2.
Obviously, comprehensive time-dependent analyses of the SARS-CoV-2 mutability are important for a better understanding of the origin of this virus and its future fate. This kind of analysis has already been conducted. For example, the time courses of emerging viral mutants and variants during the SARS-CoV-2 pandemic in ten countries reporting high numbers of COVID-19 cases and fatalities (United Kingdom, South Africa, Brazil, United States, India, Russia, France, Spain, Germany, and China) were analyzed by considering 383,500 complete SARS-CoV-2 nucleotide sequences in GISAID (Global Initiative of Sharing All Influenza Data) [123]. It was found that viral mutants and variants had different fates, where some of the previously reported mutations waned and some of them increased in prevalence over time [123]. Similar analyses were also conducted for several individual SARS-CoV-2 proteins. Troyano-Hernaez et al. studied the evolution of SARS-CoV-2 E, M, N, and S structural proteins from the beginning of the pandemic to September 2020 by looking at the 105,276 complete and partial sequences of SARS-CoV-2 from 117 countries available in the GISAID [124]. This analysis revealed that the evolution of mutations in these proteins differed across geographic regions and epidemiological weeks (epiweeks). Some illustrative examples are given below. It was shown that the D614G mutation in the S protein was the most prevalent change, followed by the R203K and G204R combination in the N protein [124]. For the first time, D614G was found in epiweek-4 in Asia and Oceania, it appeared in Europe and North America in epiweek-5 and was detected in Africa in epiweek-9. It expanded very fast, and more than half of the total sequences showed this change in epiweek-10, whereas by epiweek-37 almost all sequences contained this mutation [124]. Another example of fast evolution is given by the S477N mutation in the S protein, whose frequency in Oceania rose from 6 % in epiweek-20 to 100 % by epiweek-31 [124]. The S68F mutation in the E protein showed different evolutionary dynamics in England, where its frequency raised from epiweek-12 (0.6 %) to epiweek-19 (3 %), decreasing to 0.2 % in the last epiweek used in the analysis [124]. Although most mutations in the M protein were characterized by a very low frequency (≤ 0.2 %), significant changes over time were observed in the following six substitutions: A2S, L17I, D209Y, H125Y, V23L, and V60L, where frequencies of A2S, D209Y, H125Y, and V23L showed an increase (typically caused by accumulation of those mutations in specific geographical locations) followed by a plateau, the time course of the frequency of the L17I amino acid change passing through a maximum, and V60L frequency showing an increase around epiweeks-27 and 28 due to European sequences, specifically from England and Switzerland, decreasing later and rising again in epiweek-34, mainly due to sequences from Scotland and Switzerland [124]. Finally, the global rate of the G204R and R203K combination in the serine/arginine-rich linker (SR-linker) of the N protein rose from 23 % in epiweek-10 to 81 % in epiweek30, dropping to 16 % in epiweek-37 [124]. Although this study produced a series of important observations, it was also pointed out that the temporal analysis performed at the regional level was limited by the uneven country and epiweek distribution of available sequences [124]. Clearly, this is a global drawback, which cannot be overcome, as there is a remarkable disparity between countries in their research and diagnostics facilities. Despite all these limitations, such temporal analyses are crucial, as they provide vital information needed for a better understanding of the SARS-CoV-2 evolution and guaranteeing the success of new diagnostic tests, therapies, and vaccines against COVID-19 [124].
In the context of the origin of SARS-CoV-2, cytidine triphosphate (CTP) plays an important role in the synthesis of the precursors of the viral envelope and protein glycosylation, which has allowed to link mutational studies to the timelines [125]. The essential function of CTP in the synthesis of the viral envelope and the translation of its genome has led to the emergence of a toxic CTP analogue synthesized by viperin possessing antiviral immunogenicity [126]. Application of a probabilistic modelling approach for investigation of the molecular evolution of the virus has allowed real-time monitoring on a daily basis. It has been possible to link the evolution of the viral genome to the progeny produced over time, in particular to follow the flow of mutations and alterations of the proofreading system (formation of “blooms”) in attempts to better understand how the virus can use the host metabolism for its own benefit.
We also observed that the reference proteins (of the SARS-CoV-2, NC 045512) contained several invariant domains across the other four different beta-coronaviruses (Table 17). Mostly in all North American geo-locations, many mutations were detected in each invariant (assumed to be evolutionary conserved) region of the S protein with regard to the reference SARS-CoV-2 S protein. Likewise, several mutations in other proteins were also noted in all seven geo-locations from North America. In the rest of the 24 geo-locations, a few mutations in some of the invariant regions of the respective proteins were detected. Therefore, in a short span of one year, the NC 045512 SARS-CoV-2 changed itself in such a manner that even the evolutionarily conserved domains (invariant regions) were altered, which might lead to the emergence of new SARS-CoV-2 variants with a different degree of virulence, infectivity, and transmissibility. These observations reopen the possibility to interrogate the SARS-CoV-2 origin. Correctly identifying the characteristics of SARS-CoV-2 would enable scientists to take appropriate measures to contain future pandemics. It could also help in the development of better diagnostics, vaccines, and therapeutic tools.
The following are the supplementary data related to this article.
SARS-CoV-2 protein variants from 24 geo-locations
Multiple sequence alignments showing mutations in the invariant residue regions of various proteins of SARS-CoV-2
CRediT authorship contribution statement
SSH conceptualized the study. SSH, VK, EMR, VNU, and KL contributed to the implementation of the research, to the analysis of the results. SSH, VNU, and EMR wrote the initial draft of the manuscript. SSH, KL, PPC, ASA, GKA, AAAA, AL, GP, TMAEA, PA, GC, DB, MT, SPS, and VNU, reviewed and edited manuscript. BDU, WBC, and NGB provided constructive reviews and suggestions. All authors read and approved final version.
Declaration of competing interest
The authors declare that there is no conflict of interest in this work.
Acknowledgements
The authors thank the researchers who generated and shared the sequencing data from NCBI SARS-CoV-2 Data Hub on which this research is based.
Data availability
Data will be made available on request.
References
- 1.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020:1–14. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuen K.-S., Ye Z.-W., Fung S.-Y., Chan C.-P., Jin D.-Y. Sars-CoV-2 and COVID-19: the most important research questions. Cell Biosci. 2020;10(1):1–5. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matheson N.J., Lehner P.J. How does SARS-CoV-2 cause COVID-19? Science. 2020;369(6503):510–511. doi: 10.1126/science.abc6156. [DOI] [PubMed] [Google Scholar]
- 4.Wu D., Wu T., Liu Q., Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020;94:44–48. doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J. Sars-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas M., Karrer U., Lucas A., Klenerman P. Viral escape mechanisms–escapology taught by viruses. Int. J. Exp. Pathol. 2001;82(5):269–286. doi: 10.1046/j.1365-2613.2001.00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam M.R., Hoque M.N., Rahman M.S., Alam A.R.U., Akther M., Puspo J.A., Akter S., Sultana M., Hossain M.A., Crandall K.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Scientific Reports. 2020;10(1):1–9. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava S., Banu S., Singh P., Sowpati D.T., Mishra R.K. Sars-CoV-2 genomics: an indian perspective on sequencing viral variants. J. Biosci. 2021;46(1):1–14. doi: 10.1007/s12038-021-00145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassan S.S., Choudhury P.P., Roy B., Jana S.S. Missense mutations in SARS-CoV2 genomes from Indian patients. Genomics. 2020;112(6):4622–4627. doi: 10.1016/j.ygeno.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. Journal of translational medicine. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.-L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79(5):710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-On Y.M., Flamholz A., Phillips R., Milo R. Science forum: Sars-CoV-2 (COVID-19) by the numbers. elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanju'an R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. Journal of virology. 2010;84(19):9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sender R., Bar-On Y.M., Gleizer S., Bernshtein B., Flamholz A., Phillips R., Milo R. The total number and mass of SARS-CoV-2 virions. Proceedings of the National Academy of Sciences. 2021;118(25) doi: 10.1073/pnas.2024815118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callaway E. The coronavirus is mutating-does it matter? Nature. 2020;585(7824):174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- 18.Luo R., Delaunay-Moisan A., Timmis K., Danchin A. SARS-CoV-2 biology and variants: anticipation of viral evolution and what needs to be done. Environ. Microbiol. 2021;23(5):2339–2363. doi: 10.1111/1462-2920.15487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut, Peacock S.J. Sars-CoV-2 variants, spike mutations and immune escape. Nature Reviews Microbiology. 2021:1–16. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amicone M., Borges V., Alves M.J., Isidro J., Ze-Ze L., Duarte S., Vieira L., Guiomar R., Gomes J.P., Gordo I. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol. Med. Public Health. 2022;10(1):142–155. doi: 10.1093/emph/eoac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325(6):529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 24.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that d614g increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seyran M., Takayama K., Uversky V.N., Lundstrom K., Sherchan S.P., Attrish D., Rezaei N., Aljabali A.A., Ghosh S., et al. The structural basis of accelerated host cell entry by SARS-CoV-2. The FEBS Journal. 2021;288(17):5010–5020. doi: 10.1111/febs.15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanju'an R., Domingo-Calap P. Mechanisms of viral mutation. Cellular and molecular life sciences. 2016;73(23):4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodish H., Zipursky S.L. Molecular cell biology. Biochem. Mol. Biol. Educ. 2001;29:126–133. [Google Scholar]
- 28.Holmes E.C. The comparative genomics of viral emergence. Proc. Natl. Acad. Sci. 2010;107(suppl 1):1742–1746. doi: 10.1073/pnas.0906193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassan S.S., Aljabali A.A., Panda P.K., Ghosh S., Attrish D., Choudhury P.P., Seyran M., Pizzol D., Adadi P., El-Aziz M.Abd. A unique view of SARS-CoV-2 through the lens of ORF8 protein. Computers in Biology and Medicine. 2021;133 doi: 10.1016/j.compbiomed.2021.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan S.S., Lundstrom K., Barth D., Silva R.J.S., Andrade B.S., Azevedo V., Pal Choudhury P., Palu H., Uhal B.D., Kandinalla R., et al. Implications derived from S-protein variants of SARS-CoV-2 from six continents. Int. J. Biol. Macromol. 2021;1991:934–955. doi: 10.1016/j.ijbiomac.2021.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bajaj V., Gadi N., Spihlman A.P., Wu S.C., Choi C.H., Moulton V.R. Aging, immunity, and COVID-19: how age influences the host immune response to coronavirus infections? Front. Physiol. 2021;11:1793. doi: 10.3389/fphys.2020.571416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouse B.T., Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat. Rev. Immunol. 2010;10(7):514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kupferschmidt K. The pandemic virus is slowly mutating. But does it matter? Science. 2020;369(6501):238–239. doi: 10.1126/science.369.6501.238. [DOI] [PubMed] [Google Scholar]
- 34.Leitner T., Kumar S. Where did SARS-CoV-2 come from? Mol. Biol. Evol. 2020;37(9):2463–2464. doi: 10.1093/molbev/msaa162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jo W.K., de Oliveira-Filho E.F., Rasche A., Greenwood A.D., Osterrieder K., Drexler J.F. Potential zoonotic sources of SARS-CoV-2 infections. Transbound. Emerg. Dis. 2021;68(4):1824–1834. doi: 10.1111/tbed.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundstrom K., Seyran M., Pizzol D., Adadi P., El-Aziz T.Mohamed Abd, Hassan S., Soares A., Kandimalla R., Tambuwala M.M., Aljabali A.A. Origin of SARS-CoV-2 viruses. 2020;12(11):1203. doi: 10.3390/v12111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar V., Pruthvishree B., Pande T., Sinha D., Singh B., Dhama K., Malik Y.S., et al. Sars-CoV-2 (COVID-19): zoonotic origin and susceptibility of domestic and wild animals. J. Pure Appl. Microbiol. 2020;14(suppl 1):741–747. [Google Scholar]
- 38.Banerjee A., Doxey A.C., Mossman K., Irving A.T. Unravelling the zoonotic origin and transmission of SARS-CoV-2. Trends Ecol. Evol. 2021;36(3):180–184. doi: 10.1016/j.tree.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sallard E., Halloy J., Casane D., Decroly E., van Helden J. Tracing the origins of SARS-CoV-2 in coronavirus phylogenies: a review. Environ. Chem. Lett. 2021:1–17. doi: 10.1007/s10311-020-01151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segreto R., Deigin Y. The genetic structure of SARS-CoV-2 does not rule out a laboratory origin: Sars-CoV-2 chimeric structure and furin cleavage site might be the result of genetic manipulation. BioEssays. 2020;2000240 doi: 10.1002/bies.202000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirotkin K., Sirotkin D. Might SARS-CoV-2 have arisen via serial passage through an animal host or cell culture? A potential explanation for much of the novel coronavirus’ distinctive genome. BioEssays. 2020;42(10):2000091. doi: 10.1002/bies.202000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seyran M., Pizzol D., Adadi P., El-Aziz T.M.A., Hassan S.S., Soares A., Kandimalla R., Lundstrom K., Tambuwala M., Aljabali A.A., et al. Questions concerning the proximal origin of SARS-CoV-2. J. Med. Virol. 2021;93(3):1204–1206. doi: 10.1002/jmv.26478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maxmen A., Mallapaty S. The COVID lab-leak hypothesis: what scientists do and don’t know. Nature. 2021;594(7863):3130315. doi: 10.1038/d41586-021-01529-3. [DOI] [PubMed] [Google Scholar]
- 44.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sievers F., Higgins D.G. Clustal omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar R.C. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., et al. Vipr: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(D1):D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E.D., Zendulka J., Brezovsky J., Damborsky J. Predictsnp: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014;10(1) doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wingfield P.T. N-terminal methionine processing. Curr. Protoc. Protein Sci. 2017;88(1):6–14. doi: 10.1002/cpps.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588(7837):327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Central Science. 2020;6(10):1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ace2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 54.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. Sars-CoV-2 variants of concern are emerging in India. Nat. Med. 2021:1–3. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 55.Walensky R.P., Walke H.T., Fauci A.S. Sars-CoV-2 variants of concern in the United states—challenges and opportunities. JAMA. 2021;325(11):1037–1038. doi: 10.1001/jama.2021.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mascola J.R., Graham B.S., Fauci A.S. Sars-CoV-2 viral variants—tackling a moving target. JAMA. 2021;325(13):1261–1262. doi: 10.1001/jama.2021.2088. [DOI] [PubMed] [Google Scholar]
- 57.Vogels C.B., Breban M.I., Ott I.M., Alpert T., Petrone M.E., Watkins A.E., Kalinich C.C., Earnest R., Rothman J.E., Goes de Jesus J., et al. Multiplex qpcr discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19(5) doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591(7849):293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021:1–11. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 60.Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020;5(1):1–3. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao Y., Yang R., Lee I., Zhang W., Sun J., Wang W., Meng X. Characterization of the SARS-CoV-2 e protein: sequence, structure, viroporin, and inhibitors. Protein Sci. 2021;30(6):1114–1130. doi: 10.1002/pro.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Omari K.El, Li S., Kotecha A., Walter T.S., Bignon E.A., Harlos K., Somerharju P., Haas F.De, Clare D.K., Molin M., et al. The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins. Nature Communications. 2019;10(1):1–11. doi: 10.1038/s41467-019-08728-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alsaadi E.A., Neuman B.W., Jones I.M. Identification of a membrane binding peptide in the envelope protein of mhv coronavirus. Viruses. 2020;12(9):1054. doi: 10.3390/v12091054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boson B., Legros V., Zhou B., Siret E., Mathieu C., Cosset F.-L., Lavillette D., Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (e) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee S., Bhattacharyya D., Bhunia A. Host-membrane interacting interface of the SARS coronavirus envelope protein: immense functional potential of c-terminal domain. Biophys. Chem. 2020;106452 doi: 10.1016/j.bpc.2020.106452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curtis K.M., Yount B., Baric R.S. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 2002;76(3):1422–1434. doi: 10.1128/JVI.76.3.1422-1434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kern D.M., Sorum B., Mali S.S., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Kotecha A., Bautista D.M., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yue Y., Nabar N.R., Shi C.-S., Kamenyeva O., Xiao X., Hwang I.-Y., Wang M., Kehrl J.H. Sars-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9(9):1–15. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nieto-Torres J.L., Diego M.L.De, Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathogens. 2014;10(5) doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nieto-Torres J.L., Verdiá-Báguena C., Castaño-Rodriguez C., Aguilella V.M., Enjuanes L. Relevance of viroporin ion channel activity on viral replication and pathogenesis. Viruses. 2015;7(7):3552–3573. doi: 10.3390/v7072786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parthasarathy K., Ng L., Lin X., Liu D.X., Pervushin K., Gong X., Torres J. Structural flexibility of the pentameric SARS coronavirus envelope protein ion channel. Biophys. J. 2008;95(6):L39–L41. doi: 10.1529/biophysj.108.133041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., et al. A structural analysis of m protein in coronavirus assembly and morphology. Journal of Structural Biology. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tseng Y.-T., Chang C.-H., Wang S.-M., Huang K.-J., Wang C.-T. Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PloS One. 2013;8(5) doi: 10.1371/journal.pone.0064013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tseng Y.-T., Wang S.-M., Huang K.-J., Amber I., Lee R., Chiang C.-C., Wang C.-T. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J. Biol. Chem. 2010;285(17):12862–12872. doi: 10.1074/jbc.M109.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7(4):1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang J.Q., Fang S., Yuan Q., Huang M., Chen R.A., Fung T.S., Liu D.X. N-linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus-induced er stress response, apoptosis and pathogenesis. Virology. 2019;531:48–56. doi: 10.1016/j.virol.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arya R., Kumari S., Pandey B., Mistry H., Bihani S.C., Das A., Prashar V., Gupta G.D., Panicker L., Kumar M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021;433(2) doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang C.-K., Chen C.-M.M., Chiang M.-H., Hsu Y.-L., Huang T.-H. Transient oligomerization of the SARS-CoV n protein– implication for virus ribonucleoprotein packaging. PloS One. 2013;8(5) doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye Q., West A.M., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020;29(9):1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giri R., Bhardwaj T., Shegane M., Gehi B.R., Kumar P., Gadhave K., Oldfield C.J., Uversky V.N. Understanding COVID-19 via comparative analysis of dark proteomes of SARS-CoV-2, human SARS and bat SARS-like coronaviruses. Cell. Mol. Life Sci. 2021;78(4):1655–1688. doi: 10.1007/s00018-020-03603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tugaeva K.V., Hawkins D.E., Smith J.L., Bayfield O.W., Ker D.-S., Sysoev A.A., Klychnikov O.I., Antson A.A., Sluchanko N.N. The mechanism of SARS-CoV-2 nucleocapsid protein recognition by the human 14-3-3 proteins. Journal of Molecular Biology. 2021;433(8) doi: 10.1016/j.jmb.2021.166875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Del Veliz S., Rivera L., Bustos D.M., Uhart M. Analysis of SARS-CoV-2 nucleocapsid phosphoprotein n variations in the binding site to human 14-3-3 proteins. Biochem. Biophys. Res. Commun. 2021;569:154–160. doi: 10.1016/j.bbrc.2021.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020;527(3):618–623. doi: 10.1016/j.bbrc.2020.04.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. Msystems. 2020;5(3) doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ostaszewski M., Mazein A., Gillespie M.E., Kuperstein I., Niarakis A., Hermjakob H., Pico A.R., Willighagen E.L., Evelo C.T., Hasenauer J., et al. Covid-19 disease map, building a computational repository of SARS-CoV-2 virus-host interaction mechanisms. Scientific Data. 2020;7(1):1–4. doi: 10.1038/s41597-020-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.-Y., Zhou W., Qiu Y., et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17(8):881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shah A. Novel coronavirus-induced nlrp3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front. Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lam J.-Y., Yuen C.-K., Ip J.D., Wong W.-M., To K.K.-W., Yuen K.-Y., Kok K.-H. Loss of ORF3b in the circulating SARS-CoV-2 strains. Emerg. Microbes Infect. 2020;9(1):2685–2696. doi: 10.1080/22221751.2020.1852892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hachim A., Gu H., Kavian O., Kwan M.Y., Yau Y.S., Chiu S.S., Tsang O.T., Hui D.S., Ma F., Chan W.-H., et al. The SARS-CoV-2 antibody landscape is lower in magnitude for structural proteins, diversified for accessory proteins and stable long-term in children. medRxiv. 2021 doi: 10.1101/2021.01.03.21249180. [DOI] [Google Scholar]
- 94.Gunalan V., Mirazimi A., Tan Y.-J. A putative diacidic motif in the SARS-CoV ORF6 protein influences its subcellular localization and suppression of expression of co-transfected expression constructs. BMC Res. Notes. 2011;4(1):1–9. doi: 10.1186/1756-0500-4-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., Ge X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type i interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumar P., Gunalan V., Liu B., Chow V.T., Druce J., Birch C., Catton M., Fielding B.C., Tan Y.-J., Lal S.K. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366(2):293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of type i interferon by SARS-CoV-2. Cell Rep. 2020;33(1) doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., Halden R.U., Hogue B.G., Scotch M., Lim E.S. An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (January to March 2020) Journal of Virology. 2020;94(14) doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus ( SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81(2):718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Neches R.Y., Kyrpides N.C., Ouzounis C.A. Atypical divergence of SARS-CoV-2 ORF8 from ORF7a within the coronavirus lineage suggests potential stealthy viral strategies in immune evasion. MBio. 2021;12(1):e03014–e03020. doi: 10.1128/mBio.03014-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Su Y.C., Anderson D.E., Young B.E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., Low J.G., Tan C.W., et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. MBio. 2020;11(4) doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Farŕe D., Engel P., Angulo A. Immunoglobulin superfamily members encoded by viruses and their multiple roles in immune evasion. European Journal of Immunology. 2017;47(5):780–796. doi: 10.1002/eji.201746984. [DOI] [PubMed] [Google Scholar]
- 105.Schuster N.A. Characterization and structural prediction of the putative ORF10 protein in SARS-CoV-2. bioRxiv. 2021 [Google Scholar]
- 106.Altincekic N., Korn S.M., Qureshi N.S., Dujardin M., Ninot-Pedrosa M., Abele R., Abi Saad M.J., Alfano C., Almeida F.C., Alshamleh I., et al. Large-scale recombinant production of the SARS-CoV-2 proteome for high-throughput and structural biology applications. Front. Mol. Biosci. 2021;8:89. doi: 10.3389/fmolb.2021.653148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mena E.L., Donahue C.J., Vaites L.P., Li J., Rona G., O’Leary C., Lignitto L., Miwatani-Minter B., Paulo J.A., Dhabaria A., et al. ORF10–cullin-2–zyg11b complex is not required for SARS-CoV-2 infection. Proceedings of the National Academy of Sciences. 2021;118(17) doi: 10.1073/pnas.2023157118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2021;2(1):99–112. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang D.-M., Lin F.-C., Tsai P.-H., Chien Y., Wang M.-L., Yang Y.-P., Chang T.-J. Pandemic analysis of infection and death correlated with genomic open reading frame 10 mutation in severe acute respiratory syndrome coronavirus 2 victims. J. Chin. Med. Assoc. 2021;84(5):478–484. doi: 10.1097/JCMA.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 111.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the d614g SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang K., Zhang Y., Hui X., Zhao Y., Gong W., Wang T., Zhang S., Yang Y., Deng F., Zhang Q., et al. Q493k and q498h substitutions in spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao R., Zu W., Liu Y., Li J., Li Z., Wen Y., Wang H., Yuan J., Cheng L., Zhang S., et al. Quasispecies of SARS-CoV-2 revealed by single nucleotide polymorphisms (snps) analysis. Virulence. 2021;12(1):1209–1226. doi: 10.1080/21505594.2021.1911477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maurin M., Fenollar F., Mediannikov O., Davoust B., Devaux C., Raoult D. Current status of putative animal sources of SARS-CoV-2 infection in humans: wildlife, domestic animals and pets. Microorganisms. 2021;9(4):868. doi: 10.3390/microorganisms9040868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Frutos R., Serra-Cobo J., Pinault L., Lopez Roig M., Devaux C.A. Emergence of bat-related betacoronaviruses: hazard and risks. Front. Microbiol. 2021;12:437. doi: 10.3389/fmicb.2021.591535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Graudenzi A., Maspero D., Angaroni F., Piazza R., Ramazzotti D. Mutational signatures and heterogeneous host response revealed via large-scale characterization of SARS-CoV-2 genomic diversity. Iscience. 2021;24(2) doi: 10.1016/j.isci.2021.102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Frutos R., Gavotte L., Devaux C.A. Understanding the origin of COVID-19 requires to change the paradigm on zoonotic emergence from the spillover model to the viral circulation model. Infect. Genet. Evol. 2021;95 doi: 10.1016/j.meegid.2021.104812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ramazzotti D., Angaroni F., Maspero D., Gambacorti-Passerini C., Antoniotti M., Graudenzi A., Piazza R. Verso: a comprehensive framework for the inference of robust phylogenies and the quantification of intra-host genomic diversity of viral samples. Patterns. 2021;2(3) doi: 10.1016/j.patter.2021.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Al Khatib H.A., Benslimane F.M., Elbashir I.E., Coyle P.V., Al Maslamani M.A., Al-Khal A., Al Thani A.A., Yassine H.M. Within-host diversity of SARS-CoV-2 in COVID-19 patients with variable disease severities, Frontiers in cellular and infection. Microbiology. 2020;10 doi: 10.3389/fcimb.2020.575613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pekar J., Worobey M., Moshiri N., Scheffler K., Wertheim J.O. Timing the SARS-CoV-2 index case in Hubei province. Science. 2021;372(6540):412–417. doi: 10.1126/science.abf8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Decroly E., Claverie J.-M., Canard B. Le rapport de la mission OMS Peine `a retracer les origines de l’´epid́emie de SARS-CoV-2. Virologie. 2021;1(1) doi: 10.1684/vir.2021.0901. [DOI] [PubMed] [Google Scholar]
- 122.Maxmen A. Who report into COVID pandemic origins zeroes in on animal markets, not labs. Nature. 2021;592(7853):173–174. doi: 10.1038/d41586-021-00865-8. [DOI] [PubMed] [Google Scholar]
- 123.Weber S., Ramirez C.M., Weiser B., Burger H., Doerfler W. SARS-CoV-2 worldwide replication drives rapid rise and selection of mutations across the viral genome: a time-course study–potential challenge for vaccines and therapies. EMBO Molecular Medicine. 2021;13(6) doi: 10.15252/emmm.202114062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Troyano-Hernáaez P., Reinosa R. Evolution of SARS-CoV-2 envelope, membrane, nucleocapsid, and spike structural proteins from the beginning of the pandemic to september 2020: a global and regional approach by epidemiological week. Viruses. 2021;13(2):243. doi: 10.3390/v13020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ou Z., Ouzounis C., Wang D., Sun W., Li J., Chen W., Marlìere P., Danchin A. A path toward SARS-CoV-2 attenuation: metabolic pressure on CTP synthesis rules the virus evolution. Genome Biology and Evolution. 2020;12(12):2467–2485. doi: 10.1093/gbe/evaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cluzel N., Lambert A., Maday Y., Turinici G., Danchin A. Biochemical and mathematical lessons from the evolution of the SARS-CoV-2 virus: paths for novel antiviral warfare. C. R. Biol. 2020;343(2):177–209. doi: 10.5802/crbiol.16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SARS-CoV-2 protein variants from 24 geo-locations
Multiple sequence alignments showing mutations in the invariant residue regions of various proteins of SARS-CoV-2
Data Availability Statement
Data will be made available on request.