Rice is one of the most important food crops and feeds more than half of the world’s population [1]. Arthropod-borne rice viruses have caused devastating epidemics in Asian countries and are a major threat to food security [2]. However, little is known about the vulnerability of rice crops to viral pathogens [3,4], especially southern rice black-streaked dwarf virus (SRBSDV), rice black-streaked dwarf virus (RBSDV) and rice gall dwarf virus (RGDV), known to induce annual outbreaks by insect transmission in some localities in Asia.
To address this question, we first investigated the susceptibility of 136 conventional and hybrid rice varieties widely cultivated or approved for release in China to SRBSDV, which has been circulating in Asian countries since its first report in 2008 [2]. We monitored symptom development and asymptomatic infection in seedlings after inoculation with viruliferous white-backed plant hoppers (Sogatella furcifera, Horváth) in a greenhouse. We found that most of the varieties examined were highly susceptible to SRBSDV and developed the characteristic disease symptoms at 45 days post-inoculation (Fig. 1A and Supplementary Table S1).
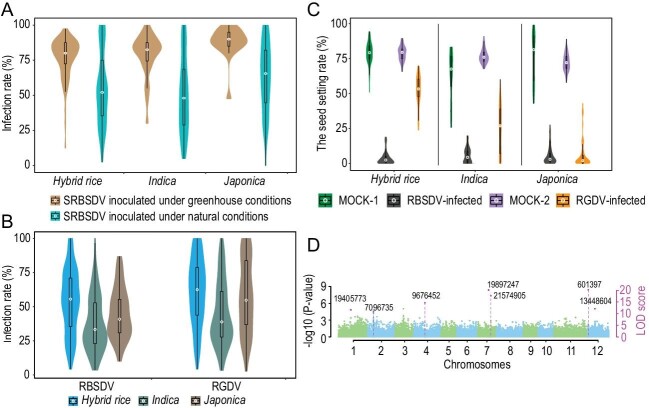
Figure 1.
Infection rate under natural and greenhouse conditions and statistical disease indicators comparison of rice varieties upon RBSDV and RGDV infection and the seven identified loci through GWAS analysis. (A) The infection rate of hybrid rice, indica type and japonica type upon SRBSDV infection under greenhouse and natural conditions. (B) The infection rate of 528 rice varieties upon RBSDV and RGDV infection under natural conditions. (C) Comparison of the seed setting rate (SSR) of 20 rice varieties between mock and RBSDV or RGDV infection, respectively. (D) The seven loci associated with rice tolerance to viral infections identified from Manhattan plots in indicate rice varieties. The left y-axis reports –log10 P-values, which are obtained from single-marker genome-wide scanning for all the markers in the first step of 3VmrMLM, and the right y-axis reports LOD scores, which are obtained from a likelihood ratio test for significant and suggested QTNs, with the threshold of LOD = 10.0 (gray dashed line), in the second step of 3VmrMLM described in Li et al. (2022). The LOD scores, along with their quantitative trait nucleotides (QTNs), are shown in points with straight lines. All the main-effect QTNs (red dots) are identified. The number on the red dot represents the chromosomal physical location of the peak of QTN, corresponding to the reference Nipponbare.
The results from greenhouse inoculation with viruliferous insect vectors predict widespread vulnerability of rice cultivars to SRBSDV in rice fields. To test this hypothesis, we planted seedlings of
528 varieties in 3 consecutive years under open field conditions at locations of Nanning and Guilin, Guangxi Province that have recorded a multiyear SRBSDV outbreak from plant hopper transmission. We found that ≥25% of the seedlings from 80%–93% of the examined hybrid or conventional indica and japonica cultivars became infected naturally with SRBSDV 60 days after seedling transplantation (Fig. 1A and Supplementary Table S2).
RBSDV and RGDV outbreaks occur annually in Kaifeng and Yunxiao of Henan and Fujian provinces, respectively. Our field evaluation of the 528 varieties at the two locations from 2016 to 2021 further revealed widespread vulnerability of these rice cultivars to both RBSDV and RGDV, which are transmitted by small brown plant hoppers (Laodelphax striatellus, Fallén) and zigzag leaf hoppers (Deltocephalus dorsalis, Motschulsky), respectively (Fig. 1B and Supplementary Table S2). A total rice category of infection rate was conducted based on 528 varieties, including different rice types of indica, japonica, hybrid indica, hybrid japonica, restorer line, sterile line and maintainer line that were infected by SRBSDV, RBSDV and RGDV, respectively (Supplementary Fig. S1 and Supplementary Table S2). These results indicate that rice varieties currently in production and promotion generally lack broad-spectrum resistance to rice viruses.
To assess the impact of viral infection, 10 seedlings from each of 20 varieties randomly selected either uninfected or infected with RBSDV or RGDV were grown to maturity for calculating seed setting rate (SSR). We found that infection with either RBSDV or RGDV drastically decreased the SSR of both conventional and hybrid cultivars (Fig. 1C and Supplementary Table S3). Together, our findings reveal that rice production and food security are dangerously vulnerable to potential epidemics caused by any of the three insect-borne viral pathogens.
We further used a recently developed methodology framework (3VmrMLM) of genome-wide association studies (GWAS) to search for QTNs (quantitative trait nucleotides) loci associated with viral tolerance based on the data sets from Nanning and Guilin and the available genomic sequences of rice varieties [5–8]. Seven QTNs located on chromosomes 1, 2, 4, 7 and 12 were found to be significantly associated with viral tolerance (Fig. 1D and Supplementary Table S4). Thus, it seems possible to investigate the genetic mechanisms of rice viral resistance using the experimental system established in this work as shown recently in Arabidopsis [9].
Major advances have been made in understanding the mechanisms of important breeding traits of rice such as high yield and rice blast disease resistance [10–15]. Current rice-breeding programs demand resistance evaluation against major fungal and bacterial pathogens. However, the direct link between the agro-ecosystem changes and the outbreaks of rice virus diseases needs attention. Our study shows that the rice varieties popularized in production generally lack antiviral function under both natural and greenhouse conditions and viral infection significantly reduce rice production. Based on our findings, we conclude that there is an urgency to develop a standardized protocol for assessing the performance of current and future rice varieties against key insect-borne viral pathogens to be incorporated into variety certification and approval guidelines.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hua-an Xie (Fujian Academy of Agricultural Sciences), Jian-long Xu (Chinese Academy of Agricultural Sciences), Yan-jun Kou (China National Rice Research Institute) and Xinhao Ouyang (Xiamen University) for providing rice variety resources; Tong Zhou (Jiangsu Academy of Agricultural Sciences), Yun-Liang Peng (Sichuan Academy of Agricultural Sciences) and Jing Huang (Zhangzhou Institute of Technology) for assistance with the RBSDV, SRBSDV and RGDV-infection assay in the field; and Suhong Bu (South China Agricultural University) for GWAS analysis.
Contributor Information
Jian-guo Wu, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Guo-yi Yang, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Shan-shan Zhao, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Shuai Zhang, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Bi-xia Qin, Institute of Plant Protection, Guangxi Academy of Agricultural Sciences, China.
Yong-sheng Zhu, Rice Research Institute, Fujian Academy of Agricultural Sciences, China.
Hui-ting Xie, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Qing Chang, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Lu Wang, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Jie Hu, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Chao Zhang, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Bao-gang Zhang, Vector-borne Virus Research Center, Fujian Province Key Laboratory of Plant Virology, Institute of Plant Virology, Fujian Agriculture and Forestry University, China.
Da-li Zeng, State Key Laboratory of Rice Biology, China National Rice Research Institute, Chinese Academy of Agricultural Sciences, China.
Jian-fu Zhang, Rice Research Institute, Fujian Academy of Agricultural Sciences, China.
Xian-bo Huang, Rice Research Institute, Sanming Academy of Agricultural Sciences, China.
Qian Qian, State Key Laboratory of Rice Biology, China National Rice Research Institute, Chinese Academy of Agricultural Sciences, China.
Shou-wei Ding, Department of Microbiology and Plant Pathology, Center for Plant Cell Biology, Institute for Integrative Genome Biology, University of California, Riverside, USA.
Yi Li, The State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, China.
FUNDING
This work was supported by the National Natural Science Foundation of China (32025031, 32090010, U1905203, 32170167 and 31871934), the National Key R&D Program of China (2021YFD1400500) and the Natural Science Foundation of Fujian Province of China (2019J01370).
AUTHOR CONTRIBUTIONS
Jian-guo Wu, Qian Qian, Shou-Wei Ding and Yi Li conceived the idea and designed the experiments for the research. Guo-yi Yang, Shan-shan Zhao, Shuai Zhang, Bi-xia Qin, Yong-sheng Zhu, Hui-ting Xie, Lu Wang, Jie Hu, Chao Zhang and Bao-gang Zhang performed the experiments. Guo-yi Yang, Qing Chang, Da-li Zeng, Jian-fu Zhang and Xian-bo Huang analysed the data. Jian-guo Wu, Shan-shan Zhao, Shou-Wei Ding and Yi Li drafted the paper. All authors worked collaboratively to edit and revise the paper.
Conflict of interest statement. None declared.
REFERENCES
- 1. Elert E. Nature 2014; 514: S50–1. 10.1038/514S50a [DOI] [PubMed] [Google Scholar]
- 2. Otuka A. Front Microbiol 2013; 4: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Q, Liu Y, He Jet al. . Nat Commun 2014; 5: 4768. 10.1038/ncomms5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Albar L, Bangratz-Reyser M, Hébrard Eet al. . Plant J 2006; 47: 417–26. 10.1111/j.1365-313X.2006.02792.x [DOI] [PubMed] [Google Scholar]
- 5. Huang XH, Yang S, Gong Jet al. . Nat communi 2015; 6: 6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang XH, Wei X, Sang Tet al. . Nat Genet 2010; 42: 961–76. 10.1038/ng.695 [DOI] [PubMed] [Google Scholar]
- 7. Qin P, Lu H, Du Het al. . Cell 2021; 184:3542–58. 10.1016/j.cell.2021.04.046 [DOI] [PubMed] [Google Scholar]
- 8. Li M, Zhang Y-W, Zhang Z-Cet al. . Mole Plant 2022; 15: 630–50. 10.1016/j.molp.2022.02.012 [DOI] [PubMed] [Google Scholar]
- 9. Liu S, Chen M, Li Ret al. . Nat communi 2022; 13: 2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu H, Lin T, Meng X. et al. Cell 2021; 184: 1156–70. 10.1016/j.cell.2021.01.013 [DOI] [PubMed] [Google Scholar]
- 11. Liu W, Liu J, Triplett Let al. . Annu Rev Phytopathol 2014; 52: 213–41. 10.1146/annurev-phyto-102313-045926 [DOI] [PubMed] [Google Scholar]
- 12. Wei X, Qiu J, Yong Ket al. . Nat Genet 2021; 53: 243–53. 10.1038/s41588-020-00769-9 [DOI] [PubMed] [Google Scholar]
- 13. Wang J, Zhou L, Shi H. et al. Science 2018; 361: 1026–8. 10.1126/science.aat7675 [DOI] [PubMed] [Google Scholar]
- 14. Wing RA, Purugganan MD, Zhang Q. Nat Rev Genet 2018; 19: 505–17. 10.1038/s41576-018-0024-z [DOI] [PubMed] [Google Scholar]
- 15. Wang W, Mauleon R, Hu Zet al. . Nature 2018; 557: 43–9. 10.1038/s41586-018-0063-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.