Abstract
Introduction
Coronavirus disease-2019 (COVID-19) is a complex infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that can cause also gastrointestinal symptoms. There are various factors that determine the host susceptibility and severity of infection, including the renin-angiotensin system, the immune response, and the gut microbiota. In this regard, we aimed to investigate the gene expression of ACE, AGTR1, ACE2, and TMPRSS2, which mediate SARS-CoV-2 pathogenesis by Akkermansia muciniphila, Faecalibacterium prausnitzii, Bacteroides thetaiotaomicron, and Bacteroides fragilis on Caco-2 cells. Also, the enrichment analysis considering the studied genes was analyzed on raw data from the microarray analysis of COVID-19 patients.
Materials and methods
Caco-2 cells were treated with live, heat-inactivated form and cell free supernatants of A. muciniphila, F. prausnitzii, B. thetaiotaomicron and B. fragilis for overnight. After RNA extraction and cDNA synthesis, the expression of studied genes was assessed by RT-qPCR. DNA methylation of studied genes was analyzed by Partek® Genomics Suite® software on the GSE174818 dataset. We used GSE164805 and GSE166552 datasets from COVID-19 patients to perform enrichment analysis by considering the mentioned genes via GEO2R, DAVID. Finally, the related microRNAs to GO terms concerned on the studied genes were identified by miRPath.
Results
The downregulation of ACE, AGTR1, and ACE2 genes by A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis in live, heat-inactivated, and cell-free supernatants was reported for the first time. These genes had hypomethylated DNA status in COVID-19 patients' raw data. The highest fold enrichment in upregulated RAS pathways and immune responses belonged to ACE, AGTR1, and ACE2 by considering the protein-protein interaction network. The common miRNAs targeting the studied genes were reported as miR-124-3p and miR-26b-5p.
Conclusion
In combination with our experimental data and bioinformatic analysis, we showed the potential of A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis and their postbiotics to reduce ACE, ATR1, and ACE2 expression, which are essential genes that drive upregulated biological processes in COVID-19 patients.
Accordingly, due to the potential of studied bacteria on the alteration of ACE, AGTR1, ACE2 genes expression, understanding their correlation with demonstrated miRNAs expression could be valuable. These findings suggest the importance of considering targeted gut microbiota intervention when designing the possible therapeutic strategy for controlling the COVID-19.
Keywords: Gut microbiota, Postbiotics, Renin angiotensin system, SARS-CoV2, COVID-19
1. Introduction
Novel beta coronaviruses named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for a coronavirus disease-2019 (COVID-19) pandemic originating from China at the end of 2019 and has been spread all over the world [1]. It has been demonstrated that coronaviruses potentially have a destructive effect on public health since the world has experienced three acute infectious coronavirus diseases, including severe acute respiratory syndrome coronavirus (SARS-CoV) the Middle East respiratory syndrome (MERS) and SARS-CoV-2. COVID-19 infection mainly occurs in the respiratory tract, with some gastrointestinal (GI) manifestations [[2], [3], [4]]. Multiple factors are involved in the susceptibility of the host cell to infection and disease development, such as the status of the renin-angiotensin system (RAS), the immune system, and the gut microbiota [5].
RAS is a complicated system with systemic and organ-specific functions that regulate blood pressure, fluid homeostasis, inflammation, and fibrosis [6]. The involvement of RAS in SARS-CoV-2 pathogenesis is caused by angiotensin converting enzyme 2 (ACE2) utilization of SARS-CoV-2 by the host cell entrance. Similar to SARS-CoV, SARS-CoV-2 employs ACE2 as a primary entrance receptor to infect the host cells by binding the receptor-binding domain (RBD) of the spike (S) protein to ACE2. Following this interaction, the protease activity of transmembrane protease serine 2 (TMPRSS2) mediates the virus entrance through priming S protein binding to ACE2 [7]. There is widely distributed ACE2 expression in the host cells, such as heart, kidney, testis, liver, gastrointestinal tract, and lung cells [8]. Interestingly, ACE2 is considered a counterbalancing player in RAS by degradation of angiotensin II (AngII), resulting in the overactivity of the classical pathway. Accumulation of Ang II, which results from degradation of AngI by ACE activity, induces devasting effects, including hypertension, inflammation, and fibrosis via activation of the angiotensin II type 1 receptor (ATR1). In contrast, ACE2 produces protective peptides from AngII degradation, Ang 1–7, to induce the MasR receptor, resulting in damping of the ACE/AngII/ATR1 arm and related complications, especially inflammation and fibrosis [9]. It is noticed that ACE2 is down-regulated after a COVID-19 infection [10]. This event could explain the lung failure and severity of SARS-CoV-2 due to loss of ACE2 activity and Dominance of ACE/AngII/ATR1 arms in acute respiratory distress syndrome (ARDS) [5]. Interestingly, the host's susceptibility to COVID-19 infection and disease development can be controlled by the GI tract and colonized microbial community called "gut microbiota" [5]. It has been proven that the establishment of lung immunity and its power to combat respiratory pathogens is governed by gut microbiota in the concept of a gut-lung axis [11].
Analysis of human transcriptomics reported the co-expression of ACE2 and TMPRSS2 in the lung alveolar cells, ileum, and colon absorptive enterocytes. Therefore, the GI tract route of the SARS-CoV-2 entrance is documented beside the respiratory tract [12]. Furthermore, RAS components are expressed in the GI and affect GI tract functions, immune response, and gut microbiota composition [13]. In this regard, several evidences demonstrate the pivotal role of ACE2 in the involvement of amino acid absorption, release of immune mediators, antimicrobial peptides, and gut microbiota composition. Also, the pathogenesis role of AngII/AGTR1 in intestinal inflammation has been reported [14].
Regarding the importance of microbiota, the composition of gut and lung microbiota has been studied in COVID-19 patients. These studies reported significant reductions and elevation of anti-inflammatory microbiota members and opportunistic pathogens in the gut and lung. The pivotal role of anti-inflammatory symbionts has been demonstrated, such as Akkermansia muciniphila, Faecalibacterium prausnitzii, Bacteroides thetaiotaomicron, and Bacteroides fragilis, based on their metabolites and direct immune cell interaction to modulate local and systemic inflammation [[15], [16], [17], [18], [19]]. This putative role is mediated by bacterial derivatives such as inactivated bacteria (paraprobiotics), metabolites, and components called postbiotics to regulate the host-microbiota crosstalk [20].
On the other hand, there is a possible link between the gut microbiota and ACE2 expression. Therefore, we can consider the gut microbiota as a potent target that can control the immune response, lung function, and ACE2 expression [5].
Considering the importance of gut microbiota members and RAS components in the determination of the susceptibility of the host to SARS-CoV-2 infection and also disease development, we studied for the first time the direct effect of gut microbiota members and their postbiotics on the expression of ACE, ACE2, AGTR1 and TMPRSS2 on Caco2 cells as a human intestinal epithelium model. Moreover, since the studied genes play essential roles in SARS-CoV-2 pathogenesis and COVID-19 development, we analyzed the status of their methylation and pathway enrichment between COVID-19 and healthy subjects to determine their importance in SARS-CoV-2 pathogenesis. Finally, the related miRNAs to GO terms concerned on the studied genes which were resulted from enrichment analysis were identified.
2. Materials and methods
2.1. Bacterial strains, growth condition and postbiotics preparation
Akkermansia muciniphila (DSM 22959), Faecalibacterium prausnitzii strain A2-165 (DSM17677), Bacteroides thetaiotaomicron (CCUG 10774) and Bacteroides fragilis (ATCC 23745) were purchased from the DSMZ institute (German collection of microorganisms and cell cultures). These bacteria were cultured in brain heart infusion (BHI) broth and agar containing with hemin (5 μg/ml), menadione (1 μg/ml), L-cystein and related supplementing (0.5% mucin for A. muciniphila, LYBHI for F. prausnitzii) and incubated at 37 °C under anaerobic conditions (80% N2, 10% Co2, and 10% H2) using an Anoxomat™ MARK II system [[21], [22], [23]]. In this study we prepared cell free supernatants (CFS) and heat inactivated (paraprobiotic) as postbiotics. To prepare CFS, bacteria were anaerobically cultured until logarithmic phase based on the optical density at 600 nm ≥ 1 [24, 25]. CFS was prepared by centrifugation at 12000g for 20 min at 4 °C and filtered through a 0.22 μm-pore-size filter (Millipore, Rockville, Maryland, USA). After harvesting fresh bacterial pellets in log phase, centrifugation at 11000 rpm for 20 min and washing with sterile PBS twice, the bacterial suspension was heat inactivated at 70 °C for 30 min. Heat inactivated bacteria were inoculated on BHI supplemented agar for at least one week at 37 °C under anaerobic conditions in order to perform the viability test [26].
2.2. Caco-2 cell culture
The human Caucasian colon adenocarcinoma, Caco-2 cell line (IBRC C10094) was purchased from the Iranian Biological Resource Center. Caco-2 cells were cultured in high-glucose Dulbecco's modifed Eagle's medium (DMEM) (Gibco Life Technologies, Paisley, UK) containing 10% heat-inactivated fetal bovine serum (FBS) (Gibco Life Technologies, Paisley,UK) 1%penicillin/streptomycin, 1% (Gibco Life Technologies, Paisley, UK) and 1% Non-essential amino acids (GibcoLife Technologies, Paisley, UK) at 37 °C under 5% CO2 atmosphere. Cells were seeded in 6-well-plates (Sorfa, Zhejiang, China) at a density of 5.0 × 105 cells per well [21]. Cells were treated with bacteria in live and heat-inactivated form at a multiplicity of infection (MOI): 50 and 100, and also 10% V/V CFS for overnight.
2.3. RNA extraction, cDNA synthesis and RT-qPCR analysis
Total RNA was extracted by RNX-Plus (CinnaGen, Iran). After assessment of RNA quantity and quality respectively evaluated by NanoDrop 2000 (Thermo Fisher Scientific, USA) and gel electrophoresis, cDNAs were synthesized by first strand cDNA synthesis kit (Thermo Scientific, USA) according to manufacturers’ instructions. RT-qPCR was performed by Rotor-Gene Q MDX (QIAGEN Hilden, Germany). The expression level of ACE, AGTR1, ACE2 and TMPRSS2 transcripts were determined using SYBR Premix Ex Taq II (Takara, China) and specific primers which were presented in Table 1 . All experiments were performed in duplicate. The relative mRNA levels in studied genes expression were normalized to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression, as internal control [21].
Table 1.
The sequence of forward and reverse primers.
| Primers | Sequence 5′→3 | Reference |
|---|---|---|
| GAPDH Forward | GAGGGCCATCCACAGTCTTCTG | [21] |
| GAPDH Reverse | CCCTTCATTGACCTCAACTACATGGT | |
| ACE Forward | GGAGGAATATGACCGGACATCC | [27] |
| ACE Reverse | TGGTTGGCTATTTGCATGTTCTT | |
| ACE2 Forward | AACCCAGATAATCCACAAGAATGC | The current study |
| ACE2 Reverse | TCATAGTCTCCTCTCCAATAATCCC | |
| AGTR1 Forward | ATTTAGCACTGGCTGACTTATGC | [28] |
| AGTR1 Reverse | CAGCGGTATTCCATAGCTGTG | |
| TMPRSS2 Forward | TCATCCTTCAGGTGTACTCATCTC | The current study |
| TMPRSS2 Reverse | TCCGCTGTCATCCACTATTCC |
2.4. DNA methylation analysis of ACE, AGTR1, ACE2 and TMPRSS2 genes on the COVID-19 and non-COVID-19 samples
We searched on Gene Expression Omnibus (GEO) database to find related GEO Series (GSE) for DNA methylation analysis between Covid and non-Covid statues. We selected “GSE174818” [29,30] for our analysis specifically to know the ACE, ACE2, AGTR1, TMPRSS2 methylation state in Covid vs non-Covid samples. Partek® Genomics Suite® software, v7.0 [31]. was used and extracted data were visualized in chromosome mapping view.
2.5. Pathways enrichment analysis of COVID-19 patients compared with healthy control
We searched on GEO database and selected 2 studies out of 342 studies of SARS-CoV-2 data (GSE164805(30) and GSE166552(32)) then we made GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) analysis between Covid and non-Covid samples. We identified differentially expressed genes (DEGs) between COVID-19 and non-COVID-19 GSMs. Gene ontology (GO) for biological processes (BP), molecular function (MF), and cellular component (CC) in addition to KEGG pathway and enrichment analyses of differential gene expressions (DEGs) were obtained using the Database for Annotation Visualization and Integrated Discovery (DAVID6.8; https://david.ncifcrf.gov) for up- and down regulated DEGs. P-values of less than 0.01 for each GO term which were obtained from GSE164805 and GSE166552 were considered statistically significant and included in the study. DAVID6.8 was also used to analyze both GO ontology and the KEGG pathway of the modules.
2.6. Construction of protein-protein interaction (PPI) network based on the considering ACE, AGTR1, ACE2, TMPRSS2 genes
To determine the interaction of studied genes (ACE, AGTR1, ACE2, TMPRSS2) with DEGs resulted from the microarray analysis of GSE164805 and GSE166552, the Search Tool for the Retrieval of Interacting Genes (STRING; version 11; string-db.org/) database was used. Visualization of the protein-protein interaction (PPI) networks was performed by Cytoscape (version 3.9, http://www.cytoscape.org/). The Cytoscape String- App was used to retrieve the functional enrichment of up- and downregulated. Key modules in the PPI network were identified by using the Cytoscape plugin Molecular Complex Detection (MCODE, version 2). A node score and node number were logFC values 4 of DEG were considered to construction of PPI network and functional enrichment analysis.
2.7. MicroRNA analysis of ACE, AGTR1, ACE2 and TMPRSS2 genes
In order to identify microRNAs regulating ACE, ACE2, AGTR1, TMPRSS2 genes expression, we used four different in silico tools. MirDB (miRDB - MicroRNA Target Prediction Database), TargetScan (Human,7.2,TargetScanHuman 7.2), miRTarBase(miRTarBase: the experimentally validated microRNA-target interactions database (cuhk.edu.cn)), miRWalk(Home - miRWalk (uni-heidelberg.de)) and multimiR package in R with default settings [33]. After gathering Data for each gene, we assumed some criteria base on each tools recommendation and reviewed literature to select target microRNAs [[34], [35], [36], [37], [38], [39], [40]]. We used venny diagram to find commons microRNAs between genes. Also, GO terms related to the studied genes which were extracted from enrichment analysis of GSE164805 and GSE166552 were examined in miRPath reverse search DIANA TOOLS - mirPath v.3 (grnet.gr) to find related microRNAs [41]. Then we used venny between our microRNAs analysis from MirDB, TargetScan, miRTarBase, miRWalk, multimiR package and the results of miRPath.
2.8. Statistics
Statistical analyses were performed by with GraphPad Prism software version 8 (GraphPad Software, Inc., CA, USA). Independent sample one-way ANOVA was used. Results are showed as means ± standard errors (SEM) and P-value<0.05. Differences were considered statistically significant when P < 0.05; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
3. Results
3.1. The effects of A. muciniphila, F. prausnitzii, B. thetaiotaomicron, B. fragilis and their postbiotics on the expression of ACE, AGTR1, ACE2 and TMPRSS2 genes
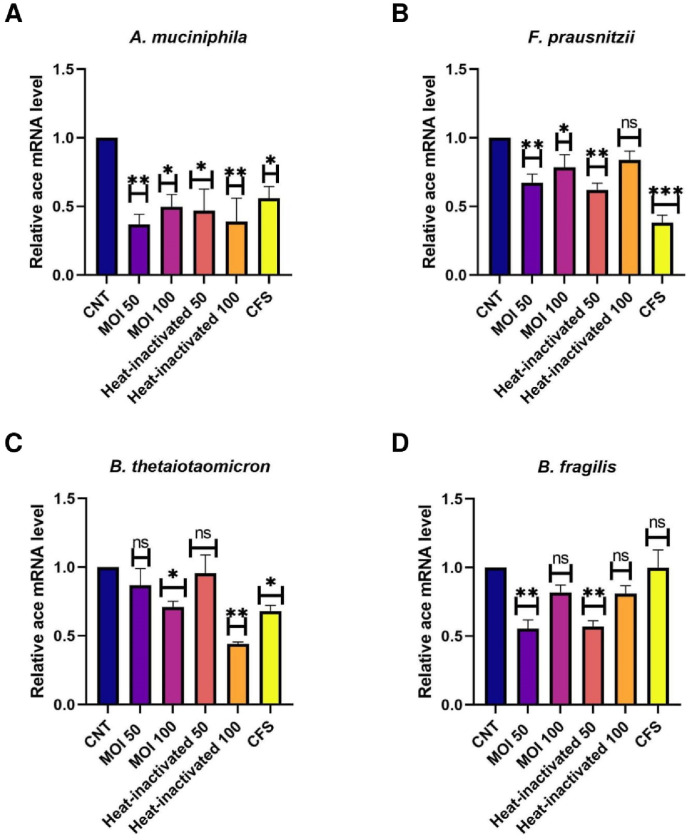
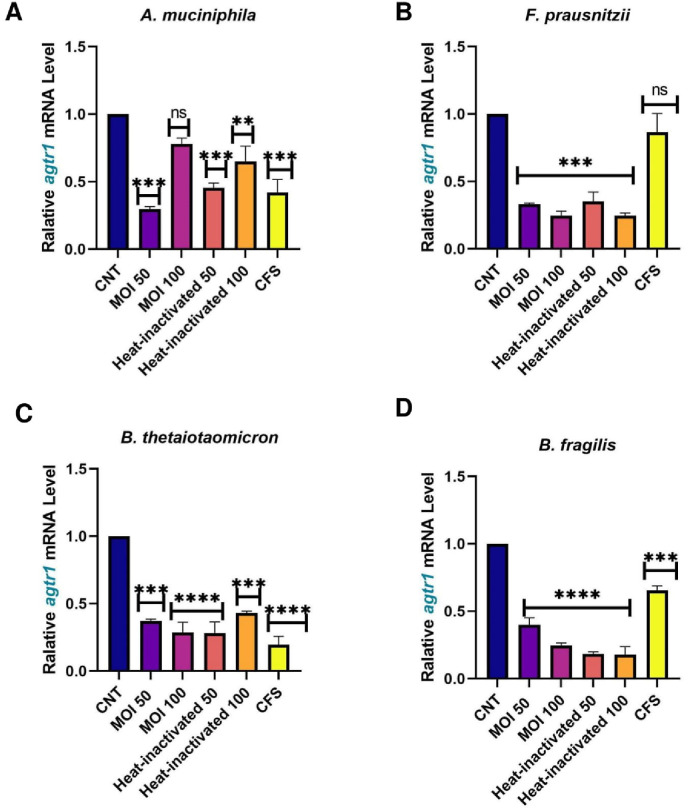
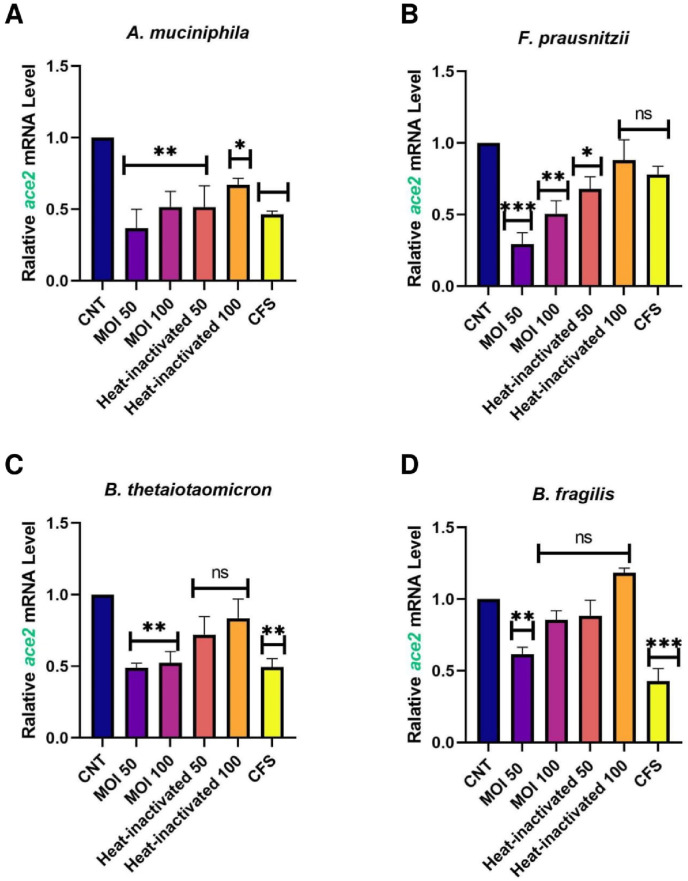
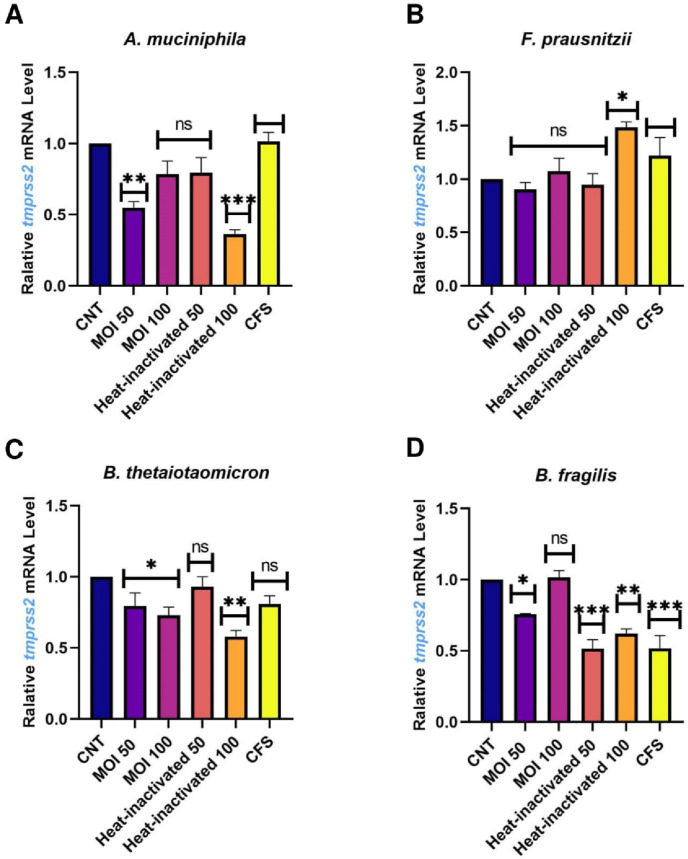
Caco-2 cells were used as a human intestinal epithelial cells model to investigate the effect of A. muciniphila, F. prausnitzii, B. thetaiotaomicron and B. fragilis, on the expression of the studied genes. Due to the importance of postbiotics in the crosstalk of gut microbiota members with the host, we also treated heat-inactivated form and CFS as postbiotics of the studied bacteria with Caco-2 cell to more elucidate the putative interaction. As shown in Fig. 1 , ACE transcript was remarkably downregulated by A. muciniphila (at MOI50-100, MOI50 heat-inactivated and CFS), F. prausnitzii (at MOI50, MOI100, MOI50 heat-inactivated and CFS), B. thetaiotaomicron (MOI100, MOI100 heat-inactivated and CFS), B. fragilis (both MOI as live and heat-inactivated form). Also, the studied bacteria and their postbiotics (unlike MOI100, CFS from A. muciniphila and CFS of F. prausnitzii) significantly reduced the gene expression of AGTR1 (Fig. 2 ). In spite of B. fragilis heat-inactivated (at MOI100) which significantly upregulated ACE2 transcripts, we identified reduction of Ace2 genes expression in the cells treated with the studied bacteria and their derivatives (Fig. 3 ). The TMRSS2 mRNA levels were reduced by A. muciniphila, F. prausnitzii, B. thetaiotaomicron, B. fragilis and their postbiotics in the cells unlike TMRSS2 transcripts significant upregulation resulted from heat-inactivated F. prausnitzii (at MOI100) (Fig. 4 ). In overall, the genes expression of studied genes was influenced by mentioned bacteria and derivatives at the same manner favored to reducing of COVID-19 infection in human intestinal epithelial cells.
Fig. 1.
Effect of A. muciniphila, F. prausnitzii, B. thetaiotaomicron and B. fragilis on ACE mRNA expression. Caco-2 cells were initially deprived of serum and then treated with A. A. muciniphila, B. F. prausnitzii, C. B. thetaiotaomicron and D. B. fragilis as live, heat-inactivated at multiplicity of infection (MOI) = 50,100 and 10% V/V CFS for overnight. phosphate buffer solution (PBS) and specific supplemented BHI were treated as control. Values of triplicate experiments are demonstrated as mean ± standard errors (SEM). Statistically Significant results are presented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Fig. 2.
Effect of A. muciniphila, F. prausnitzii, B. thetaiotaomicron and B. fragilis on AGTR1 mRNA expression. Caco-2 cells were initially deprived of serum and then treated with A. A. muciniphila, B. F. prausnitzii, C. B. thetaiotaomicron and D. B. fragilis as live, heat-inactivated at multiplicity of infection (MOI) = 50,100 and 10% V/V CFS for overnight. phosphate buffer solution (PBS) and specific supplemented BHI were treated as control. Values of triplicate experiments are demonstrated as mean ± standard errors (SEM). Statistically Significant results are presented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Fig. 3.
Effect of A. muciniphila, F. prausnitzii, B. thetaiotaomicron and B. fragilis on ACE2 mRNA expression. Caco-2 cells were initially deprived of serum and then treated with A. A. muciniphila, B. F. prausnitzii, C. B. thetaiotaomicron and D. B. fragilis as live, heat-inactivated at multiplicity of infection (MOI) = 50,100 and 10% V/V CFS for overnight. phosphate buffer solution (PBS) and specific supplemented BHI were treated as control. Values of triplicate experiments are demonstrated as mean ± standard errors (SEM). Statistically Significant results are presented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Fig. 4.
Effect of A. muciniphila, F. prausnitzii, B. thetaiotaomicron and B. fragilis on TMPRSS2 mRNA expression. Caco-2 cells were initially deprived of serum and then treated with A. A. muciniphila, B. F. prausnitzii, C. B. thetaiotaomicron and D. B. fragilis as live, heat-inactivated at multiplicity of infection (MOI) = 50,100 and 10% V/V CFS for overnight. phosphate buffer solution (PBS) and specific supplemented BHI were treated as control. Values of triplicate experiments are demonstrated as mean ± standard errors (SEM). Statistically Significant results are presented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
3.2. DNA methylation state of ACE, AGTR1, ACE2 and TMPRSS2 genes on the COVID-19 patients compared with healthy subjects
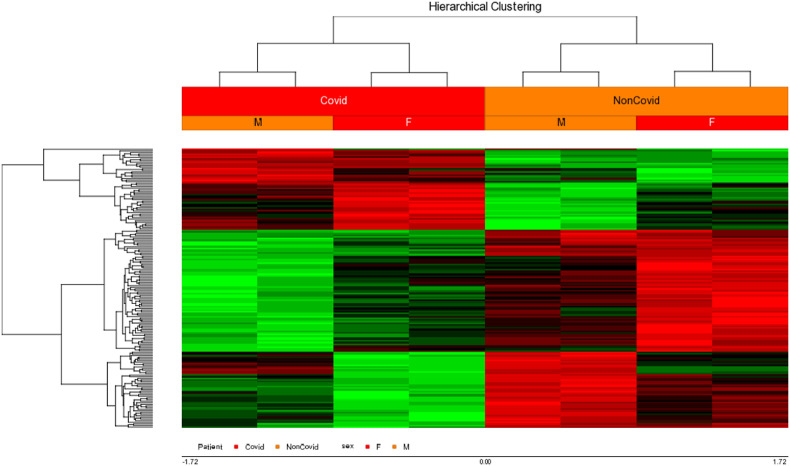
DNA methylation pattern was investigated on GSE174818 in COVID-19 patients in comparison to healthy controls (Fig. 5 ).
Fig. 5.
DNA methylation analysis on COVID-19 patients compared with healthy control from GSE174818. Hierarchical clustering heatmap of the significantly differentially methylated CPG sites, containing a hypermethylated (red color) versus hypomethylated (green color) signature. X axis labels: number of differentially methylated CPG positions (DMPs) from each gene.
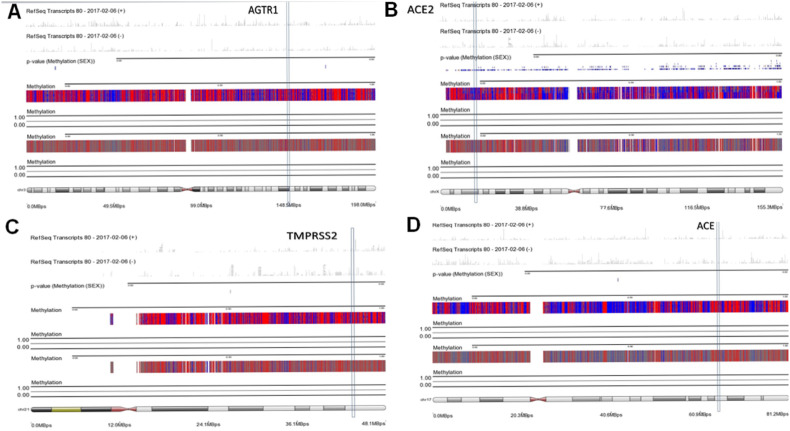
According to the importance of ACE, AGTR1, ACE2 and TMPRSS2 genes on the COVID-19 infection and development, we also specifically checked the DNA methylation of the mentioned genes. It was identified that all of them were hypomethylated in both groups (Fig. 6 ).
Fig. 6.
Chromosomal methylation mapping view of ACE, ACE2, TMPRSS2, AGTR1.
3.3. Pathways enrichment analysis of COVID-19 patients compared with healthy control
Two microarray datasets, GSE164805 [42] and GSE166552 [32], were considered for construction of GEO2R in order to identify DEGs between COVID-19 and non-COVID-19 samples. Based on our analysis, 45 upregulated and 154 downregulated DEGs derived from GSE166552 and 2094 upregulated and 1921 downregulated DEGs resulted from GSE164805 (by taking the DEGs adjusted on P-values of less than 0.01). To find upregulated and downregulated DEGs based on the GO between COVID-19 and healthy control, BP, CC, MF and KEGG enrichment analysis was performed by DAVID platform. We demonstrated in detail, BP, CC, MP and KEGG-up regulated and down regulated DEGs in COVID-19 compared with non-COVID-19 samples in supplementary tables 1-8.
It was determined that multicellular organism development, immune response, cell-cell signaling, response to virus, DNA damage checkpoint, regulation of systemic blood pressure, regulation of vasodilation, regulation of vasoconstriction and angiotensin maturation were BP-upregulated DEGs in COVID-19 samples. in contrast, translation, viral transcription, signal transduction, immune response, cell-cell signaling were BP-downregulated DEGs in COVID-19 samples. Based on the KEGG pathway enrichment analysis RAS system was upregulated in COVID-19 patients compared with healthy control.
3.4. PPI network construction based on the interaction of ACE, AGTR1, ACE2 and TMPRSS2 with DEGs derived from COVID-19 patients
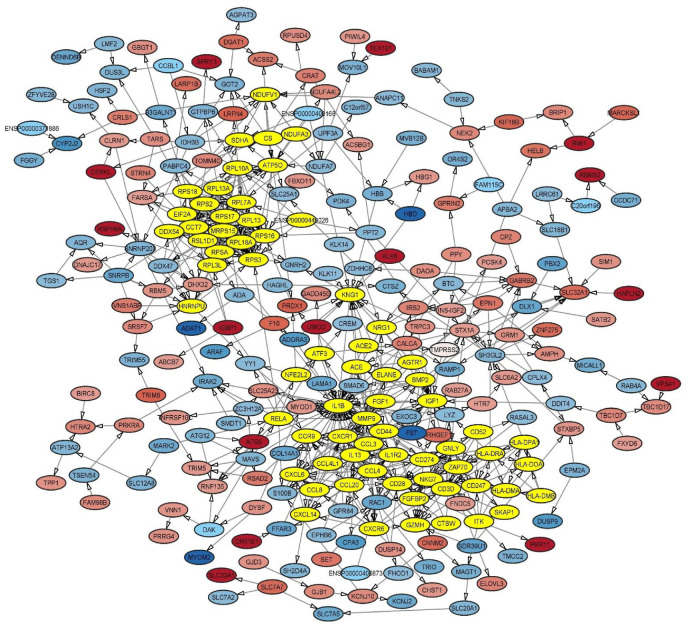
According to the importance of ACE, AGTR1, ACE2 and TMPRSS2 genes in the determination of the host susceptibility to SARS-CoV-2 infection and the COVID-19 severity (based on the review literature), we aimed to determination the interaction of these genes with DEGs derived from COVID-19 samples adjusted on the LogFC . After PPI construction with MCOD, it was determined that ACE, AGTR1 and ACE2 are being as hub nodes by considering both node score and node number (Fig. 7 ).
Fig. 7.
PPI network of DEGs from GSE164805 and GSE166552 enrichment analysis with MCODE.
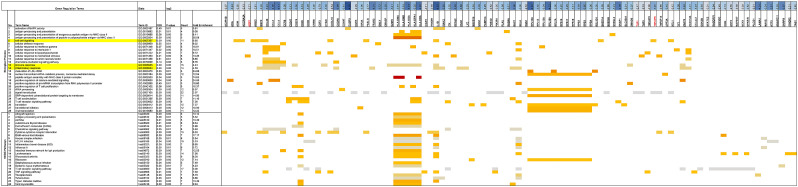
The enrichment analysis DEGs derived from COVID-19 patients compared with healthy control, ACE, ACE2 and AGTR1 had highest fold enrichment compared with another DEGs in the up regulation of systemic arterial blood pressure by renin-angiotensin biological process and RAS system. Also, regulation of vasoconstriction and regulation of vasodilation were BP-upregulated which highest fold enrichment belong to ACE, ACE2 and AGTR1. Our enrichment analysis demonstrated the pivotal role of ACE, ACE2 and AGTR1 genes in the upregulated process in COVID-19 patients. TMPRSS2 is not located as a hub node and has the moderate fold enrichment which participated on the upregulation of protein auto processing in COVID-19 patients (Fig. 8, Fig. 9 ).
Fig. 8.
Up-regulated terms by considering the interaction of ACE, ACE2, TMPRSS2, AGTR1 in COVID-19 patents in compared with healthy control.
Fig. 9.
Down-regulated terms by considering the interaction of ACE, ACE2, TMPRSS2, AGTR1 in COVID-19 patents in compared with healthy control.
3.5. Network analysis of microRNs targeting ACE, AGTR1, ACE2 and TMPRSS2 genes
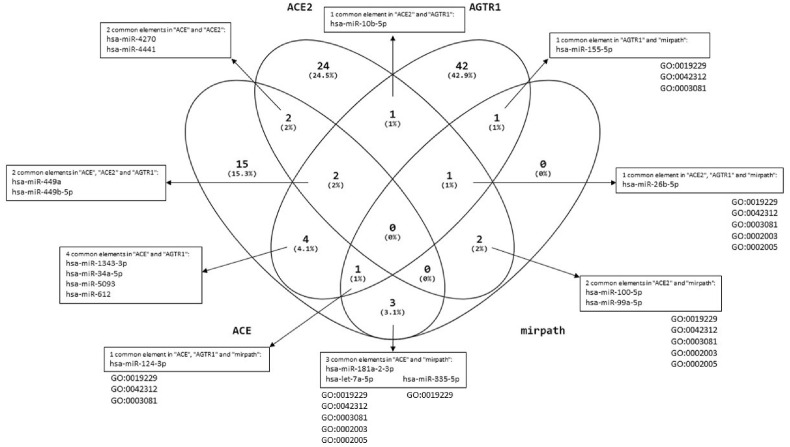
We extracted miRNAs targeting ACE, AGTR1, ACE2 and TMPRSS2 using MirDB, TargetScan, miRTarBase and miRWalk, multimiR package in R (Fig. 10 ). Since, we reported nearly the same reduction of the studied genes, we aimed to determine common miRNAs between the genes. In this way, the shared miRNA between these genes were determined by Venn diagram as shown in Fig. 10. Based on our analysis, hsa-miR-27a-3p, hsa-miR-130a-3p and hsa-miR-3908 were shared miRNAs between ACE2 and TMPRSS2 genes. Also, hsa -miR-10b-5p and hsa -miR-26b-5p were common between ACE2 and AGTR1. ACE had shared regulating miRNAs with TMPRSS2 (hsa -miR5001-5p and hsa -miR-let-7a-5p) and AGTR1 (hsa -miR-5093, hsa -miR-1343-3p, has-miR-612, has-miR-124-39). hsa -miR-let-7b-5p was a common miRNA which can regulate TMPRSS2 and AGTR1 genes. ACE, ACE2 and AGTR1 could be regulated by hsa -miR-449b-5p and hsa -miR-449a. Also, hsa -miR-4441 and hsa -miR-4270 were common regulating miRNAs between ACE, ACE2 and TMPRSS2 (Fig. 10).
Fig. 10.
miRNAs targeting ACE, ACE2, AGTR1 and TMPRSS2 and common agents between them.
Also, we evaluated functional miRNAs related to GO terms from enrichment analysis (related to the studied genes) which were derived from two studied GSE164805 and GSE166552 between COVID-19 and non-COVID-19 samples. Related microRNAs with GO:0003081, GO:0042312, GO:0019229, GO:0002003 and hsa04614 were extracted by miRPath. Finally, we used Venn diagrams between our microRNAs analysis from MirDB, TargetScan, miRTarBase, miRWalk, multimiR package and the results of miRPath (Fig. 11 ).
Fig. 11.
microRNAs results related to GOs based on enrichment analysis of “GSE164805, GSE166552” between COVID-19 patients and healthy control in mirPath v.3 from DIANAtools.
4. Discussion
Various factors determine the host susceptibility to SARS-CoV-2 and the severity of COVID-19 infection, including the RAS system, immune response, and gut microbiota. These agents have an essential role in preserving host homeostasis, lung function, and systemic immunity [5]. Despite the proven role of gut microbiota on the regulation of lung and systemic immunity and the pathogenesis of respiratory pathogens, there is no study on the direct and specific gut microbiota members and correlation with genes involved in SARS-CoV-2 pathogenesis. Furthermore, there is a lack of studies concerning their correlation between RAS and human gut microbiota members. Accordingly, we aimed to study the specific correlation between A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis as influential regulating host functions on the genes involved in SARS-CoV-2 entrance and COVID-19 severity, ACE, AGTR1, ACE2, and TMPRSS2 on the Caco-2 cell line for the first time. To determine the role of studied genes, we analyzed the state of DNA methylation of the studied genes on the raw data of COVID-19 patients. We analyzed enrichment pathways/PPI networks by considering the impact of the studied genes. Finally, the targeting miRNAs of these genes were identified from related GO terms from the enrichment analysis.
Because gut microbiota and the majority of immune cells reside in the GI tract, it plays a role in health and disease. Gut microbiota demonstrated an establishing and regulating effect on immunity in the gut and other organs such as the lung by establishing and regulating the gut-lung immune system by governing the gut-lung axis [43]. In this way, the frequency of anti-inflammatory gut microbiota members could influence the lung immunity against respiratory pathogens through various routes, such as regulation of gut barrier function, short chain fatty acids (SCFAs) production, hematopoiesis, and migration of sensitized immune cells to the lungs [5,44]. There is growing attention to understanding the gut microbiota's role in COVID-19 [[45], [46], [47], [48], [49]]. Based on the studies, it has been suggested that the entrance of SARS-CoV-2 to alveolar cells could disrupt the gut-lung axis, which in turn results in gut microbiota dysbiosis, dysfunction of gut barrier, and penetration of the virus into ACE-2 expressing enterocytes. These events explain the GI manifestations in COVID-19 and the presence of SARS-CoV-2 RNA in the fecal samples of patients [50]. Several studies report the depletion of anti-inflammatory bacteria in the gut microbiota composition of patients compared to healthy controls [[51], [52], [53], [54]]. For example, Zuo et al. analyzed the gut microbiota composition of COVID-19 and the fecal shedding of SARS-CoV-2 patients during hospitalization in association with disease severity. They reported a significant alteration in these patients with reduced F. prausnitzii frequency, which is inversely correlated with COVID-19 severity. Also, the inverse association between fecal shedding of the virus and Bacteroides spp. such as B. thetaiotaomicron has been described [55].
The bioinformatic analysis based on the single-cell transcriptome reported as potential target for SARS-CoV-2 due to ACE2 expression in the ileum and colon enterocytes [12]. The RAS components, especially ACE2, ACE, and AGTR1, is expressed in GI tract and influences immunity and gut microbiota dysbiosis [56]. Therefore, it is possible that there is a correlation between gut microbiota members and the gene expression of ACE, AGTR1, and ACE2. In this regard, we treated A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis, anti-inflammatory bacteria, and their derivatives as postbiotics on Caco-2 cells to investigate the effect on the putative genes. Also, there are reports that ACE2 and TMPRSS2, which mediate the viral entrance, are expressed in the GI tract. Therefore, we studied the alteration of TMPRSS2 gene expression in this study.
We reported for the first time that studied bacteria and their derivatives were able to downregulate ACE, ACE2, and AGTR1 on Caco-2 cells. ACE and AGTR1 belong to the deleterious arm of RAS, whose activation results in increased inflammation, fibrosis, and hypertension [57]. Our obtained results could be a complementary explanation for the anti-inflammatory features of these bacteria. The relative abundance of A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis is significantly decreased during GI inflammation such as metabolic syndrome, inflammatory bowel diseases (IBD), and irritable bowel syndrome (IBS) [[58], [59], [60]]. The gut barrier function and tuning of the immune system are disrupted due to a dysbiotic gut microbiota and its metabolites, which are caused by inflammatory disorders. Interestingly, the disrupted gut barrier function and increased inflammatory mediators (due to a dysbiotic gut microbiota composition) may be involved in the induction of a cytokine storm in severe COVID-19 patients [61].
On the other hand, the presence of ACE2 could facilitate the increased susceptibility to GI infection by SARS-CoV-2. Zhang et al. also reported the co-expression of two main vital elements, ACE2 and TMPRSS2, driving the viral entrance into lung alveolar and intestinal enterocytes [12]. By studying bacteria and their postbiotics, we reported the downregulation of ACE2 and TMPRSS2 in Caco-2 cells. Our results parallel the only existing study that reported the negative correlation between Bacteroides spp. such as B. dorei, B. thetaiotaomicron, B. massiliensis, and B. ovatus and ACE2 expression in mouse colonocytes [62]. It was noted that the ACE2 function favored the RAS independent role in the GI tract, including mediation of amino acid uptake and gut microbiota composition. Furthermore, the increased ACE2 expression and poor outcome of inflammatory intestinal diseases such as Crohn's disease (CD) have been suggested [63]. Conversely, a report shows that reduced ACE2 is accompanied by a worthwhile effect in CD patients [27]. Suárez-Fariñas et al. emphasized a different distribution of ACE2 expression across the intestine with high expression of ACE2 in colonocytes in inflammatory bowel diseases (IBD). Also, they found overlapping genes in IBD and COVID-19, which suggested the potential effect of IBD medication during the COVID-19 pandemic [64]. Therefore, based on our data reduction of A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis due to loss of inhibitory effect on ACE2 expression may increase the GI tract manifestations and possible oral-fecal route of viral transmission. It can also explain how diabetic and hypertensive patients are in the high-risk group for infection with SARS-CoV-2 due to a dysbiotic gut microbiota with a decrease in A. muciniphila and F. prausnitzii. Taken together, it was noticed that ACE2 has a dual function in SARS-CoV-2 pathogenesis. Although higher levels of ACE2 facilitate viral entrance, preserving expression could dampen inflammation and fibrosis (resulting from disease development) by counterbalancing ACE/AngII/AGTR1 activity. Therefore, it is necessary to more elucidate the interaction between gut microbiota members and the putative genes after disease development in various infected groups with different historical GI complications.
Due to the dominant role of ACE2-TMPRSS2 in the early events of SARS-CoV-2 pathogenesis and the harmful effect of ACE-AGTR1 overactivity in developing infection to a severe state, we determined the DNA methylation of COVID-19 vs. healthy controls. We analyzed the specific DNA methylation of studied genes on data related to genome-wide analysis of circulating blood DNA CpG methylation in COVID-19 [29]. Balnis et al. reported that the global mean methylation profile did not alter in COVID-19 subjects compared with control subjects [29]. Our obtained results showed that the studied genes' DNA methylation was hypomethylated the same as control subjects. Furthermore, we investigated the genes roles and their interactions with other DEGs, obtained from two microarray analyses of COVID-19(32, 42). Our analysis demonstrated the upregulation of RAS with the highest fold enrichment of ACE, ATR1, and ACE2. Also, our pathway enrichment analysis demonstrated the upregulation of AGTR1 accompanied by increased CCL8, CXCL6 and CXCL14, which mediate cell chemotaxis. Khalil et al. reviewed the elevated monocyte-attractant chemokines such as CCL8 and CXCL6 to induce lung inflammation and poor outcome in COVID-19 patients [65]. Since the studied genes are located as hub nodes to control various pathways' gene expression, especially immune response and RAS activity, their downregulation could affect the mentioned pathways. In combination with our experimental data and bioinformatic analysis, we showed the potential of A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis and postbiotics to reduce ACE, ATR1, and ACE2 expression, which are essential genes that drive upregulated biological processes in COVID-19 patients.
Finally, we identified the shared miRNA which target these genes to control expression. We obtained 17 microRNAs in common between the four target genes analyzed, but we believe that the most important microRNAs are miR-124-3p and miR-26b-5p related to GO terms from enrichment analysis. In this case the miR-124-3p was reported to inhibit the Ang II-induced apoptosis and ROS production in HUVECs cells by down-regulating early growth response factor 1 (EGR1) reducing the vascular constriction [66]. Moreover, miR-124-3p was observed to regulate strongly ACE2 gene expression which is correlated to respiratory and cardiovascular disfunction particularly in SARS-CoV-2 patients [34]. A correlation with ACE gene was predicted for miR-26b-5p [67]that it often dysregulated in cardiovascular disease like heart failure [68], myocardial infarction [69] or vascular modifications [70]. We predicted that ACE/ACE2/AGTR1 axis could be targeted by both hsa-miR-449a and hsa-miR-449b that could act as vasodilator or vasoconstriction regulator, as already observed in the pulmonary arterial hypertension (PAH), where has-miR-449a regulates the pulmonary arterial smooth muscle cell proliferation via Myc inhibition [71]. Moreover, we predicted that hsa-miR-4441 and hsa-miR-4270 are common regulators of ACE, ACE2 and TMPRSS2, suggesting their role as new interfering agents in the SARS-CoV-2 infection mechanism. All of these evidences strengthen the idea to use microRNAs for personalized diagnosis and prognosis in COVID-19 disease.
5. Conclusion
In conclusion, we investigated the possible interaction between gut microbiota members and genes involved in SARS-CoV-2 pathogenesis in the Caco-2 cells. Our experimental data showed the significant downregulation of ACE, ATR1, and ACE2 by A. muciniphila, F. prausnitzii, B. thetaiotaomicron, and B. fragilis and postbiotics. Therefore, these bacteria could have a protective role against infection with SARS-CoV-2 besides exerting an anti-inflammatory role. This potential of studied gut microbiota members to reduce the expression of considered genes could be interpreted in two different ways: determination of the host susceptibility to infection (ACE2/TMPRSS2) and disease development by dominancy of inflammation and fibrosis (ACE/AGTR1), which are beneficial against SARS-CoV-2 infection in healthy subjects. Furthermore, the upregulated RAS pathways due to high fold enrichment of ACE, ATR1, and ACE2 in COVID-19 patients confirmed the pivotal role of these putative genes in COVID-19. According to the potential of studied bacteria on the alteration of ACE, AGTR1, ACE2 and TMPRSS2 genes expression, understanding their correlation with demonstrated miRNAs expression could be valuable. The combination of our experimental data and in silico analyses suggested the possible therapeutic potential of targeted gut microbiota intervention to manage the COVID-19 infection.
Data availability
The data used to support the findings in this study are available from the corresponding author upon reasonable request.
Funding statement
This project was supported by a grant [1771] from thePasteur Institute of Iran.
CRediT authorship contribution statement
Sara Ahmadi Badi: Writing – review & editing, Writing – original draft, Validation, Supervision, Project administration, Methodology, Investigation, Data curation, Conceptualization. Amin Malek: Software, Methodology. Alessandro Paolini: Writing – original draft, Software. Mahya Rouhollahi Masoumi: Software, Methodology. Seyed Amirhesam Seyedi: Methodology. Amir Amanzadeh: Methodology. Andrea Masotti: Writing – review & editing, Writing – original draft, Visualization, Validation. Shohreh Khatami: Writing – review & editing, Supervision. Seyed Davar Siadat: Validation, Supervision.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors would like to thank Mycobacteriology and Pulmonary Research Department, Pasteur Institute of Iran Pasteur Institute of Iran.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micpath.2022.105798.
Glossary
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- COVID-19
coronavirus disease-2019
- SARS-CoV
severe acute respiratory syndrome coronavirus
- MERS
Middle East respiratory syndrome
- GI
gastrointestinal
- RAS
renin-angiotensin system
- ACE2
angiotensin converting enzyme 2
- TMPRSS2
transmembrane protease serine 2
- AngII
angiotensin II
- AGTR1
angiotensin II type 1 receptor
- ARDS
acute respiratory distress syndrome
- CFS
cell free supernatants
- Paraprobiotic
heat inactivated
- SCFAs
short chain fatty acids
- IBD
inflammatory bowel diseases
- IBS
irritable bowel syndrome
- CD
Crohn's disease
- MOI
multiplicity of infection
- GEO
Gene Expression Omnibus
- DEGs
differential gene expressions
- GO
Gene ontology
- BP
biological processes
- MF
molecular function
- CC
cellular component
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Rohani P., Badi S.A., Moshiri A., Siadat S.D. Coronavirus disease 2019 (COVID-19) and pediatric gastroenterology. Gastroenterol. Hepatol. Bed to Bench. 2020;13(4):351. [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza a respiratory viruses. Front. Immunol. 2020;11:2309. doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liya G., Yuguang W., Jian L., Huaiping Y., Xue H., Jianwei H., et al. Studies on viral pneumonia related to novel coronavirus SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV: a literature review. Apmis. 2020;128(6):423–432. doi: 10.1111/apm.13047. [DOI] [PubMed] [Google Scholar]
- 4.Villapol S. Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl. Res. 2020;226:57–69. doi: 10.1016/j.trsl.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmadi Badi S., Tarashi S., Fateh A., Rohani P., Masotti A., Siadat S.D. Mediators of Inflammation; 2021. From the Role of Microbiota in Gut-Lung Axis to SARS-CoV-2 Pathogenesis; p. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fyhrquist F., Saijonmaa O. Renin‐angiotensin system revisited. J. Intern. Med. 2008;264(3):224–236. doi: 10.1111/j.1365-2796.2008.01981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Zhou W., Yang L., You R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong M., Zhang J., Ma X., Tan J., Chen L., Liu S., et al. Biomedicine & Pharmacotherapy; 2020. ACE2, TMPRSS2 Distribution and Extrapulmonary Organ Injury in Patients with COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng J., Xiao G., Zhang J., He X., Ou M., Bi J., et al. Renin-angiotensin system inhibitors improve the clinical outcomes of COVID-19 patients with hypertension. Emerg. Microb. Infect. 2020;9(1):757–760. doi: 10.1080/22221751.2020.1746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leach D., Mohr A., Giotis E., Cil E., Isac A., Yates L., et al. The antiandrogen enzalutamide downregulates TMPRSS2 and reduces cellular entry of SARS-CoV-2 in human lung cells. Nat. Commun. 2021;12(1):1–12. doi: 10.1038/s41467-021-24342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumas A., Bernard L., Poquet Y., Lugo‐Villarino G., Neyrolles O. The role of the lung microbiota and the gut–lung axis in respiratory infectious diseases. Cell Microbiol. 2018;20(12) doi: 10.1111/cmi.12966. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., et al. Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69(6):1010–1018. [Google Scholar]
- 13.Garg M., Angus P.W., Burrell L.M., Herath C., Gibson P.R., Lubel J.S. The pathophysiological roles of the renin–angiotensin system in the gastrointestinal tract. Aliment Pharmacol. Therapeut. 2012;35(4):414–428. doi: 10.1111/j.1365-2036.2011.04971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katada K., Yoshida N., Suzuki T., Okuda T., Mizushima K., Takagi T., et al. Dextran sulfate sodium-induced acute colonic inflammation in angiotensin II type 1a receptor deficient mice. Inflamm. Res. 2008;57(2):84–91. doi: 10.1007/s00011-007-7098-y. [DOI] [PubMed] [Google Scholar]
- 15.Derrien M., Belzer C., de Vos W.M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Zhai Q., Feng S., Arjan N., Chen W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2019;59(19):3227–3236. doi: 10.1080/10408398.2018.1517725. [DOI] [PubMed] [Google Scholar]
- 17.He X., Zhao S., Li Y. Faecalibacterium prausnitzii: a next-generation probiotic in gut disease improvement. Can. J. Infect Dis. Med. Microbiol. 2021:2021. [Google Scholar]
- 18.Hiippala K., Kainulainen V., Suutarinen M., Heini T., Bowers J.R., Jasso-Selles D., et al. Isolation of anti-inflammatory and epithelium reinforcing Bacteroides and Parabacteroides spp. from a healthy fecal donor. Nutrients. 2020;12(4):935. doi: 10.3390/nu12040935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blandford L.E., Johnston E.L., Sanderson J.D., Wade W.G., Lax A.J. Promoter orientation of the immunomodulatory Bacteroides fragilis capsular polysaccharide A (PSA) is off in individuals with inflammatory bowel disease (IBD) Gut Microb. 2019;10(5):569–577. doi: 10.1080/19490976.2018.1560755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuevas-González P., Liceaga A., Aguilar-Toalá J. Postbiotics and paraprobiotics: from concepts to applications. Food Res. Int. 2020 doi: 10.1016/j.foodres.2020.109502. [DOI] [PubMed] [Google Scholar]
- 21.Badi S.A., Khatami S., Irani S., Siadat S.D. Induction effects of bacteroides fragilis derived outer membrane vesicles on toll like receptor 2, toll like receptor 4 genes expression and cytokines concentration in human intestinal epithelial cells. Cell J. (Yakhteh) 2019;21(1):57. doi: 10.22074/cellj.2019.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabiei N., Badi S.A., Marvasti F.E., Sattari T.N., Vaziri F., Siadat S.D. Induction effects of Faecalibacterium prausnitzii and its extracellular vesicles on toll-like receptor signaling pathway gene expression and cytokine level in human intestinal epithelial cells. Cytokine. 2019;121 doi: 10.1016/j.cyto.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Ashrafian F., Shahriary A., Behrouzi A., Moradi H.R., Keshavarz Azizi Raftar S., Lari A., et al. Akkermansia muciniphila-derived extracellular vesicles as a mucosal delivery vector for amelioration of obesity in mice. Front. Microbiol. 2019;10:2155. doi: 10.3389/fmicb.2019.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antunes L.C.M., Ferreira L.Q., Ferreira E.O., Miranda K.R., Avelar K.E.S., Domingues R.M.C.P., et al. Bacteroides species produce Vibrio harveyi autoinducer 2-related molecules. Anaerobe. 2005;11(5):295–301. doi: 10.1016/j.anaerobe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Jafari B., Nejad R.A.K., Vaziri F., Siadat S.D. Evaluation of the effects of extracellular vesicles derived from Faecalibacterium prausnitzii on lung cancer cell line. Biologia. 2019;74(7):889–898. [Google Scholar]
- 26.Ashrafian F., Keshavarz Azizi Raftar S., Shahryari A., Behrouzi A., Yaghoubfar R., Lari A., et al. Comparative effects of alive and pasteurized Akkermansia muciniphila on normal diet-fed mice. Sci. Rep. 2021;11(1):1–13. doi: 10.1038/s41598-021-95738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang H., Xu C., Lu A., Zou C.-J., Xie S., Chen Y., et al. (Pro) renin receptor mediates albumin-induced cellular responses: role of site-1 protease-derived soluble (pro) renin receptor in renal epithelial cells. Am. J. Physiol. Cell Physiol. 2017;313(6):C632–C643. doi: 10.1152/ajpcell.00006.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee D.K., Nevo O. Microvascular endothelial cells from preeclamptic women exhibit altered expression of angiogenic and vasopressor factors. Am. J. Physiol. Heart Circ. Physiol. 2016;310(11):H1834–H1841. doi: 10.1152/ajpheart.00083.2016. [DOI] [PubMed] [Google Scholar]
- 29.Balnis J., Madrid A., Hogan K.J., Drake L.A., Chieng H.C., Tiwari A., et al. Blood DNA methylation and COVID-19 outcomes. Clin. Epigenet. 2021;13(1):1–16. doi: 10.1186/s13148-021-01102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corley M.J., Pang A.P., Dody K., Mudd P.A., Patterson B.K., Seethamraju H., et al. Genome‐wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID‐19. J. Leukoc. Biol. 2021 doi: 10.1002/JLB.5HI0720-466R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Zhou W., McReynolds L.J., Katki H.A., Griffiths E.A., Thota S., et al. Prognostic impact of pre-transplant chromosomal aberrations in peripheral blood of patients undergoing unrelated donor hematopoietic cell transplant for acute myeloid leukemia. Sci. Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-94539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y., Zhao T., Deng R., Xia X., Li B., Wang X. A study of differential circRNA and lncRNA expressions in COVID-19-infected peripheral blood. Sci. Rep. 2021;11(1):1–14. doi: 10.1038/s41598-021-86134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ru Y., Kechris K.J., Tabakoff B., Hoffman P., Radcliffe R.A., Bowler R., et al. The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42(17):e133–e. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wicik Z., Eyileten C., Jakubik D., Simões S.N., Martins D.C., Pavão R., et al. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors. J. Clin. Med. 2020;9(11):3743. doi: 10.3390/jcm9113743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-coding RNA res. 2020;5(4):153–166. doi: 10.1016/j.ncrna.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roganović J.R. microRNA‐146a and‐155, upregulated by periodontitis and type 2 diabetes in oral fluids, are predicted to regulate SARS‐CoV‐2 oral receptor genes. J. Periodontol. 2021;92(7):e35–e43. doi: 10.1002/JPER.20-0623. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay D., Mussa B.M. Identification of novel hypothalamic microRNAs as promising therapeutics for SARS-CoV-2 by regulating ACE2 and TMPRSS2 expression: an in silico analysis. Brain Sci. 2020;10(10):666. doi: 10.3390/brainsci10100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce J.B., Simion V., Icli B., Pérez-Cremades D., Cheng H.S., Feinberg M.W. Computational analysis of targeting SARS-CoV-2, viral entry proteins ACE2 and TMPRSS2, and interferon genes by host MicroRNAs. Genes. 2020;11(11):1354. doi: 10.3390/genes11111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sevgin O., Sevgin K. Systematic review of microRNAs in the SARS-CoV-2 infection: are microRNAs potential therapy for COVID-19. J. Genet. Genom. Res. 2021;8 053. [Google Scholar]
- 40.Kaur T., Kapila S., Kapila R., Kumar S., Upadhyay D., Kaur M., et al. Tmprss2 specific miRNAs as promising regulators for SARS-CoV-2 entry checkpoint. Virus Res. 2021;294 doi: 10.1016/j.virusres.2020.198275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., et al. DIANA miRPath v. 2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40(W1):W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q., Meng Y., Wang K., Zhang X., Chen W., Sheng J., et al. Inflammation and antiviral immune response associated with severe progression of COVID-19. Front. Immunol. 2021;12:135. doi: 10.3389/fimmu.2021.631226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12(4):843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 44.Chan C.K., Tao J., Chan O.S., Li H.-B., Pang H. Preventing respiratory tract infections by synbiotic interventions: a systematic review and meta-analysis of randomized controlled trials. Adv. Nutr. 2020;11(4):979–988. doi: 10.1093/advances/nmaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal J.P., Mak J.W., Mullish B.H., Alexander J.L., Ng S.C., Marchesi J.R. The gut microbiome: an under-recognised contributor to the COVID-19 pandemic? Therapeut. Adv. Gastroenterol. 2020;13 doi: 10.1177/1756284820974914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuo T., Wu X., Wen W., Lan P. 2021. Gut Microbiome Alterations in COVID-19. Genomics, Proteomics & Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janda L., Mihalčin M., Šťastná M. Is a healthy microbiome responsible for lower mortality in COVID-19? Biologia. 2021;76(2):819–829. doi: 10.2478/s11756-020-00614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soffritti I., D'Accolti M., Fabbri C., Passaro A., Manfredini R., Zuliani G., et al. Oral microbiome dysbiosis is associated with symptoms severity and local immune/inflammatory response in COVID-19 patients: a cross-sectional study. Front. Microbiol. 2021;12:1397. doi: 10.3389/fmicb.2021.687513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roudbary M., Kumar S., Kumar A., Černáková L., Nikoomanesh F., Rodrigues C.F. Overview on the prevalence of fungal infections, immune response, and microbiome role in COVID-19 patients. J. Fungi. 2021;7(9):720. doi: 10.3390/jof7090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain I., Cher G.L.Y., Abid M.A., Abid M.B. Role of gut microbiome in COVID-19: an insight into pathogenesis and therapeutic potential. Front. Immunol. 2021:4164. doi: 10.3389/fimmu.2021.765965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020;71(10):2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao W., Zhang G., Wang X., Guo M., Zeng W., Xu Z., et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 2020;5 doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y., Gu S., Chen Y., Lu H., Shi D., Guo J., et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022;71(1):222–225. doi: 10.1136/gutjnl-2021-324090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeoh Y.K., Zuo T., Lui G.C.-Y., Zhang F., Liu Q., Li A.Y., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70(4):698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuo T., Zhang F., Lui G.C., Yeoh Y.K., Li A.Y., Zhan H., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159(3):944–955. doi: 10.1053/j.gastro.2020.05.048. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perlot T., Penninger J.M. ACE2–From the renin–angiotensin system to gut microbiota and malnutrition. Microb. Infect. 2013;15(13):866–873. doi: 10.1016/j.micinf.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D., Chai X-q, Magnussen C.G., Zosky G.R., Shu S-h, Wei X., et al. Renin-angiotensin-system, a potential pharmacological candidate, in acute respiratory distress syndrome during mechanical ventilation. Pulm. Pharmacol. Therapeut. 2019;58 doi: 10.1016/j.pupt.2019.101833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez-Siles M., Enrich-Capó N., Aldeguer X., Sabat-Mir M., Duncan S.H., Garcia-Gil L.J., et al. Alterations in the abundance and co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the colonic mucosa of inflammatory bowel disease subjects. Front. Cell. Infect. Microbiol. 2018;8:281. doi: 10.3389/fcimb.2018.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pittayanon R., Lau J.T., Yuan Y., Leontiadis G.I., Tse F., Surette M., et al. Gut microbiota in patients with irritable bowel syndrome—a systematic review. Gastroenterology. 2019;157(1):97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 60.Velasquez M.T. Altered gut microbiota: a link between diet and the metabolic syndrome. Metab. Syndr. Relat. Disord. 2018;16(7):321–328. doi: 10.1089/met.2017.0163. [DOI] [PubMed] [Google Scholar]
- 61.Ferreira C., Viana S.D., Reis F. Is gut microbiota dysbiosis a predictor of increased susceptibility to poor outcome of COVID-19 patients? An update. Microorganisms. 2021;9(1):53. doi: 10.3390/microorganisms9010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T.G., Ortiz-Lopez A., et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168(5):928–943. doi: 10.1016/j.cell.2017.01.022. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toyonaga T., Araba K.C., Kennedy M.M., Keith B.P., Wolber E.A., Beasley C., et al. Increased colonic expression of ACE2 associates with poor prognosis in Crohn's disease. Sci. Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-92979-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suárez-Fariñas M., Tokuyama M., Wei G., Huang R., Livanos A., Jha D., et al. Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-CoV-2–related disease. Gastroenterology. 2021;160(1):287–301. doi: 10.1053/j.gastro.2020.09.029. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khalil B.A., Elemam N.M., Maghazachi A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021 doi: 10.1016/j.csbj.2021.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lv L., Shen J., Xu J., Wu X., Zeng C., Lin L., et al. MiR-124-3p reduces angiotensin II-dependent hypertension by down-regulating EGR1. J. Hum. Hypertens. 2021;35(8):696–708. doi: 10.1038/s41371-020-0381-x. [DOI] [PubMed] [Google Scholar]
- 67.Teodori L., Sestili P., Madiai V., Coppari S., Fraternale D., Rocchi M.B.L., et al. MicroRNAs bioinformatics analyses identifying HDAC pathway as a putative target for existing anti‐COVID‐19 therapeutics. Front. Pharmacol. 2020;11:1729. doi: 10.3389/fphar.2020.582003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vegter E.L., Ovchinnikova E.S., Silljé H.H., Meems L.M., van der Pol A., van der Velde A.R., et al. Rodent heart failure models do not reflect the human circulating microRNA signature in heart failure. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eyyupkoca F., Ercan K., Kiziltunc E., Ugurlu I.B., Kocak A., Eyerci N. Determination of microRNAs associated with adverse left ventricular remodeling after myocardial infarction. Mol. Cell. Biochem. 2022:1–11. doi: 10.1007/s11010-021-04330-y. [DOI] [PubMed] [Google Scholar]
- 70.Chang Z., Yan G., Zheng J., Liu Z. The lncRNA GAS5 inhibits the osteogenic differentiation and calcification of human vascular smooth muscle cells. Calcif. Tissue Int. 2020;107(1):86–95. doi: 10.1007/s00223-020-00696-1. [DOI] [PubMed] [Google Scholar]
- 71.Zhang C., Ma C., Zhang L., Zhang L., Zhang F., Ma M., et al. MiR-449a-5p mediates mitochondrial dysfunction and phenotypic transition by targeting Myc in pulmonary arterial smooth muscle cells. J. Mol. Med. 2019;97(3):409–422. doi: 10.1007/s00109-019-01751-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings in this study are available from the corresponding author upon reasonable request.