Abstract
Linear plasmids were unknown in mycobacteria until recently. Here, we report the complete nucleotide sequence of 23-kb linear plasmid pCLP from Mycobacterium celatum, an opportunistic pathogen. The sequence of pCLP revealed at least 19 putative open reading frames (ORFs). Expression of pCLP genes in exponential-phase cultures was determined by reverse transcriptase PCR (RT-PCR). Twelve ORFs were expressed, whereas no transcription of the 7 other ORFs of pCLP was detected. Five of the 12 transcribed ORFs detected by RT-PCR are of unknown function. Sequence analysis revealed similar loci in both M. celatum pCLP and the Mycobacterium tuberculosis chromosome, including transposase-related sequences. This result suggests horizontal transfer between these two organisms. pCLP also contains ORFs that are similar to genes of bacterial circular plasmids involved in partition (par operon) and postsegregational (pem operon) mechanisms. Functional analysis of these ORFs suggests that they probably carry out similar maintenance roles in pCLP.
Since Hayakawa et al. (13) discovered the first bacterial linear plasmid in Streptomyces rochei, many linear double-stranded DNA plasmids of various sizes (from 12 to 1,700 kb) have been isolated from other Streptomyces spp. Linear plasmids in other Actinomycetales, including Rhodococcus spp. (7, 8, 17, 19, 29), Mycobacterium spp. (24, 25, 29), and Planobispora rosea (26), have also been described. All of these linear replicons belong to a class of genetic elements called invertrons (30), which have terminal inverted repeats (IRs) with their 5′ ends covalently linked to a terminal protein. Another type of bacterial linear plasmid, with covalently closed hairpin loops at each end, in Borrelia spp. and in prophage of coliphage N15 has been characterized (14). Information pertaining to the genetic organization of linear plasmids with invertron structures is limited. Indeed, only linear plasmid pSCL1 from Streptomyces clavuligerus has been completely sequenced (36). Plasmid pSCL1 is 12 kb in length and contains eight possible open reading frames (ORFs), two of which encode proteins with significant sequence similarity to replication and regulatory proteins; other ORFs have no significant matches with databases (36). Rhodococcus and Streptomyces linear plasmids also encode enzymes for some catabolic pathways and carry genes involved in antibiotic biosynthesis (18). Although most of the extensively studied linear replicons in Streptomyces have been found to be transmissible plasmids (18), genetic information for conjugational transfer on these plasmids has not yet been identified.
pCLP, a 23-kb linear plasmid from opportunistic pathogen Mycobacterium celatum, has been previously cloned, and its telomeres have been sequenced. The telomeres have both covalently attached proteins (24, 25) and sequence features similar to those of other Actinomycetales linear plasmids (25). Recently, we identified the internal origin of replication of pCLP (23), which is similar to those of many bacterial circular plasmids in that it harbors putative replication and partitioning genes, iteron sequences, and an AT-rich region. The replication region of pCLP was used to construct an Escherichia coli-mycobacterium shuttle vector which is able to replicate in both slow- and fast-growing mycobacteria; this shuttle vector is also compatible with other mycobacterial vectors (23).
To further characterize the genetic organization of these atypical plasmids and to illuminate the origin of these linear structures, we used a direct approach and sequenced pCLP. The complete nucleotide sequence of pCLP reveals transposase-related sequences similar to those in the M. tuberculosis chromosome. This suggests possible genetic exchange between mycobacteria mediated by mobile elements. We also studied the transcription of all the ORFs and analyzed in more detail those encoding homologs to proteins involved in partition and postsegregational killing mechanisms.
(This work was part of a doctoral thesis by C. Le Dantec.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The M. celatum strain 4 used in this study is a clinical isolate which contains two linear replicons, one of about 23 kb designated pCLP and another of 320 kb (24). We also used Mycobacterium smegmatis mc2155 (32) and Mycobacterium tuberculosis H37Rv strain Pasteur. Mycobacteria were grown in 7H9 Middlebrook liquid and solid media at 37°C, with antibiotics added to the media as required.
Sequencing of pCLP.
pCLP, a 23-kb plasmid, was cloned in five 4- to 5-kb fragments into a Km pUC19 derivative called pPV8 (23). Each insert was then subcloned in smaller fragments for sequencing. Plasmid constructs were introduced into E. coli DH5α by electroporation (gene pulser unit; Bio-Rad, Richmond, Calif.), and transformants were selected on solid Luria-Bertani medium supplemented with 20 μg of kanamycin/ml. Double-stranded plasmid DNA was recovered using a Midi kit (Qiagen, Hilden, Germany) and sequenced by the dideoxy chain termination method (31) using a Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Perkin-Elmer Corp., Foster City, Calif.), a model 9600 GenAmp PCR system (Perkin-Elmer), and a model 373 stretch DNA analysis system (Applied Biosystems). We used universal forward and reverse primers and a DNA-walking strategy to sequence the fragments of the linear plasmid. Nucleotide sequences were analyzed using the GCG package (Genetics Computer Group, University of Wisconsin, Madison), and we searched for sequence similarities using the BLAST algorithm (1) (Table 1).
TABLE 1.
Linear plasmid pCLP gene analysis
| ORF no.a | Predicted protein size (aa) | Identity (%) | Closest homolog (species and strain or plasmid) | Known or putative function | Accession no. |
|---|---|---|---|---|---|
| 1 | 424 | 86 | Rv3128c (M. tuberculosis H37Rv) | Unknown | C70990 |
| 2 | 98 | 70 | pemI (M. morganii R446b) | Maintenance | AAC82515 |
| 71 | pemI (E. coli R100) | Maintenance | BAA78897 | ||
| 3 | 84 | 87 | pemK (M. morganii R446b) | Maintenance | AAC82516 |
| 86 | pemK (E. coli R100) | Maintenance | BAA78898 | ||
| 8 | 271 | 80 | Rv2813 (M. tuberculosis H37Rv) | Secretion | H70690 |
| 9b | 318 | 86 | Rv2812 (M. tuberculosis H37Rv) | Transposase (IS1604) | G70690 |
| 11 | 215 | 57 | parA (P. alcaligenes) | Partition | AAD40334 |
| 14 | 351 | 65 | rep (M. fortuitum) | Replication | CAB43095 |
| 16 | 195 | 71 | Rv0921 (M. tuberculosis H37Rv) | Resolvase (IS1535) | A70583 |
| 63 | Rv2792c (M. tuberculosis H37Rv) | Resolvase (IS1602) | G70884 | ||
| 17 | 486 | 58 | Rv0922 (M. tuberculosis H37Rv) | Transposase (IS1535) | B70583 |
| 53 | Rv2791c (M. tuberculosis H37Rv) | Transposase (IS1602) | F70884 |
ORF4 to -7, -10, -12, -13, -15, -18, and -19 have no significant similarities in the databases.
No valid start codon.
mRNA detection with RT-PCR.
M. celatum was grown to the exponential phase in 40 ml of 7H9 Tween medium at 37°C and resuspended in 1 ml of TRIzol reagent (Life Technologies). Cells were shaken for 2 min with 0.1-mm glass beads (PolyLabo), and then 0.2 ml of isoamyl chloroform was added. After centrifugation (12,000 × g for 15 min at 4°C), the supernatant was recovered and 0.5 ml of isopropyl alcohol was added. To precipitate RNA, tubes were incubated for 10 min on ice and then centrifuged and nucleic acids were washed three times with 75% ethanol. Pellets were dissolved in 50 μl of diethyl pyrocarbonate-treated water and stored at −70°C. To remove DNA contamination, samples were treated with RNase-free DNase I (Roche Diagnostics). PCR primer pairs were designed to amplify the transcripts corresponding to each of the 19 ORFs (Table 2). All primer pairs were 18 to 22 nucleotides long and produced amplicons of the expected sizes (from 102 to 424 bp) when tested on M. celatum genomic DNA (Table 2). Reverse transcription of RNA was carried out as described by the manufacturer (Superscript; Gibco-BRL) by using an antisense primer at a final concentration of 1 pmol/μl (Table 2) plus 2 U of RNasin (Amersham Pharmacia Biotech). After 50 min at 42°C, reverse transcriptase (RT) was inactivated by incubation at 70°C for 15 min. PCR was performed in a Perkin-Elmer model 480 thermal cycler in a 50-μl reaction volume containing 2 μl of cDNA, 1× Taq DNA buffer (Perkin-Elmer), 2.5 mM MgCl2, 0.5 U of Taq DNA polymerase (Perkin-Elmer), 0.2 mM deoxynucleoside triphosphates, and 10 ng of a given primer pair (Table 2). The PCR conditions were 94°C for 5 min, followed by 35 cycles at 94°C for 1 min, 55°C for 1 min, and 70°C for 1 min. RNA samples were tested in the presence and absence of RT to test for amplification of contaminant genomic DNA. Each ORF was tested at least three times. Amplicons were detected by electrophoresis in 2% agarose gels, followed by ethidium bromide staining.
TABLE 2.
Primers used to amplify the internal fragments of the given gene of pCLP
| ORFa no. (homolog) | Sequence of:
|
Size of product (bp) | |
|---|---|---|---|
| Forward primer (5′–3′) | Antisense primer (5′–3′)b | ||
| 1 (Rv3128c) | CCTGGGCGCCCTCAAATCCG | GCAGCTCTGATTCTTGGGCC | 424 |
| 2 (pemI) | CCCGGATGGAGGTGGGCG | GCTTCACCGTTGAGCC | 255 |
| 3 (pemK) | ATTGCTCTGACGGAACGCGG | CCCGCCGCGTGTTATAGGTAGC | 152 |
| 2 and 3 (pem operon) | GGACCCCCAGGTGCGACCGC | CCGTTCGGGCAAAGTTCCCG | 152 |
| 4 | CCGGCAAGACCTCGTCAGCC | GCCAATGAGTAGTCCAGCGG | 277 |
| 5 | GGACTACTCGTGTGGCGACAGC | GCTGACGCGGCACAGGAACC | 272 |
| 6 | GGTCGCCCGTGAATACCGG | GCATCTTGCGCCCAATCGCC | 293 |
| 7 | CGAGCGACGGTGATCCGTACCG | GCGTCTATTGATGCCGAACACC | 316 |
| 8 (Rv2813) | CGTCATCTATCTGCCTGACC | CGCGGCCGGCCAGCTTCAGG | 413 |
| 9 (Rv2812) | GGCAGACCCCGCTGGCCCGC | CGGCGCGCCGGCCAGCCGC | 200 |
| 10 | CGCAGCCCGCGCGGGACAGC | GGCGTCGAGTGCGCTCCGCC | 223 |
| 11 (parA) | GCCGTTGCCGGTTTCCCTCG | CGGTTGATCGCCCACACCGC | 253 |
| 12 | CCGCGCCGGCCTAGCGCCG | CGGCGGTGTCGGCGTGCTGG | 102 |
| 13 | GCGCGCAGGCACGCGGACG | CCCCCAGGTGGGGGCGACCC | 288 |
| 14 (rep) | GCCGGGCGTGGATCGAACGG | ATCCGGCCGCCCCAAGAACC | 301 |
| 15 | GCCGGCGTCGGTGGTGGCG | GCTCGACCCACCGGCAGCCGG | 264 |
| 16 (Rv0921) | GGGGAATCCGGCCACACGG | CGGCGCGCCGGCCAGCCGC | 310 |
| 17 (Rv0922) | GGCGCCGAAGTCGTTGACCG | GGGCGGCATCAGCCAGGGC | 316 |
| 18 | CGGCCCCGCCGACGCACCGC | GCCCGCGACGAAGCGTCGGG | 277 |
| 19 | CCGCCAGCGGCGTCATTGG | GGTCCTGCCGACCCGCGGCG | 268 |
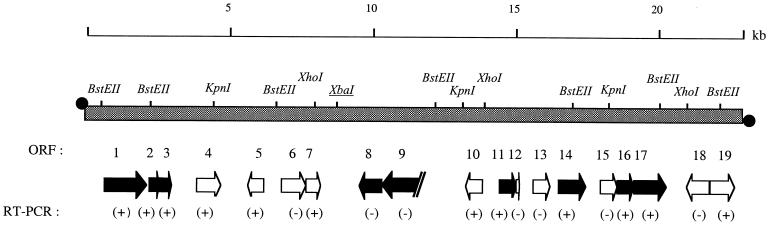
See Fig. 1 for location of ORFs on pCLP.
These primers were also used to prime cDNA.
Southern blotting.
Genomic DNA of M. celatum and M. tuberculosis H37Rv was extracted as described previously (23), digested with PvuII, subjected to electrophoresis overnight in a 1% agarose gel, and transferred onto nylon membranes. Membranes were hybridized overnight at 50°C in Rapid hybridization buffer (Amersham International, Amersham, United Kingdom) with Rv2812 or Rv2813 homolog probes. Probes were amplified from M. celatum pCLP by PCR (with primers P1, 5′-GCC AAG CGA TCC AGA TGG C-3′, and P2, 5′-CGG CGC CGG ACA GGT CGGCG-3′, for the Rv2812 homolog and primers P3, 5′-GGT CTT GAG TTC ATC ACG GC-3′, and P4, 5′-CGC GTG ACC AAG ACC GCG-3′, for the Rv2813 homolog) and radiolabeled with [α-32P]dCTP using a commercial kit (Megaprime; Amersham). The membranes were then washed at 50°C as previously described (23).
Expression of the pCLP pemK homolog.
Plasmid pMIP12 is an E. coli-mycobacterium shuttle vector. It carries the pAL5000 origin of replication (20) and a kanamycin resistance gene derived from Tn903. An expression cassette which allows expression of heterologous genes in mycobacteria has been cloned into pMIP12. This cassette consists of the up-regulated pBlaF* promoter derived from Mycobacterium fortuitum (33), an optimized Shine-Dalgarno sequence (Mega SD) that allows ribosomal attachment, and an ATG translation start codon followed by a multiple cloning site (MCS). A stretch of six histidine codons downstream from the MCS allows recombinant proteins synthesized in mycobacteria and transformed with pMIP12 derivatives to be purified by use of nickel columns. A transcription terminator derived from the ESAT-6 operon (3), downstream from the stop codon, favors the stability of mRNAs transcribed from pBlaF*. Bacterial, viral, eukaryotic, and parasitic genes have been successfully expressed in mycobacteria using this vector (N. Winter, unpublished results). The pemK homolog (ORF3) from M. celatum was amplified by PCR with primers P5, 5′-CGG GAT CCA TTG CTC TGA CGG AAC GCG G-3′, and P6, 5′-CGC TGCAGG GCG ACG GGC GGC GGC GGC-3′. The 373-bp PCR product was introduced into pMIP12 so that it was under the control of the pBlaF* promoter; the resulting vector was called pCK12. EcoRV, which cuts once in pCK12 (in the middle of the pemK homolog), was used to digest pCK12 DNA, and the samples were treated with T4 DNA polymerase to introduce a deletion into the pemK homolog; the linearized plasmid was then religated, resulting in pCKM12. The pCKM12 point mutation was confirmed by sequencing (one nucleotide was missing in the locus corresponding to the EcoRV site). M. smegmatis mc2155 was transformed by electroporation with pCK12 and pCKM12 as previously described (22), and cells were selected on solid 7H11 medium supplemented with 20 μg of kanamycin/ml.
Stability study in the presence or absence of the pCLP par operon.
We studied the stability of recombinant plasmid pCL4D (23), which contains the intact par operon from the pCLP replication region, and recombinant plasmid pLB2 (23), which lacks the par operon, in M. smegmatis. M. smegmatis cells carrying pCL4D or pLB2 (both encoding kanamycin resistance) were grown in liquid medium with no antibiotic selection pressure for 24 h at 37°C. The culture was then diluted 1:100, and the bacteria were grown in fresh antibiotic-free medium for a further 24 h; this procedure was repeated three times. After each dilution, aliquots of the cells were plated out on agar plates with or without kanamycin (20 μg/ml), and the proportion of resistant cells was determined to estimate the number of cells carrying the plasmid.
Nucleotide sequence accession number.
The GenBank accession number for the pCLP nucleotide and amino acid sequences is AF312688.
RESULTS
Complete nucleotide sequence of pCLP, a 23-kb linear plasmid.
We have previously demonstrated that pCLP of M. celatum strain 4 is a linear plasmid with an invertron terminal structure, i.e., containing terminal IRs with ends covalently linked to a terminal protein (25). Various restriction fragments of pCLP were ligated into pUC19 derivatives (23) to obtain a library covering the complete sequence of pCLP, and clones were sequenced. The fully assembled linear DNA sequence of pCLP was 22,691 bp long. The GC content of the plasmid is 65.6%, which is typical for a Mycobacterium sp. A four-enzyme (BstEII, KpnI, XbaI, and XhoI) restriction map of pCLP (23) was compared with the map predicted from the complete nucleotide sequence. All restriction sites found in the pCLP nucleotide sequence had been previously identified by restriction analysis. PCR was used to link the different restriction fragments, further confirming that the pCLP sequence was accurate and well assembled. We determined the locations of putative ORFs by using a combination of the GCG software, the BLAST algorithm, the known codon usage for Mycobacterium genes (2), and inspection of the sequences by eye. We thus identified 19 ORFs (designated ORF1 to -19) that seem likely to be expressed (Fig. 1). ORF9 was found to be similar to an M. tuberculosis gene (Rv2812) but to have a truncated 5′ terminus relative to its homolog (Fig. 2A); in addition, no valid start codon was found to replace the missing ATG start codon in the homolog. However, ORF9 is included among the ORFs likely to be expressed solely on the basis of its high degree of homology to M. tuberculosis Rv2812 (Table 1). We identified nine ORFs (including ORF9) with significant homologies to genes in the databases. Of these, five have similarities with M. tuberculosis chromosomal genes and three are similar to maintenance and partitioning genes of bacterial circular plasmids (Table 1). Although the minimum ORF size in genomic annotation is often considered to be 300 nucleotides, ORF12 (144 bp) was selected as a putative gene. Indeed, a previous study indicates that ORF12 (downstream from the parA homolog, ORF11) could be the pCLP parB counterpart of a par operon (11). The other 10 ORFs have no homologs in databases, but they were selected as putative ORFs due to their good coding potential. To confirm that they are genuine ORFs, we studied the transcription of all 19 ORFs.
FIG. 1.
Physical and genetic map of pCLP linear plasmid. Arrows indicate predicted genes (ORFs are numbered 1 through 19), with their direction indicating the direction of transcription (for ORF9, no valid start codon was identified); black arrows, genes similar to known genes. Amplification (+) or lack of amplification (−) of a product corresponding to the size predicted from the primer locations by RT-PCR (see Table 1) is indicated. black circles, terminal proteins covalently linked to DNA ends.
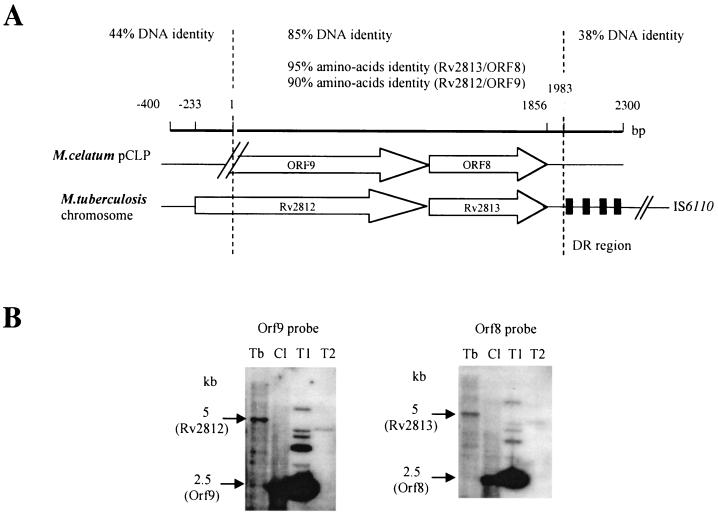
FIG. 2.
Conserved region of the M. tuberculosis chromosome and M. celatum pCLP. (A) Genetic organization of the cluster comprising Rv2812 and Rv2813 in the M. tuberculosis complex and the corresponding homologs in pCLP. Arrows, ORFs. Percentages of nucleotide and amino acid sequence identity are indicated. Locations of the DR region and IS6110 are indicated. (B) Southern blot analysis of genomic DNA of M. tuberculosis H37Rv and M. celatum. DNA in all lanes was digested with PvuII and probed with ORF9 and ORF8 (Rv2812 and Rv2813 homologs, respectively) from pCLP. Hybridization was carried out at 50°C. Lanes: Tb, M. tuberculosis H37Rv; Cl, M. celatum; T1, plasmid construct with the left end of pCLP; T2, plasmid construct with the right end of pCLP. Left and right ends correspond to the two XbaI fragments of pCLP (Fig. 1) (25).
Transcription study of putative ORFs.
We determined the presence or absence of mRNA corresponding to the 19 ORFs of pCLP using RT-PCR. As expected, the rep gene (ORF14) of pCLP, which had been previously identified (23), was transcribed and was therefore used as the RT-PCR positive control. Each gene was tested at least three times to ensure the reproducibility of the experiment. The genes tested, as well as the presence or absence of a PCR product of the expected size (Table 2), are shown in Fig. 1. Twelve ORFs were found to be transcribed during the exponential phase, whereas 7 ORFs were not.
Among the ORFs that were not transcribed, ORF8 and ORF9 have homologs in M. tuberculosis (Rv2813 and Rv2812, respectively). For ORF9, no valid start codon was identified and the 5′ terminus was truncated compared to that of Rv2812 (Fig. 2A); therefore ORF9 may be a pseudogene. The ORF8 DNA and encoded protein sequences are 85 and 90% identical, respectively, to those of Rv2813 of M. tuberculosis, whose expression and putative function (secretion) in M. tuberculosis have not been determined. None of other nontranscribed ORFs (ORF6, ORF12, ORF13, ORF15, and ORF18) are similar to previously determined protein-coding sequences. ORF13 contains direct repeats and an AT-rich region that we previously identified as the possible origin of replication in pCLP (23), and therefore ORF13 may not be part of a coding sequence. Despite several attempts, we failed to amplify transcripts for ORF12, whose organization suggests that it may constitute the parB counterpart of the par operon (11).
The majority of the transcribed ORFs have homologs in the databases. In addition to ORF14 (rep homolog), involved in the initiation of pCLP replication, homologs to genes implicated in maintenance (ORF2 and ORF3) and partitioning systems (ORF11) of bacterial circular plasmids were identified (Fig. 1). pCLP ORF3 and ORF4 are very similar to the pemI and pemK genes of E. coli plasmid R100 and Morganella morganii plasmid R446b (Table 1). Transcription between ORF2 and ORF3 was also detected, suggesting that these two ORFs are cotranscribed and constitute an operon. The ORF11 protein exhibits 57% identity to Pseudomonas alcaligenes ParA and shows a strong RT-PCR signal. As described above, genetic organization of par systems (11) led us to suspect the existence of a second ORF downstream from parA; however, in our hands, neither ORF12 nor ORF13 was found to be transcribed. Other transcribed ORFs include ORF1, homologous to M. tuberculosis Rv3128c, which is of unknown function. Products of transcribed ORF16 and ORF17 are similar to the putative resolvase (Rv0921 homolog) and transposase (Rv0922 homolog) of M. tuberculosis. The other ORFs with positive results (ORF4, ORF5, ORF7, ORF10, and ORF19) do not have homologs in the databases and therefore could constitute new genes.
Sequence similarity with M. tuberculosis chromosomal loci and identification of transposase-like sequences.
The complete nucleotide sequence of the M. tuberculosis chromosome (6) provided a wealth of data for mycobacterial genomics. Sequence analysis of linear plasmid pCLP revealed several regions with sequence homologies (in both DNA and amino acid sequences) with the M. tuberculosis chromosome. Many of these regions are mobile elements or related sequences. Sequence alignment of Rv2812 and Rv2813 from M. tuberculosis and their pCLP homologs shows that they have a high degree of nucleotide sequence identity (90%) (Fig. 2A). This nucleotide sequence identity continues beyond Rv2813 but stops 10 bp before the DR region of IS6110 of M. tuberculosis. The ends of the fragment homologous between the two organisms are clearly defined (the sequence identity drops from 85 to 44% within a few base pairs) (Fig. 2A). M. tuberculosis Rv2812 encodes the putative transposase of IS1604, which did not contain IRs or DRs (12). The region surrounding ORF8 and ORF9 was searched, unsuccessfully, for DNA sequence features typical of IRs and/or DRs suggestive of a past event of homologous recombination and/or the presence of mobile elements. To confirm the presence of M. tuberculosis Rv2812 and Rv2813 homologs in pCLP, the digested genomic DNA of M. celatum and M. tuberculosis H37Rv was hybridized with labeled probes corresponding to these pCLP homologs (Fig. 2B). Rv2812- and Rv2813-related sequences were both found on the same restriction fragments in the two species: only in a plasmid region for M. celatum and in a chromosomal region for M. tuberculosis (Fig. 2B). Other regions of homology between pCLP and M. tuberculosis include Rv0921 (ORF16) and Rv0922 (ORF17), both from M. tuberculosis IS1535 (12). However, in this case, the sequence identity was not homogeneous throughout the homologous fragment (data not shown). Again, no appropriate IRs or DRs could be found for the IS1535 homolog of pCLP; this was also the case for other M. tuberculosis members of the IS1535 family (12).
Maintenance and partitioning functions of pCLP: identification of pem and par operon homologs.
Previous pCLP sequence analysis and cloning experiments (23) revealed a single replication region (positions 14292 to 17277 bp) consisting of a rep gene and additional sequence elements characteristic of plasmid replicons that use iteron-based replication initiation (9). The region upstream from the pCLP rep coding region contains a parA homolog that may be part of the partitioning locus. Partitioning genes are required for reliable plasmid segregation upon cell division. ParA is an ATPase stimulated by ParB, which is a DNA-binding protein that recognizes the cis-acting parS site. These three components are required for plasmid stability. We searched for an ORF downstream from parA that might function as parB. No homology with the coding sequence for ParB was found in the origin region or anywhere on the plasmid. A recent phylogenetic analysis (11) of the par loci of bacterial plasmids and chromosomes separated the par operon into three distinct subgroups on the basis of the ParA sequence and the par genetic organization. In many plasmids, the partitioning locus is organized like the par and sop loci of E. coli plasmids P1 and F (11). However, the par loci from plasmids of gram-positive origin are distinct from those of P1 and F and are similar to those of the Agrobacterium tumefaciens pTAR plasmid. In this case, the par locus encodes a small ParA (208 to 227 amino acids [aa]) and very small ParB (46 to 113 aa). In pCLP, a very small ORF, ORF12 (encoding a putative protein of 47 aa), is present 10 bp downstream from the putative parA TGA codon (pCLP parA encodes a 214-aa protein) and therefore may constitute the parB of the pCLP par locus. However, RT-PCR assays for ORF12 (and even ORF13) did not detect transcripts. We also failed to identify a centromere analog, parS, which may be upstream of or downstream from the par operon (11). To determine whether the par-homologous region of pCLP can act as a partition locus, we compared the segregational stability of an M. smegmatis-E. coli shuttle vector (23) carrying the intact pCLP par region with that of a similar vector with the pCLP par region deleted. The stabilities of the two constructs (with or without the putative par operon) were determined by measuring the rates at which plasmid-free cells accumulated during 80 generations of growth in the absence of antibiotic selection (Fig. 3). The plasmid with the putative par operon, called pCL4D, was significantly more stable than plasmid pLB2, which lacks ORF11 and ORF12 (Fig. 3). This result indicates that ORF11 and/or ORF12 confer partition functions to pCLP.
FIG. 3.
Stability studies of pCLP derivatives in M. smegmatis mc2155. Constructs pCL4D and pLB2 contain the replication region of pCLP with or without the putative par operon, respectively. Percentages of cells resistant to kanamycin are shown. These values correspond to the percentages of cells retaining the plasmid when grown in the absence of the antibiotic.
The transcribed ORF2 and ORF3 map between nucleotide positions 1919 and 2545 (Fig. 1). They encode putative proteins of 98 and 84 amino acids with 71 and 86% identity with PemI and PemK of E. coli plasmid R100, respectively (Table 1). The pem system (for plasmid emergency maintenance) uses a killer protein (PemK) and a regulatory protein (PemI) to eliminate plasmid-free segregants from the population. PemK inhibits the growth of the host cell, causing cell death, whereas PemI suppresses the growth inhibition caused by PemK (10). PemI and PemK are autoregulated by binding to the promoter region of the pem operon upstream from pemI (34). The genetic organization of the pem homolog operon of pCLP is similar to that in E. coli plasmid R100. Although E. coli and M. morganii are both enterobacteria, pem promoters of plasmids R100 and R446b exhibit a high degree of homology to the upstream region of the pemI homolog of pCLP (Fig. 4A). This homology includes the putative pem −10 promoter region of plasmid R100 (34) and IRs corresponding to the specific DNA binding sites of the Pem proteins encoded by R100 (34) (Fig. 4A). To analyze the putative pem operon, we tested the killing function of the pCLP-encoded PemK homolog. We constructed plasmid pCK12, which carries a 373-bp fragment containing the putative pCLP pemK gene cloned into expression vector pMIP12. Plasmid pMIP12 is an E. coli-mycobacterium shuttle vector carrying the pAL5000 origin of replication (20) and containing an expression cassette which has been optimized for the expression of heterologous genes in mycobacteria (Fig. 5A). M. smegmatis transformed with pCK12 was not able to grow on selective medium (Fig. 5B); this is probably due to the production of the PemK homolog in the absence of PemI. To confirm that this was the case, a point mutation was introduced into the pCK12 pemK homolog, resulting in pCKM12. We found that the mutated allele restored the capacity of M. smegmatis transformants to grow on selective media (Fig. 5C). This shows that the pMIP12 promoter allows the efficient expression of the pemK homolog and that the PemK homolog alone causes cell death in M. smegmatis.
FIG. 4.
Sequence alignment of the pem operon of pCLP. (A) Nucleotide sequence alignment of the pem promoters of E. coli plasmid R100, M. morganii plasmid R446b, and M. celatum plasmid pCLP. IRs a and b correspond to binding sites of the R100-encoded Pem proteins (34). The −10 (consensus sequence is in boldface) and −35 regions of the R100 pem promoter are also indicated. (B and C) Alignment of the deduced amino acid sequences encoded by ORF2 and ORF3 of pCLP with PemI (B) and PemK (C) encoded by R100, respectively. Conserved residues are boxed.
FIG. 5.
Expression of pCLP pemK in M. smegmatis. (A) Schematic diagram of expression cassette Mega SD of pMIP12. Thick arrow, pBlaF* promoter. Relevant restriction sites are indicated. ATG, pMIP12 translation start codon. Stop codons are underlined (B and C) Transformation of M. smegmatis mc2155 with replicative plasmids pCK12 (B) and pCKM12 (C). Plasmid pCK12 contains the pCLP pemK homolog alone cloned into expressing vector pMIP12. For pCKM12, a point mutation was introduced into the pemK homolog. Plates were incubated for 5 days at 37°C.
DISCUSSION
In bacteria, linear plasmids with invertron structures have only been found in the gram-positive group Actinomycetales. The few linear plasmids of this class to have been studied in detail were all isolated from streptomycetes. The complete nucleotide sequence of linear plasmid pCLP from M. celatum reveals at least 19 putative ORFs. The functions of ORF2, ORF3, ORF11, and ORF14 have been demonstrated (rep, par, and pem systems); ORF8, ORF9, ORF16, and ORF17 have been assigned putative functions based on their database matches (transposase, resolvase, secretion protein); and ORF1 has a good match to an M. tuberculosis hypothetical gene (Rv3128c). The remaining ORFs (ORF4, ORF5, ORF6, ORF7, ORF10, ORF12, ORF13, ORF15, ORF18, and ORF19) have only poor matches to database entries. Despite the accumulation of sequence information in databases, the pCLP sequence contains at least 10 uncharacterized ORFs, i.e., ORFs without any resemblance to previously determined protein coding sequences. Amplicons were detected for 12 of the 19 ORFs assayed by RT-PCR in exponential-phase cultures of M. celatum (Fig. 1). In some cases, the transcripts may have been present at levels below the detection threshold of the technique or may have been unstable or only present in other growth conditions. A high percentage of plasmid DNA seems to be composed of noncoding sequences, and at least 5 of the 10 positive ORFs detected by RT-PCR have unknown functions; it is likely that at least some of these ORFs with unknown functions have biological relevance.
Evidence for horizontal transfer.
The overall organization and genes of pCLP show similarities to those of bacterial circular plasmids or of mycobacterial origin. Although it is plausible that Actinomycetales linear plasmids could have evolved from phages or eucaryotic viruses with the same terminal structure (14), no sequence features suggestive of a phage or virus origin were identified in pCLP. In addition, no pCLP region had a G+C content significantly different from those of surrounding regions of DNA, suggesting the absence of recent horizontal transfer from organisms of low G+C content.
pCLP and M. tuberculosis loci show a high degree of nucleotide sequence identity. For example, the region between nucleotide positions 8951 and 10934 (containing ORF8 and ORF9) exhibits sequence identity with the M. tuberculosis region containing Rv2812 and Rv2813; beyond these two points essentially no general homology exists. This suggests that this fragment has been transferred from M. tuberculosis to linear plasmid pCLP as a single fragment. The high conservation of the DNA sequence in this region and the fact that ORF8 and ORF9 were not transcribed (and therefore there can have been no pressure to conserve the native coding sequence) suggest that this fragment has been acquired relatively late in pCLP evolution. Interestingly, the analogous region in M. tuberculosis is located near insertion sequence IS6110, and M. tuberculosis Rv2812 may encode a transposase. Another region conserved between pCLP and the M. tuberculosis chromosome contains Rv0921 and Rv0922, which encode a putative resolvase and transposase, respectively, of IS1535 (12). In this case, the Rv0921 and Rv0922 homologs were transcribed in pCLP and therefore could still be mobile elements. Such mobile element-like sequences may promote illegitimate recombination and genetic plasticity. It is therefore very likely that pCLP acquired DNA fragments from an M. tuberculosis-like organism through recombination events that originated from mobile elements. Note that linear plasmid pCLP can replicate within most Mycobacterium species, including the M. tuberculosis complex (23), so it can easily spread between species and promote gene transfer between mycobacteria. A promiscuous lifestyle involving both M. celatum and M. tuberculosis may have contributed to the evolution of pCLP. Similarly, the sequences and functions of the rep, pem, and par systems of some circular plasmids isolated from phylogenetically remote bacteria were found to be conserved in pCLP. Sequence homology studies did not identify a conjugation system in pCLP. Although such a conjugation system has not been identified at the molecular level in mycobacteria, naturally occurring conjugation has been demonstrated for mycobacteria (21) and some Streptomyces linear plasmids (18). Future studies will include investigation of the possible conjugational transfer of pCLP in mycobacterial species.
A genetic organization for pCLP typical of a circular plasmid.
The pCLP sequence reveals a backbone similar to that of a typical bacterial circular plasmid. Little is known about the replication of bacterial linear plasmids. We previously identified an internal origin of replication in mycobacterial linear plasmid pCLP similar to those of mycobacterial circular plasmids (23). Previous studies on Streptomyces linear plasmids also revealed an internal origin of replication, but their genetic organization showed similarities with that of phages and archaeal plasmid replication regions (4, 15, 28, 36). This difference suggests that mycobacterial and Streptomyces linear plasmids do not have a common ancestor. It has been shown that bidirectional replication of Streptomyces linear plasmids is initiated from the internal origin of replication and continues toward the ends of plasmids, leaving single-strand gaps at the 3′ ends (5). A second replication mechanism to fill in the recessed 5′ ends may therefore be involved. Sequence analysis of the ends of linear plasmids with invertron structures revealed palindromes which could form secondary structures that may be recognized and used by terminal proteins to complete DNA synthesis (5, 16, 25, 27). Although terminal proteins for bacterial linear plasmids have not yet been identified in Actinomycetales, it is noteworthy that ORF16 of pCLP encodes a protein similar to resolvases. Proteins of the resolvase family promote strand exchange by making and rejoining DNA ends and therefore could form a covalent DNA-protein linkage with 5′ ends of linear plasmids. Qin and Cohen (27) suggested that telomere replication of linear plasmids may require an endonucleolytic processing step, also carried out by resolvases.
Two loci responsible for plasmid maintenance were identified on linear plasmid pCLP. Both loci exhibit high sequence similarity to the maintenance genes (pem and par operons) of bacterial circular plasmids. It seems very likely that these genes carry out similar roles in pCLP. Indeed, the stability of the minimal plasmid replicon of pCLP is affected similarly to that of a circular plasmid when the putative par operon is not present. The pCLP par operon may therefore govern the accurate partitioning of plasmid copies into daughter cells, and its deletion may destabilize pCLP maintenance, as in circular plasmids. Interestingly, no par systems have been identified so far in circular mycobacterial plasmids and in other Actinomycetales linear plasmids, but we have identified a gene encoding a ParA homolog within the sequence of linear plasmid pSCL1 from S. clavuligerus (36). ORF3 may also be responsible for pCLP plasmid maintenance. A Blast search revealed strong amino acid sequence similarity between the product of ORF3 and PemK encoded by E. coli plasmid R100 and M. morganii plasmid R446b. PemI (also called Kis for killing suppressor) and PemK (also called Kid for killing determinant) are encoded by the pem operon, which is similar in its genetic organization and function to the components of several other toxin-antitoxin-encoding loci carried on plasmids such as the ccd and parDE operons of plasmids F and RK2, respectively (10). We expressed pCLP pemK in M. smegmatis using pMIP12, an expression vector for heterologous genes in mycobacteria. Expression of the pemK gene under pBlaF* control in this vector caused death of the transformed M. smegmatis, evidencing the killing function of PemK. The cotranscribed ORF2 and ORF3 may therefore constitute the pCLP pem operon. The region containing recognition motifs for the Pem proteins encoded by R100 and R446b was conserved in the pCLP pem operon. Sequence analysis of the M. tuberculosis chromosome has revealed several toxin-antitoxin-encoding loci (6, 10, 35). However, the PemK homolog encoded by pCLP was found to be more closely related to the PemK encoded by E. coli plasmid R100 than to toxin homologs of M. tuberculosis. Chromosomally encoded toxin-antitoxin modules are thought to be involved in the stringent response by suppressing growth under certain conditions (10). In conclusion, pCLP has two different systems that are responsible for its stable maintenance: the par system, which may use a mitosis-like apparatus to bring about a plasmid centromere-like site movement to partition replicons prior to cell division, and the pem system, a postsegregational killing system which inhibits the initiation of DNA replication in cells that have lost the pem+ plasmids. Neither of these two maintenance systems has been described so far for mycobacterial plasmids. Plasmid pCLP is therefore an interesting candidate for mycobacterial genetic studies which require long-term vector stability, especially for use in vaccination or in other in vivo studies.
The complete sequence of linear plasmid pCLP provides an interesting model for the study of replication and evolution of linear plasmids. Further characterization of the pCLP ORFs should provide insight into the replication of linear plasmids and into the transcribed genes of unknown functions, in addition to the involvement of linear plasmids in the spread of genes.
ACKNOWLEDGMENTS
This work received support from Novotech (Lyonnaise des Eaux), Sagep, and the European Commission (contract no. BMH4-CT97-2167). M.P. thanks the Fondation de France (prix Jacques Monod) for financial support.
We thank J. Rauzier for help with sequencing and I. Saint Girons for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson G E, Sharp P M. Codon usage in the Mycobacterium tuberculosiscomplex. Microbiology. 1996;142:915–925. doi: 10.1099/00221287-142-4-915. [DOI] [PubMed] [Google Scholar]
- 3.Berthet F X, Rasmussen P B, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosisoperon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10) Microbiology. 1998;144:3195–3203. doi: 10.1099/00221287-144-11-3195. [DOI] [PubMed] [Google Scholar]
- 4.Chang P C, Kim E S, Cohen S N. Streptomyceslinear plasmids that contain a phage-like, centrally located, replication origin. Mol Microbiol. 1996;22:789–800. doi: 10.1046/j.1365-2958.1996.01526.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen C W. Complications and implications of linear bacterial chromosomes. Trends Genet. 1996;12:192–196. doi: 10.1016/0168-9525(96)30014-0. [DOI] [PubMed] [Google Scholar]
- 6.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosisfrom the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Crespi M, Messens E, Caplan A B, van Montagu M, Desomer J. Fasciation induction by the phytopathogen Rhodococcus fasciansdepends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 1992;11:795–804. doi: 10.1002/j.1460-2075.1992.tb05116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dabrock B, Kesseler M, Averhoff B, Gottschalk G. Identification and characterization of a transmissible linear plasmid from Rhodococcus erythropolisBD2 that encodes isopropylbenzene and trichloroethene catabolism. Appl Environ Microbiol. 1994;60:853–860. doi: 10.1128/aem.60.3.853-860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Solar G, Giraldo R, Ruiz-Echevarria M J, Espinosa M, Diaz Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes K, Moller-Jensen J, Jensen R B. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 12.Gordon S V, Heym B, Parkhill J, Barrell B, Cole S T. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosisH37Rv. Microbiology. 1999;145:881–892. doi: 10.1099/13500872-145-4-881. [DOI] [PubMed] [Google Scholar]
- 13.Hayakawa T, Otake N, Yonehara H, Tanaka T, Sakaguchi K. Isolation and characterization of plasmids from Streptomyces. J Antibiot (Tokyo) 1979;32:1348–1350. doi: 10.7164/antibiotics.32.1348. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch J, Tilly K. Linear plasmids and chromosomes in bacteria. Mol Microbiol. 1993;10:917–922. doi: 10.1111/j.1365-2958.1993.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 15.Hiratsu K, Mochizuki S, Kinashi H. Cloning and analysis of the replication origin and the telomeres of the large linear plasmid pSLA2-L in Streptomyces rochei. Mol Gen Genet. 2000;263:1015–1021. doi: 10.1007/pl00008689. [DOI] [PubMed] [Google Scholar]
- 16.Huang C H, Lin Y S, Yang Y L, Huang S W, Chen C W. The telomeres of Streptomyceschromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol Microbiol. 1998;28:905–916. doi: 10.1046/j.1365-2958.1998.00856.x. [DOI] [PubMed] [Google Scholar]
- 17.Kalkus J, Dorrie C, Fischer D, Reh M, Schlegel H G. The giant linear plasmid pHG207 from Rhodococcussp. encoding hydrogen autotrophy: characterization of the plasmid and its termini. J Gen Microbiol. 1993;139:2055–2065. doi: 10.1099/00221287-139-9-2055. [DOI] [PubMed] [Google Scholar]
- 18.Kinashi H, Mori E, Hatani A, Nimi O. Isolation and characterization of linear plasmids from lankacidin-producing Streptomycesspecies. J Antibiot (Tokyo) 1994;47:1447–1455. doi: 10.7164/antibiotics.47.1447. [DOI] [PubMed] [Google Scholar]
- 19.Kosono S, Maeda M, Fuji F, Arai H, Kudo T. Three of the seven bphC genes of Rhodococcus erythropolisTA421, isolated from a termite ecosystem, are located on an indigenous plasmid associated with biphenyl degradation. Appl Environ Microbiol. 1997;63:3282–3285. doi: 10.1128/aem.63.8.3282-3285.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labidi A, David H L, Roulland-Dussoix D. Restriction endonuclease mapping and cloning of Mycobacterium fortuitumvar. fortuitum plasmid pAL5000. Ann Inst Pasteur Microbiol. 1985;136B:209–215. doi: 10.1016/s0769-2609(85)80045-4. [DOI] [PubMed] [Google Scholar]
- 21.Parsons L M, Jankowski C S, Derbyshire K M. Conjugal transfer of chromosomal DNA in Mycobacterium smegmatis. Mol Microbiol. 1998;28:571–582. doi: 10.1046/j.1365-2958.1998.00818.x. [DOI] [PubMed] [Google Scholar]
- 22.Pelicic V, Reyrat J M, Gicquel B. Positive selection of allelic exchange mutants in Mycobacterium bovisBCG. FEMS Microbiol Lett. 1996;144:161–166. doi: 10.1111/j.1574-6968.1996.tb08524.x. [DOI] [PubMed] [Google Scholar]
- 23.Picardeau M, Le Dantec C, Vincent V. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology. 2000;146:305–313. doi: 10.1099/00221287-146-2-305. [DOI] [PubMed] [Google Scholar]
- 24.Picardeau M, Vincent V. Characterization of large linear plasmids in mycobacteria. J Bacteriol. 1997;179:2753–2756. doi: 10.1128/jb.179.8.2753-2756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picardeau M, Vincent V. Mycobacterial linear plasmids have an invertron-like structure related to other linear replicons in actinomycetes. Microbiology. 1998;144:1981–1988. doi: 10.1099/00221287-144-7-1981. [DOI] [PubMed] [Google Scholar]
- 26.Polo S, Guerini O, Sosio M, Deho G. Identification of two linear plasmids in the actinomycete Planobispora rosea. Microbiology. 1998;144:2819–2825. doi: 10.1099/00221287-144-10-2819. [DOI] [PubMed] [Google Scholar]
- 27.Qin Z, Cohen S N. Replication at the telomeres of the Streptomyceslinear plasmid pSLA2. Mol Microbiol. 1998;28:893–903. doi: 10.1046/j.1365-2958.1998.00838.x. [DOI] [PubMed] [Google Scholar]
- 28.Redenbach M, Bibb M, Gust B, Seitz B, Spychaj A. The linear plasmid SCP1 of Streptomyces coelicolor A3(2) possesses a centrally located replication origin and shows significant homology to the transposon Tn4811. Plasmid. 1999;42:174–185. doi: 10.1006/plas.1999.1419. [DOI] [PubMed] [Google Scholar]
- 29.Saeki H, Akira M, Furuhashi K, Averhoff B, Gottschalk G. Degradation of trichloroethene by a linear-plasmid-encoded alkene monooxygenase in Rhodococcus corallinus (Nocardia corallina) B-276. Microbiology. 1999;145:1721–1730. doi: 10.1099/13500872-145-7-1721. [DOI] [PubMed] [Google Scholar]
- 30.Sakaguchi K. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol Rev. 1990;54:66–74. doi: 10.1128/mr.54.1.66-74.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 33.Timm J, Lim E M, Gicquel B. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J Bacteriol. 1994;176:6749–6753. doi: 10.1128/jb.176.21.6749-6753.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuchimoto S, Ohtsubo E. Autoregulation by cooperative binding of the PemI and PemK proteins to the promoter region of the pemoperon. Mol Gen Genet. 1993;237:81–88. doi: 10.1007/BF00282787. [DOI] [PubMed] [Google Scholar]
- 35.Tyagi J S, Das T K, Kinger A K. An M. tuberculosis DNA fragment contains genes encoding cell division proteins ftsX and ftsE, a basic protein and homologues of PemK and small protein B. Gene. 1996;177:59–67. doi: 10.1016/0378-1119(96)00271-5. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Roy K L. Complete nucleotide sequence of a linear plasmid from Streptomyces clavuligerusand characterization of its RNA transcripts. J Bacteriol. 1993;175:37–52. doi: 10.1128/jb.175.1.37-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]