Abstract
Replication arrests due to the lack or the inhibition of replicative helicases are processed by recombination proteins. Consequently, cells deficient in the Rep helicase, in which replication pauses are frequent, require the RecBCD recombination complex for growth. rep recA mutants are viable and display no growth defect at 37 or 42°C. The putative role of chaperone proteins in rep and rep recA mutants was investigated by testing the effects of dnaK mutations. dnaK756 and dnaK306 mutations, which allow growth of otherwise wild-type Escherichia coli cells at 40°C, are lethal in rep recA mutants at this temperature. Furthermore, they affect the growth of rep mutants, and to a lesser extent, that of recA mutants. We conclude that both rep and recA mutants require DnaK for optimal growth, leading to low viability of the triple (rep recA dnaK) mutant. rep recA mutant cells form colonies at low efficiency when grown to exponential phase at 30°C. Although the plating defect is not observed at a high temperature, it is not suppressed by overexpression of heat shock proteins at 30°C. The plating defect of rep recA mutant cells is suppressed by the presence of catalase in the plates. The cryosensitivity of rep recA mutants therefore results from an increased sensitivity to oxidative damage upon propagation at low temperatures.
Interconnections between DNA replication and homologous recombination have been observed in a number of organisms and are likely to play an important role in the maintenance of genome integrity (reviewed in references 19, 21, and 29). An additional link was found with the observation that recombination enzymes act in Escherichia coli to rescue blocked replication forks (40). rep mutants were used to study the fate of replication forks upon blockage. The rep mutation causes a slow progression of chromosomal replication forks which suggests the occurrence of frequent pauses (6, 23). Because the Rep helicase is able to displace a DNA-bound protein in vitro, it was proposed that in vivo Rep could facilitate chromosomal replication by dislodging DNA-bound proteins from the path of the replication forks (28, 46).
rep mutants require the recombination complex RecBCD for viability (43), suggesting a link between replication fork arrest and homologous recombination. RecBCD initiates homologous recombination of linear DNA and is therefore essential for the repair of DNA double-strand breaks. It binds to DNA double-strand ends and opens while simultaneously degrading the DNA. Upon encounter with a specific sequence named CHI, RecBCD promotes the formation of single-stranded DNA recognized by RecA (reviewed in references 20, 25, and 33). rep recBC mutant lethality results from the occurrence of RuvABC-dependent DNA double-strand breaks (30, 40). The RuvAB proteins bind to Holliday junctions and catalyze branch migration. The RuvAB-bound DNA is cleaved by RuvC, which resolves the recombination intermediates by introducing nicks in strands of opposite polarity (reviewed in reference 44). To account for the action of RuvABC at blocked replication forks, a model was proposed in which, upon replication arrest, a Holliday junction forms by annealing of the two nascent strands (40). In the absence of RecBCD, resolution of the RuvAB-bound DNA by RuvC leads to chromosomal breakage. In cells proficient for homologous recombination, reincorporation of the double-strand tail formed by replication fork reversal into the chromosome allows replication restart from a recombination intermediate. However, rep recA mutants defective for homologous recombination are viable. The viability of rep recA mutants depends on the exonuclease V activity of RecBCD (40, 43); therefore, we proposed that in rep recA mutants RecBCD may degrade the double-strand tail formed by replication fork reversal, allowing replication restart from a Y-structure.
In this work we further analyzed the properties of the rep recA double mutant. We tested the effects of dnaK mutations on the viability of rep recA mutants. DnaK is a member of the Hsp70 family of stress-induced proteins which are highly conserved in procaryotes and eucaryotes (1). It is one of the major chaperone proteins induced by a shift to a high temperature in E. coli. Like several other chaperone proteins, DnaK is involved in processes that protect cells against various stresses and plays a role in DNA replication (reviewed in reference 12). We report that dnaK mutations that do not affect the viability of wild-type strains affect the growth of rep mutants and are lethal in rep recA double mutants. This indicates that rep recA mutants depend on DnaK for growth.
A peculiar property of the rep recA mutants remains unexplained. Liquid cultures grown to exponential phase at 30°C exhibit a defect in plating efficiency (43). A normal plating efficiency is spontaneously recovered when cells reach the end of exponential phase or if cells are grown at 37 or 42°C. The best characterized origin of double-strand breaks is the presence of oxidative compounds, a natural consequence of aerobic growth. DNA is the primary site of lethal damages (16, 17, 24). As a first line of defense against oxidative stress, bacterially encoded catalases and superoxide dismutases prevent the accumulation of reactive oxygen species (reviewed in references 8 and 10). A second line of defense is a set of DNA repair enzymes. DNA damages include mainly base modifications and DNA single-strand and double-strand breaks (24; reviewed in reference 17). Consequently, cells that lack enzymes required for recombinational or base-excision DNA repair pathways (RecA, RecB, PolA, Xth) are killed by low doses of H2O2 (16). In contrast, the rep mutants are not more sensitive to H2O2 than wild-type strains (16). We explored the reason for the plating defect of rep recA mutant cells grown at 30°C. The plating defect was not suppressed by overexpression of heat shock proteins at 30°C, whereas it was suppressed by the presence of catalase in plates, suggesting that it results from oxidative damage.
MATERIALS AND METHODS
Strains and media.
Strains used in this work are described in Table 1. dnaK756 carries three mutations in dnaK (31). dnaK306 is a single mutant (45). Strains were constructed by P1 transduction. rep, recA, and rep recA derivatives of the dnaK306 and dnaK756 mutants were constructed at 30°C, and the isogenic rep recA double mutants were constructed at 37°C (with an MC1061 background for the dnaK306 mutant and a C600 background for the dnaK756 mutant, in contrast with the AB1157 background used for Fig. 3). Transductants obtained by introduction of the Δrep::kan allele in the dnaK756 mutant originally exhibited variable plating efficiencies at 40°C, but acquired during propagation at 30°C the capacity to form about 100% small colonies at 40°C in 24 to 48 h. One such clone, which was a rep (defective for M13 replication) and dnaK756 (defective for λ growth) mutant but which may have acquired a compensatory mutation facilitating its propagation at 30°C, was used to perform the experiments reported below. All other double mutants were obtained with the expected efficiency and had the expected phenotype (see below). P1 transduction of the Δ(recA-srl)::Tn10 mutation in the rep dnaK306 mutant strain led to very few clones, and only one Δ(recA-srl)::Tn10 clone was obtained with the rep dnaK756 mutant strain. This may result from the poor plating efficiency of these strains (see below). Since rep recA dnaK mutants exhibited the expected phenotype (UV sensitive, with an M13 replication defect and a λ growth defect), they were used for further experiments. The Rep− phenotype was verified by transformation of CaCl2 competent cells with M13mp2 DNA on a lawn of Hfr indicator strain, and recA mutants were verified by measuring their UV sensitivity. The dnaK756 mutation was verified as preventing growth of wild-type λ phages and replication of the mini-F plasmid. The dnaK306 mutation was verified as preventing mini-F replication. Plasmids pMob45 (dnaKJ carried by pMob vector; McMacken Laboratory) and pNRK416 (dnaK under lacUV5 promoter control; C. Gross Laboratory) were provided by Marie-Agnes Petit, and pCG179 (ptac12HrpoH+) was provided by Philippe Bouloc (42). Plasmid pDWS2, a pBR322 derivative carrying the recBCD region (35), was provided by G. Smith. Cells were grown in LBT medium (Luria broth [LB] supplemented with 25 mg of thymidine per ml). Sigma catalase from bovine liver was used at a final concentration of 150 U per ml.
TABLE 1.
Strains
| Strain | Relevant genotype | Construction or origin |
|---|---|---|
| CAG9270 | Wild type (C600) | C. Gross |
| CAG9271 | dnaK756 derivative of CAG9270 | C. Gross |
| GY9701 | recA938::cat miniF-kan-recA+ | R. Devoret |
| K1019 | rep71 ilv::Tn10 | N. Zinder |
| MC1061 | Wild type | M. Casadaban |
| TS154 | dnaK306 derivative of MC1061 | C. Gross |
| JJC16 | Δ(recA-srl)::Tn10 | Laboratory stock |
| JJC40 | Wild type (AB1157 hsdR) | Laboratory stock |
| JJC213 | Δrep::kan | Reference 38 |
| JJC356 | Δ(recA-srl)::Tn10 Δrep::kan | JJC213 ∗ P1 (JJC16) |
| JJC489 | rep71 ilv::Tn10 | JJC40 ∗ P1 (K1019) |
| JJC502 | recA938::cat rep71 ilv::Tn10 | JJC489 ∗ P1 (GY9701) |
| JJC526 | Δrep::kan | MC1061 ∗ P1(JJC213) |
| JJC527 | Δ(recA-srl)::Tn10 | MC1061 ∗ P1(JJC16) |
| JJC528 | dnaK306 Δrep::kan | TS154 ∗ P1(JJC213) |
| JJC529 | dnaK306 Δ(recA-srl)::Tn10 | TS154 ∗ P1(JJC16) |
| JJC530 | Δrep::kan Δ(recA-srl)::Tn10 | JJC526 ∗ P1(JJC16) |
| JJC531 | dnaK306 Δrep::kan Δ(recA-srl)::Tn10 | JJC528 ∗ P1(JJC16) |
| JJC546 | dnaK756 Δ(recA-srl)::Tn10 | CAG9271 ∗ P1(JJC16) |
| JJC547 | dnaK756 Δrep::kan | CAG9271 ∗ P1(JJC213) |
| JJC633 | Δ(recA-srl)::Tn10 | C600 ∗ P1(JJC16) |
| JJC634 | Δrep::kan | C600 ∗ P1(JJC213) |
| JJC636 | Δrep::kan Δ(recA-srl)::Tn10 | JJC634 ∗ P1(JJC16) |
| JJC677 | dnaK756 Δrep::kan Δ(recA-srl)::Tn10 | JJC547 ∗ P1(JJC16) |
FIG. 3.
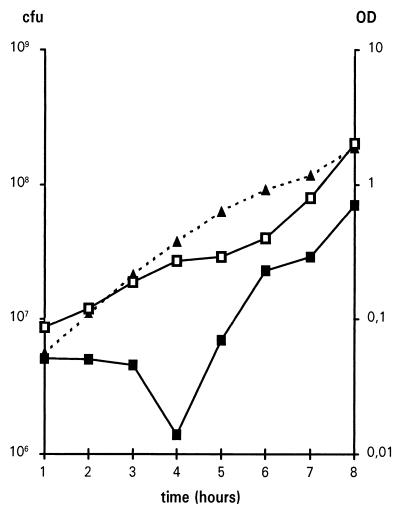
Growth curves of rep recA mutant (JJC502) at 30°C. Overnight cultures grown at 37°C were diluted to an OD of 0.03, and cells were incubated in LB at 30°C with shaking. Every hour, the OD at 650 nm was measured, and 100 μl of an appropriate dilution was plated on LBAT plates with or without catalase to determine the number of CFU/ml. The defect of plating was variable, from a plateau to a 100-fold decrease in CFU, while the recovery on catalase-containing plates was always observed. An experiment in which the decrease was of intermediate level is shown. Triangles, OD at 650 nm; closed squares, CFU on LBAT; open squares, CFU on LBAT containing 150 U of catalase per ml.
Micrographs of bacteria.
Cell samples were fixed with 4% paraformaldehyde phenylindole (Sigma, St. Louis, Mo.), deposited on the slides, dried, colored with 4,6-diamidino-2-phenylindole (DAPI; Sigma) (2.5 μg/ml), and added directly to glycerol–phosphate-buffered saline mountant (Citifluor Ltd., Canterbury, United Kingdom). Photographs of cells were taken using an epifluorescence microscope (Nikon) with Sensia 400 film (Fuji). The slides were scanned and processed with the Adobe Illustrator, 7.0, program (Edinburgh, Scotland).
RESULTS
dnaK mutations decrease the plating efficiencies of rep recA mutants at 30°C.
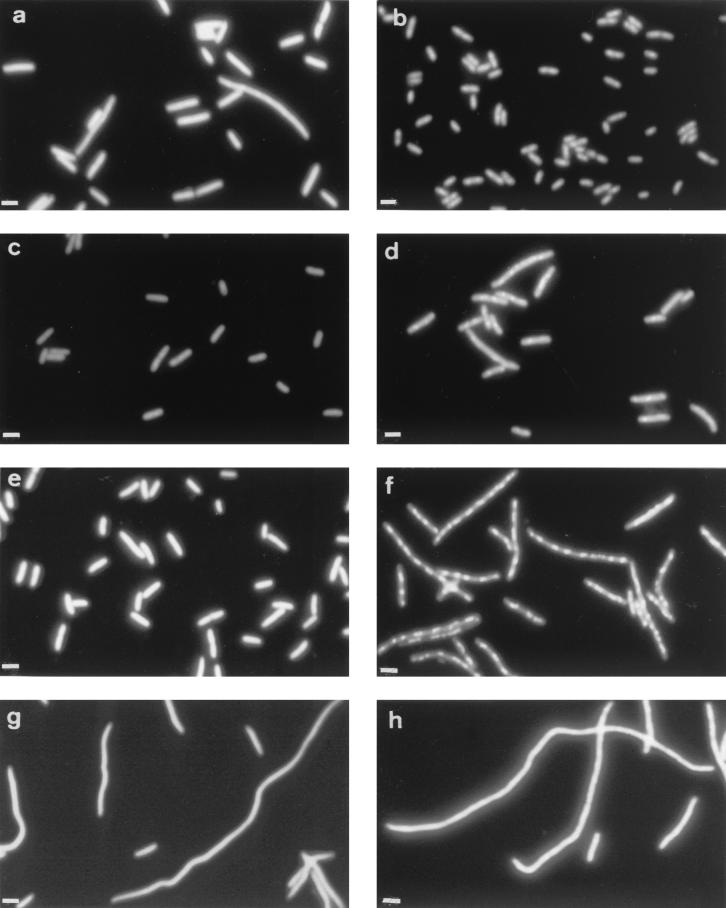
We studied the effects of mutations of the major heat-induced chaperone protein, DnaK, in rep, recA, and rep recA mutants. Two dnaK mutants, TS214 (dnaK306) and CAG9271 (dnaK756), that grow at 40°C were used (45). Either rep, recA, or both rep and recA null mutations were introduced in these two strains as well as in isogenic dnaK+ strains (see Material and Methods; Table 1). Cells were grown at 30°C to exponential phase and plated at the same temperature (Table 2). dnaK306 and dnaK756 mutations decreased the plating efficiency of rep mutant cells three- to fivefold in exponential and stationary phase. The term plating efficiency is used here as the ratio of CFU to optical density (OD). This ratio depends on (i) the average size of the cells (OD measures cell mass per ml) and (ii) the ability of individual cells to give rise to a colony. To further analyze the contribution of cell filamentation to the plating defect of these mutants, cells in exponential growth were examined by fluorescent microscopy and DAPI staining. Microscopic observations indicated that filamentation participates in the loss of plating efficiency for rep dnaK756 mutant cells (Fig. 1, compare panel g with panels a and c), whereas rep dnaK306 mutant cells were not significantly more elongated than dnaK306 single mutants (data not shown). recA single mutants presented a 20 to 50% plating defect compared to isogenic recA+ cells, as expected (3) (Table 2). In recA mutant cells, the dnaK306 mutation decreased the plating efficiency twofold without inducing significant filamentation (Table 2; data not shown), whereas the dnaK756 mutation had little effect (Table 2; Fig. 1e).
TABLE 2.
rep recA dnaK mutant cells present a plating defect at 30°Ca
| Strain | Mutant genotype | Exponential cultures
|
Overnight cultures
|
||
|---|---|---|---|---|---|
| CFU/ml | Relative value | CFU/ml | Relative value | ||
| MC1061 | Wild type | 1.1 × 108 ± 3.5 × 107 | 1 | 3.8 × 109 ± 7 × 107 | 1 |
| JJC526 | rep | 1.3 × 108 ± 1.1 × 107 | 1.2 | 2.8 × 109 ± 4.7 × 108 | 0.74 |
| JJC527 | recA | 4.7 × 107 ± 1.5 × 107 | 0.43 | 2.4 × 109 ± 2.5 × 108 | 0.63 |
| TS154 | dnaK306 | 1.4 × 108 ± 5.8 × 107 | 1.3 | 2 × 109 ± 2 × 108 | 0.5 |
| JJC529 | dnaK306 recA | 2 × 107 ± 1.3 × 107 | 0.18 | 1.2 × 109 ± 8 × 108 | 0.3 |
| JJC528 | dnaK306 rep | 4 × 107 ± 3.7 × 107 | 0.36 | 6.6 × 108 ± 2.5 × 108 | 0.17 |
| JJC530 | rep recA | 2.4 × 107 ± 1.8 × 106 | 0.23 | 8.7 × 108 ± 3.7 × 108 | 0.22 |
| JJC531 | dnaK306 rep recA | 3.4 × 106 ± 4.4 × 106 | 0.031 | 2.9 × 108 ± 1.4 × 108 | 0.07 |
| CAG9270 | Wild type | 1.3 × 108 ± 1.8 × 107 | 1 | 1.8 × 109 ± 8 × 108 | 1 |
| JJC634 | rep | 8 × 107 ± 3.4 × 107 | 0.61 | 1.6 × 109 ± 1.4 × 108 | 0.9 |
| JJC633 | recA | 1 × 108 ± 2.3 × 107 | 0.77 | 2.5 × 109 ± 2 × 108 | 1.4 |
| CAG9271 | dnaK756 | 1.4 × 108 ± 4.5 × 107 | 1.1 | 1.3 × 109 ± 2 × 108 | 0.72 |
| JJC546 | dnaK756 recA | 7 × 107 ± 3.4 × 107 | 0.53 | 9.5 × 108 ± 3.4 × 108 | 0.52 |
| JJC547 | dnaK756 rep | 2.8 × 107 ± 2.3 × 107 | 0.21 | 5 × 108 ± 5 × 107 | 0.28 |
| JJC636 | rep recA | 4.5 × 106 ± 2 × 106 | 0.03 | 4 × 108 ± 3 × 108 | 0.22 |
| JJC677 | dnaK756 rep recA | 1.2 × 106 ± 1.2 × 106 | 0.009 | 1.4 × 107 ± 9 × 106 | 0.008 |
Results are the average of three or four independent experiments. Cells were grown at 30°C and plated at the same temperature. Cells in exponential phase were harvested at an OD of 0.3 to 0.6 (most often at 0.4 to 0.5). Saturated cultures reached an OD of 1.5 to 4, depending on the strain. In order to compare the different cultures, results normalized for an OD of 0.4 (exponential cultures) or 2 (overnight cultures) were calculated.
FIG. 1.
Morphology of dnaK756 derivatives. Cells were grown overnight at 30°C, diluted 100-fold, and grown for 3 h at either 30°C (a, c, e, g) or 42°C (b, d, f, h). JJC213 (rep mutant; a, b), CAG9271 (dnaK756 mutants; c, d), JJC546 (dnaK756 recA mutant; e, f), and JJC547 (dnaK756 rep mutant; g, h) are shown. Wild-type cells were similar to JJC213 at 42°C and to CAG9271 at 30°C (data not shown). Bars, 1 μm.
The combination of rep and recA mutations decreased the plating efficiency of C600 cells in exponential phase and not in stationary phase, as previously observed in an AB1157 background (43) (compare CAG9270 and JJC636 in Table 2). The plating efficiency of the MC1061 rep recA mutant (JJC530) was higher (23% of that of the isogenic wild-type strain MC1061 [Table 2]), suggesting that the plating defect of rep recA mutants grown to exponential phase may depend on the cellular background. The plating efficiency of rep recA mutant cells was further decreased three- to sevenfold by dnaK306 or dnaK756 mutations, in part because of filamentation, since dnaK756 induced in rep recA mutant cells a level of filamentation similar to that observed in rep mutant cells (Fig. 1g) and dnaK306 induced a lower level (data not shown). In rep recA double mutants, the defect in the ability to form colonies is not accompanied by filamentation (data not shown). Interestingly, plating efficiency was not improved in overnight cultures of the rep recA dnaK756 mutant, suggesting that DnaK may participate in the recovery of plating efficiency of the rep recA mutant upon saturation (compare JJC636 and JJC677 overnight cultures in Table 2).
Taken together, these results show that (i) growth of rep, and to a lesser extent, recA mutant cells is affected at 30°C by dnaK756 or dnaK306 mutations (Fig. 1g; Table 2, compare recA with recA dnaK306 mutants) and (ii) both dnaK306 and dnaK756 mutations affect the growth of rep recA mutant cells at 30°C.
rep recA dnaK mutants are thermosensitive for growth.
To measure the capacity of the various strains to form colonies at a high temperature, overnight cultures were plated at 30 and 40°C (Table 3). For all strains the number of colonies was similar at both temperatures, with the exception of the two rep recA dnaK mutants, which did not form colonies at 40°C (Table 3). This shows that in the two dnaK mutants tested, the rep recA combination of mutations renders cells thermosensitive for growth.
TABLE 3.
rep recA dnaK306 and rep recA dnaK756 mutants are thermosensitive for growth
| Strain | Mutant genotype | Plating efficiencya |
|---|---|---|
| MC1061 | Wild type | 0.9 ± 0.1 |
| TS154 | dnaK306 | 0.9 ± 0.1 |
| JJC529 | dnaK306 recA | 0.6 ± 0.09 |
| JJC528 | dnaK306 rep | 0.7 ± 0.3 |
| JJC530 | rep recA | 1.1 ± 0.5 |
| JJC531 | dnaK306 rep recA | ≤0.00046 ± 0.00012b |
| CAG9270 | Wild type | 0.8 ± 0.4 |
| CAG9271 | dnaK756 | 0.9 ± 0.09 |
| JJC546 | dnaK756 recA | 1.1 ± 0.14 |
| JJC547 | dnaK756 rep | 0.95 ± 0.03 |
| JJC636 | rep recA | 1.1 ± 0.5 |
| JJC677 | dnaK756 rep recA | ≤0.046 ± 0.046b |
Overnight cultures grown at 30°C were plated at 30 or 40°C. For each culture, the ratio of clones obtained at 40°C to those obtained at 30°C was calculated, and the results are expressed as the average of the ratios.
A few clones were obtained at 40°C that were no longer thermosensitive, indicating that they probably have acquired a compensatory mutation that allows growth at a high temperature. In contrast, rep recA dnaK mutant clones obtained at 30°C were still thermosensitive.
The plating efficiency of rep dnaK and recA dnaK mutants was not affected by temperature (Table 3). However, at 40°C colonies were smaller than those of single dnaK mutants (Fig. 2). Microscopic observations showed that dnaK756 mutant cells were mildly elongated at 42°C (Fig. 1d). The recA mutation increased the number and the length of the filaments. For unknown reasons, it also increased the condensation of nucleoids (Fig. 1f). The rep dnaK756 mutant was found to be strongly filamentous (Fig. 1h). In contrast, dnaK306 single mutants (TS154) were more elongated at 42°C than dnaK756 mutants (data not shown), and when introduced in the dnaK306 mutant, the rep and recA mutations had a mild effect on cell morphology. In conclusion, our results indicate that both recA and rep mutants require a physiological concentration of chaperone proteins for normal growth and that this requirement is additive in rep recA mutant cells.
FIG. 2.
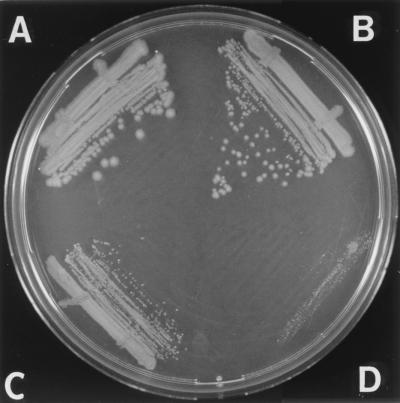
Colony growth of dnaK756 derivatives. CAG9271 (dnaK756 mutant; A), JJC546 (dnaK756 recA mutant; B), JJC547 (dnaK756 rep mutant; C), and JJC677 (dnaK756 rep recA mutant; D) are shown. Overnight cultures grown at 30°C were streaked on LB agar thymine (LBAT), and plates were incubated overnight at 40°C. The size of dnaK756 colonies was not significantly different from that of wild-type colonies at this temperature (data not shown).
Loss of plating of the rep recA mutants at 30°C results from oxidative damage.
The only noticeable growth defect of the rep recA mutant in laboratory conditions is a variable but significant defect in colony forming efficiency when cells in exponential phase at 30°C are plated on rich medium (43) (Table 2; Fig. 3). The plating defect is observed for liquid cultures propagated at 30°C regardless of the temperature at which the plates are then incubated. The low plating efficiency of the rep recA mutants is not observed when cells are grown at 37 or 42°C, suggesting that a higher temperature protects rep recA mutant cells. During growth at 42°C, the steady-state level of heat shock proteins is about twice as high as that at 30°C (12), which could be sufficient to restore a normal plating efficiency in rep recA mutants. In order to test whether the defect in the plating efficiency of AB1157 rep recA mutants at 30°C could be restored by an increased concentration in heat shock proteins, plasmids overexpressing DnaK (pNRK416) or DnaK and DnaJ (pMob-dnaKJ) were introduced in the rep recA mutant JJC356. Plating efficiency was not restored by the presence of either of these plasmids (data not shown). Hence, an increased level of DnaK and DnaJ is not sufficient to restore full viability of rep recA mutants. However, DnaK controls the expression of several other heat shock proteins (reviewed in reference 12). These proteins are repressed by overexpression of DnaK, and some of them may be required in rep recA mutant cells. The hypothesis that loss of colony-forming ability of rep recA mutant cells at 30°C is because of insufficient heat shock protein expression was therefore tested by overproduction of ς32, the rpoH gene product, which governs the heat shock response. The ptac12HrpoH+ plasmid was used (42). Overexpression of ς32 did not restore the plasmid efficiency of the rep recA mutant (data not shown). In conclusion, increasing the level of heat shock proteins did not restore the plating efficiency of the rep recA mutant, suggesting that heat shock proteins are not at a limiting concentration at 30°C in the rep recA mutant cells.
The defect in plating of rep recA mutant cells could result from a limiting amount of an enzymatic activity essential for the viability of rep recA mutant cells other than that of heat shock proteins. RecBCD is a good candidate because (i) it is present in only 10 to 20 copies per cell (20), (ii) it is essential in rep mutants, and (iii) it is titrated out in cells defective for RecA due to the extensive DNA degradation that occurs in these cells (22, 41). A decrease in the amount of available RecBCD enzyme in vivo can be detected with the use of T4 phages deficient for the gene 2 product that protects linear T4 molecules against exonuclease V-mediated degradation upon infection (22, 38). As previously reported (22), we observed that infection by T4 2− was increased by the presence of a recA mutation in the recipient cells. However, we found that it was not further modified by inactivation of rep and was not significantly influenced by temperature (data not shown). This suggests that rep recA mutant cells are not more deficient in exonuclease V activity at 30°C than at 42°C and are not more deficient than recA single mutants. To check more directly whether a decrease in the concentration of free RecBCD complexes could be responsible for the plating defect of rep recA mutants, we measured the plating efficiency of rep recA mutants containing the plasmid pDWS2, which carries the recBCD genes (35). Overexpression of RecBCD from this plasmid prevented growth of T4 2− mutant phages on rep recA mutants as expected, while it did not revert the plating defect of the strain (data not shown). We conclude that a lack of RecBCD cannot be the reason for this plating defect.
Interestingly, exogenous catalase has been shown to improve the recovery of E. coli cells under various stress conditions, for example, heat-injured DNA-repair mutants (27). In addition, bacterial catalase does not protect isolated organisms but favors the survival of high-density and colonial E. coli (26). To test whether the low plating efficiency of the rep recA mutant could result from a defect in the repair of oxidative damage that occurs upon plating (26), we measured the capability of this strain to form colonies on catalase-containing plates (Fig. 3). The number of CFU was restored by the presence of 150 U of catalase/ml in the LB plates. This result indicates that the defect in colony formation results from oxidative damage due to the presence of compounds degraded by catalase. The oxidative damage occurred upon plating and not during exponential growth in liquid medium, as the presence of catalase in the culture did not rescue the cells plated on catalase-lacking plates and did not further enhance the plating efficiency on catalase-containing plates (data not shown). We conclude that the low plating efficiency of the rep recA mutant propagated at low temperature could result from the additive effects of replication pauses due to the rep mutation and lesions due to the aerobic environment.
DISCUSSION
In this work, we studied the effects of dnaK mutations in rep, recA, and rep recA mutants and showed that two different dnaK mutations that do not prevent the growth of wild-type cells significantly impair the growth of rep and rep recA mutants. We also investigated the reasons for the cryosensitivity of rep recA mutants. We found that the presence of catalase in the plates relieves the plating defect of the strain, indicating that the defect in colony formation results from oxidative damage.
Role of DnaK in rep mutants.
The combination of rep and dnaK mutations is sufficient to impair growth. dnaK306 and dnaK756 mutations decrease the plating efficiency of rep mutants three- to fivefold independently of the growth phase (Table 2) and independently of the temperature (Table 3). The strong filamentation induced by the combination of rep and dnaK756 mutations is observed at low and high temperatures (Fig. 1g and h). It can be noted that the rep mutant cells are slightly elongated at 30°C, which may reflect their requirement for a high level of DnaK. The rep mutation affects the propagation of replication forks in E. coli, and the chaperone proteins DnaK and DnaJ play a role in the replication of several replicons. They are required for the initiation of F, P1, and lambda (reviewed in reference 5). In F and P1, they control the multimerization of the specific initiator protein encoded by the replicon. In λ, they appear to act by dissociating DnaB protein from the λ P protein, thereby allowing the helicase to act. They are also involved in E. coli chromosomal replication, where their role is less well understood. Deletion of DnaK causes temperature sensitivity for cell growth, abnormal cell division, and a reduced rate of replication at 30°C (2). dnaK, dnaJ, and grpE mutations cause a dramatic decrease in the level of epsilon, the proofreading subunit of DNA polymerase III, and a decrease in the apparent level of RNase H1 (9). DnaK and DnaJ are also required for the proper folding of UmuC, one of the subunits of DNA polymerase V (PolV) involved in replication restart from a lesion and lesion bypass (34, 37). The properties of the dnaK756 and dnaK306 mutants used in this work have been characterized (34, 45). These dnaK mutations prevent proteolysis (measured by degradation of the pyromycyl fragment) and mini-F plasmid replication. They differ in that the dnaK756 mutant is deficient for λ growth and proficient for UmuC folding, whereas the dnaK306 mutant has the opposite properties (34, 45). In rep mutants, chromosome replication is slowed down, which is compensated for by more replication forks per chromosome (6, 23). Replication pauses due to the Rep defect may be more frequent or longer in dnaK mutants, due to a role of chaperone proteins in the release of the obstacles that block replication forks in rep mutants. Alternatively, DnaK could facilitate replication restart after restoration of a replication fork by homologous recombination. In this case, as the defect due to the rep dnaK combination is also observed in recA mutants, DnaK would also participate in the restart of replication forks from arrested forks that have not recombined. Comparison of [3H]thymidine incorporation under various conditions did not allow us to detect a defect in nucleotide incorporation in the rep dnaK756 double mutant compared to the parental strains (data not shown). This suggests that the replication defect which causes the filamentation of the rep dnaK756 mutant is, as in rep mutants, compensated by overinitiation at the origin. Altogether, this work supports the hypothesis that chaperone proteins play a role in the normal propagation of replication forks.
Plating defect and oxidative damage.
The plating defect of the rep recA mutant indicates that lethal lesions occur in these cells upon cell isolation, and the recovery of plating efficiency upon addition of catalase in the plates shows that these lesions are caused by oxidative compounds. This is consistent with the observation that isolated cells are not protected against oxidative damage by their own catalase (26). This transient inactivation of the first line of defense, mediated by enzymes that destroy oxygen radicals, renders essential the action of the second line of defense, mediated by enzymes that repair lesions. Homologous recombination plays an essential role in this process. Oxidative lesions lead to the formation of single-strand and double-strand DNA breaks, known to be repaired by homologous recombination (4, 16). recA mutations also inactivate the induction of the SOS response, a set of genes involved in DNA repair. However, inactivation of SOS induction only in rep mutants has no effect on plating efficiency (43), suggesting that the defect in homologous recombination plays an essential role in the cryosensitivity of the rep recA mutant. In contrast with cells defective in homologous recombination, rep mutants are not more sensitive to H2O2 than wild-type strains (16); nevertheless, here rep mutations increase the sensitivity of recA mutants to oxidative stress. This suggests an additive effect of oxidative lesions and replication pauses due to the rep mutation. The rep recA mutant requires exonuclease V-mediated degradation for viability; however, we show here that the plating defect of rep recA mutant cells does not result from a lack of RecBCD. The plating defect was suppressed at 42°C; however, it was not suppressed by overexpression of heat shock proteins. The Rep helicase is very similar to the repair helicase UvrD (11), and rep uvrD double mutants are lethal. Interestingly, a mutation has been described in the uvrD gene (uvrD307) (47) that also specifically decreases the plating efficiency of certain wild-type strains of E. coli at 30°C. Whether the cryosensitivities of the uvrD307 mutant and of rep recA mutants have a common origin remains to be determined.
DnaK and oxidative damage.
A protective effect of heat shock proteins against oxidative damage has been previously documented in several studies. Heat shock proteins, including DnaK, are induced by H2O2 treatment (32; reviewed in reference 8). A dnaK null mutation increases the sensitivity of E. coli to H2O2 (7). We report here that rep recA mutant cells require the following for full viability: (i) DnaK and (ii) protection against oxidative damage. However, no direct link between these two observations could be found. If the low level of heat shock proteins at 30°C participates in the plating defect of rep recA mutant cells, it is not the only cause, since overproduction of heat shock proteins does not restore the plating efficiency. Conversely, the growth defect of rep recA dnaK mutant cells at high temperature does not result only from oxidative damage: plating on catalase-containing plates of either rep recA dnaK mutant increased 10- to 100-fold the number of colonies at 40°C (data not shown); however, all colonies tested had acquired a suppressor mutation, suggesting that catalase only permits some residual growth that facilitates the acquisition of suppressor mutations, without restoring viability. Therefore, the observation that rep mutants are handicapped by either recA or dnaK mutations and that these handicaps are additive suggests that RecA and DnaK do different things that increase survival.
ACKNOWLEDGMENTS
We thank I. Matic, M. Radman, and F. Taddei for suggesting to us the use of catalase. We are very grateful to S. Kulaskauskas and to J. Tremblay for the bacterial micrographs. We thank M. A. Petit for careful reading of the manuscript and an anonymous referee for insightful suggestions.
B.M. is on the CNRS staff. This work is supported in part by the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses.
REFERENCES
- 1.Ang D, Liberek K, Skowyra D, Zylicz M, Georgopoulos C. Biological role and regulation of the universally conserved heat shock proteins. J Biol Chem. 1991;266:24233–24236. [PubMed] [Google Scholar]
- 2.Bukau B, Walker G C. ΔdnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J Bacteriol. 1989;171:6030–6038. doi: 10.1128/jb.171.11.6030-6038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capaldo F N, Barbour S D. Isolation of nonviable cells produced during normal growth of recombination deficient strains of Escherichia coli K12. J Bacteriol. 1973;115:928–936. doi: 10.1128/jb.115.3.928-936.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlsson J, Carpenter V S. The recA+ gene product is more important than catalase and superoxide dismutase in protecting Escherichia coli against hydrogen peroxide toxicity. J Bacteriol. 1980;142:319–321. doi: 10.1128/jb.142.1.319-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chattoraj D K. Role of molecular chaperones in the initiation of plasmid DNA replication. Genet Eng (NY) 1995;17:81–98. [PubMed] [Google Scholar]
- 6.Colasanti J, Denhardt D T. The Escherichia coli rep mutation. X. Consequences of increased and decreased Rep protein levels. Mol Gen Genet. 1987;209:382–390. doi: 10.1007/BF00329669. [DOI] [PubMed] [Google Scholar]
- 7.Delaney J M. Requirement of the Escherichia coli dnaK gene for thermotolerance and protection against H2O2. J Gen Microbiol. 1990;136:2113–2118. doi: 10.1099/00221287-136-10-2113. [DOI] [PubMed] [Google Scholar]
- 8.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster P L, Marinus M G. Levels of epsilon, an essential replication subunit of Escherichia coli DNA polymerase III, are controlled by heat shock proteins. J Bacteriol. 1992;174:7509–7516. doi: 10.1128/jb.174.23.7509-7516.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 11.Gilchrist C A, Denhardt D T. Escherichia coli rep gene: sequence of the gene, the encoded helicase, and its homology with uvrD. Nucleic Acids Res. 1987;15:465–475. doi: 10.1093/nar/15.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross C. Function and regulation of heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umberger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 13.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umberger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1497–1512. [Google Scholar]
- 14.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 15.Imlay J A, Chin S M, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 16.Imlay J A, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol. 1986;166:519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 18.Jenkins D E, Schultz J E, Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1988;170:3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuzminov A, Stahl F W. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J Bacteriol. 1997;179:880–888. doi: 10.1128/jb.179.3.880-888.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane H E, Denhardt D T. The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J Mol Biol. 1975;97:99–112. doi: 10.1016/s0022-2836(75)80025-8. [DOI] [PubMed] [Google Scholar]
- 24.Lesko S A, Lorentzen R J, Ts'o P O. Role of superoxide in deoxyribonucleic acid strand scission. Biochemistry. 1980;19:3023–3028. doi: 10.1021/bi00554a029. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umberger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 2236–2255. [Google Scholar]
- 26.Ma M, Eaton J W. Multicellular oxidant defense in unicellular organisms. Proc Natl Acad Sci USA. 1992;89:7924–7928. doi: 10.1073/pnas.89.17.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackey B M, Seymour D A. The effect of catalase on recovery of heat-injured DNA-repair mutants of Escherichia coli. J Gen Microbiol. 1987;133:1601–1610. doi: 10.1099/00221287-133-6-1601. [DOI] [PubMed] [Google Scholar]
- 28.Matson S W, Bean D W, George J W. DNA helicases—enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 29.Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 30.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki T, Tanaka S, Fujita H, Itikawa H. DNA sequence analysis of the dnaK gene of Escherichia coli B and of two dnaK genes carrying the temperature-sensitive mutations dnaK7(Ts) and dnaK756(Ts) J Bacteriol. 1992;174:3715–3722. doi: 10.1128/jb.174.11.3715-3722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan R W, Christman M F, Jacobson F S, Storz G, Ames B N. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci USA. 1986;83:8059–8063. doi: 10.1073/pnas.83.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers R S, Stahl F W. chi and the RecBCD enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 34.Petit M A, Bedale W, Osipiuk J, Lu C, Rajagopalan M, Mcinerney P, Goodman M F, Echols H. Sequential folding of UmuC by the Hsp70 and Hsp60 chaperone complexes of Escherichia coli. J Biol Chem. 1994;269:23824–23829. [PubMed] [Google Scholar]
- 35.Ponticelli A S, Schultz D W, Taylor A F, Smith G R. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985;41:145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- 36.Privalle C T, Fridovich I. Induction of superoxide dismutase in Escherichia coli by heat shock. Proc Natl Acad Sci USA. 1987;84:2723–2726. doi: 10.1073/pnas.84.9.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 38.Rinken R, Thoms B, Wackernagel W. Evidence that RecBC-dependent degradation of duplex DNA in Escherichia coli recD mutants involves DNA unwinding. J Bacteriol. 1992;174:5424–5429. doi: 10.1128/jb.174.16.5424-5429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rockabrand D, Livers K, Austin T, Kaiser R, Jensen D, Burgess R, Blum P. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J Bacteriol. 1998;180:846–854. doi: 10.1128/jb.180.4.846-854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 41.Skarstad K, Boye E. Degradation of individual chromosomes in recA mutants of Escherichia coli. J Bacteriol. 1993;175:5505–5509. doi: 10.1128/jb.175.17.5505-5509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilly K, Pierce J, Georgopoulos C. Modulation of stability of the Escherichia coli heat shock regulatory factor ς32. J Bacteriol. 1989;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uzest M, Ehrlich S D, Michel B. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol Microbiol. 1995;17:1177–1188. doi: 10.1111/j.1365-2958.1995.mmi_17061177.x. [DOI] [PubMed] [Google Scholar]
- 44.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 45.Wild J, Kamath-Loeb A, Ziegelhoffer E, Lonetto M, Kawasaki Y, Gross C A. Partial loss of function mutations in DnaK, the Escherichia coli homologue of the 70-kDa heat shock proteins, affect highly conserved amino acids implicated in ATP binding and hydrolysis. Proc Natl Acad Sci USA. 1992;89:7139–7143. doi: 10.1073/pnas.89.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yancey-Wrona J E, Matson S W. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992;20:6713–6721. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang G, Deng E X, Baugh L R, Hamilton C M, Maples V F, Kushner S R. Conserved motifs II to VI of DNA helicase II from Escherichia coli are all required for biological activity. J Bacteriol. 1997;179:7544–7550. doi: 10.1128/jb.179.23.7544-7550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]