Abstract
The biologically critical, exquisite specificity and efficiency of nucleases, such as those acting in DNA repair and replication, often emerge in the context of multiple other macromolecules. The evolved complexity also makes biologically relevant nuclease assays challenging and low-throughput. Meiotic recombination 11 homolog 1 (MRE11) is an exemplary nuclease that initiates DNA double-strand break (DSB) repair and processes stalled DNA replication forks. Thus, DNA resection by MRE11 nuclease activity is critical for multiple DSB repair pathways as well as in replication. Traditionally, in vitro nuclease activity of purified enzymes is studied either through gel-based assays or fluorescence-based assays like fluorescence resonance energy transfer (FRET). However, adapting these methods for a high-throughput application such as inhibitor screening can be challenging. Gel-based approaches are slow, and FRET assays can suffer from interference and distance limitations. Here we describe an alternative methodology to monitor nuclease activity by measuring the small-angle X-ray scattering (SAXS) interference pattern from gold nanoparticles (Au NPs) conjugated to 5′-ends of dsDNA using X-ray scattering interferometry (XSI). In addition to reporting on the enzyme activity, XSI can provide insight into DNA-protein interactions, aiding in the development of inhibitors that trap enzymes on the DNA substrate. Enabled by efficient access to synchrotron beamlines, sample preparation, and the feasibility of high-throughput XSI data collection and processing pipelines, this method allows for far greater speeds with less sample consumption than conventional SAXS techniques. The reported metrics and methods can be generalized to monitor not only other nucleases but also most other DNA-protein interactions.
Keywords: DNA repair, Small-angle X-ray scattering, Gold nanoparticles, Nuclease assay, MRE11A, X-ray scattering interferometry
1. Introduction
Meiotic recombination 11 homolog 1 (MRE11) is a critical nuclease that initiates DNA double-strand break (DSB) repair and processes stalled DNA replication forks [1–3]. Thus, DNA resection by MRE11 nuclease activity is important for multiple DSB repair pathways as well as in replication. The crystal structures of MRE11 show that the active site of MRE11 contains two Mn2+ ions with the protein forming a dimer both in an apo state and in DNA-bound states [4–6]. MRE11 is one of the first proteins to respond to DNA damage causing DSBs [1–3]. The DSBs are mainly repaired through either homologous-recombination (HR) that repairs the DSB in an error-free fashion or through non-homologous end-joining (NHEJ) pathway which may result in deletions or insertions in the repaired DNA. The first step in the HR repair pathway is MRE11-mediated resection (in the 3′–5′ direction) of the DSB leading to 3′ ssDNA overhangs [1]. These ssDNA overhangs inhibit NHEJ which requires very little processing of the broken DNA ends. Thus, MRE11 activity is the key determinant of whether DSBs are resected through HR or NHEJ [7].
In general, in vitro nuclease activity of purified enzymes is studied either through gel-based assays or fluorescence-based assays like fluorescence resonance energy transfer (FRET). However, adapting these methods for a high-throughput application such as inhibitor screening can be challenging. Gel-based approaches are slow, and FRET assays can suffer from interference and distance limitations (~1–10 nm) [8]. In the current method, we are combining our expertise in small-angle X-ray scattering (SAXS) with the scattering power of gold nanoparticles (Au NPs) conjugated to dsDNA substrates (Au-dsDNA). As will be demonstrated, when Au NPs are held at fixed distances on the 5′-ends of dsDNA, they act as molecular rulers through X-ray scattering interferometry (XSI) and can be used as a high-throughput technique to measure the binding and nuclease activity of MRE11 or other proteins that interact with DNA.
In biological research, SAXS is empowering for structural characterization of biomacromolecules at near physiological conditions [9]. Molecular assemblies, conformational changes, and flexibility can be robustly analyzed from a properly performed SAXS experiment [10]. SAXS is generally performed in solution with modest sample requirements, probing sub-nm distances and microsecond time-scales at many synchrotrons around the world. Coupled with sample handling robotics or microfluidics, impactful measurements can be made in high throughput. Although directional information is lost due to orientational averaging of the macromolecules relative to the probing beam, SAXS provides critical dynamics information to complement atomic resolution techniques like macromolecule X-ray crystallography (MX), nuclear magnetic resonance (NMR), and cryogenic-electron microscopy (cryo-EM) where both the distances and direction between atoms can be recorded [11]. Yet, high-resolution structures from MX or cryo-EM can typically only be attained on specific constructs that are sufficiently homogeneous in conformation and assembly. Fortunately, using these experimental models and SAXS data, models of the full-length or alternate conformations can be determined along with information on flexibility, assembly, and conformational states [12]. SAXS is the right balance between information and throughput for many biological systems [9].
In a typical SAXS experiment, X-ray scattering from the biomacromolecule is measured in a buffer solution. The particle scattering intensity I(q) is a function of momentum transfer q = (4π sin θ)/λ, where 2θ is the scattering angle and λ is the wavelength of the incident X-ray beam. I(q) can be derived from the electron distribution within the biomacromolecule as:
where r is the distance between election pairs within the macromolecule which leads to a statistical distribution of electron pair distances, or pair-distribution function, P(r), where the maximal dimension, Dmax, of a molecule is found as the function goes to zero [13, 14].
SAXS is inherently a contrast measurement technique where the signal is derived from differences in electron density Δρ(r) between biomolecule ρ(r) and that of the bulk solvent ρ(s) [14] as:
The approximate values for electron density of protein, DNA, and bulk solvent (pure water) are 0.43, 0.55, and 0.33 e−/Å3, respectively [15]. Given that the differences in the electron density between the biomolecules of interest and the buffer are already small, very minor fluctuations in buffer composition used for subtraction can greatly affect the results. Thus, a reasonable concentration of the analyte and a careful buffer subtraction are essential for obtaining useful information in SAXS experiments. To overcome the challenges in producing the large amounts of protein required for large-scale assays and the sensitivity of buffer fluctuations, high-throughput XSI can be used. This technique expands upon all of the same physical phenomenon of conventional SAXS by utilizing the interference pattern generated not between atom pairs but rather between heavy atom clusters (e.g., Au NPs) held at fixed distances by biomolecules (e.g., dsDNA). These Au NPs function as slits in reciprocal space to the atomic scale wavelengths of hard X-rays in an analogous way as the physical slits in Young’s experiments with visible light from classical physics [16]. Importantly, Au NPs having significantly more electron density (5-nm NP ~4.6 e−/Å3) scatter X-rays with ~200-fold greater intensity as compared to a 172-kDa protein or 5400-fold higher than that of a 31-bp dsDNA since scattering intensity on an absolute scale, I(0), (where q = 0) is proportional to the square of the number of electrons (m) in a particle [17] as:
where N is the number of particles and ψ is ratio between the particle volume and its number of electrons. The original idea of measuring scattering from heavy metals in a biomolecule was proposed in as early as the late 1940s and successfully performed in 1980 [18]. In 2008, Mathew-Fenn et al. were the first group to use Au-dsDNA as molecular rulers via XSI [19], applying this technique to measuring the double helix with exceptional accuracy [20]. The P(r) functions derived from these experiments can be divided into two major peak regions. One corresponding to intra-Au and another for the inter-Au distances [17, 20, 21]. Using this approach, inter-particle distances between 2 Au NPs separated by up to 100-bp have been accurately measured [21], and greater distances are presumed possible. Others have followed this technique, studying Au NPs conjugated to DNA, RNA, and even proteins using XSI [22–27].
Our group has applied the XSI technique to probe the mismatch repair of MutS/L, demonstrating that the technique can be used to study damage-specific structural changes in the DNA caused by MutS/L [17]. This study focused on the qualitative changes in the inter-particle distances providing information on the DNA-protein interactions. In our study of MRE11 nuclease activity, we sought to observe such DNA-protein interactions as well as develop a more quantitative assay towards the future of high-throughput XSI experiments. As such we have designed Au-DNA substrates of two different lengths (37-bp and 57-bp) conjugated to 10 nm Au NPs via a Trithiol (TrT) (Letsinger’s type) linker on the 5′-end annealed to a shorter (9-bp) duplex forming ssDNA oligo leaving a long stretch of ssDNA available for MRE11 binding. In both substrate cases, the inter-Au distance distributions are shifted to lower mean values compared to the substrate alone for samples where nuclease activity was not observed indicating structural changes in the DNA associated with MRE11 binding. As expected, MRE11 nuclease activity decreases the population of doubly Au Nps-labelled dsDNA as observed by the decrease in the amplitude of P(r) corresponding only to inter-Au distances. The intra-Au regions remain unperturbed as the amount of Au NPs is not changing. The P(r) functions are normalized to the intra-Au peak during analysis to account for any minor fluctuations in Au NP concentration. From these XSI assays, we observe that MRE11 is not active when the active site metal (Mn2+) is not present in the reaction buffer or a nuclease-dead mutant (H129N) is used in the reaction instead of the wild-type (WT) enzyme.
Prior to analysis, we validated that the substrate is cleaved by MRE11 via gel-based assays using the same substrates as in the XSI experiments except with Fluorescein (6-FAM) substituted for the TrT linkers and Au NPs on the 5′-ends. In general, it is useful (but not essential) to have an independent assay for protein-DNA interactions. For MRE11, the gel-based activity assay data agreed with our XSI assays showing that the nuclease activity is only observed in the reaction with the WT enzyme in the presence of MnCl2. Additionally, these gel-based assays indicate that MRE11 can cut on both strands, and on the longer strand, it can chew all the way to the 5′-end of the DNA.
Since SAXS probes all molecules in a solution, homogeneous samples are often used [9]. In DNA repair and damage responses, there is a need to examine enzyme activities where their active states may be in complexes that are transient and dynamic. To address this challenge, we combined the efficiency of SAXS with the high contrast of Au and present here a SAXS method with Au-labeled DNA as a robust prototypic assay on DNA processing. More specifically, these experiments can be carried out in a variety of solution conditions, in high throughput, provide sub-nm resolution at low concentrations, and have the inherent potential to categorize sub-millisecond reaction steps. Furthermore, many DNA repair processes have longer DNA footprints than are comfortably assayed using FRET. This method and the approach defined here for MRE11 can complement and extend more traditional, fluorescent-based assays. High-throughput XSI has a robust ability to test combinations and additives including other macromolecules without loss of signal. These protocols offer strategic and tactical advantages for studies to identify novel inhibitors from screening chemical libraries with the expectation that 1000 experiments can be done weekly and with batched compounds 10,000 compounds can be screened in 1 week.
In the following sections, we describe our XSI method to conjugate DNA substrate to the Au NPs for XSI experiments and data analysis protocol. We employ MRE11 as an example, but by changing the design of the DNA substrate, this method can be applied for many other enzymes that are known to cause structural changes in DNA including major types of DNA damage responses. For example, some DNA repair proteins of biological interest bind DNA without making any chemical alterations to control pathway selection [28]; however, if these bend DNA or otherwise alter the distance between DNA ends as they typically do, then XSI will provide a sensitive high-throughput measure of their interactions. We therefore expect XSI will be able to interrogate the impacts of proteins and RNA binding to DNA repair and replication complexes, ranging from scaffold proteins such as XRCC1 that is essential to micro-homology-mediated end joining [29] to RNA that can act in efficient DSB repair machines [30], to PAR clouds at DNA damage controlled by poly(ADP-ribose) polymerase (PARP1) and poly(ADP-ribose) glycohydrolase (PARG) whose inhibitors are actively being pursued for cancer therapy [31, 32], and even to G-quadruplex, repetitive sequence elements, and other non-B DNA sequences associated with DNA instability and mutation sites [33–35]. We demonstrate how to leverage this technique for use at a researcher’s home institution as well as how to take advantage of the mail-in user program of the SIBYLS beamline at the Advanced Light Source (ALS) at Lawrence Berkeley National Laboratory (LBNL) helping to design and carry out experiments like those mentioned herein.
2. Materials
2.1. Preparation of BSPP Protected Au Nanoparticles Via BSPP-Citrate Exchange (Au-BSPP)
15 mg/mL BSPP solution: Dissolve 375 mg bis(p-sulfonatophenyl)phenylphosphine (BSPP) in 25 mL ddH2O.
5 M NaCl solution: Dissolve 146 g sodium chloride (NaCl) in 400 mL ddH2O, and then add ddH2O until total volume equals 500 mL.
100 mM Phosphate buffer (PBS) pH 7: Dissolve 7.744 g of sodium phosphate dibasic heptahydrate (Na2HPO4·7H2O) and 2.913 g of sodium phosphate monobasic monohydrate (NaH2PO4·H2O) in 400 mL ddH2O. Adjust pH to 7 using HCl or NaOH, and then add ddH2O until total volume equals 500 mL.
Au-BSPP storage buffer (15 mM PBS, 1 mg/mL BSPP, 1 mM TCEP, pH 6.4): Dissolve 50 mg BSPP and 14 mg Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 30 mL ddH2O. Add 15 mL 100 mM phosphate buffer (PBS) pH 7 to solution. Adjust pH to 6.4 using HCl or NaOH, and then add ddH2O until total volume equals 50 mL.
10 nm Au NP: Purchased from Ted Pella.
2.2. Au-ssDNA Conjugation, Anion Exchange Chromatography, and Au-dsDNA Annealing
5′ Tri-thiolated ssDNA in solution: Purified/lyophilized ssDNA sequences with a Trithiol (Letsinger’s type) modification to the 5′-end are purchased from Fidelity Oligos at ~100 nmole scale (Table 1) and are re-hydrated in 0.5 mL ddH2O.
SH-PEG solution: Thiolated poly(ethylene glycol) (SH-PEG), MW = 356.5 was purchased from PolyPure (Oslo, Norway), and 20 μL SH-PEG is added to 480 μL ddH2O.
High salt FPLC buffer (15 mM Tris, 1 M NaCl, pH 8): Dissolve 58.44 g NaCl and 1.82 g tris(hydroxymethyl)-aminomethane (Tris) in 900 mL ddH2O. Adjust pH to 8 using HCl or NaOH, and then add ddH2O until total volume equals 1 L.
No salt FPLC buffer (15 mM Tris, pH 8): Dissolve 1.82 g Tris in 990 mL ddH2O. Adjust pH to 8 using HCl or NaOH, and then add ddH2O until total volume equals 1 L.
FPLC AKTA purifier for an automated anion-exchange chromatography.
Table 1.
Table showing DNA substrate sequences

|
2.3. Protein Expression and Purification
The catalytic domain of human MRE11 nuclease (1–411) selected is based on the previous report [6] and is cloned into pET series expression vector with an N-terminus His-tag (Addgene#29653). Surface-exposed methionines (M26, M84, M157, M309, M343) are modified to leucines for improving the protein stability; the modified MRE11 construct maintains the nuclease activity as the parental construct.
DH5α chemical competent cells (Thermo Fisher).
Rosetta™ chemical competent cells (Novagen).
BD Difco™ LB Broth, Miller (Luria-Bertani) media.
BD Difco™ LB Agar, Miller (Luria-Bertani) media.
Kanamycin sulfate UPS grade (Teknova).
37 °C incubator and refrigerated shaker.
Lysis buffer (50 mM Tris (pH = 7.5), 500 mM KCl, 5% glycerol, 0.5% T-20, 1 mM TCEP, protease inhibitors).
Buffer A (25 mM Tris (pH = 7.5), 300 mM NaCl, 2.5% glycerol, 1 mM TCEP, 20 mM imidazole).
Buffer B (25 mM Tris (pH = 7.5), 300 mM NaCl, 2.5% glycerol, 1 mM TCEP, 500 mM imidazole).
SEC buffer (20 mM Tris (pH = 8.0), 200 mM NaCl, 0.1 mM EDTA, 5 mM DTT).
HisTrap FF Crude pre-packed 5 mL column (GE/Cytiva).
Hi Load™ 16/600 Superdex200 pg (GE/Cytiva).
FPLC AKTA Pure system (GE/Cytiva) for automated affinity and size-exclusion chromatography.
Thermo Scientific NanoDrop 2000 Spectrophotometer.
2.4. DNA Substrate Preparation for the Fluorescence-Based Nuclease Reaction
DNA sequences (from IDT) used in the gel-based assay are given in Table 1 and annealed using a PCR machine.
2.5. Fluorescence-Based Nuclease Reaction to Validate Substrates and the Activity
Nuclease reaction buffer for FAM-based detection (25 mM HEPES (pH = 7), 50 mM KCl, ±1 mM MnCl2).
PCR machine.
To make 3× Stop Buffer, mix 0.5 mL formamide, 0.12 mL of 0.5 M EDTA (pH = 8), 0.25 mL of 100% glycerol, and 0.15 mL of 10% SDS.
4–20% Mini-PROTEAN® TGX Stain-Free protein gels (Bio-Rad).
15% Mini-PROTEAN® TBE-Urea gel (Bio-Rad).
Mini-PROTEAN® Tetra Vertical electrophoresis cell and PowerPac™ power supply.
2.6. Sample Preparation of XSI Experiments
Nuclease reaction buffer for SAXS-based detection (25 mM MOPS, 60 mM KCl, 0.2% T-20, pH 7): Dissolve 2.89 g (3-(N-morpholino)propanesulfonic acid) (MOPS), 2.24 g potassium chloride (KCl), and 1.095 g Tween-20 in 980 mL ddH2O. Adjust pH to 7 using HCl or KOH, and then add ddH2O until total volume equals 1 L.
20 mM MnCl2 solution: Dissolve 125.8 mg in 50 mL ddH2O.
PCR machine.
3. Methods
3.1. Preparation of BSPP Protected Au Nanoparticles Via BSPP-Citrate Exchange (Au-BSPP)
Add 25 mL of 15 mg/mL BSPP to 400 mL of either purchased 10 nm colloidal Au NPs, and filter solution through 0.22 μm filter.
Stir 400 mL citrate-stabilized colloidal Au NPs with BSPP overnight.
Add 5 M NaCl until the solution turns from red to dark red/-purple (~75 mL).
Pour into Beckman 100 mL polypropylene bottles w/cap assembly.
Spin in Beckman centrifuge in JA-18 rotor @ 12,000 G for 10 min.
Decant slowly, or pipette off supernatant (see Note 1).
Use 0.5 M NaCl solution to wash NPs, sonicate, and repeat step 6.
Repeat step 7 twice.
Resuspend in 25 mL Au-BSPP storage buffer.
3.2. Au-ssDNA Conjugation, Anion Exchange Chromatography, and Au-dsDNA Annealing
One limitation of Au NP conjugation to DNA is feasible only in the 5′ end of the DNA; thus, we designed a substrate (Table 1) that would be cut by MRE11 as well as leads to separation of paired Au NPs upon the nuclease reaction.
For the conjugation, measure the concentrations of Au-BSPP and ssDNA solutions (diluted appropriately; see Note 2) using Thermo Scientific NanoDrop 2000 Spectrophotometer at 520 and 260 nm, respectively.
Calculate concentration using Beer’s law and the appropriate extinction coefficients for Au NPs and ssDNA (see calculation Note 3).
Colloidal Au-BSPP and selected ssDNA solutions are mixing at a mole ratio of 3:1 and shaken gently at room temperature (RT) overnight.
SH-PEG solution is added to final to mixture at v/v% ratio of 10% (i.e., 100 μL added to 1000 μL solution), and mixture is shaken gently at RT for 2 h.
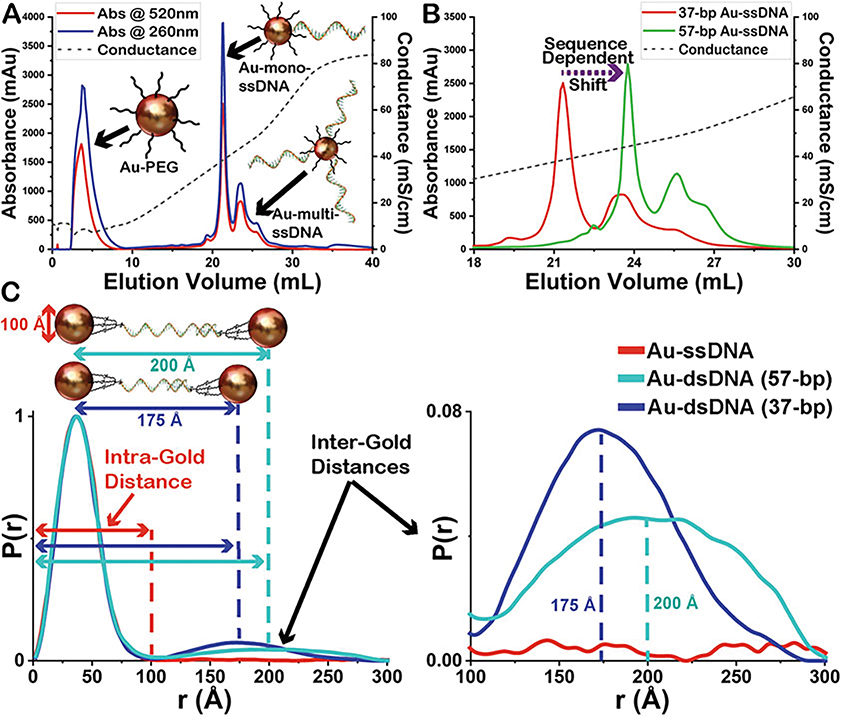
Separate and collected mono-conjugated Au-ssDNA from multi-conjugated using a Dionex DNA-Pac PA100 anion exchange column on an AKTA series fast protein liquid chromatography (FPLC) (see FPLC Method Note 4, Fig. 1a, b).
Complementary Au-ssDNA conjugates are annealed by heating at 94 °C for 3 min and allowing to cool to RT slowly to form final Au-dsDNA substrates (Fig. 1c).
Final Au-dsDNA substrates are observed via XSI to ensure inter-particle signal only seen from the properly annealed substrate (Fig. 1c).
Fig. 1.
Anion exchange chromatograms showing (a) the separation of mono-conjugated Au-ssDNA from un-conjugated Au-PEG and multi-conjugated Au-ssDNA using fast protein liquid chromatography (FPLC) and (b) the sequence-dependent shift in the elution volume. Both as measured by diode array detector (DAD) at 520 and 280 nm for Au NPs and ssDNA, respectively, as well as measurement of conductance showing salt gradient conditions. (c) Demonstration of the two Au-dsDNA substrates used and the normalized electron-pair distance distribution P(r) functions from these experiments showing the peak regions corresponding to the intra-Au and inter-Au distances as well as the disappearance of the inter-Au distances in the Au-ssDNA sample. The P(r) functions are normalized to the intra-Au peak to compensate for fluctuations in concentration (Au NP conc 200 nM ± 10)
3.3. Protein Expression and Purification
After expression plasmids are verified through DNA sequencing, plasmids are amplified by transforming into DH5α cells, and cells are grown on LB-agar plates with kanamycin selection (50 μg/mL) overnight at 37 °C and are extracted using Qiagen® miniprep kit as per the manufacturer protocol.
For protein expression, extracted plasmids are transformed into Rosetta™ competent cells in a similar fashion as above (see Note 5).
Expression-plasmid transformed Rosetta™ cells are inoculated into a small LB culture medium (200 mL) supplemented with kanamycin (50 μg/mL) and grown overnight at 37 °C in a shaker.
Overnight culture is further utilized to inoculate large-scale (6 L) LB media (1.5 L/flask) supplemented with kanamycin (50 μg/mL), and protein expression is induced with 0.75 mM IPTG at 16 °C overnight.
Cells are harvested and stored in −80 °C deep freezer until further use.
Cell pellets are thawed and resuspended in the lysis buffer and homogenized using a Dounce homogenizer.
Homogenized cells are lysed by sonication.
Lysed cells are clarified by centrifugation at 39,191 × g for 45 min.
Automated affinity purification is performed on an FPLC system (e.g., AKTA Pure). The clarified lysate is loaded onto a prepacked 5 mL HisTrap column. Prior to loading the lysate, the column is pre-equilibrated with Buffer A. The following steps are used for the automated affinity purification: column wash, 100 mL of Buffer A; second wash, 25 mL of 10% Buffer B; and elution, 50 mL of 60% Buffer B, second elution, 50 mL of 100% Buffer B. Protein eluted with 60% Buffer B is used for downstream activity assays.
Eluted protein fractions are verified by protein gel electrophoresis, and protein-containing fractions are pooled and concentrated and loaded onto pre-equilibrated (with SEC buffer) Superdex 200 16/600 column mounted on an AKTA pure machine for further purification by size-exclusion chromatography.
Protein fractions are verified by gel electrophoresis, and protein-containing fractions are pooled and concentrated and quantified by NanoDrop.
Protein is distributed into 20–30 μL fractions and flash frozen in the liquid nitrogen and stored in −80 °C deep freezer until further use.
Plasmid for the nuclease-dead version of the enzyme (H129N) was generated through mutagenesis and purified exactly as the wild-type (WT) enzyme.
Given the composition of SEC buffer contains 0.1 mM EDTA, the purified proteins at the end are in a metal-free state.
3.4. DNA Substrate Preparation for the Fluorescence-Based Nuclease Reaction
Identical DNA substrates are used in Au-SAXS and gel-based nuclease reactions (see Note 6 and Table 1).
All 5′-Fluorescein (FAM) labelled DNA oligos are purchased from IDT with HPLC purification.
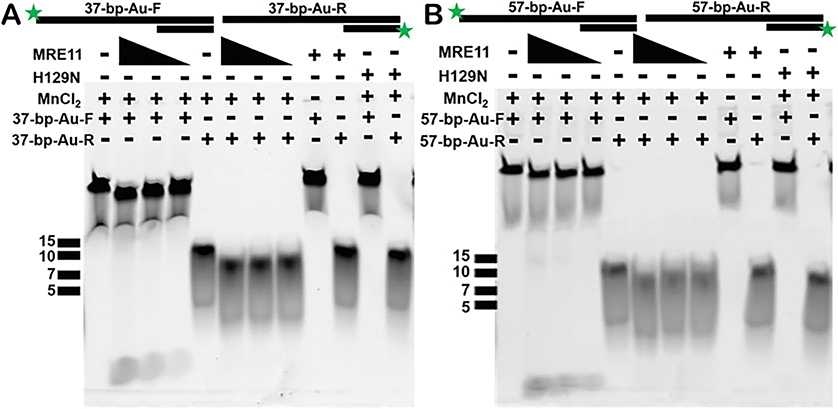
We verified that MRE11 cuts our substrate through monitoring the cleavage in a fluorescence-based nuclease assay (Fig. 2).
To monitor how MRE11 cuts the DNA substrates on both strands, both 37-bp and 57-bp substrates are labelled with FAM at 5′ individually resulting in four different substrates: (1) duplex with a 5′-FAM on longer strand of 37-bp, (2) duplex with 5′-FAM on shorter strand of 37-bp, (3) duplex with a 5′-FAM on longer strand of 57-bp, and (4) duplex with a 5′-FAM on shorter strand of 57-bp.
DNA substrates (in Table 1) used in gel-based nuclease reaction are prepared by annealing complementary non-labelled strand with fluorescently labelled oligo (in 1.3:1 ratio) and by heating at 95 °C for 5 min followed by gradual cooling to room temperature for the duplex formation.
Substrates are stored at −20 °C until further use (@ 1 μM stock concentration).
Fig. 2.
MRE11 nuclease activity as monitored in gel-based assays. Both substrates (37-bp and 57-bp) used in the XSI experiments were used in fluorescence-based nuclease assay. To monitor the nuclease activity of MRE11 on both strands of the duplex substrate, a 5′-FAM label is added on either end resulting in four substrates as shown above (37-bp-Au-F, 37-bp-Au-R, 57-bp-Au-F, and 57-bp-Au-R). (a) MRE11 shows nuclease activity (at 2, 1, and 0.5 μM concentration) on both strands of 37-bp substrate, and the activity is dependent on the presence of MnCl2 in the reaction buffer. As expected, the nuclease-dead mutant H129N is not active even in the presence of MnCl2 at 2 μM enzyme concentration. (b) MRE11 shows nuclease activity (at 2, 1, and 0.5 μM concentration) on both strands of 57-bp substrate, and the activity is dependent on the presence of MnCl2 in the reaction buffer. As expected, the nuclease-dead mutant H129N is not active even in the presence of MnCl2 at 2 μM enzyme concentration. ssDNA markers are indicated for each gel
3.5. Fluorescence-Based Nuclease Reaction to Validate Substrates and the Activity (Fig. 2)
Proteins (WT or H129N) are diluted to the desired concentration in the nuclease reaction buffer with or without MnCl2.
Nuclease reaction is initiated by adding the substrate to the reaction mixture and incubating at 37 °C for 1 h.
Nuclease reaction is stopped by adding a stop buffer and incubated further at 37 °C for 15 min (see Note 7).
For each substrate, a non-labelled version of the cleaved FAM-labelled strand is added (100–200-fold excess) to the reaction mixture to visualize only FAM-labelled ssDNA product.
Reaction mixture is run on a denaturing TBE-UREA gel for 50 min at 185 V (see Note 8).
Gel can be imaged with FAM excitation/emission filter on any gel imager (Fig. 2).
3.6. Sample Preparation of XSI Experiments
Dialyze Au-dsDNA substrates overnight at 4 °C in 1 L reaction buffer using 4 kDa dialysis membranes. Be cautious of strong reducing agents in the buffer (see Note 9).
Measure the concentrations of Au-dsDNA (diluted appropriately; see Note 2) using Thermo Scientific NanoDrop 2000 Spectrophotometer at 520 nm. Adjust concentration if needed (see Note 10).
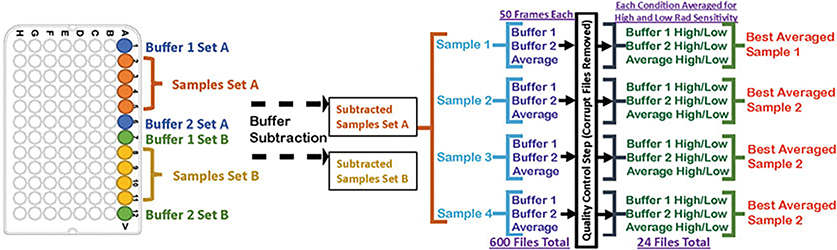
Combine enzymes with Au-dsDNA in an Axygen 96-well Polypropylene PCR Microplate at a final molar ratio of 10:1 (MRE11 2 μM and Au NPs 200 nM) in nuclease reaction buffer (with or without 2 mM MnCl2), and then bracket the samples with a blank buffer sample on either end for buffer subtraction (see Note 11, Fig. 3).
Incubate plate containing samples at 37 °C for 1 h for the reaction to take place.
Fig. 3.
Exemplary demonstration of how to set up a 96-well plate for high-throughput XSI assay and the subsequent data processing pipeline
3.7. XSI Data Collection at the SIBYLS Beamline (See Note 12)
XSI data is collected at the SIBYLS beamline (BL12.3.1), at the Advanced Light Source at Lawrence Berkeley National Laboratory, Berkeley, California [36]. To send samples for collection, see Note 12.
Load 96-well sample plate onto cooled 10 °C sampling position.
Samples are transferred from a 96-well plate via a Tecan Evo liquid handling robot with modified pipetting needles acting as sample cells to the X-ray beam as described previously [9].
X-ray wavelength is set at λ = 1.24 Å, and the sample-to-detector distance is 2.1 m, resulting in scattering vector q, ranging from 0.01 to 0.45 Å−1. The scattering vector is defined as q = 4π sin θ/λ, where 2θ is the scattering angle. Data is collected using a Dectris PILATUS3X 2M detector at 20 °C and processed as previously described [37]. Samples are exposed to X-ray synchrotron radiation for a total of 10 s at a frame rate of 0.2 s for a total of 50 images.
For each sample collected, two sample-free buffer samples are also collected to reduce error in subtraction (Fig. 3).
Each collected image is circularly integrated and normalized for beam intensity to generate a one-dimensional scattering profile by beamline specific software (Fig. 3).
Buffer subtraction is performed for the one-dimensional scattering profile of each sample by using each of the two corresponding buffers, producing two sets of buffer-subtracted sample profiles to ensure the subtraction process was not subject to instrument variations (Fig. 3).
3.8. Setting up XSI Data Processing Pipeline
- Once data collection has been completed, you will receive your data back with the following file hierarchy:
Username_Date: Results Subtracted A2_results Average 50 Integrated Curves (.dat files) Buffer1 50 Integrated Curves (.dat files) Buffer2 50 Integrated Curves (.dat files) Unsubtracted A1b_results A2_results A3b_results Scattering profiles over the 10 s exposure (50 frames total) should be sequentially averaged to eliminate any radiation damage affects. This can be done either manually, for each sample using our web-based beamline software FrameSlice (sibyls.als.lbl.gov/ran), or by batch processing using our XSI data processing pipeline (Fig. 3) which is recommended for large data sets.
To set up your system for running the frame averaging pipeline, we have recommended Bash terminal environment (see Note 13), but it should be able to run on any platform with Python 3 and pip (both required).
- Check your versions of python and pip.
- To check your python version from terminal:
$ python3 --version
-
If no version of python 3 (see Note 13).To check your pip version from terminal:
$ python3 -m pip --version
- If no version of pip type:
$ python3 get-pip.py
- In a new bash terminal clone our gitlab repository (see Note 14):
$ git clone https://git.bl1231.als.lbl.gov/djrosenberg/frame_averaging_pipeline.git
- Go to the folder called frame_averaging_pipeline, and install:
$ cd frame_averaging_pipeline $ pip install .
- To make sure pip has installed frame_averaging_pipeline:
$ pip list
- Note the location of the repository folder, frame_averaging_pipeline:
$ pwd
-
Example Output: folder_path/frame_averaging_pipeline.This output we will call folder_path (needed in step 7 to run the main script).
Start Xserver if on Windows or Mac, and leave it running in the background (see Note 15).
- If your data is local, cd to the folder containing the Results folder you would like to process (called Username_Date in the file hierarchy example above), and run the xsi_batch_processing shell script.
- Here we use the example of the Test_Data included in the frame_averaging_pipeline folder:
$ cd tests/Test_Data
- Run xsi_batch_processing.sh using the path from step 4:
$ folder_path/frame_averaging_pipeline /xsi_batch_processing.sh
When asked “Is your data on your local machine and are you in the folder containing your Results folder,” answer “y” or “yes.”
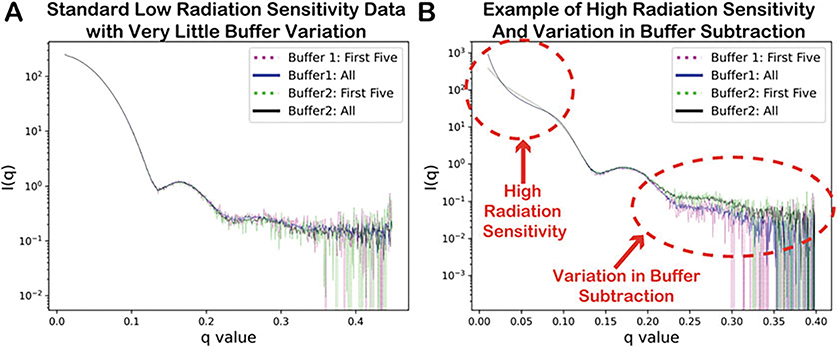
The data processing pipeline should start. Once complete you are asked to “Please Review Output In” the folder. Scroll through the .png images in the Xserver window or preferred image viewer if prompted (see Note 15), and decide whether buffer subtraction one, two, or the average should be used, and enter 1, 2, or A, respectively (if buffers match closely, use average). Then select whether the samples show high or low sensitivity to radiation, and enter either H or L, respectively (see Fig. 4).
Make sure the desired data has been selected and the output directory is correct, and enter “Y” to continue or “N” to repeat selection step 9.
To pull your data directly from the SIBYLS beamline database (must have an account; see Note 12), answer “n” or “no” when asked “Is your data on your local machine and are you in the folder containing your Results folder.”
When asked, “Please Enter Your SIBYLS User Name: (this is caps sensitive).”
When asked, “Is Current_Year the correct year of your data collection?,” answer “Y” to continue or “N” to enter the year of your data collection as YYYY.
Enter user password.
When asked, “Please Select Data Folder Name You’d Like to Work on (this is caps sensitive) (For example: 2020_02_25_username_results).”
Make sure the folder and path are correct, and answer “Y” to continue or “N” to repeat data selection step 14.
Enter user password.
Batch data processing will start automatically. Follow steps 9 and 10.
- Once XSI batch processing is completed, the following file hierarchy is output:
Username_Date: Best_Curves (best selected averaged curves) Results (original subtracted/unsubtracted data) Username_Date _Ave_Curves Full_Ave (full frame averaged curves (low radiation)) Region_Ave (Regionally averaged curves (high radiation)) Username_Date _Ave_Graphs (.png images of averaged curves) logs (logs for debugging code)
Fig. 4.
Output from the xsi_batch_processing.sh script with examples of (a) standard low radiation sensitivity data and (b) high radiation sensitivity with variations in buffer subtraction
3.9. XSI Data Analysis and Interpretation
Once you have your best frame averaged XSI curves (best_curves in the above file hierarchy), you can use those for analysis.
The simplest analysis is to generate pair-distribution functions, P(r), from the inverse Fourier transformation of the best averaged XSI profiles [14, 38]. For this we recommend using SCÅTTER [39] (see Note 17), but there are many other options [40, 41].
P(r) functions were normalized to the intra-Au peak to account for variations in concentration.
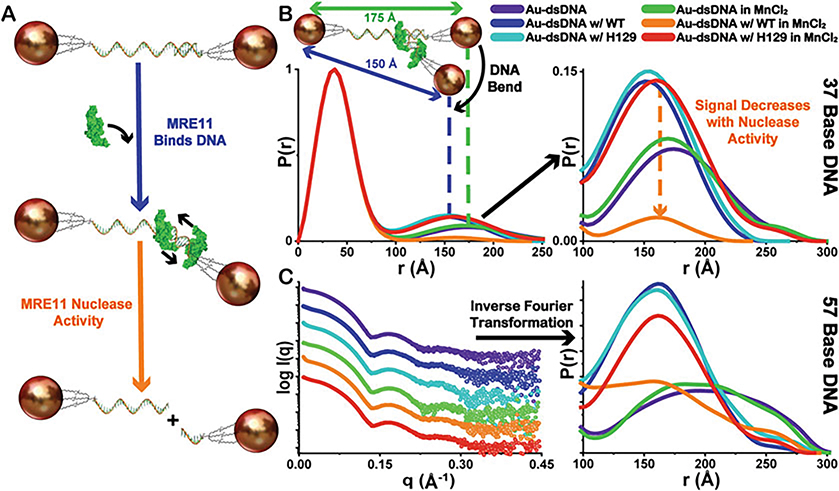
A shift in the peak maximum in the inter-Au region indicates a highly accurate change in inter-particle distance suggesting a change in the substrate. For the example of MRE11, the inter-Au distance distributions are shifted to lower mean values compared to the substrate alone for samples where nuclease activity was not observed indicating structural changes in the DNA associated with MRE11 binding (Fig. 5). These finding are consistent with both Au-dsDNA substrate lengths.
The integration of the inter-Au peaks is used to estimate the relative changes in concentration for intact Au-dsDNA substrates after the enzymatic reaction takes place. As expected, MRE11 nuclease activity decreases the population of doubly Au-NP-labelled dsDNA as observed by the decrease in the amplitude of P(r) corresponding only to inter-Au distances. From these XSI assays, we observe that MRE11 is not active when the active site metal (Mn2+) is not present in the reaction buffer or a nuclease-dead mutant (H129N) is used in the reaction instead of the wild-type (WT) enzyme (Fig. 5). These findings are consistent with both Au-dsDNA substrate lengths.
Fig. 5.
Demonstration of overall XSI assay scheme. (a) The proposed mechanism of MRE11 interaction with intact Au-dsDNA substrates and the subsequent nuclease activity leading to separation of the fixed inter-particle distances as Au-ssDNA. (b) Demonstration of the shifts in the distribution of inter-Au electron-pair distances, seen in the normalized P(r) functions (37-bp DNA), to lower mean values compared to the substrate alone representing the structural changes in the Au-dsDNA substrates associated with MRE11 binding. Additionally, a decrease in the amplitude of P(r) corresponding only in the inter-Au regions is observed only with WT-MRE11 in the presence of MnCl2 (orange) suggesting increased MRE11 nuclease activity (Au-dsDNA to Au-ssDNA). Legend for sample identity shown. (c) Exemplary experimental XSI curves and derived P(r) functions for 57-bp DNA colored as in the Panel (b). Curves have scaled I(q) for visualization purposes. P(r) function plot is scaled to show inter-Au distance region. All P(r) functions are normalized to the intra-Au peak to compensate for fluctuations in concentration as depicted in Fig. 1c
4. Notes
Au NP pellets can be disturbed easily if not careful. It is recommended to decant with particles held at side closest to the floor if decanting or to use a pipette.
To measure concentrations accurately within the confines of Beer’s law, analytes should be diluted so that the absorbance range is with 0.1 and 1 mAu.
- Extinction Coefficient Au @ 520 nm = 9.69 × 106 L/mol cm, Path Length NanoDrop 0.1 = cm, Extinction Coefficient ssDNA @ 260 nm sequence dependent.
Using no salt and high salt FPLC buffers, a salt gradient from 10 to 1000 mM is created over a period of 50 min (Fig. 1a, b). Sample elution monitored UV-Vis absorption at the Au plasmon maximum of 520 nm. Typical final concentrations for collected conjugates were 0.1–0.2 μM.
The catalytic domain of MRE11 used (with Met-to-Leu mutations) in this chapter is not super-soluble when expressed in E. coli; however, soluble fraction of the purified enzyme is active in the nuclease reaction.
Prior knowledge of enzyme-DNA substrate reaction can be quite useful in designing the substrates. By carefully modifying the substrate, this method can be adapted for other DNA nucleases.
Enzymes can be removed from the substrate if desired so by adding proteinase K at the end of the reaction.
It is recommended to pre-run the TBE-Urea gel (@200 V for 60 min) prior to running the nuclease reaction products on the gel.
DNA is conjugated to Au NPs through Au-S interaction. Strong reducing agents in the reaction buffer can disrupt this interact and cause aggregation of the Au NPs, especially when exposed to strong synchrotron X-ray radiation. Thus, the reaction conditions need to be optimized accordingly.
Samples can be diluted with reaction buffer or concentrated by using 4 kDa centrifuge concentrator tubes and spinning at 10,000 × g. Generally, concentrations >100 nM give great scattering signal.
SAXS is a contrast measurement, as such the buffer used for subtraction must be as close to the buffer containing the sample as possible. Dialyze samples and use the dialysis buffer for best subtraction.
While it should be possible to leverage this technique for use at a researcher’s home institution, we also offer a mail-in user program at the SIBYLS beamline where we can help to design and carry out experiments like those mentioned herein. To obtain XSI data collection time at the SIBYLS beamline, please follow the directions on our website https://bl1231.als.lbl.gov/htsaxs/instructions/htsaxs and/or contact us.
The use of Python 3 is required. The desired version of Python 3 can be installed by following the instruction on https://www.python.org/downloads/, and any version should work. We also recommend running bash terminal in a Conda, Python 3.7+ environment as it may streamline the setup of the code, but Conda is not required. To set up your own conda environment, follow the instructions on https://docs.conda.io/projects/conda/en/latest/user-guide/install/index.html for setting up miniconda on your system if desired.
This code under active development and the newest setup and usage information can be found in the README file at our gitlab for frame_averaging_pipeline at: https://git.bl1231.als.lbl.gov/djrosenberg/frame_averaging_pipeline.git.
If running xsi_bacth_processing.sh on Windows or Mac, you will likely need an Xserver (https://kb.thayer.dartmouth.edu/article/336-x11-for-windows-and-mac) to run graphical interfaces, and you will also need Eye of Gnome (eog) installed in your terminal if you don’t already have it (see Note 16 below). Alternatively, just open the .png images in your preferred image viewer, and ignore Notes 15 and 16.
- The xsi_bacth_processing.sh will prompt you if “Eye of Gnome (eog) could not be found. Please use your prefer image viewer to view .png files in: username_date_Ave_Graphs folder.” To install eog:
$ eog --version # first check if you have eog installed $ sudo apt-get install eog #if you don’t have it (ubuntu) $ sudo yum install eog #if you don’t have it (centos/redhat)
The SAXS analysis software SCÅTTER can be downloaded from http://www.bioisis.net/tutorials, and the website includes tutorials for its use.
Acknowledgments
The work is supported by R35 CA220430, by the Cancer Prevention Research Institute of Texas (CPRIT-grant#RP180813), and by a Robert A. Welch Chemistry Chair. Efforts to apply SAXS to characterize eukaryotic pathways relevant to human cancers and merge nano- to mesoscale structures are supported in part by National Cancer Institute grants Structural Biology of DNA Repair (SBDR) CA092584. SAXS data was collected at the Advanced Light Source (ALS) beamline SIBYLS which is supported by the DOE-BER IDAT DE-AC02-05CH11231 and NIGMS ALS-ENABLE (P30 GM124169 and S10OD018483).
References
- 1.Syed A, Tainer JA (2018) The MRE11–R-AD50–NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu Rev Biochem 87:263–294. 10.1146/annurev-biochem-062917-012415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paull TT (2018) 20 years of Mre11 biology: no end in sight. Mol Cell 71:419–427. 10.1016/j.molcel.2018.06.033 [DOI] [PubMed] [Google Scholar]
- 3.Stracker TH, Petrini JHJ (2011) The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 12:90–103. 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopfner K-P, Karcher A, Craig L et al. (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105:473–485. 10.1016/S0092-8674(01)00335-X [DOI] [PubMed] [Google Scholar]
- 5.Williams RS, Moncalian G, Williams JS et al. (2008) Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135:97–109. 10.1016/j.cell.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park YB, Chae J, Kim YC, Cho Y (2011) Crystal structure of human Mre11: understanding tumorigenic mutations. Structure 19:1591–1602. 10.1016/j.str.2011.09.010 [DOI] [PubMed] [Google Scholar]
- 7.Shibata A, Moiani D, Arvai AS et al. (2014) DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 53:7–18. 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Hildebrandt N (2020) Resonance energy transfer to gold nanoparticles: NSET defeats FRET. TrAC Trends Anal Chem 123:115748. 10.1016/j.trac.2019.115748 [DOI] [Google Scholar]
- 9.Hura GL, Menon AL, Hammel M et al. (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6:606–612. 10.1038/nmeth.1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosey CA, Tainer JA (2019) Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr Opin Struct Biol 58:197–213. 10.1016/j.sbi.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang HYH, Tainer JA, Hura GL (2017) High resolution distance distributions determined by X-ray and neutron scattering. In: Chaudhuri B, Muñoz IG, Qian S, Urban VS (eds) Biological small angle scattering: techniques, strategies and tips. Springer, Singapore, pp 167–181 [Google Scholar]
- 12.Rambo RP, Tainer JA (2013) Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu Rev Biophys 42:415–441. 10.1146/annurev-biophys-083012-130301 [DOI] [PubMed] [Google Scholar]
- 13.Rambo RP, Tainer JA (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496:477–481. 10.1038/nature12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putnam CD, Hammel M, Hura GL, Tainer JA (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40:191–285. 10.1017/S0033583507004635 [DOI] [PubMed] [Google Scholar]
- 15.Rambo RP, Tainer JA (2011) Characterizing flexible and instrinsically unstructured biological macromolecules by SAS using the Porod-Debye Law. Biopolymers 95:559–571. 10.1002/bip.21638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young T (1804) I. The Bakerian Lecture. Experiments and calculations relative to physical optics. Philos Trans R Soc Lond 94:1–16. 10.1098/rstl.1804.0001 [DOI] [Google Scholar]
- 17.Hura GL, Tsai C-L, Claridge SA et al. (2013) DNA conformations in mismatch repair probed in solution by X-ray scattering from gold nanocrystals. Proc Natl Acad Sci 110:17308–17313. 10.1073/pnas.1308595110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vainshtein BK, Feigin LA, Lvov YM et al. (1980) Determination of the distance between heavy-atom markers in haemoglobin and histidine decarboxylase in solution by small-angle X-ray scattering. FEBS Lett 116:107–110. 10.1016/0014-5793(80)80539-4 [DOI] [PubMed] [Google Scholar]
- 19.Mathew-Fenn RS, Das R, Silverman JA et al. (2008) A molecular ruler for measuring quantitative distance distributions. PLoS One 3:e3229. 10.1371/journal.pone.0003229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathew-Fenn RS, Das R, Harbury PAB (2008) Remeasuring the double helix. Science 322:446–449. 10.1126/science.1158881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mastroianni AJ, Sivak DA, Geissler PL, Alivisatos AP (2009) Probing the conformational distributions of subpersistence length DNA. Biophys J 97:1408–1417. 10.1016/j.bpj.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X, Herschlag D, Harbury PAB (2013) Structural ensemble and microscopic elasticity of freely diffusing DNA by direct measurement of fluctuations. Proc Natl Acad Sci 110:E1444–E1451. 10.1073/pnas.1218830110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X, Beauchamp KA, Harbury PB, Herschlag D (2014) From a structural average to the conformational ensemble of a DNA bulge. Proc Natl Acad Sci 111:E1473–E1480. 10.1073/pnas.1317032111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X, Huang L, Lilley DMJ et al. (2016) The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat Chem Biol 12:146–152. 10.1038/nchembio.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zettl T, Mathew RS, Shi X et al. (2018) Gold nanocrystal labels provide a sequence–to–3D structure map in SAXS reconstructions. Sci Adv 4:eaar4418. 10.1126/sciadv.aar4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zettl T, Mathew RS, Seifert S et al. (2016) Absolute intramolecular distance measurements with angstrom-resolution using anomalous small-angle X-ray scattering. Nano Lett 16:5353–5357. 10.1021/acs.nanolett.6b01160 [DOI] [PubMed] [Google Scholar]
- 27.Claridge SA, Mastroianni AJ, Au YB et al. (2008) Enzymatic ligation creates discrete multinanoparticle building blocks for self-assembly. J Am Chem Soc 130:9598–9605. 10.1021/ja8026746 [DOI] [PubMed] [Google Scholar]
- 28.Tubbs JL, Latypov V, Kanugula S et al. (2009) Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature 459:808–813. 10.1038/nature08076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckelmann BJ, Bacolla A, Wang H et al. (2020) XRCC1 promotes replication restart, nascent fork degradation and mutagenic DNA repair in BRCA2-deficient cells. NAR Cancer 2:zcaa013. 10.1093/narcan/zcaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thapar R, Wang JL, Hammel M et al. (2020) Mechanism of efficient double-strand break repair by a long non-coding RNA. Nucleic Acids Res 48:10953–10972. 10.1093/nar/gkaa784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zandarashvili L, Langelier M-F, Velagapudi UK et al. (2020) Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368:eaax6367. 10.1126/science.aax6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houl JH, Ye Z, Brosey CA et al. (2019) Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat Commun 10:5654. 10.1038/s41467-019-13508-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacolla A, Tainer JA, Vasquez KM, Cooper DN (2016) Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res 44:5673–5688. 10.1093/nar/gkw261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacolla A, Ye Z, Ahmed Z, Tainer JA (2019) Cancer mutational burden is shaped by G4 DNA, replication stress and mitochondrial dysfunction. Prog Biophys Mol Biol 147:47–61. 10.1016/j.pbiomolbio.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo SH, Bacolla A, Yoo D et al. (2020) Replication-based rearrangements are a common mechanism for SNCA duplication in Parkinson’s disease. Mov Disord 35:868–876. 10.1002/mds.27998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Classen S, Hura GL, Holton JM et al. (2013) Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. J Appl Crystallogr 46:1–13. 10.1107/S0021889812048698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyer KN, Hammel M, Rambo RP et al. (2014) High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol Biol 1091:245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glatter O (1977) A new method for the evaluation of small-angle scattering data. J Appl Crystallogr 10:415–421. 10.1107/S0021889877013879 [DOI] [Google Scholar]
- 39.Tully MD, Tarbouriech N, Rambo RP, Hutin S (2021) Analysis of SEC-SAXS data via EFA deconvolution and scatter. J Vis Exp (167):e61578. 10.3791/61578 [DOI] [PubMed] [Google Scholar]
- 40.Hopkins JB, Gillilan RE, Skou S (2017) BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J Appl Crystallogr 50:1545–1553. 10.1107/S1600576717011438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manalastas-Cantos K, Konarev PV, Hajizadeh NR, et al. (2021) ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J Appl Crystallogr 54:343–355. 10.1107/S1600576720013412 [DOI] [PMC free article] [PubMed] [Google Scholar]