Abstract
By informing timely targeted treatments, rapid whole-genome sequencing can improve the outcomes of seriously ill children with genetic diseases, particularly infants in neonatal and pediatric intensive care units (ICUs). The need for highly qualified professionals to decipher results, however, precludes widespread implementation. We describe a platform for population-scale, provisional diagnosis of genetic diseases with automated phenotyping and interpretation. Genome sequencing was expedited by bead-based genome library preparation directly from blood samples and sequencing of paired 100-nt reads in 15.5 hours. Clinical natural language processing (CNLP) automatically extracted children’s deep phenomes from electronic health records with 80% precision and 93% recall. In 101 children with 105 genetic diseases, a mean of 4.3 CNLP-extracted phenotypic features matched the expected phenotypic features of those diseases, compared with a match of 0.9 phenotypic features used in manual interpretation. We automated provisional diagnosis by combining the ranking of the similarity of a patient’s CNLP phenome with respect to the expected phenotypic features of all genetic diseases, together with the ranking of the pathogenicity of all of the patient’s genomic variants. Automated, retrospective diagnoses concurred well with expert manual interpretation (97% recall and 99% precision in 95 children with 97 genetic diseases). Prospectively, our platform correctly diagnosed three of seven seriously ill ICU infants (100% precision and recall) with a mean time saving of 22:19 hours. In each case, the diagnosis affected treatment. Genome sequencing with automated phenotyping and interpretation in a median of 20:10 hours may increase adoption in ICUs and, thereby, timely implementation of precise treatments.
One-sentence summary:
Automated phenotyping and interpretation of rapid whole-genome sequencing improve time to diagnosis of genetic diseases in hospitalized children.
Editor’s Summary:
A streamlined genetic diagnosis pipeline
When treating seriously ill children, time is of the essence. Clark et al. built an automated pipeline to analyze EHR data and genome sequencing data from dried blood spots to deliver a potential diagnosis for hospitalized, often critically ill, children with suspected genetic diseases. Their pipeline required minimal user intervention, increasing usability and shortening time to diagnosis, delivering a provisional finding in a median time of less than 24 hours. Although this pipeline would need to be adapted for use at different hospital systems, such an automated tool could aid clinicians to expedite an accurate genetic disease diagnosis, potentially hastening lifesaving changes to patient care.
INTRODUCTION
Genetic diseases are the leading cause of infant mortality in the United States, particularly among about 15% of infants admitted to neonatal, pediatric, and cardiovascular intensive care units (ICUs) (1-11). As disease progression in infants is rapid, etiologic diagnosis must be equally fast to inform interventions that can lessen suffering, morbidity, and mortality (12, 13). Unfortunately, this is rarely the case. More than 13,000 genetic diseases are known (14, 15), and their presentations often overlap in seriously ill infants and are typically abridged with respect to classical descriptions (14, 15). Standard genome sequencing takes weeks to return results, which is too slow to guide inpatient management. Rapid whole-genome sequencing (rWGS) provides faster diagnosis, enabling precision medicine interventions in time to decrease the morbidity and mortality of infants with genetic diseases (12, 13). Furthermore, in genetic diseases with uniformly dismal prognosis, rapid diagnosis facilitates end-of-life care decisions that can alleviate suffering and aid the grieving process.
Clinical studies are starting to substantiate the diagnostic and clinical utility and cost effectiveness of rWGS in seriously ill infants in ICUs, with reported rates of diagnosis of 42 to 57%, changes in medical management in 30 to 72% of cases, and altered outcomes in 24 to 34% of cases (12, 14, 16-30). This evidence has led to calls for accelerated implementation in national health care systems as the new standard of care (31-33). The National Health Service of the United Kingdom, for example, will offer whole-genome sequencing as part of care for all seriously ill children from 2019 (34). The major impediments to universal implementation in ICUs are absence of reimbursement outside the United Kingdom, lack of knowledge of genomic medicine by pediatricians, and the high capital and labor intensity of current clinical rWGS and interpretation.
We previously described diagnosis by rWGS in 26 hours in a research setting (16, 17). In the clinical studies reported to date, however, the fastest genetic diagnosis by genome sequencing was 37 hours, the mean time to diagnosis was 16 days, and the largest cohort comprised only 63 patients (8, 16-30). The small cohort size and longer time to diagnosis in those clinical studies substantiate the limitations of current methods of rWGS. Here, we report methods for clinical diagnosis of genetic diseases in a median of 20:10 hours that can be scaled to 30 patients per week per genome sequencing instrument, with automated provisional diagnosis.
RESULTS
rWGS for genetic disease diagnosis
In light of the limitations of current methods of rWGS, we developed an automated platform for rapid, high-throughput, provisional diagnosis of genetic diseases with genome sequencing by automating and accelerating our conventional workflow (Fig. 1). Conventional clinical genome sequencing requires preparatory steps of manual purification of genomic DNA from blood samples, DNA quality assessment, normalization of DNA concentration, sequencing library preparation, and library quality assessment (Fig. 1A). Instead, we manually prepared sequencing libraries directly from blood samples or dried blood spots using microbeads to which transposons were attached (Nextera DNA Flex Library Prep Kit, Illumina Inc.; Fig. 1B) (35), because this method was both faster and less labor intensive. Dried blood spots are the sample type used in mandatory newborn screening worldwide. In four timed runs with retrospective samples, manual Nextera library preparation from dried blood spots took a mean of 2 hours and 45 min, compared with at least 10 hours by conventional DNA purification and library preparation (TruSeq DNA PCR-free Library Prep Kit, Illumina Inc.; Table 1). As with standard methods, Nextera Flex allowed samples to be prepared in batches and was amenable to automation with liquid-handling robots.
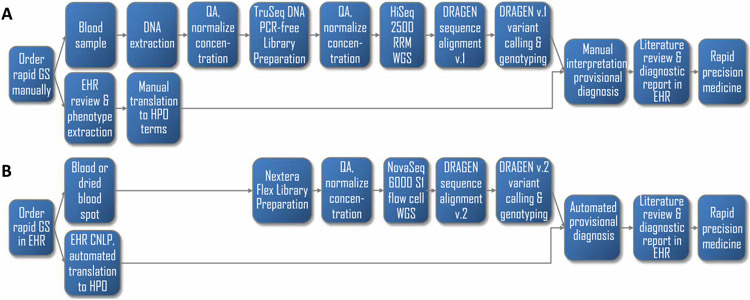
Fig. 1. Flow diagrams of the diagnosis of genetic diseases by standard genome sequencing and rWGS.
(A) Steps in conventional clinical diagnosis of a single patient by genome sequencing (GS) with manual analysis and interpretation in a minimum of 26 hours but with a mean time to diagnosis of 16 days (8, 16-30). Genome sequencing was requested manually. We manually extracted genomic DNA from blood samples, assessed the DNA quality (QA), and manually normalized the DNA concentration. We then manually prepared TruSeq PCR-free DNA sequencing libraries, performed the QA again, and manually normalized the library concentration. Genome sequencing was performed on the HiSeq 2500 system (Illumina) in rapid run mode (RRM). Sequences were manually transferred to the DRAGEN Platform version 1 (Illumina) for alignment and variant calling. Phenotypic features were identified by manual review of the electronic health record (EHR). Variant files and phenotypic features were manually loaded into Opal software (Fabric), and interpretation was performed manually. (B) Steps in autonomous diagnosis of up to six patients concurrently in a minimum of 19 hours (fig. S3). Steps included (i) automation of order entry from the EHR with a portal; (ii) manual or robotic preparation of Nextera DNA Flex sequencing libraries directly from the blood in 2.5 hours; (iii) rapid 40-fold coverage genome sequencing in 15.5 hours with the NovaSeq 6000 system and S1 flow cell (Illumina); (iv) automation of sequence transfer, alignment, and variant calling in 1 hour with the DRAGEN platform, version 2 (Illumina); (v) automated extraction of patient phenomes from the EHR by clinical natural language processing (CNLP) and translation to Human Phenotype Ontology (HPO) terms in 20 s; and (vi) automated transfer of variant and phenotype files and automated Bayesian comparison of the CNLP phenome with those of all genetic diseases (MOON, Diploid) combined with automated assessment of the pathogenicity of their genomic variants based on aggregated literature knowledge and in silico predictive tools (InterVar) and with automated display of the highest-ranked provisional diagnosis(es).
Table 1. Duration and metrics for the major steps in the diagnosis of genetic diseases by genome sequencing with rapid standard methods and a rapid, autonomous platform.
Primary (1°) and secondary (2°) analyses: Conversion of raw data from base call to FASTQ format, read alignment to the reference genomes, and variant calling. Tertiary (3°) analysis processing: Time to process variants and phenotypic features and make them available for manual interpretation in Opal interpretation software (Fabric Genomics) or to display a provisional, automated diagnosis(es) in MOON interpretation software (Diploid). Std., rapid standard methods; auto., rapid, autonomous platform; dev. delay, global developmental delay; PPHN, persistent pulmonary hypertension of the newborn; HIE, hypoxic ischemic encephalopathy; n.a., not applicable. Patients 263, 6124, and 3003 were retrospectively analyzed by the autonomous system. Patient 263 was analyzed two times by the autonomous system. Patients 6194, 290, 352, 362, 412, and 7072 were prospectively analyzed by both autonomous and standard diagnostic methods.
| Use type | Retrospective patients | Prospective patients | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject ID | 263 | 6124 | 3003 | 6194 | 290 | 352 | 362 | 374 | 7052 | 412 | ||||||||
| Age | 8 days | 14 years | 1 year | 5 days | 3 days | 7 weeks | 4 weeks | 2 days | 17 months | 3 days | ||||||||
| Sex | ♀ | ♂ | ♀ | ♀ | ♂ | ♀ | ♂ | ♂ | ♂ | ♂ | ||||||||
| Abbreviated presentation | Neonatal seizures | Rhabdomyolysis | Dystonia, dev. delay | Hypoglycemia, seizures | Pulmonary hemorrhage, PPHN | Diabetic ketoacidosis | Neonatal seizures | HIE, anemia | Pseudomonal septic shock | Neonatal seizures | ||||||||
| Method | Auto. | Auto. | Auto. | Auto. | Auto. | Std. | Auto. | Std. | Auto. | Std. | Auto. | Std. | Auto. | Std. | Auto. | Std. | Auto. | Std. |
| Number of phenotypic features | 51 | 115 | 148 | 14 | 2 | 257 | 4 | 103 | 4 | 65 | 1 | 112 | 6 | 124 | 3 | 33 | 1 | |
| Molecular diagnosis | Early infantile epileptic encephalopathy 7 | Glycogen storage disease V | Dopa-responsive dystonia | None | None | None | None | Permanent neonatal diabetes mellitus | None | None | None | None | X-linked agammaglobulinemia 1 | Benign familial neonatal seizures 1 | ||||
| Gene and causative variant(s) | KCNQ2 c.727C > G | PYGM c.2262delA c.1726C > T | TH c.785C > G c.541C > T | n.a. | n.a. | n.a. | n.a. | INS c.26C > G | n.a. | n.a. | n.a. | n.a. | BTK c.974 + 2 T > C | KCNQ2 c.1051C > G | ||||
| Sample/Library Prep (hours) | 3:20 | 2:55 | 2:24 | 2:22 | 2:10 | 23:54 | 2:12 | 22:05 | 2:13 | 15:42 | 2:31 | 18:30 | 3:30 | 10:10 | 4:30 | 12:10 | 3:05 | 23:50 |
| NovaSeq loading (hours) | 0:20 | 0:17 | 0:16 | 0:20 | 1:38* | 0:20 | 0:29 | 0:22 | 0:30 | 0:53 | 0:15 | 2:30 | 0:45 | 0:35 | 1:00 | 1:00 | 0:20 | 0:53 |
| 2 × 101 nt sequencing (hours) | 15:36 | 15:31 | 15:34 | 15:27 | 15:26 | 24:13 | 15:25 | 24:08 | 15:21 | 22:44 | 15:17 | 33:36 | 15:17 | 21:07 | 15:19 | 22:46 | 15:58 | 21:00 |
| 1° & 2° analysis (hours) | 1:03 | 1:02 | 0:59 | 0:59 | 1:07 | 3:05 | 1:00 | 1:57 | 1:01 | 2:30 | 1:02 | 2:30 | 1:02 | 2:30 | 1:09 | 2:25 | 1:24 | 2:24 |
| 3° analysis processing (hours) | 0:06 | 0:05 | 0:07 | 0:05 | 0:06 | 0:15 | 0:08 | 0:14 | 0:06 | 0:15 | 0:05 | 0:15 | 10:28† | 0:16 | 0:06 | 0:16 | 0:06 | 0:16 |
| Total (hours) | 20:25 | 19:56 | 19:20 | 19:14 | 20:42 * | 56:03 | 19:29 | 48:46 | 19:11 | 42:04 | 19:10 | 57:21 | 31:02 † | 34:38 | 22:04 | 38:37 | 20:53 | 48:23 |
Included time to thaw a second set of NovaSeq reagents.
Included 10:20 hours of downtime due to data center relocation.
Following the preparatory steps, our previous method performed rWGS with the HiSeq 2500 sequencer (Illumina) in rapid run mode, with one sample sequenced per sequencing instrument [~120 gigabases (Gb) of 2 × 101 nucleotides (nt)] in ~25 hours (Fig. 1A) (16, 17). Here, we instead performed rWGS with the NovaSeq 6000 sequencer and S1 flow cell (Illumina) (Fig. 1B), as this instrument was faster and less labor intensive, requiring fewer steps to set up a sequencing run and automatically washing the instrument after a run. In four timed runs with retrospective samples, genome sequencing of 2 × 101 nt took a mean 15:32 hours and yielded 404 to 537 Gb per flow cell, sufficient for two to three 40× genome sequences (Table 1 and table S1).
Dynamic Read Analysis for GENomics (DRAGEN, Illumina) is a hardware and software platform for alignment and variant calling that has been highly optimized for speed, sensitivity, and accuracy (16). We wrote scripts to automate the transfer of files from the sequencer to the DRAGEN platform. The DRAGEN platform then automatically aligned the reads to the reference genome and identified and genotyped nucleotide variants. Alignment and variant calling took a median of 1 hour for 150 Gb of 101-nt paired-end sequences (primary and secondary analyses; Table 1). Analytic performance of this new method, from blood sample receipt to output of genomic variant genotypes, was similar to standard clinical methods with reference human genome samples, retrospective patient samples, and prospective patient samples, except for lower sensitivity in the detection of nucleotide insertions and deletions (tables S1 and S2). The new method did not assess structural variations.
CNLP of EHRs
Genetic disease diagnosis requires determination of a differential diagnosis based on the overlap of the observed clinical features of a child’s illness (phenotypic features) with the expected features of all genetic diseases. However, a comprehensive EHR review can take hours. In addition, manual phenotypic feature selection can be sparse and subjective (36, 37), and even expert reviewers can carry an unwritten bias into interpretation (Fig. 1A). We sought automated, complete phenotypic feature extraction from EHRs, unbiased by expert opinion. The simplest approach would be to extract universal, structured phenotypic features, such as International Classification of Diseases (ICD) medical diagnosis codes or diagnosis-related group (DRG) codes. However, these are sparse and lack sufficient specificity (38, 39). Instead, we extracted clinical features from unstructured text in patient EHRs by CNLP that we optimized for identification of patients with orphan diseases (CLiX ENRICH, Clinithink Ltd.) (Figs. 1B and 2A). We then iteratively optimized the protocol for the Rady Children’s Hospital Epic EHRs using a training set of 16 children who had received genome sequencing for genetic disease diagnosis (table S3). The standard output from CLiX ENRICH is in the form of Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT). However, our automated methods required phenotypic features described in the HPO, a hierarchical reference vocabulary designed for description of the clinical features of genetic diseases (Fig. 2B). For this reason, we mapped 7706 (60%) of 12,786 HPO terms (13,685 including synonyms) and 75.4% of Orphanet Rare Disease HPO terms (released in June 2018) to SNOMED CT by lexical and logical methods and then manually verified them (data file S1). This enabled automated translation of phenotypic features extracted from the EHR by CNLP from SNOMED CT concepts to HPO terms (Fig. 1B). In contrast, Dhombres and Bodenreider (40) mapped 92% of HPO terms to SNOMED CT, but only 49% were shown to be ontologically valid and clinically relevant.
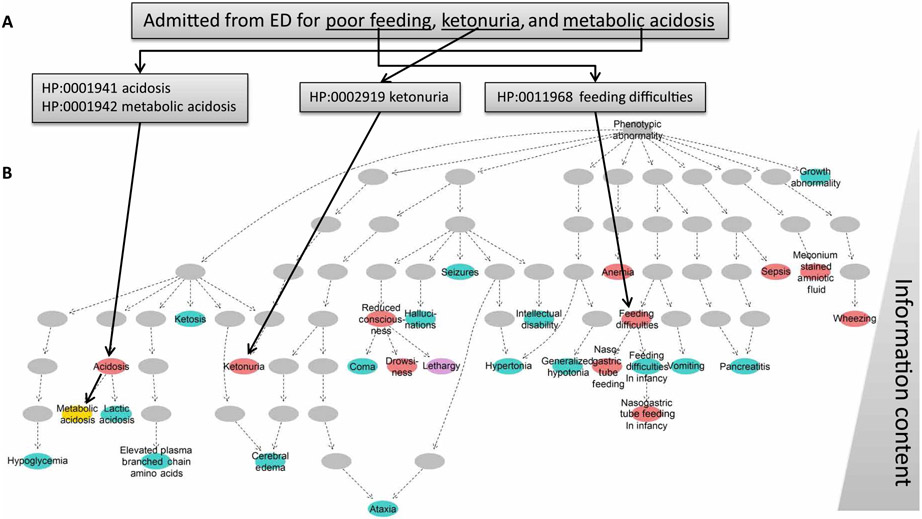
Fig. 2. CNLP can extract a more detailed phenome than manual EHR review or OMIM clinical synopsis.
(A) Example CNLP of a sentence from the EHR of an 8-day-old baby (patient 341) with maple syrup urine disease, showing four extracted HPO terms. ED, emergency department. (B) Hierarchical display of HPO phenotypic features extracted by manual review of the EHR of neonate 341 and by CNLP (red) and expected phenotypic features (from the OMIM Clinical Synopsis; blue). Yellow circles: Phenotypic features extracted by both CNLP and expert review. Purple circles: Phenotypic overlap between CNLP and OMIM. Gray circles: The location of parent terms of identified phenotypic features within the HPO hierarchy. The information content (IC) was defined by IC(phenotype) = −log(pphenotype), where pphenotype was the probability of observing the exact term or one of its subclasses across all diseases in OMIM. IC increases from top (general) to bottom (specific).
The performance of the optimized CNLP was tested with the EHRs of 10 test children who had received genome sequencing for genetic disease diagnosis. The training and test sets did not overlap. Both exact EHR phenotypic feature matches and their hierarchical root terms were extracted from the first record until time of enrollment for genome sequencing. CNLP identified a mean of 86.7 phenotypic features (SD, 32.8; range, 26 to 158) (table S4) in about 20 s per patient. A detailed manual review of the EHR was performed to identify all true-positive, false-positive, and false-negative CNLP phenotypic features in the test children. The precision (positive predictive value) of CNLP was 80% and the recall (sensitivity) was 93% (table S4), which were superior to previous CNLP-based extraction of HPO terms (36, 41). The principal reasons for false positives were as follows: (i) incorrect CLiX encoding (n = 89, 38% of 237 phenotypic features) due to misinterpreted context (n = 31), unrecognized headings (n = 23), incorrect acronym expansion (n = 21), incorrect interpretation of a clinical word (n = 8), or incorrectly attributed finding site for disease (n = 6); (ii) ambiguity of source text (unrecognized or incorrect syntax, abbreviations, acronyms, or terminology; n = 46, 19% of 237); (iii) incongruity among SNOMED CT, HPO, and clinical acumen (n = 20, 8%); (iv) failure to recognize a pasted citation as nonclinical text (n = 68, 29%); and (v) incorrect query logic (n = 14, 6%) (tables S5 to S14).
Characterization of the CNLP-derived phenomes of children with suspected genetic diseases
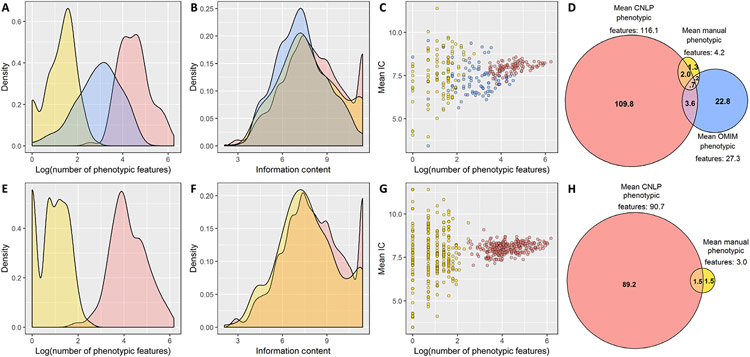
Development of an autonomous diagnostic system has been hindered by a dearth of knowledge of the topography of the phenomes of children with suspected genetic diseases (36, 42-44). Therefore, we compared EHR CNLP-derived phenomes with the comparatively sparse phenotypic features selected by experts during manual interpretation of the first 375 symptomatic children to receive genome sequencing for diagnosis of genetic diseases at Rady Children’s Hospital [101 children diagnosed with genome sequencing (Fig. 3, A to D) and 274 children who were not diagnosed (Fig. 3, E to H); data files S3 and S4]. In 101 of these children, who had received genomic diagnoses of 105 genetic diseases (four had dual diagnoses), we also compared the observed phenotypic features with the expected phenotypic features for those diseases, obtained from the Clinical Synopsis field of Online Mendelian Inheritance in Man (OMIM) (table S15) (18, 22-24, 41). In the 101 diagnosed children, CNLP identified 27-fold more phenotypic features (mean, 116.1; SD, 93.6; range, 13 to 521) than expert manual selection at interpretation (mean, 4.2; SD, 2.6; range, 1 to 16) and 4-fold more than OMIM (mean, 27.3; SD, 22.8; range, 1 to 100) (Fig. 3, A and D, and data files S3 and S4) (45, 46). Similarly, previous studies demonstrated 2-fold more phenotypic features extracted by CNLP than comprehensive, expert manual extraction (36) and 18-fold more phenotypic features extracted by CNLP than Orphanet HPO terms for those diseases (47). CNLP extracted more phenotypic features in the 101 diagnosed children than in the 274 undiagnosed children (mean, 116.1 versus 90.7, respectively; P = 0.0004, Mann-Whitney U test; Fig. 3, A, D, E, and H). This suggested the possibility that undiagnosed children, in part, did not have enough detail in their medical records to make a molecular diagnosis. In addition, there was greater overlap between CNLP and manually extracted phenotypic features in diagnosed children (mean, 2.74 terms; SD, 1.7; range, 0 to 9) than in undiagnosed children (mean, 1.52 terms; SD, 1.48; range, 0 to 7; P < 0.0001, Mann-Whitney U test) (Fig. 3, D and H). This suggested that undiagnosed children, in part, had less consistent information on phenotypic features.
Fig. 3. Comparison of observed and expected phenotypic features of 375 children with suspected genetic diseases.
(A to D) One hundred one children diagnosed with 105 genetic diseases. (E to H) Two hundred seventy-four children with suspected genetic diseases that were not diagnosed by genome sequencing. Phenotypic features identified by manual EHR review are in yellow, those identified by CNLP are in red, and the expected phenotypic features, derived from the OMIM Clinical Synopsis, are in blue. (A) Frequency distribution of the number of phenotypic features (log-transformed) in 101 children with genetic diseases. The mean number of features detected per patient was 4.2 (SD, 2.6; range, 1 to 16) for manual review, 116.1 (SD, 93.6; range, 13 to 521) for CNLP, and 27.3 (SD, 22.8; range, 1 to 100) for OMIM (OMIM versus manual, P < .0001; CNLP versus OMIM, P < .0001; CNLP versus manual, P < 0.0001; paired Wilcoxon tests). (B) Frequency distribution of IC for each phenotypic feature set in 101 diagnosed patients. The mean IC was 7.8 (SD, 2.0; range, 2.1 to 11.4) for manual review, 8.1 (SD, 2.0; range, 2.6 to 11.4) for CNLP, and 7.3 (SD, 1.7; range, 3.2 to 11.4) for OMIM (manual versus OMIM, P < .0001; CNLP versus OMIM, P < .0001; manual versus CNLP, P = 0.003; Mann-Whitney U tests). (C) Correlation of the mean IC of phenotypic terms with the number of phenotypic terms in each patient. Spearman’s rank correlation coefficient (rs) was 0.24 for manually extracted phenotypic features (P = 0.02), 0.44 for CNLP (P < 0.0001), and −0.001 for OMIM (P > 0.05). (D) Venn diagram showing overlap of phenotypic terms by the three methods for diagnosed patients. Phenotypic features extracted by CNLP overlapped expected OMIM phenotypic features (mean, 4.31 terms; SD, 4.59; range, 0 to 32) significantly more than manually (mean, 0.92 terms; SD, 1.02; range, 0 to 4; P < 0.0001, paired Wilcoxon test for the difference in the number of terms that overlap with OMIM). (E) Frequency distribution of the number of phenotypic features (log-transformed) in 274 children with suspected genetic diseases that were not diagnosed by genome sequencing. The mean number of features was 3.0 (SD, 1.9; range, 1 to 12) for manual review and 90.7 (SD, 81.1; range, 6 to 482) for CNLP (CNLP versus manual, P < 0.0001; paired Wilcoxon test). (F) Frequency distribution IC for each phenotypic feature set in 274 undiagnosed patients. The mean IC was 7.7 (SD, 2.1; range, 2.1 to 11.4) for manual review and 8.1 (SD, 2.0; range, 2.6 to 11.4) for CNLP (manual versus CNLP, P < 0.0001; Mann-Whitney U test). (G) Correlation of the mean IC of phenotypic terms with the number of phenotypic terms in each patient. rs was 0.02 for manually extracted phenotypic features (P > 0.05) and 0.30 for CNLP (P < 0.0001). (H) Venn diagram showing overlap of phenotypic terms for undiagnosed patients by CNLP and manual methods.
In the 101 diagnosed children, phenotypic features extracted by CNLP overlapped expected OMIM phenotypic features (mean, 4.31 terms; SD, 4.59; range, 0 to 32) significantly more than the manually extracted phenotypic features (mean, 0.92 terms; SD, 1.02; range, 0 to 4; P < 0.0001, paired Wilcoxon test) (Fig. 3B). Although the cohort included eight genetic diseases that were incidental findings, their exclusion did not materially change these results (table S15 and fig. S1). Thus, the recall of OMIM phenotypic features by CNLP, although small (mean, 0.20; SD, 0.16; range, 0 to 0.67), was substantially greater than the sparse expert manual phenotypic features used in expert manual interpretation (mean, 0.04; SD, 0.06; range, 0 to 0.25) (fig. S2). However, the much larger number of phenotypic features extracted by CNLP was associated with lower precision (mean, 0.04; SD, 0.03; range, 0 to 0.15) than manual extraction (mean, 0.25; SD, 0.30; range, 0 to 1) when compared with OMIM, indicating that, by design, an autonomous diagnostic system should not penalize false-positive phenotypic features. Recall and F1 values increased when phenotypic features with one degree of hierarchical separation to those extracted were included [(mean CNLP recall with inexact matches, 0.29; SD, 0.22; range, 0 to 1), (mean CNLP F1 with inexact matches, 0.12; SD, 0.08; range, 0 to 0.38), and (mean CNLP F1 with exact matches, 0.06; SD, 0.05; range, 0 to 0.23)], indicating that, by design, an autonomous system should include hierarchical parents of extracted terms (fig. S2).
Traditionally, genetic diseases have been clinically diagnosed by the identification of one or more pathognomonic phenotypic features. Such phenotypic features have high IC (the logarithm of the probability of that phenotypic feature being observed in all OMIM diseases; Fig. 2) (48). A potential concern was that phenotypic features extracted by CNLP would have less IC than those prioritized manually by experts during interpretation. However, among the 101 children, the mean IC of CNLP phenotypic features (8.1; SD, 2.0; range, 2.6 to 11.4) was significantly higher than manual (7.8; SD, 2.0; range, 2.1 to 11.4; P = 0.003, Mann-Whitney U test) or OMIM phenotypic features (7.3; SD, 1.7; range, 3.2 to 11.4; P < 0.0001, Mann-Whitney U test) (Fig. 3E). We note that the mean IC correlated significantly with the number of phenotypic features extracted manually and by CNLP [Spearman’s rho, 0.24 (P = 0.02) and 0.44 (P < 0.0001), respectively; Fig. 3C]. The mean IC of CNLP phenotypic features was higher than manual phenotypic features (Fig. 3F), and the mean IC correlated significantly with the number of phenotypic features extracted by CNLP [Spearman’s rho, 0.30 (P < 0.0001); Fig. 3G].
Retrospective performance of an autonomous system for diagnosis of childhood genetic diseases
The remaining step in automated diagnosis of genetic diseases was to combine the automated ranking of the patient’s CNLP phenome with respect to all genetic diseases, together with the automated ranking of the pathogenicity of all their genomic variants based on literature knowledge and in silico tools (Fig. 1 and fig. S3). We wrote scripts to automatically transfer the patient’s CNLP-derived phenotypic features and genomic variants to autonomous interpretation software (MOON, Diploid). MOON identified the phenotypic features associated with each genetic disease by natural language processing of the medical literature. Typically, this was a larger set of phenotypic features than those listed in the OMIM Clinical Synopsis. MOON then compared the patient’s phenotypic features with those associated with each genetic disease and rank-ordered the genetic diseases on the basis of their likelihood of causing the child’s illness.
We also wrote scripts to automatically transfer a patient’s nucleotide and structural variants (SVs) from the DRAGEN platform to MOON as soon as it finished, without user intervention. For rWGS, there was a mean of 4,742,595 nucleotide variants and 19.3 SVs, and rapid whole-exome sequencing (rWES) had a mean of 39,066 nucleotide variants and 10.3 SVs per patient (table S16). Of these, MOON retained 67,589 nucleotide variants and 12 SVs and 791 nucleotide variants and 4.5 SVs for rWGS and rWES, respectively, that had allele frequencies of <2% and affected known disease genes (table S17). A Bayesian framework and probabilistic model in MOON ranked the pathogenicity of these variants with 15 in silico prediction tools, ClinVar assertions, and inheritance pattern–based allele frequencies. In singleton and family trio analyses, on average five and three provisional diagnoses were ranked, respectively (table S18). Because MOON was optimized for sensitivity, it shortlisted a median of six nucleotide variants per diagnosed subject (range, 2 to 24) and often shortlisted false-positive diagnoses in cases considered negative by manual interpretation. Both were largely remedied, however, by processing the MOON output in InterVar software and retaining only pathogenic and likely pathogenic variants (49). InterVar classified variants with regard to 18 of the 28 consensus pathogenicity recommendations (50), specifically triaging variants of uncertain significance (VUS). Automated interpretation took a median of 5 min from transfer of variants and HPO terms to display of the provisional diagnosis and supporting evidence, including patient phenotypic features matching that disorder, for laboratory director review. In four timed runs, the time from blood samples or blood spot receipt to display of the correct diagnosis as the top-ranked variant was 19:14 to 20:25 hours (median, 19:38 hours; Table 1, retrospective cases). This conformed well to a daily clinical operation cycle: Sample receipt in the morning enabled library preparation in the afternoon, genome sequencing overnight, and provisional reporting early the following morning for laboratory director review.
We retrospectively examined the concordance between the autonomous system and previous, team-based, manual expert interpretation in 95 of the 101 children, diagnosed with 97 of the 105 genetic diseases (table S15). We excluded eight findings that had been reported but that were considered incidental (without current evidence of any of the expected phenotypic features). This cohort was diverse in race and ancestry. Eleven diagnoses were associated with SVs, and 86 were associated with nucleotide variants. No training patients were included in the test set. In two patients, a revised clinical report was issued of a new diagnosis (infant 6007, EIEE9, Xp22 del, and patient 6033, Cockayne syndrome B, ERCC6 p.Gly528Glu and c.-15 + 3G > T, which was validated by functional studies). Therefore, initial expert manual interpretation had a recall of 98% (95 of 97). Although we did not re-analyze manual diagnoses, none of them had been demoted in the period since initially reported clinically. The autonomous diagnostic system had a precision of 99% (93 of 94) and a recall of 97% (94 of 97). For nucleotide variants and SVs, the median rank of the correct diagnosis was first (range, 1 to 4 for nucleotide variants; range, 1 to 13 for SVs) (table S18).
The three false-negative autonomous diagnoses comprised the following cases:
Infant 6159, with autosomal dominant Alport syndrome (COL4A4 c.4715C > T, p.Pro1572Leu), had hematuria, nephrotic syndrome, glomerulonephritis, hypertension, and anasarca. OMIM indicated that COL4A4-associated Alport syndrome (CAS) was autosomal recessive, and p.Pro1572Leu was recorded as pathogenic in ClinVar for autosomal recessive Alport syndrome. There are, however, a large number of reports of autosomal dominant CAS. The variant was maternally inherited. Because the infant’s mother was asymptomatic, we assumed that she exhibited incomplete penetrance of autosomal dominant CAS, as has been reported (51, 52). The autonomous system classified the infant as a carrier for autosomal recessive CAS.
Infant 253 had autosomal dominant optic atrophy plus syndrome (OPA1 c.556 + 1G > A). The autonomous system did not rank this variant because of insufficient overlap of the 70 CNLP phenotypic features with the MOON disease phenotypic feature model. Recent reports indicate that OPA1 can be associated with complex, severe multisystem mitochondrial disorders, similar to infant 253.
Neonate 213 had dextrocardia and transposition of the great vessels. He received singleton genome sequencing and was diagnosed manually with autosomal dominant visceral heterotaxy type 5 associated with a likely pathogenic variant in NODAL (c.778G > A; p.Gly260Arg). This variant was filtered out by the autonomous system based on classification as a VUS by InterVar (based on PM1-PP3-PP5) and the presence of conflicting interpretations in ClinVar, including a “likely benign” assertion.
When the relatively sparse phenotypic features selected by experts during manual interpretation were substituted for phenotypic features identified by CNLP, the recall of the autonomous system decreased (88%; 85 of 97).
Prospective performance of an autonomous system for diagnosis of childhood genetic diseases
We prospectively compared the performance of the autonomous diagnostic system with the fastest manual methods in seven seriously ill infants in ICUs and three previously diagnosed infants (Table 1). The median time from blood sample to diagnosis with the autonomous platform was 19:56 hours (range, 19:10 to 31:02 hours), compared with the median manual time of 48:23 hours (range, 34:38 to 56:03 hours). This included two automated runs that were delayed by operator error or data center downtime. The autonomous system coupled with InterVar post-processing made three diagnoses and no false-positive diagnoses. All three diagnoses were confirmed by manual methods and Sanger sequencing. The first was for patient 352, a 7-week-old female, admitted to the pediatric ICU with diabetic ketoacidosis. rWGS was performed on the singleton proband. In 19:11 hours, the autonomous system identified a previously unreported, heterozygous missense variant in the insulin gene (INS c.26C > G, pPro9Arg), which is associated with autosomal dominant permanent neonatal diabetes mellitus (OMIM disease record 606176). According to American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) pathogenicity criteria, the variant was of uncertain significance (VUS). After 42:04 hours, parent-child trio sequencing with the fastest manual methods confirmed the result and showed the variant to be de novo, which changed the variant classification to likely pathogenic.
The second diagnosis was made in patient 7052, a previously healthy 17-month-old boy admitted to the pediatric ICU with pseudomonal septic shock, metabolic acidosis, ecthyma gangrenosum, and hypogammaglobulinemia. Singleton, proband, rapid sequencing, and automated interpretation identified a pathogenic hemizygous variant in the Bruton tyrosine kinase gene (BTK c.974 + 2 T > C) associated with X-linked agammaglobulinemia 1 (OMIM #300755) in 22:04 hours. This was 16:33 hours earlier than a concurrent trio run with the fastest manual methods. The provisional result provided confidence in treatment with high-dose intravenous immunoglobulin (to maintain serum immunoglobulin G concentration of >600 mg/dl) and 6 weeks of antibiotic treatment. This provisional diagnosis was verbally conveyed to the clinical team upon review of the autonomous result by a laboratory director. Clinical whole-genome sequencing subsequently returned the same result and showed the variant to be maternally inherited.
The third diagnosis was made in patient 412, a 3-day-old boy admitted to the neonatal ICU with seizures and a strong family history of infantile seizures responsive to phenobarbital. The autonomous system identified a likely pathogenic, heterozygous variant in the potassium voltage-gated channel, KQT-like subfamily, member 2 gene (KCNQ2 c.1051C > G). This gene is associated with autosomal dominant benign familial neonatal seizures 1 (OMIM disease record 121200). The diagnosis was made in 20:53 hours, which was 27:30 hours earlier than a concurrent run with the fastest manual methods. A verbal provisional result was conveyed to the clinical team upon review of the result by a laboratory director as the diagnosis provided confidence in treatment with phenobarbital and changed the prognosis. For the remaining four patients, no diagnosis was evident with either the manual or autonomous method.
DISCUSSION
Previously, the fastest time to diagnosis by genome sequencing in clinical practice was 37 hours (8, 15-26) . The protocol was, however, extremely labor and capital intensive and was limited to one sample at a time. Here, we described a prototypic, autonomous system for genetic disease diagnosis in a median of 20:10 hours requiring decreased user intervention and a throughput of up to two parent-child trios or six probands per run. Most decision-making in ICUs is made deliberatively in morning rounds attended by a multidisciplinary health care team. Thus, a potential 20-hour diagnosis would return results to the on-call physician who had ordered testing in time for morning rounds. This would simplify information transfer during rounds and facilitate management decisions. A 20-hour diagnosis is important in seriously ill infants because most timely genomic diagnoses result in changes in ICU management (16-25).
Our autonomous platform for potential 20-hour diagnosis of genetic diseases was designed to meet the needs of acutely ill infants in ICUs with diseases of unknown etiology. It has been estimated that 10 to 12% of infants admitted to regional ICUs may benefit from same-day diagnosis and implementation of targeted treatments (8, 16-30). In 2014, the U.S. Food and Drug Administration (FDA) permitted provisional reporting in seriously ill children when the diagnosis indicated changes in management that could improve outcomes and where a delay in reporting until confirmation of results by Sanger sequencing could result in avoidable morbidity or mortality (18, 20, 21). In our previous experience, provisional diagnoses were reported in 17% (114 of 684) of genome sequencing cases, with a mean time to report of 3.6 days. Presentations in which 20-hour diagnoses were likely to be associated with improved outcomes included neonatal epileptic encephalopathies, metabolic diseases (as in patient 352), septic shock possibly associated with immunodeficiency (as in patient 7052), organ failure, and extracorporeal membrane oxygenation that is considered in the absence of a known disease etiology (18-24, 28). Thus, a circumscribed application of an autonomous diagnostic system is to identify provisional diagnoses for laboratory director review, earlier than standard rapid testing, in a subset of neonatal and pediatric ICU admissions in which morbidity or mortality is likely to be avoided by early institution of targeted treatment. It will be important to evaluate the proportion of seriously ill patients and extent of urgent health care settings in which a potential 20-hour diagnosis would inform acute interventions and for which a longer time to result would not be effective.
This paper demonstrated the automated extraction of a deep, digital phenome from the EHR. The analytic performance of the extraction of phenotypic features from the EHRs of children with genetic diseases by CNLP herein was considerably better than previous reports and appeared adequate for replacement of expert manual EHR review (36, 41). CNLP extracted 27-fold more phenotypic features from the EHR than those selected by experts during manual interpretation, consistent with previous reports (36, 41, 47). In addition, the mean IC of the CNLP phenome was greater than that of the phenotypic features selected by experts during manual interpretation. The superiority of deep CNLP phenomes was shown by substantially greater overlap with the expected (OMIM) clinical features than by those selected by experts during manual interpretation. Phenotypic features selected by experts during manual interpretation had poorer diagnostic utility than CNLP-based phenotypic features when used in the autonomous diagnostic system. This concurred with two recent reports of genome sequencing of cohorts of patients in which the rate of diagnosis was greater when more than 15 phenotypic features were used at time of interpretation than when one to five features were used (53, 54).
Here, we described fully automated interpretation of sequencing results. In 95 seriously ill children, the automated system had 97% recall and 99% precision in recapitulating 97 genetic disease diagnoses made by a team of experts. Where the system suggested more than one diagnosis, the median rank of a variant associated with the correct diagnosis was first. The three false-negative automated results had explanations that either can be addressed by parameter adjustments or were of types that cause assessments of variant pathogenicity to vary between laboratories (55). Prospectively, molecular laboratory directors determined that the automated system made correct provisional diagnoses in three of seven seriously ill ICU infants (100% precision and recall) with an average time saving of 22:19 hours. In light of insufficient expert analysts, molecular laboratory directors, medical geneticists, and genetic counselors to expand genomic diagnosis to regional ICU infants worldwide, such diagnostic performance was sufficient to suggest several, high-throughput clinical applications (31-33). Supervised autonomous systems may provide effective first-tier, provisional diagnoses, allowing valuable cognitive resources to be reserved for unsolved or difficult cases, manual curation of variants, and clinical report generation that includes a summary of medical management literature. Second, in the roughly 67% of cases where manual interpretation fails to provide a diagnosis, it is difficult to know when analysis should be considered complete. With further development, autonomous diagnostic systems could provide an independent, objective analysis in such cases. Third, autonomous systems could reanalyze unsolved cases periodically. This is burdensome to perform manually because 250 new gene-disease associations and 9200 new variant-disease associations are reported annually. However, reanalysis yields up to 8 to 10% new diagnoses per annum (56-60). Automated reanalysis could include updated CNLP of the EHR, which would be useful when the phenotype evolves with time. A known risk of genetic testing is overtreatment as a result of overdiagnosis (61). Periodic, autonomous reanalysis would also detect cases where the diagnosis is changed as a result of reclassification of the causality of the gene or pathogenicity of the variant and/or where phenome overlap was minimal. An autonomous system, akin to an autopilot, can decrease the labor intensity of genome interpretation. One hundred six years after the invention of the autopilot, however, two pilots are still employed in cockpits of commercial aircraft. Likewise, a skilled team will still be required to curate the literature and make tough decisions/classifications for the foreseeable future.
The automated system has several limitations. First, system performance is partly predicated on the quality of the history and physical examination and on the completeness of the write-up in EHR notes. The performance of the autonomous diagnostic system, although acceptable, is anticipated to improve with additional training, increased mapping of HPO terms associated with genetic diseases in OMIM, Orphanet, and the literature to SNOMED CT (the native language of the CNLP), inclusion of phenotypes from structured EHR fields, measurements of phenotype severity (such as phenotype term frequency in EHR documents), and material-negative phenotypes (pathognomonic phenotypes whose absence rules out a specific diagnosis). As part of this, a quantitative data model is needed for improved multivariate matching of nonindependent phenotypes that appropriately weights related, inexact phenotype matches. Although possible, the automated system did not take advantage of commercial variant database annotations, such as the Human Gene Mutation Database, and did not eliminate the labor-intensive literature curation that is the current standard for variant reporting. Diagnosis of genetic diseases due to SVs requires standard library preparation and additional software steps that add several hours to turnaround time. Because the autonomous system uses the same knowledge of allele and disease frequencies as manual interpretation, which underrepresent minority races or ethnicities, pathogenicity assertions in the latter groups are less certain. Likewise, because the autonomous system uses the same consensus guidelines for variant pathogenicity determination as manual interpretation, it is subject to the same general limitations of assertions of pathogenicity (55-61).
The major barriers to widespread adoption of genomic medicine for seriously ill infants with disorders of unknown etiology are an untrained medical workforce and substantial shortage of domain experts, including medical geneticists, molecular laboratory directors, and genetic counselors. Manual genome analysis and interpretation are very labor intensive. In addition, the extreme number of rare genetic diseases precludes easy domain mastery by nonexperts. Thus, pediatric genomic medicine may be one of the first clinical areas where artificial intelligence is necessary for its general adoption (62). Diagnosis of seriously ill infants with diseases of unknown etiology represents an early application of autonomous diagnostic systems because such cases are abundant in ICUs and a faster time to result is critical for optimal outcomes.
MATERIALS AND METHODS
Study design
This study was designed to furnish training and test datasets to assist in the development of a prototypic, autonomous system for very rapid, population-scale, provisional diagnoses of genetic diseases by genome sequencing and to separate datasets to test the analytic and diagnostic performance of the resultant system both retrospectively and prospectively. The 401 subjects analyzed herein were a convenience sample of the first symptomatic children who were enrolled in four studies that examined the diagnostic rate, time to diagnosis, clinical utility of diagnosis, outcomes, and health care utilization of rWGS between 26 July 2016 and 25 September 2018 at Rady Children’s Hospital, San Diego, USA (ClinicalTrials.gov identifiers: NCT03211039, NCT02917460, and NCT03385876) (18, 22-24, 28, 30). One of the studies was a randomized controlled trial of genome and exome sequencing (NCT03211039); the others were cohort studies. All subjects had a symptomatic illness of unknown etiology in which a genetic disorder was suspected. All subjects had a Rady Children’s Hospital Epic EHR and a genome sequence (genome or exome) that had been interpreted manually for diagnosis of a genetic disease. They included five groups, namely, 16 children tested for genetic diseases by rWGS whose EHRs were used to train CNLP (table S3), 10 children with genetic diseases diagnosed by rWGS whose EHRs were used to test the performance of CNLP (table S4), 101 children with genetic diseases diagnosed by rWGS whose genome sequences and EHRs were used to test the retrospective performance of the autonomous diagnostic system (table S15), 7 seriously ill children with suspected genetic diseases whose DNA samples and EHRs were used to test the prospective performance of the autonomous diagnostic system (Table 1), and 274 control children in whom rWGS did not disclose a genetic disease diagnosis. The studies were approved by the institutional review board at Rady Children’s Hospital, San Diego, USA. The studies were designated to be of “nonsignificant risk” by the FDA in response to an investigational device exemption presubmission inquiry in April 2014. The studies were performed in accordance with the Declaration of Helsinki. Informed consent was obtained from at least one parent or guardian.
Standard clinical rWGS and rWES, analysis, and interpretation
Standard clinical rWGS and rWES were performed in laboratories accredited by the College of American Pathologists (CAP) and certified through Clinical Laboratory Improvement Amendments (CLIA). Experts selected key clinical features representative of each child’s illness from the Epic EHR and mapped them to genetic diagnoses with Phenomizer or Phenolyzer (16, 18, 20-24, 45, 63). Trio EDTA-blood samples were obtained where possible. Genomic DNA was isolated with an EZ1 Advanced XL robot and the EZ1 DSP DNA Blood Kit (Qiagen). DNA quality was assessed with the Quant-iT Picogreen dsDNA Assay Kit (Thermo Fisher Scientific) with the Gemini EM Microplate Reader (Molecular Devices). Genomic DNA was fragmented by sonication (Covaris), and bar-coded, paired-end, polymerase chain reaction (PCR)–free libraries were prepared for rWGS with TruSeq DNA LT kits (Illumina) or Hyper kits (KAPA Biosystems). Sequencing libraries were analyzed with the Library Quantification Kit (KAPA Biosystems) and High Sensitivity NGS Fragment Analysis Kit (Advanced Analytical), respectively. One hundred one–nucleotide paired-end rWGS was performed to 45-fold coverage with Illumina HiSeq 2500 (rapid run mode), HiSeq 4000, or NovaSeq 6000 (S2 flow cell) instruments, as described (16). rWES was performed by GeneDx. Exome enrichment was with the xGen Exome Research Panel v1.0 (Integrated DNA Technologies), and amplification was performed using the Herculase II Fusion DNA Polymerase (Agilent) (18, 64). Sequences were aligned to human genome assembly GRCh37 (hg19), and variants were identified with the DRAGEN Platform (v.2.5.1, Illumina, San Diego; table S16) (16). SVs were identified with Manta and CNVnator (using DNAnexus), a combination that provided the highest sensitivity and precision in 21 samples with known SVs (table S18) (18, 65, 66). SVs were filtered to retain those affecting coding regions of known disease genes and with allele frequencies of <2% in the Rady Children’s Institute for Genomic Medicine (RCIGM) database. Nucleotide variants and SVs were annotated, analyzed, and interpreted by clinical molecular geneticists with Opal Clinical (Fabric Genomics), according to standard guidelines (50, 67). Opal annotated variants with respect to pathogenicity, generated a rank-ordered differential diagnosis based on the disease-gene algorithm VAAST (Variant Annotation, Analysis, and Search Tool; a gene burden test) and the algorithm PHEVOR (Phenotype Driven Variant Ontological Reranking), which combined the observed HPO phenotype terms from patients, and re-ranked disease genes based on the phenotypic match and the gene score (68-70). Automatically generated, ranked results were manually interpreted through iterative Opal searches. Initially, variants were filtered to retain those with allele frequencies of <1% in the Exome Variant Server, 1000 Genomes Samples, and Exome Aggregation Consortium database (71). Variants were further filtered for de novo, recessive, and dominant inheritance patterns. The evidence supporting a diagnosis was then manually evaluated by comparison with the published literature. Analysis, interpretation, and reporting required an average of 6 hours of expert effort. If rWGS or rWES established a provisional diagnosis for which a specific treatment was available to prevent morbidity or mortality, then this was immediately conveyed to the clinical team, as described. All causative variants were confirmed by Sanger sequencing or chromosomal microarray, as appropriate. Secondary findings were not reported, but medically actionable incidental findings were reported if families consented to receiving this information.
Natural language processing and phenotype extraction
Extraction of HPO terms from the EHR entailed the following four steps:
(1) Clinical records were exported from the EHR data warehouse, transformed into a compatible format (JSON), and loaded into CLiX ENRICH.
(2) A semi-automated query map was created, with HPO terms (and their synonyms) as the input and CLiX queries as the output. The HPO terms were passed through the CLiX encoding engine, resulting in creation of CLiX post-coordinated SNOMED CT expressions for each recognized HPO term or synonym. Where matches were not exact, manual review was used to validate the generated CLiX queries. Where there was no match or incorrect matches, new content was added to the Clinithink SNOMED CT extension and terminology files to ensure appropriate matches between phenotypes in HPO and those in SNOMED CT. This was an iterative process that resulted in a CLiX query set that covered 60% (7706) of 12,786 HPO terms (9 October 2017, HPO build).
(3) EHR documents containing unstructured data were passed through the CNLP engine. The natural language processing engine read the unstructured text and encoded it in structured format as post-coordinated SNOMED CT expressions. These expressions were more complex than simple SNOMED CT codes, and examples of their processing are included in Supplementary Materials and Methods.
(4) These encoded data were then interrogated by the CLiX query technology (abstraction). To trigger an HPO query, the encoded data had to contain either an exact match or one of its logical descendants (exploiting the parent-child hierarchy of the SNOMED CT ontology), resulting in a list of HPO terms for each patient.
Rapid whole-genome sequencing
Sequencing libraries were prepared from 10 μl of EDTA blood or five 3-mm punches from a Nucleic-Card Matrix dried blood spot (Thermo Fisher Scientific) with Nextera DNA Flex Library Prep kits (Illumina) and five cycles of PCR, as described (35). For SV analysis, libraries were prepared by Hyper kits (KAPA Biosystems), as described above. Libraries were quantified with Quant-iT PicoGreen dsDNA assays (Thermo Fisher Scientific). Libraries were sequenced (2 × 101 nt) without indexing on the S1 FC with NovaSeq 6000 S1 reagent kits (Illumina). Sequences were aligned to human genome assembly GRCh37 (hg19), and nucleotide variants were identified with the DRAGEN Platform (v.2.5.1, Illumina; table S16) (16).
Automated tertiary analysis
Automated variant interpretation was performed with MOON (Diploid) (72). Data sources and versions were ClinVar (2018-04-29), dbNSFP (3.5), dbSNP (150), dbscSNV (1.1), Apollo (2018-07-20), Ensembl (37), gnomAD (2.0.1), HPO (2017-10-05), Database of Genomic Variants (DGV; 2016-03-01), dbVar (2018-06-24), and MOON (2.0.5). MOON generated a list of potential provisional diagnoses by sequentially filtering and ranking variants with decision trees, Bayesian models, neural networks, and natural language processing. MOON was iteratively trained with thousands of previous patient samples uploaded by previous investigators. No samples analyzed in this study were used in training of MOON.
The filtering pipeline was designed to minimize false negatives. For single-nucleotide variant analysis, MOON excluded low-quality and common variants [>2% in Genome Aggregation Database (gnomAD)] and known likely benign/benign variants in ClinVar. We retained only variants in coding and splice site regions and known pathogenic variants in noncoding regions. A disease annotation was added to the remaining variants on the basis of a proprietary disorder model (72). The disorder model performs natural language processing of the genetics literature to automatically extract associations between diseases, disease genes, inheritance patterns, specific clinical features, and other metadata on an ongoing basis.
Subsequent steps included filtering on variant frequency, with variable frequency thresholds depending on the inheritance pattern of the associated disease, known pathogenicity of the variant, and typical age of onset range of the annotated disease. In family analyses (duo and trio analyses), cosegregation of the variant with the phenotype, according to autosomal dominant, autosomal recessive, X-linked dominant, or X-linked recessive inheritance patterns, was taken into account. Parent-child variant segregation was not applied as a strict filter criterion, thereby also ensuring that causal mutations following non-Mendelian inheritance (e.g., with incomplete penetrance) were identified in family analyses. For proband-only analyses, only variants for which the zygosity of the called variant fit the inheritance pattern of the annotated disease were retained. In a final filter step, the phenotype overlap was scored between the input HPO terms describing the patient’s phenotype and known disease manifestations of the disorder annotated from the published literature. Variants in genes for which the phenotype match with the annotated disease was considered too limited on the basis of Apollo were removed from the analysis. The final rank of variants was based on proprietary algorithms that took phenotype match and variant effect into account. In addition, MOON provided all metadata supporting the pathogenicity of ranked variants. MOON also returned an annotated list of all rare variants (<2% in gnomAD) and carrier status for recessive disorders.
For SV analysis, MOON removed known benign SVs on the basis of the DGV. SVs overlapping pathogenic SVs listed in dbVar were retained for analysis. From the remaining variants, MOON discarded SV that did not overlap with coding regions of known disease genes (Apollo). If a family analysis was performed, then segregation of the SV was taken into account, although non-Mendelian inheritance patterns (e.g., incomplete penetrance) were also supported. In a final filter step, only SVs for which there was a phenotype overlap between the input HPO terms and known disease presentations of at least one of the genes affected by the SV were retained. MOON then reported a ranked list of candidate SVs, where ranking was mostly based on the phenotype overlap.
Statistical analysis
To assess the complexity of phenomes associated with childhood genetic diseases, we compared phenotypes identified by manual review and by CNLP and listed for each patient’s diagnosis in OMIM. All analyses were conducted in R v3.3.3 (73). When applying CNLP to a patient’s EHR, the list of HPO terms produced contained both terms that had an exact match to a phenotype in the clinical notes and terms that were superclasses (ancestor terms) of exact matches. The R package ontologyIndex v2.4 was used to load the October 2017 build of HPO into R and to calculate the IC of each HPO term in the entire OMIM corpus (74). The IC for term ‘phenotype’, which reflects its clinical specificity, is given by IC(phenotype) = −log(pphenotype), where pphenotype was the probability of observing the exact term or one of its subclasses across all diseases in OMIM. Because phenotypes that were extracted manually and by CNLP were restricted to subclasses of “phenotypic abnormality” (HP:0000118), OMIM terms that were subclasses of “clinical modifier” (HP:0012823), “frequency” (HP:0040279), “mode of inheritance” (HP:0000005), and “mortality/aging” (HP:0040006) were not included in the analyses. Phenotype sets were first compared visually by plotting the HPO graph for each patient with the R package hpoPlot v2.4 (75). Summary statistics for outcomes of interest include the means, SD, and range. Before testing for significant differences, outcome variables were tested for normality with the Shapiro-Wilk test. Because of deviations from normality, differences in phenotype counts and IC were evaluated with two-sided Mann-Whitney U tests and, when the data were paired, Wilcoxon signed-rank tests. Correlation was assessed with Spearman’s rank correlation coefficient (rs). Precision and recall were given by tp/(tp + fp) and tp/(tp + fn), respectively, where tp was true positives, fp was false positives, and fn was false negatives. The number of true positives, tp, was defined in two ways. First, tp was set to the number of HPO terms that overlapped between sets of phenotypes. Second, tp was calculated on the basis of terms that were up to one degree of separation apart within the HPO hierarchy (parent-child terms) between sets of phenotypes, allowing for inexact, but similar, matches. Additional graphics were produced with packages ggplot2 v2.2.1 and eulerr v4.0.0 (76, 77). A significance cutoff of P < 0.05 was used for all analyses.
Supplementary Material
Fig. S1. Venn diagram showing overlap of observed and expected patient phenotypic features in 95 children diagnosed with 97 genetic diseases.
Fig. S2. Precision, recall, and F1 score of phenotypic features identified manually and by CNLP and OMIM.
Fig. S3. Flow diagram of the software components of the autonomous system for provisional diagnosis of genetic diseases by rWGS.
Table S1. Comparison of the analytic performance of standard and new library preparation and genome sequencing in retrospective samples.
Table S2. Comparison of the analytic performance of standard and rapid library preparation and genome sequencing methods in seven matched prospective samples.
Table S3. Characteristics of 16 children with genetic diseases used to train CNLP.
Table S4. Precision and recall of phenotypic features extracted by CNLP from EHRs in 10 children with genetic diseases.
Table S18. Number of SVs shortlisted by MOON and rank of the causal variant in MOON in 11 children with genetic diseases.
Table S19. Summary statistics of provisional diagnoses reported for clinical rWGS.
Table S5. Precision and recall of 26 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 201.
Table S7. Precision and recall of 95 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 213.
Table S6. Precision and recall of 96 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 205.
Table S8. Precision and recall of 158 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 233.
Table S10. Precision and recall of 90 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 6094.
Table S9. Precision of 85 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 243.
Table S11. Precision and recall of 96 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 6098.
Table S12. Precision and recall of 83 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 6108.
Table S13. Precision and recall of 44 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 7003.
Table S14. Precision and recall of 71 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 7004.
Table S15. The test cohort diagnosed manually by rWGS or rWES and interpreted retrospectively with an autonomous system.
Table S17. Number of nucleotide variants shortlisted by MOON and rank of the causal variant in MOON in 84 children with 86 genetic diseases.
Table S16. Variant characteristics in rWGS or rWES of the 101 children with 105 genetic diseases.
Data file S2. Phenotypic features of 101 children with genetic diseases that were manually extracted by experts from the EHR.
Data file S3. Phenotypic features of 101 children with genetic diseases that were automatically extracted from the EHR by CNLP.
Data file S1. Mapping of HPO terms to SNOMED CT expressions.
Acknowledgments:
The autonomous diagnostic system was made possible by previous advances by many groups with regard to data extraction from EHRs, CNLP, standardized genetic disease and phenotype vocabularies and databases, algorithms for ranking differential diagnosis based on observed phenotype, protocols and algorithms for variant pathogenicity assessment, databases of pathogenic variants and variant allele frequencies, and previous methods for rWGS. We acknowledge informatics effort by G. Concepcion, C. Sepulveda, L. Leonard, M. Wallis, D. O’Hagan, J. Kohrumel, J. Ambrose, T. Elmer, and E. Jones at Rady Children’s Hospital. We acknowledge effort by the RCIGM investigators J. Barea, G. Chiang, C. Cohenmeyer, N. G. Coufal, M. Evans, J. Honold, F. B. Imam, A. Kimball, B. Lane, C. Le, S. Leibel, L. Moyer, P. Ordonez, R. Wechsler-Reya, M. Speziale, D. Suttner, C. Sauer, R. Song, and A. Wise. We acknowledge valuable feedback and discussions from K. Lamoreaux (Cogenticity) and from P. Billings and B. Stache-Crain (Fabric Genomics).
Funding:
This study was supported by grant U19HD077693 from NICHD and NHGRI to S.F.K., grant U01TR002271 from NCATS to J. M. Davis and J. L. Maron (with sub-award to S.F.K.), and grant UL1TR002550 from NCATS to E. J. Topol (with sub-award to S.F.K.).
Footnotes
Competing interests: M.G.R. is an employee and stockholder of Fabric Genomics Inc. J.R., T.D., P.D.M., and M.B. are employees of Alexion Pharmaceuticals Inc., and each have both equity holdings and stock options. C. Yacoubian, A.F., R.G., and B.W. are employees of Clinithink Ltd. K.H. and H.G. are employees of Illumina Inc. C.K. and P.S. are employees of Diploid Inc. J. Gore is an employee of Tessella Inc. D.D. received funding from BioMarin (consultant for Pegvaliase trials), Audentes Therapeutics (Scientific Advisory Board), and Ichorion Therapeutics (consultant for mitochondrial disease drugs). M.B. owns stock in Codified Genomics and is a consultant for Baebies Inc. S.F.K. consults for AB2BIO. S.F.K. filed a provisional patent application filing number 62/659,495, entitled “Method and system for rapid genetic analysis” with the U.S. Patent and Trademark Office. J.F.’s spouse is the founder and principal of Friedman Bioventure, which holds a variety of publicly traded and private biotechnology interests.
Data and materials availability: All data associated with this study are present in the paper and/or the Supplementary Materials or are available at the Longitudinal Pediatric Data Resource under a material transfer agreement (MTA) or data use agreement, as appropriate, and are subject to the limitations of the informed consent documents for each subject (accession number nbs000003.v1.p; www.nbstrn.org/research-tools/longitudinal-pediatric-data-resource). InterVar is available at Github (https://github.com/WGLab/InterVar). CLiX ENRICH is available from CliniThink (info@clinithink.com). Moon is available from Diploid (info@diploid.com). The DRAGEN Platform is available from Illumina [S. Mehtalia (smehtalia@illumina.com)]. OPAL is available from Fabric Genomics (info@fabricgenomics.com). The RCIGM GEMS portal and pipeline are available under an MTA from R. Veeraraghavan (nveeraraghavan@rchsd.org).
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Khokha MK, Mitchell LE, Wallingford JB, White paper on the study of birth defects. Birth Defects Res. 109, 180–185 (2017). [DOI] [PubMed] [Google Scholar]
- 2.March of Dimes Foundation Data Book for Policy Makers: Maternal, Infant and Child Health in the United States 2016 (March of Dimes, 2016); www.marchofdimes.org/March-of-Dimes-2016-Databook.pdf. [Google Scholar]
- 3.Murphy SL, Xu J, Kochanek KD, Arias E, Mortality in the United States, 2017. NCHS Data Brief, 1–8 (2018). [PubMed] [Google Scholar]
- 4.Yoon PW, Olney RS, Khoury MJ, Sappenfield WM, Chavez GF, Taylor D, Contribution of birth defects and genetic diseases to pediatric hospitalizations. A population-based study. Arch. Pediatr. Adolesc. Med 151, 1096–1103 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Arth AC, Tinker SC, Simeone RM, Ailes EC, Cragan JD, Grosse SD, Inpatient hospitalization costs associated with birth defects among persons of all ages—United States, 2013. MMWR Morb. Mortal. Wkly Rep 66, 41–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry MA, Shah PS, Brouillette RT, Hellmann J, Predictors of mortality and length of stay for neonates admitted to children's hospital neonatal intensive care units. J. Perinatol 28, 297–302 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Committee on Approaching Death: Addressing Key End of Life Issues; Institute of Medicine, in Dying in America: Improving Quality and Honoring individual Preferences Near the End of Life (National Academies Press, 2015), chap. Appendix F Pediatric End-of-Life and Palliative Care: Epidemiology and Health Service Use. [PubMed] [Google Scholar]
- 8.Daoud H, Luco SM, Li R, Bareke E, Beaulieu C, Jarinova O, Carson N, Nikkel SM, Graham GE, Richer J, Armour C, Bulman DE, Chakraborty P, Geraghty M, Lines MA, Lacaze-Masmonteil T, Majewski J, Boycott KM, Dyment DA, Next-generation sequencing for diagnosis of rare diseases in the neonatal intensive care unit. CMAJ 188, E254–E260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malam F, Hartley T, Gillespie MK, Armour CM, Bariciak E, Graham GE, Nikkel SM, Richer J, Sawyer SL, Boycott KM, Dyment DA, Benchmarking outcomes in the neonatal intensive care unit: Cytogenetic and molecular diagnostic rates in a retrospective cohort. Am. J. Med. Genet. A 173, 1839–1847 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, Jiang Y-H, Xie P, Need A, Goldstein DB, The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet. Med 16, 176–182 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Weiner J, Sharma J, Lantos J, Kilbride H, How infants die in the neonatal intensive care unit: Trends from 1999 through 2008. Arch. Pediatr. Adolesc. Med 165, 630–634 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Petrikin JE, Willig LK, Smith LD, Kingsmore SF, Rapid whole genome sequencing and precision neonatology. Semin. Perinatol 39, 623–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith LD, Willig LK, Kingsmore SF, Whole-exome sequencing and whole-genome sequencing in critically ill neonates suspected to have single-gene disorders. Cold Spring Harb. Perspect. Med 6, a023168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.OMIM Entry Statistics (Johns Hopkins University, 2018); www.omim.org/statistics/geneMap. [Google Scholar]
- 15.National Center for Biotechnology Information, National Library of Medicine, Database of Single Nucleotide Polymorphisms (dbSNP) (2018); www.ncbi.nlm.nih.gov/dbvar?term=(%22clin%20pathogenic%22%5BFilter%5D)%20AND%20homo%20sapiens%5BOrganism%5D.
- 16.Miller NA, Farrow EG, Gibson M, Willig LK, Twist G, Yoo B, Marrs T, Corder S, Krivohlavek L, Walter A, Petrikin JE, Saunders CJ, Thiffault I, Soden SE, Smith LD, Dinwiddie DL, Herd S, Cakici JA, Catreux S, Ruehle M, Kingsmore SF, A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 7, 100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF, Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci. Transl. Med 4, 154ra135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, Cakici JA, Benson W, Kaplan RH, Kronick R, Bainbridge MN, Friedman J, Gold JJ, Ding Y, Veeraraghavan N, Dimmock D, Kingsmore SF, Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med 3, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, Zhang J, He W, Dharmadhikari AV, Qu C, Ward P, Braxton A, Narayanan S, Ge X, Tokita MJ, Santiago-Sim T, Dai H, Chiang T, Smith H, Azamian MS, Robak L, Bostwick BL, Schaaf CP, Potocki L, Scaglia F, Bacino CA, Hanchard NA, Wangler MF, Scott D, Brown C, Hu J, Belmont JW, Burrage LC, Graham BH, Sutton VR, Craigen WJ, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Muzny DM, Miller MJ, Wang X, Leduc MS, Xiao R, Liu P, Shaw C, Walkiewicz M, Bi W, Xia F, Lee B, Eng CM, Yang Y, Lalani SR, Use of exome sequencing for infants in intensive care units: Ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 171, e173438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, Saunders CJ, Thiffault I, Miller NA, Zellmer L, Herd SM, Holmes AM, Batalov S, Veeraraghavan N, Smith LD, Dimmock DP, Leeder JS, Kingsmore SF, The NSIGHT1-randomized controlled trial: Rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom. Med 3, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willig LK, Petrikin JE, Smith LD, Saunders CJ, Thiffault I, Miller NA, Soden SE, Cakici JA, Herd SM, Twist G, Noll A, Creed M, Alba PM, Carpenter SL, Clements MA, Fischer RT, Hays JA, Kilbride H, McDonough RJ, Rosterman JL, Tsai SL, Zellmer L, Farrow EG, Kingsmore SF, Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: A retrospective analysis of diagnostic and clinical findings. Lancet Respir. Med 3, 377–387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnaes L, Nahas SA, Chowdhury S, Nelson J, Batalov S, Dimmock DM, Kingsmore SF; RCIGM Investigators, Rapid whole-genome sequencing identifies a novel GABRA1variant associated with west syndrome. Cold Spring Harb. Mol. Case Stud 3, a001776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hildreth A, Wigby K, Chowdhury S, Nahas S, Barea J, Ordonez P, Batalov S, Dimmock D, Kingsmore S; RCIGM Investigators, Rapid whole-genome sequencing identifies a novel homozygous NPC1 variant associated with Niemann-Pick type C1 disease in a 7-week-old male with cholestasis. Cold Spring Harb. Mol. Case Stud 3, a001966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanford E, Watkins K, Nahas S, Gottschalk M, Coufal N, Farnaes L, Dimmock D, Kingsmore S; RCIGM Investigators, Rapid whole-genome sequencing identifies a novel AIRE variant associated with autoimmune polyendocrine syndrome type 1. Cold Spring Harb. Mol. Case Stud 4, a002485 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen DY, Chowdhury S, Farnaes L, Friedman JR, Honold J, Dimmock DP; RCIGM Investigators, Gold JJ, Rapid diagnosis of KCNQ2-associated early infantile epileptic encephalopathy improved outcome. Pediatr. Neurol 86, 69–70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark Z, Lunke S, Brett GR, Tan NB, Stapleton R, Kumble S, Yeung A, Phelan DG, Chong B, Fanjul-Fernandez M, Marum JE, Hunter M, Jarmolowicz A, Prawer Y, Riseley JR, Regan M, Elliott J, Martyn M, Best S, Tan TY, Gaff CL, White SM; Melbourne Genomics Health Alliance, Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet. Med 20, 1554–1563 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Mestek-Boukhibar L, Clement E, Jones WD, Drury S, Ocaka L, Gagunashvili A, Le Quesne Stabej P, Bacchelli C, Jani N, Rahman S, Jenkins L, Hurst JA, Bitner-Glindzicz M, Peters M, Beales PL, Williams HJ, Rapid Paediatric Sequencing (RaPS): Comprehensive real-life workflow for rapid diagnosis of critically ill children. J. Med. Genet 55, 721–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanford E, Farnaes L, Batalov S, Bainbridge M, Laubach S, Worthen HM, Tokita M, Kingsmore SF, Bradley J, Concomitant diagnosis of immune deficiency and Pseudomonas sepsis in a 19 month old with ecthyma gangrenosum by host whole-genome sequencing. Cold Spring Harb. Mol. Case Stud 4, a003244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, LePichon J-B, Miller NA, Thiffault I, Dinwiddie DL, Twist G, Noll A, Heese BA, Zellmer L, Atherton AM, Abdelmoity AT, Safina N, Nyp SS, Zuccarelli B, Larson IA, Modrcin A, Herd S, Creed M, Ye Z, Yuan X, Brodsky RA, Kingsmore SF, Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci. Transl. Med 6, 265ra168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briggs B, James KN, Chowdhury S, Thornburg C, Farnaes L, Dimmock D, Kingsmore SF; RCIGM Investigators, Novel factor XIII variant identified through whole-genome sequencing in a child with intracranial hemorrhage. Cold Spring Harb. Mol. Case Stud 4, a003525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stark Z, Dolman L, Manolio TA, Ozenberger B, Hill SL, Caulfied MJ, Levy Y, Glazer D, Wilson J, Lawler M, Boughtwood T, Braithwaite J, Goodhand P, Birney E, North KN, Integrating genomics into healthcare: A global responsibility. Am. J. Hum. Genet 104, 13–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman JM, Bombard Y, Cornel MC, Fernandez CV, Junker AK, Plon SE, Stark Z, Knoppers BM; Paediatric Task Team of the Global Alliance for Genomics and Health Regulatory and Ethics Work Stream, Genome-wide sequencing in acutely ill infants: Genomic medicine's critical application? Genet. Med 21, 498–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borghesi A, Mencarelli MA, Memo L, Ferrero GB, Bartuli A, Genuardi M, Stronati M, Villani A, Renieri A, Corsello G; Scientific Societies, Intersociety policy statement on the use of whole-exome sequencing in the critically ill newborn infant. ital. J. Pediatr 43, 100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.U.K. Department of Health and Social Care, Matt Hancock announces ambition to map 5 million genomes (2018); www.gov.uk/government/news/matt-hancock-announces-ambition-to-map-5-million-genomes.
- 35.Illumina Inc., Nextera DNA Flex Library Prep Reference Guide (Document # 1000000025416 v00, 2017); https://support.illumina.com/downloads/nextera-dna-flex-library-prep-reference-guide-1000000025416.html.
- 36.Son JH, Xie G, Yuan C, Ena L, Li Z, Goldstein A, Huang L, Wang L, Shen F, Liu H, Mehl K, Groopman EE, Marasa M, Kiryluk K, Gharavi AG, Chung WK, Hripcsak G, Friedman C, Weng C, Wang K, Deep phenotyping on electronic health records facilitates genetic diagnosis by clinical exomes. Am. J. Hum. Genet 103, 58–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hripcsak G, Albers DJ, High-fidelity phenotyping: Richness and freedom from bias. J. Am. Med. inform. Assoc 25, 289–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei W-Q, Denny JC, Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 7, 41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campillo-Gimenez B, Garcelon N, Jarno P, Chapplain JM, Cuggia M, Full-text automated detection of surgical site infections secondary to neurosurgery in Rennes, France. Stud. Health Technol. inform 192, 572–575 (2013). [PubMed] [Google Scholar]
- 40.Dhombres F, Bodenreider O, Interoperability between phenotypes in research and healthcare terminologies—Investigating partial mappings between HPO and SNOMED CT. J. Biomed. Semantics 7, 3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandel HL, “Performance evaluation of a natural language processing tool to extract infectious disease problems,” thesis, University of Washington Seattle, WA; (2013); http://bime.uw.edu/wordpress/wp-content/uploads/2016/11/Mandel-Hannah-L.-2013-MS.pdf. [Google Scholar]
- 42.Garcelon N, Neuraz A, Salomon R, Faour H, Benoit V, Delapalme A, Munnich A, Burgun A, Rance B, A clinician friendly data warehouse oriented toward narrative reports: Dr. Warehouse. J. Biomed. inform 80, 52–63 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Garcelon N, Neuraz A, Benoit V, Salomon R, Kracker S, Suarez F, Bahi-Buisson N, Hadj-Rabia S, Fischer A, Munnich A, Burgun A, Finding patients using similarity measures in a rare diseases-oriented clinical data warehouse: Dr. Warehouse and the needle in the needle stack. J. Biomed. Inform 73, 51–61 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Carlsson G, Topology and data. Bull. Am. Math. Soc 46, 255–308 (2009). [Google Scholar]
- 45.Köhler S, Schulz MH, Krawitz P, Bauer S, Dölken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN, Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet 85, 457–464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The Human Phenotype Ontology, Build #1246 (2017); http://compbio.charite.de/jenkins/job/hpo.annotations/1246/.
- 47.Garcelon N, Neuraz A, Salomon R, Bahi-Buisson N, Amiel J, Picard C, Mahlaoui N, Benoit V, Burgun A, Rance B, Next generation phenotyping using narrative reports in a rare disease clinical data warehouse. Orphanet J. Rare Dis 13, 85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Resnik P, Semantic similarity in a taxonomy: An information-based measure and its application to problems of ambiguity in natural language. J. Artif. Int. Res 11, 95–130 (1999). [Google Scholar]
- 49.Li Q, Wang K, InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet 100, 267–280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee, Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med 17, 405–423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kharrat M, Makni S, Makni K, Kammoun K, Charfeddine K, Azaeiz H, Jarraya F, Ben Hmida M, Gubler MC, Ayadi H, Hachicha J, Autosomal dominant Alport's syndrome: Study of a large Tunisian family. Saudi J. Kidney Dis. Transpl 17, 320–325 (2006). [PubMed] [Google Scholar]
- 52.Pescucci C, Mari F, Longo I, Vogiatzi P, Caselli R, Scala E, Abaterusso C, Gusmano R, Seri M, Miglietti N, Bresin E, Renieri A, Autosomal-dominant Alport syndrome: Natural history of a disease due to COL4A3 or COL4A4 gene. Kidney Int. 65, 1598–1603 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, Kingsmore SF, Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genome Med. 3, 16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss MER, Köster J, Marais A, Paknia O, Schröder R, Garcia-Aznar JM, Werber M, Brandau O, Calvo Del Castillo M, Baldi C, Wessel K, Kishore S, Nahavandi N, Eyaid W, Al Rifai MT, Al-Rumayyan A, Al-Twaijri W, Alothaim A, Alhashem A, Al-Sannaa N, Al-Balwi M, Alfadhel M, Rolfs A, Abou Jamra R, Clinical exome sequencing: Results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet 25, 176–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, Berg JS, Biswas S, Bowling KM, Conlin LK, Cooper GM, Dorschner MO, Dulik MC, Ghazani AA, Ghosh R, Green RC, Hart R, Horton C, Johnston JJ, Lebo MS, Milosavljevic A, Ou J, Pak CM, Patel RY, Punj S, Richards CS, Salama J, Strande NT, Yang Y, Plon SE, Biesecker LG, Rehm HL, Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet 98, 1067–1076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wenger AM, Guturu H, Bernstein JA, Bejerano G, Systematic reanalysis of clinical exome data yields additional diagnoses: Implications for providers. Genet. Med 19, 209–214 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Williams E, Retterer K, Cho M, Richard G, Juusola J, paper presented at the ACMG 2016, Tampa, FL, 2016. [Google Scholar]
- 58.Costain G, Jobling R, Walker S, Reuter MS, Snell M, Bowdin S, Cohn RD, Dupuis L, Hewson S, Mercimek-Andrews S, Shuman C, Sondheimer N, Weksberg R, Yoon G, Meyn MS, Stavropoulos DJ, Scherer SW, Mendoza-Londono R, Marshall CR, Periodic reanalysis of whole-genome sequencing data enhances the diagnostic advantage over standard clinical genetic testing. Eur. J. Hum. Genet 26, 740–744 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nambot S, Thevenon J, Kuentz P, Duffourd Y, Tisserant E, Bruel A-L, Mosca-Boidron A-L, Masurel-Paulet A, Lehalle D, Jean-Marçais N, Lefebvre M, Vabres P, El Chehadeh-Djebbar S, Philippe C, Tran Mau-Them F, St-Onge J, Jouan T, Chevarin M, Poé C, Carmignac V, Vitobello A, Callier P, Rivière J-B, Faivre L, Thauvin-Robinet C; Orphanomix Physicians' Group, Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: Substantial interest of prospective annual reanalysis. Genet. Med 20, 645–654 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Wright CF, McRae JF, Clayton S, Gallone G, Aitken S, FitzGerald TW, Jones P, Prigmore E, Rajan D, Lord J, Sifrim A, Kelsell R, Parker MJ, Barrett JC, Hurles ME, FitzPatrick DR, Firth HV; DDD Study, Making new genetic diagnoses with old data: Iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet. Med 20, 1216–1223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS, Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med 375, 655–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang H, Tsui BY, Ni H, Valentim CCS, Baxter SL, Liu G, Cai W, Kermany DS, Sun X, Chen J, He L, Zhu J, Tian P, Shao H, Zheng L, Hou R, Hewett S, Li G, Liang P, Zang X, Zhang Z, Pan L, Cai H, Ling R, Li S, Cui Y, Tang S, Ye H, Huang X, He W, Liang W, Zhang Q, Jiang J, Yu W, Gao J, Ou W, Deng Y, Hou Q, Wang B, Yao C, Liang Y, Zhang S, Duan Y, Zhang R, Gibson S, Zhang CL, Li O, Zhang ED, Karin G, Nguyen N, Wu X, Wen C, Xu J, Xu W, Wang B, Wang W, Li J, Pizzato B, Bao C, Xiang D, He W, He S, Zhou Y, Haw W, Goldbaum M, Tremoulet A, Hsu C-N, Carter H, Zhu L, Zhang K, Xia H, Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med 25, 433–438 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, Baynam G, Bello SM, Boerkoel CF, Boycott KM, Brudno M, Buske OJ, Chinnery PF, Cipriani V, Connell LE, Dawkins HJS, DeMare LE, Devereau AD, de Vries BBA, Firth HV, Freson K, Greene D, Hamosh A, Helbig I, Hum C, Jähn JA, James R, Krause R, Laulederkind SJF, Lochmüller H, Lyon GJ, Ogishima S, Olry A, Ouwehand WH, Pontikos N, Rath A, Schaefer F, Scott RH, Segal M, Sergouniotis PI, Sever R, Smith CL, Straub V, Thompson R, Turner C, Turro E, Veltman MWM, Vulliamy T, Yu J, von Ziegenweidt J, Zankl A, Züchner S, Zemojtel T, Jacobsen JOB, Groza T, Smedley D, Mungall CJ, Haendel M, Robinson PN, The Human Phenotype Ontology in 2017. Nucleic Acids Res. 45, D865–D876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Powis Z, Hart A, Cherny S, Petrik I, Palmaer E, Tang S, Jones C, Clinical diagnostic exome evaluation for an infant with a lethal disorder: Genetic diagnosis of TARP syndrome and expansion of the phenotype in a patient with a newly reported RBM10 alteration. BMC Med. Genet 18, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abyzov A, Urban AE, Snyder M, Gerstein M, CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21, 974–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT, Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Coonrod EM, Margraf RL, Russell A, Voelkerding KV, Reese MG, Clinical analysis of genome next-generation sequencing data using the Omicia platform. Expert Rev. Mol. Diagn 13, 529–540 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu H, Huff CD, Moore B, Flygare S, Reese MG, Yandell M, VAAST 2.0: Improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet. Epidemiol 37, 622–634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL, Durtschi JD, Tavtigian SV, Shankaracharya A, Wu W, Scheet P, Wang S, Xing J, Glusman G, Hubley R, Li H, Garg V, Moore B, Hood L, Galas DJ, Srivastava D, Reese MG, Jorde LB, Yandell M, Huff CD, A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nat. Biotechnol 32, 663–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singleton MV, Guthery SL, Voelkerding KV, Chen K, Kennedy B, Margraf RL, Durtschi J, Eilbeck K, Reese MG, Jorde LB, Huff CD, Yandell M, Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Genet 94, 599–610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha KE, Cummings BB, Birnbaum D; Exome Aggregation Consortium, Daly MJ, MacArthur DG, The ExAC browser: Displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 45, D840–D845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lumaka A, Race V, Peeters H, Corveleyn A, Coban-Akdemir Z, Jhangiani SN, Song X, Mubungu G, Posey J, Lupski JR, Vermeesch JR, Lukusa P, Devriendt K, A comprehensive clinical and genetic study in 127 patients with ID in Kinshasa, DR Congo. Am. J. Med. Genet. A 176, 1897–1909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R Development Core Team, R: A language and environment for statistical computing (2017); www.R-project.org/.
- 74.Greene D, Richardson S, Turro E, ontologyX: A suite of R packages for working with ontological data. Bioinformatics 33, 1104–1106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greene D, hpoPlot: Functions for Plotting HPO Terms. R package version 2.4 (2015); https://CRAN.R-project.org/package=hpoPlot. [Google Scholar]
- 76.Wickham H, ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag New York, 2009), pp. 216. [Google Scholar]
- 77.Larsson J, eulerr: Area-Proportional Euler and Venn Diagrams with Ellipses. R package version 4.0.0 (2018); https://cran.r-project.org/package=eulerr. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Venn diagram showing overlap of observed and expected patient phenotypic features in 95 children diagnosed with 97 genetic diseases.
Fig. S2. Precision, recall, and F1 score of phenotypic features identified manually and by CNLP and OMIM.
Fig. S3. Flow diagram of the software components of the autonomous system for provisional diagnosis of genetic diseases by rWGS.
Table S1. Comparison of the analytic performance of standard and new library preparation and genome sequencing in retrospective samples.
Table S2. Comparison of the analytic performance of standard and rapid library preparation and genome sequencing methods in seven matched prospective samples.
Table S3. Characteristics of 16 children with genetic diseases used to train CNLP.
Table S4. Precision and recall of phenotypic features extracted by CNLP from EHRs in 10 children with genetic diseases.
Table S18. Number of SVs shortlisted by MOON and rank of the causal variant in MOON in 11 children with genetic diseases.
Table S19. Summary statistics of provisional diagnoses reported for clinical rWGS.
Table S5. Precision and recall of 26 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 201.
Table S7. Precision and recall of 95 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 213.
Table S6. Precision and recall of 96 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 205.
Table S8. Precision and recall of 158 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 233.
Table S10. Precision and recall of 90 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 6094.
Table S9. Precision of 85 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 243.
Table S11. Precision and recall of 96 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 6098.
Table S12. Precision and recall of 83 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 6108.
Table S13. Precision and recall of 44 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 7003.
Table S14. Precision and recall of 71 phenotypic features extracted and proportion of OMIM clinical features detected by CNLP from the EHR of patient 7004.
Table S15. The test cohort diagnosed manually by rWGS or rWES and interpreted retrospectively with an autonomous system.
Table S17. Number of nucleotide variants shortlisted by MOON and rank of the causal variant in MOON in 84 children with 86 genetic diseases.
Table S16. Variant characteristics in rWGS or rWES of the 101 children with 105 genetic diseases.
Data file S2. Phenotypic features of 101 children with genetic diseases that were manually extracted by experts from the EHR.
Data file S3. Phenotypic features of 101 children with genetic diseases that were automatically extracted from the EHR by CNLP.
Data file S1. Mapping of HPO terms to SNOMED CT expressions.