Abstract
Objective
The incidence of postmenopausal endometrial cancer (EC) is rising, and the uterine microbiota has recently been suggested to be an etiology of EC. However, the differences in microbiota profiles in paired EC and the adjacent non-EC endometrium, and the functional microbiota of clinical relevance remain largely unknown. Therefore, we examined the differences in microbiota profiles between EC and non-EC endometrium and investigated their clinical relevance to EC.
Methods
Twenty-eight EC-affected postmenopausal women undergoing hysterectomy were enrolled. Endometrial microbiome from paired EC and adjacent non-EC tissue samples were detected using 16S rRNA sequencing, and the data were analyzed using R language software.
Results
The α diversity and evenness of the endometrial bacterial community significantly increased in EC tissues than those in pericancer tissues (P < 0.05 for all variables). Lactobacillus and Gardnerella were the main bacterial genera present in both EC and adjacent non–EC-invading endometrium, whereas Prevotella, Atopobium, Anaerococcus, Dialister, Porphyromonas, and Peptoniphilus were more commonly enriched in the EC endometrium (corrected P < 0.05 for all variables). Finally, the abundance of some observed endometrial bacteria was associated with clinical aspects, particularly the vaginal pH, vaginal Lactobacillus abundance, and EC clinical stage.
Conclusions
Paired EC and adjacent non-EC endometrium harbor different endometrial microbiota, and the functional bacteria residing in the endometrium are clinically relevant but require further investigation.
Key Words: 16S rRNA sequencing, Clinical relevance, Endometrial cancer, Tissue microbiota
Endometrial cancer (EC), which originates in the endometrium, has become the most common gynecological tumor in developed or high-income countries because of the increase in risk factors, such as obesity.1,2 Histopathological studies suggest that endometrioid adenocarcinoma with estrogen dependence (type I EC) is the most frequent subtype.3 The removal of the uterus with or without uterine appendages through surgery followed by adjuvant therapy is the primary treatment strategy.4 Recently, several studies have focused on elucidating the mechanisms underlying endometrial carcinogenesis by identifying potential causative factors and validating them at the molecular or cellular level.5,6 To date, the disparate roles of host genetic alterations, hereditary predisposition, and identified environmental factors for EC development, including obesity, diabetes, inflammation, gonadal hormones, and menopausal status, have advanced the understanding of EC's biological heterogeneity.1,2 Despite these advances, further studies are necessary to identify previously unrecognized tumorigenic mechanisms.
Emerging evidence demonstrates that the uterus harbors its own microbiota, which can modify the uterine functions in health and disease.7 Through next-generation sequencing techniques, several cross-sectional studies have presented and compared endometrial microbiota profiles in healthy women and women with EC.8-11 The endometrial microbial community exhibits considerable heterogeneity, with contributing factors including procedures for obtaining endometrial samples, gonadal hormones, menopausal status, and vaginal microbial communities.12,13 However, the uterine microbial composition and its role in EC progression or development remain poorly elucidated. In particular, differences in tissue microbiota from EC and paired pericancer (PC) tissues have not yet been addressed.
In this cohort study, we compared the tissue microbiota in EC and paired PC tissues in postmenopausal women using 16S rRNA sequencing and attempted to clarify its correlation with clinicopathological EC profiles. We hope that our results will provide definitive evidence for preventing EC occurrence and contribute to improving related therapy by modifying the uterine microbial composition.
METHODS
Participant Enrollment
This investigation was conducted in the Department of Obstetrics and Gynecology of a comprehensive tertiary hospital in Taiyuan, China, from October 2020 to December 2021. Menopausal women histopathologically diagnosed with endometrioid adenocarcinoma and who underwent hysterectomy using the open surgical approach were enrolled voluntarily. The exclusion criteria were as follows: (a) nonmenopausal women; (b) individuals with current autoimmune diseases and gastrointestinal disorders, or a history of gastrointestinal surgery; (c) individuals with a history of genital tract infection and/or application of any antimicrobial treatments to the genital area within the past 3 months; (d) individuals receiving preoperative chemotherapy or radiotherapy; (e) use of systemic antibiotics, corticosteroids, or any other immunosuppressive therapy in the past 6 months; (f) smokers with smoking index >400; and (g) drinkers with daily ethanol intake ≥20 g in the past 5 years or ≥80 g in the past 2 weeks. The study was approved by the Medical Ethics Committee of the First Hospital of Shanxi Medical University, and written informed consent was obtained from all participants during recruitment.
Participant Profiles
A structured questionnaire was used to acquire data on general demographic variables (age, age of menarche and menopause, ethnicity, height, and weight), surgical history, previous or current medications, smoking habits, and alcohol intake. A worksheet was used to collect clinicopathological profiles including diabetes, fertility, number of abortions, intrauterine device (IUD) use, vaginal pH, vaginal Lactobacillus abundance, estrogen level, necrosis in EC, clinical stage, and EC differentiation grade, from electronic medical records. Specifically, semiquantitative vaginal Lactobacillus determination (Lactobacillus presence in average five to eight microscopic fields) was performed under an oil microscope: 0, no Lactobacillus; 1+, 1 Lactobacillus; 2+, 2 to 5 Lactobacillus; 3+, 6 to 30 Lactobacillus; and 4+, >30 Lactobacillus. Histopathological assessment of the EC differentiation degree was conducted referring to the International Federation of Gynecology and Obstetrics grading system of EC. Briefly, an endometrial glandular component area corresponding to >95%, 50% to 94%, and <50% neoplasm was classified as well-, moderate-, poor-differentiated EC, respectively, with corresponding cellular atypia.14
Tissue Sample Processing, DNA Extraction, and Sequencing
Once removed, any necrotic or eroded uterine surface tissue was immediately scraped using sterile scalpels before macrodissecting the residual endometrium and cancer tissues. In detail, the visible cancerous tissue and normal tissue 2 cm away from the cancerous tissue were defined as EC and PC groups (confirmed by histopathology), respectively. Notably, the tissue specimens obtained were rinsed three to five times in sterile saline (0.85% NaCl) for removing the microbiota with unstable adhesions or tissue fluid adherence. To homogenize the samples, the tissues collected in a sterile plastic cup were first frozen in liquid nitrogen for 24 hours and then stored at −80°C until DNA extraction. DNeasy PowerSoil kit (Qiagen, Hilden, Germany) was used to isolate bacterial DNA from the endometrial samples according to the manufacturer's instructions. A NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and 1.0% agarose gel electrophoresis were used to determine the DNA concentration and integrity, respectively. The entire process was conducted by an experienced gynecologist and a laboratory technician under strict aseptic procedures. In addition, a negative control group including six tubes of prepared diethyl pyrocarbonate water was set to avoid potential bacterial contamination of molecular biology reagents. The processes of DNA extraction from water specimens to polymerase chain reaction (PCR) were completely synchronized with those from tissues. Our PCR assays failed to obtain positive results from the negative control group.
To analyze the bacterial colonies in tissue, the V3-V4 region of the 16S rRNA gene was amplified using PCR with bacterial universal primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GGACTACHVGGGTATCTAAT-3′). All PCRs were performed in triplicate using a 50 μL mixture containing 1 × Phanta Flash Master Mix (#P520; Vazyme, Wuhan, China), 10 μM primers, and 100 ng template DNA. The amplification conditions were as follows: 98°C for 30 seconds (1 cycle); followed by 98°C for 10 seconds, 56°C for 5 seconds, 72°C for 5 seconds (30 cycles); and 72°C for 1 minute (1 cycle). The amplicons were then recovered from 2% agarose gels and further purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, San Francisco, CA) and quantified using QuantiFluor-ST (Promega, Madison, WI) according to corresponding protocols. According to the instruction, the purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, CA). The raw reads were deposited into the NCBI Sequence Read Archive database (accession number, PRJNA847034).
Bioinformatic Analysis and Statistical Tests
Paired-end raw reads with overlap were merged to tags, and tags were clustered to operational taxonomic units (OTUs) at 97% sequence similarity using UPARSE (version 7.1, http://drive5.com/uparse/). The taxonomy of each 16S rRNA gene sequence was analyzed using the RDP Classifier Algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU128) 16S rRNA database, with 70% confidence threshold. QIIME and R packages (v3.2.0) from the free online Majorbio I-Sanger Cloud Platform (www.i-sanger.com) were used to analyze the sequencing data. Species diversity in a single sample was determined by calculating the α diversity indices (Shannon and Simpson index), evenness indices (Shannon even index), and richness estimators (Sobs and Chao index) at OTU and genus level using Mothur v.1.30.2. The differences in these indices between the groups were further assessed using the Wilcoxon rank-sum test. β Diversity, representing the microbial community heterogeneity between groups, was first shown on the principal coordinates analysis (PCoA) conducted using the R package at the genus level. Then, rank of distance, representing the microbial distribution difference between groups, was determined using the analysis of similarities package in R-vegan. Statistically, significant differences in the relative abundance of taxa were determined by Wilcoxon rank-sum tests, whereas linear discriminant analysis was used for assessing taxa responsible for the differences between the groups. Pearson correlation was calculated between the differential abundance at the genus level and clinical characteristics using the VGAM package. Before the correlation analysis, the variance inflation factor (VIF) was analyzed to assess the existing multicollinearity between clinical factors, and the factors with VIF >10 were usually removed because of their high multicollinearity. The continuous and categorical variables are presented as mean ± SD and percentage (%), respectively. The χ2 test or Student t test was conducted to compare the demographic information and clinical characteristics between the two groups. P < 0.05 (two sides) was accepted as the cutoff for statistical significance, and a false discovery rate was considered for correcting the P value.
RESULTS
Participant Characteristics
During the time of study, 134 women were diagnosed with EC and underwent hysterectomy in our department. After excluding cases with nonmenopausal status (57 cases) and the other exclusion conditions mentioned before (36 cases), 41 EC samples with corresponding PC samples were further used for bacterial DNA extraction. However, bacterial DNA was undetectable in 3 EC tissues, 6 PC tissues, and 4 EC with PC tissues; therefore, only 28 cases were included. The demographic and clinical characteristics of the included participants are summarized in Table 1. Specifically, 3 women had a nonhormone-containing IUD in place during surgery, and 14 women had a history of IUD use (the average interval between the time of IUD removal and the time of surgery was 4.88 ± 4.38 years).
Table 1.
Demographic and clinical characteristics of the 28 included participants
| Items | Valuations |
|---|---|
| Age, y | 60.41 ± 5.22 |
| Age of menarche, y | 14.20 ± 1.31 |
| Age of menopause, y | 54.36 ± 2.69 |
| BMI, kg/m2 | 25.68 ± 1.97 |
| Diabetes, n (%) | 13 (46.43) |
| Childless, n (%) | 5 (17.86) |
| No. abortions (−/1/2) | 8/13/7 |
| IUD history or presence, n (%) | 17 (60.71) |
| Vaginal pH | 4.76 ± 1.06 |
| Vaginal Lactobacillus (0/1+/2+/3+/4+), n | 4/11/4/8/1 |
| Estrogen level, pg/mL | 23.51 ± 10.49 |
| Necrosis in EC, n (%) | 7 (25.00) |
| Clinical stage (I/II/III),a n | 14/12/2 |
| EC differentiation (well/moderate/poor), n | 11/10/7 |
Data are presented as mean ± SD unless otherwise is indicated.
BMI, body mass index; EC, endometrial cancer; IUD, intrauterine device; pH, pondus hydrogenii.
aStaging by International Federation of Gynecology and Obstetrics.
Sequencing Data
We obtained total 9,331,057 raw reads with 2,332,764,250 bases from the 56 tissue samples. After quality filtering and trimming, 171,488 (ranging from 81,218 to 299,622; median, 168,918) valid sequences with a mean length of 422 bp and 161,763 (ranging from 89,791 to 211,375; median, 164,717) valid sequences with a mean length of 426 bp were screened from 28 tissue samples in EC and PC cohorts, respectively. The reads from the two cohorts involved 1,472 OTUs, including 25 phyla, 60 classes, 127 orders, 204 families, 367 genera, and 547 species. The value of the good's coverage estimator was 99.99%. Clear asymptotes observed in refraction curve analyses with Shannon or Sobs index at the OTU level indicate a near-complete sampling of the bacterial communities.
Bacterial Diversity Analysis
The EC group had higher index values for Sobs and Chao at the OTU and genus levels than the PC group (Table 2), demonstrating increased bacterial community richness in the EC tissues (corrected P > 0.05 for all variables). In comparison with those in the PC group, the Simpson estimator and index values for Shannon or Shannoneven at OTU and genus levels in the EC group were remarkably lower and higher, respectively (corrected P < 0.01 for all variables), which implies increased α diversity and evenness of the endometrial bacterial community in EC tissues. At the genus level, distinguished cohorts were visualized by hierarchical clustering tree, and β diversity analyzed using the PCoA based on unweighted UniFrac metrics also showed significant distinct clusters among the two cohorts (R = 0.149, P = 0.001, Fig. 1A, B).
Table 2.
α Diversity analysis in the two cohorts at OTU and genus levels
| EC (OTU) | PC (OTU) | EC (Genus) | PC (Genus) | ||
|---|---|---|---|---|---|
| Sobs | 118.82 ± 128.41a | 57.11 ± 29.94 | 45.85 ± 33.81b | 24.85 ± 8.09 | |
| Chao | 154.34 ± 117.72a | 113.84 ± 63.25 | 57.62 ± 36.63a | 41.81 ± 34.64 | |
| Shannon | 1.17 ± 0.94b | 0.42 ± 0.36 | 0.84 ± 0.89b | 0.15 ± 0.23 | |
| Simpson | 0.54 ± 0.32b | 0.77 ± 0.22 | 0.66 ± 0.35b | 0.91 ± 0.16 | |
| Shannoneven | 0.25 ± 0.19b | 0.11 ± 0.01 | 0.21 ± 0.22b | 0.05 ± 0.07 | |
| Good coverage | 0.99 ± 0.00 | 0.99 ± 0.00 | 0.99 ± 0.00 | 0.99 ± 0.00 | |
Wilcox rank-sum test was used for statistical differences between the groups.
EC, endometrial cancer; OTU, operational taxonomic units; PC, pericancer.
aCorrected P < 0.05.
bCorrected P < 0.01.
FIG. 1.
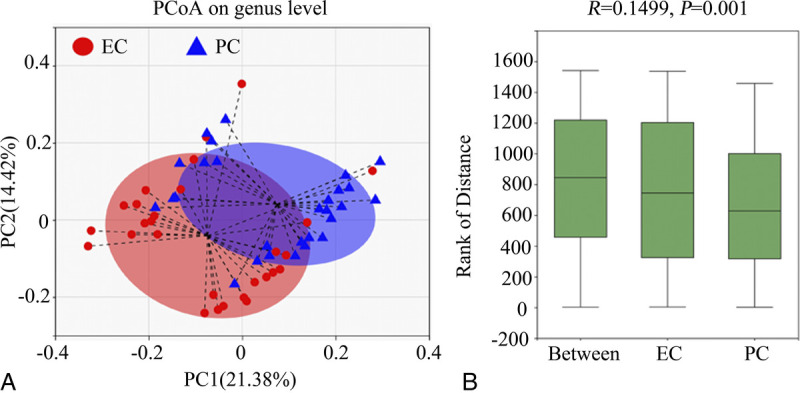
β Diversity analysis of the gut microbial structure. At the genus level, (A) principal co-ordinate analysis and (B) comparison of distance rank by analysis of similarities were performed as specified in the Methods. EC, endometrial cancer; PC, pericancer; PCoA, principal coordinates analysis.
Overall Distribution of Endometrial Microbiota
The groups shared 157 genera, with 178 and 32 unique genera detected in the EC and PC groups, respectively. More than 65% endometrial bacteria in both groups belonged to the phyla Firmicutes and Actinobacteriota (Fig. 2A). The genera Lactobacillus (85.24%) and Gardnerella (12.26%) occupied the vast majority of the bacteria in the PC tissues, whereas the top eight genera identified in EC tissues were Lactobacillus (53.79%), Gardnerella (12.19%), Prevotella (8.79%), Atopobium (3.58%), Bifidobacterium (3.48%), Streptococcus (3.15%), Sneathia (2.08%), and Proteus (1.36%) (Fig. 2B).
FIG. 2.
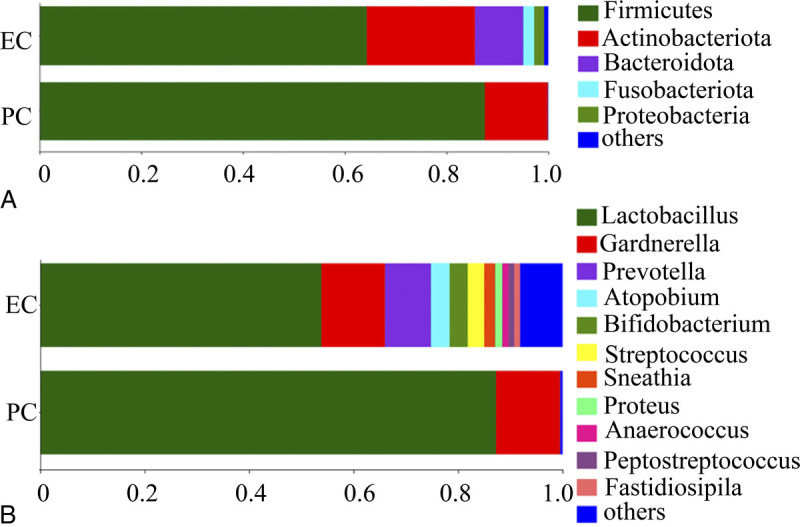
Composition and relative proportion of endometrial bacteria in the two separate tissues at (A) phylum and (B) genus levels. EC, endometrial cancer; PC, pericancer.
Community Structure Differences
At the phylum level, EC tissues had significantly lower relative abundance of Firmicutes (64.27 ± 38.91 vs 87.51 ± 26.85, corrected P < 0.05) and higher relative abundance of Bacteroidota (9.49 ± 17.19 vs 0.01 ± 0.03, corrected P < 0.000), Fusobacteriota (2.15 ± 6.05 vs 0.002 ± 0.004, corrected P < 0.05), and Campilobacterota (0.07 ± 0.21 vs 0.005 ± 0.001, corrected P < 0.05) than the PC tissues. Differences in the relative abundance of the top 20 genera in the endometrial microbiota in the two cohorts were further analyzed using the Wilcox rank-sum test. The results showed that Prevotella (8.79 ± 16.69 vs 0.01 ± 0.03, corrected P < 0.000), Atopobium (5.58 ± 6.49 vs 0.003 ± 0.007, corrected P < 0.01), Anaerococcus (1.18 ± 3.64 vs 0.009 ± 0.017, corrected P < 0.01), Dialister (0.59 ± 0.97 vs 0.002 ± 0.007, corrected P < 0.000), Porphyromonas (0.57 ± 1.69 vs 0.005 ± 0.000, corrected P < 0.000), and Peptoniphilus (0.56 ± 1.35 vs 0.004 ± 0.006, corrected P < 0.000) abundance significantly increased in the EC cohort than that in the PC cohort (Fig. 3).
FIG. 3.
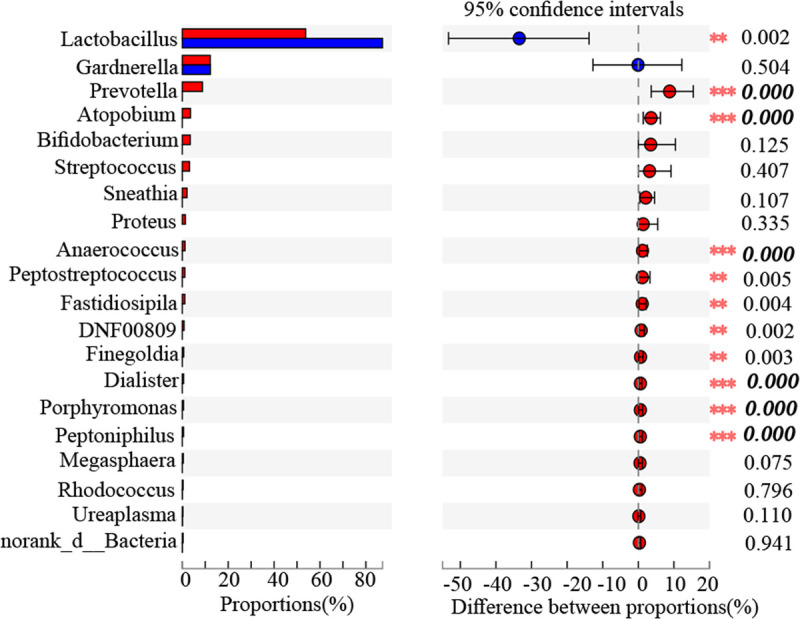
Wilcoxon rank-sum test bar plot at genus level. The bolded and italicized values indicate corrected P < 0.05.
In addition, the application of the linear discriminant analysis effect size method identified 31 features with significantly different abundance from the phylum to genus levels between the EC and PC cohorts (linear discriminant analysis score >3, P < 0.05; Fig. 4A). Specifically, the endometrial microbiota of EC tissues was differently enriched in the genera Prevotella, Atopobium, Peptostreptococcus, Anaerococcus, Fastidiosipila, Finegoldia, DNF00809, Dialister, Peptoniphilus, Porphyromonas, Anaeroglobus, and Criibacterium (P < 0.05 for all variables), whereas that of PC tissues was enriched only in genera Lactobacillus (P < 0.05; Fig. 4B).
FIG. 4.
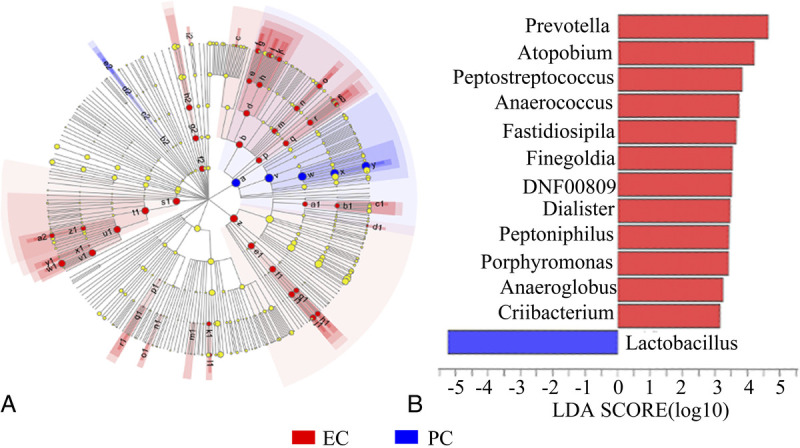
Differently abundant taxa identified using LEfSe analysis among cohorts. (A) Distribution of differently abundant taxa at the phylum (the innermost circle), class (the second circle), order (the third circle), family (the fourth circle), and genus (the outermost circle) levels between the EC and PC cohorts. The effect of the microbial taxa significantly enriched in the EC and PC cohorts on the differences between the cohorts are expressed as red and blue solid circles, respectively. Large solid circles indicate a high degree of influence of the species on the intergroup. Light yellow circles indicate that the microbial taxa had no significant effects on the differences between groups. (B) Linear discriminant analysis showing the impact of different species on the difference between EC and PC cohorts and visualization of only taxa meeting an LDA ≥3. EC, endometrial cancer; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; PC, pericancer.
Relationship with Clinical Characteristics
Finally, we analyzed the relationship between the observed genera in the participants and the clinical characteristics. A clinical indicator with VIF >10 was considered to influence subsequent correlation analysis because of the existence of multicollinearity between clinical factors. The VIF of age during enrollment, menarche age, menopause age, body mass index, diabetes status, childless status, number of abortions, IUD use, vaginal pH, vaginal Lactobacillus abundance, estrogen level, necrosis in EC, pathological stage, and EC differentiation were 4.75, 2.13, 4.11, 1.51, 1.78, 2.71, 1.81, 1.27, 2.51, 2.27, 1.96, 1.95, 1.57, and 1.71, respectively. As the correlation heatmap graph demonstrated (Fig. 5), vaginal pH is moderately positively and negatively correlated with the abundance of the endometrial genera Lactobacillus (R = 0.540, P < 0.01) and Atopobium (R = −0.491, P < 0.01), respectively. Vaginal Lactobacillus abundance is strongly positively associated with endometrial genera Prevotella (R = 0.603, P < 0.001); moderately with Gardnerella (R = 0.487, P < 0.01), Atopobium (R = 0.533, P < 0.01) and Dialister (R = 0.555, P < 0.01); and strongly negatively with Lactobacillus (R = −0.785, P < 0.001) abundance. In addition, endometrial genera Gardnerella (R = 0.661, P < 0.001), Atopobium (R = 0.782, P < 0.001), Sneathia (R = 0.518, P < 0.01), and Fastidiosipila (R = 0.488, P < 0.01) abundance was strongly-to-moderately positively correlated with EC clinical stage.
FIG. 5.
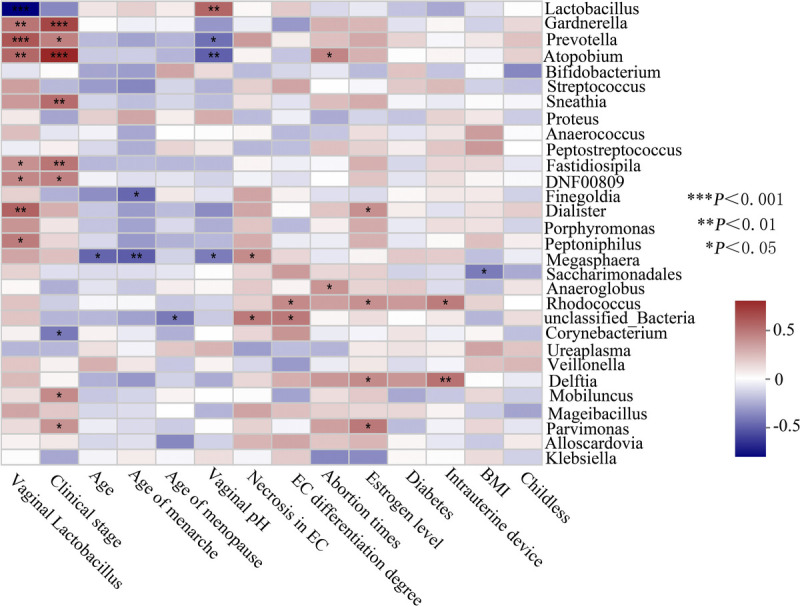
Correlation analysis of endometrial bacteria and related clinical indicators. BMI, body mass index; EC, endometrial cancer.
DISCUSSION
The incidence of EC is rising, and the estimated cumulative risk of developing EC by the age of 75 years is 1%. Endometrial cancer, the fifth most common cancer in women, is the 14th leading cause of death in cancers.1,2,15 Clarifying the potential causative events and pathophysiology of EC is a continuing need, and efforts are in progress. Available evidence has shown that only 10% to 20% EC cases can be attributed to host genetic alterations or hereditary factors.8 However, all EC cases are likely to be the combined result from genetic and environmental factors, such as long-term estrogen exposure, early menarche, late-onset menopause, nulliparity, and obesity.16 The microbiota is significantly associated with a wide range of diseases. The effect of gut microbiota dysbiosis on gastrointestinal tumors has been extensively explored, and it is supposed to affect EC carcinogenesis through regulating the risk factors for EC occurrence, including estrogen metabolism, obesity, and gut–vaginal microbiome axis.17-19 However, the relationship between endometrial bacteria and EC development is of greater concern than that between gut microbiota and EC development because tissue microbiota may directly affect tumor behavior.20,21 About tissue microbiota, more enigmas wait to be urgently clarified.
The uterus was long considered to be sterile, but current knowledge suggests that the uterus in healthy or diseased conditions harbors its own microbiota.22-25 Endometrial microbiota has been preliminarily proven to be involved in several gynecological disorders, including chronic endometritis, infertility, endometriosis, dysfunctional menstrual bleeding, and EC.26-31 Indeed, many clinical factors may independently influence the endometrial microbiome structure. Of particular relevance, postmenopausal status is the main driver of an EC-associated polymicrobial network.32 This study is the first attempt to characterize the tissue microbiota from EC and paired PC tissues in postmenopausal women and determine its correlation with clinicopathological EC profiles. The endometrial microbiota profiles in EC participants and individuals with benign uterine lesions have been recently characterized,10,11 and decreased α diversity of the bacterial community was supposed to be of great importance for EC development. However, in this study, the increased α diversity and evenness of endometrial bacterial community in the EC tissues compared with those in the adjacent nontumor endometrial tissues further underlines some unclear connections between endometrial microbiota shift and EC progression. The analysis of β diversity measured with spatial distance among EC and corresponding PC tissues using PCoA showed that the samples from different locations in uterine tissues could be clustered independently, which is to be expected given that they are tumor and non–tumor-affected samples.
Several studies also uncovered potential alterations in endometrial microbiota composition and function in EC tissues.8-11 The genera Anaerostipes, Dialister, Peptoniphilus, Ruminococcus, Anaerotruncus, Bacteroides, Arthrospira, Atopobium, and Porphyromonas were significantly enriched in EC tissues than those in endometrial hyperplasia tissues; moreover, relevance analysis showed that the coexistence of Atopobium and Porphyromonas with vaginal pH >4.5 was strongly associated with EC severity.8 In China, a study enrolling endometrial tissue samples from 30 EC participants and 10 healthy individuals showed that Pelomonas, Prevotella, Nocardioides, and Muribaculum were enriched and Oscillibacter was not enriched in the EC endometrium. Of particular interest, the increasing abundance of endometrial Prevotella was positively correlated with elevated serum D-dimer and fibrin degradation products, which implies a high tumor burden.10 Compared with that in benign uterine lesions, the genera Micrococcus was also more abundant in EC tissues and positively correlated with endometrial interleukin 6 and interleukin 17 messenger RNA levels, highlighting the clear role of microbiota-inflammation crosstalk in endometrium for EC development.11 Chen et al9 recently observed that the abundance of 17 bacterial species in EC tissues is significantly different than that in the normal endometrium, and further interaction analysis suggested the involvement of the functionally activated endometrial bacteria in the tumor metabolic process, including the 6-sulfo-sialyl Lewis x epitope and N-acetyl-β-glucosaminyl, as well as contributions to tumor migration. Differences in the number of participants, sexual and physical habits, menopausal status, contamination of endometrial samples, and methodological considerations, such as PCR primers, reference database, and analysis software used among studies, may lead to partially inconsistent conclusions.33
To minimize the heterogeneity among samples, paired EC and adjacent non-EC endometrium were chosen in this study. In contrast to the case of gut bacteria, endometrial bacteria were detectable in both EC and paired PC tissues in only 68.29% (28 of 41) enrolled cases, which highlights that bacterial invasion of or residency in the endometrium tissue may only occur under specific conditions. Figure 4 showed the increased richness of the genera Prevotella, Atopobium, Anaerococcus, Dialister, Porphyromonas, and Peptoniphilus in the EC endometrium, which partly corroborates the endometrial microbiota composition observed in previous findings.8-11 The result suggests that uncovering the variability of endometrial flora by subclassifying the endometrium according to EC affection is warranted. The distributions and function of endometrium-resident microbiota remain largely unknown. The tumor-resident intracellular microbiota was recently proved to promote metastatic colonization in breast cancer.34 Moreover, the endometrial Porphyromonas presence has been suggested to be the most predictive microbial marker of EC in high-risk, asymptomatic women. Crooks et al35 have presented evidence for intracellular invasion of Porphyromonas in EC cells in vitro, which revealed the possible activities and distributions of the uterine microbiome, underscoring the potential role of intracellular Porphyromonas in EC progression. However, it is noteworthy that the living bacteria, short bacterial DNA fragments, or microbial components may act synergistically. Furthermore, the origin of endometrium-resident microbiota has not been established yet. The evidence presented by the correlation analysis showed that the abundance of endometrial genera Lactobacillus was strongly inversely linked with that of vaginal Lactobacillus and moderately positively with vaginal pH, which may lend some support to the supposed bacterial redistribution hypothesis that Lactobacillus in the lower reproductive tract may partially migrate and colonize the endometrium, which should be further studied in future studies focusing on the clarification of bacterial transmission. Based on this assumption, the EC clinical stage was demonstrated to be strongly-to-moderately positive with endometrial non-Lactobacillus abundance, which may clue that non-Lactobacillus species easily colonize the disrupted tissue with EC progression.
The limitations of the current study include the following: (1) the causal relationships among specific endometrial bacteria and EC, particularly whether EC predisposes an area of the uterus to microbial invasion/colonization or the endometrium-resident microbiota predisposes the endometrium to EC occurrence or progression, are not adequately explained as with all case-controlled clinical studies; (2) isolation of bacteria invasive in the endometrium or adherent to the endometrial surface by removing endometrium luminal surface was unsuccessful because the cancer-unaffected endometrium from the postmenopausal uterus is very thin; (3) within-subject verification of specific endometrial bacteria in another newly enrolled cohort of larger study populations from multicenter is lacking, including the women with EC in clinical stages III and IV, if the bottlenecks in specimens acquisition are overcome is promising; (4) uterine microbial composition should be modified in clinical settings to confirm the possibilities of bacterial dysbiosis correction for EC interventions; and (5) premenopausal or perimenopausal women were not included in this study.
CONCLUSIONS
Overall, this study indicates that EC and adjacent EC-unaffected endometrium in postmenopausal individuals have significantly different microbiota, and some observed bacteria are strongly clinically relevant to the tumor's biological behavior.
Acknowledgments: We thank the key research and development projects in Shanxi Province (number 201803D31111).
Footnotes
This study was approved by the Medical Ethics Committee of the First Hospital of Shanxi Medical University.
All data in this study are available upon reasonable request made to the corresponding author.
Funding/support: This study was funded by key research and development projects in Shanxi Province (no. 201803D31111).
Financial disclosure/conflicts of interest: None reported.
Author Contributions: S.Z. contributed in the project development. L.W. contributed in the manuscript writing. L.W., J.Y. and H.S. contributed in the tissue sample collection. L.S. and B.C. contributed in the data analysis. B.C. contributed in the manuscript revision.
Contributor Information
Lili Wang, Email: w15513051926@163.com.
Jiaolin Yang, Email: yangjiaolin@163.com.
Huancheng Su, Email: 18734890406@163.com.
Liuming Shi, Email: shiliuming228@163.com.
Bangtao Chen, Email: medisci@163.com.
REFERENCES
- 1.Lu KH, Broaddus RR. Endometrial cancer. N Engl J Med 2020;383:2053–2064. doi: 10.1056/NEJMra1514010 [DOI] [PubMed] [Google Scholar]
- 2.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0 [DOI] [PubMed] [Google Scholar]
- 3.Akhtar M, Al Hyassat S, Elaiwy O, Rashid S, Al-Nabet ADMH. Classification of endometrial carcinoma: new perspectives beyond morphology. Adv Anat Pathol 2019;26:421–427. doi: 10.1097/PAP.0000000000000251 [DOI] [PubMed] [Google Scholar]
- 4.van den Heerik ASVM, Horeweg N, de Boer SM, Bosse T, Creutzberg CL. Adjuvant therapy for endometrial cancer in the era of molecular classification: radiotherapy, chemoradiation and novel targets for therapy. Int J Gynecol Cancer 2021;31:594–604. doi: 10.1136/ijgc-2020-001822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang Z, Talukdar S, Mullany SA, Winterhoff B. Molecular characterization of endometrial cancer and therapeutic implications. Curr Opin Obstet Gynecol 2019;31:24–30. doi: 10.1097/GCO.0000000000000508 [DOI] [PubMed] [Google Scholar]
- 6.Brooks RA Fleming GF Lastra RR, et al. Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin 2019;69:258–279. doi: 10.3322/caac.21561 [DOI] [PubMed] [Google Scholar]
- 7.Giudice LC. Challenging dogma: the endometrium has a microbiome with functional consequences! Am J Obstet Gynecol 2016;215:682–683. doi: 10.1016/j.ajog.2016.09.085 [DOI] [PubMed] [Google Scholar]
- 8.Walther-António MR Chen J Multinu F, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med 2016;8:122. doi: 10.1186/s13073-016-0368-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P Guo Y Jia L, et al. Interaction between functionally activate endometrial microbiota and host gene regulation in endometrial cancer. Front Cell Dev Biol 2021;9:727286. doi: 10.3389/fcell.2021.727286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C Gu Y He Q, et al. Integrated analysis of microbiome and transcriptome data reveals the interplay between commensal bacteria and fibrin degradation in endometrial cancer. Front Cell Infect Microbiol 2021;11:748558. doi: 10.3389/fcimb.2021.748558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu W He F Lin Z, et al. Dysbiosis of the endometrial microbiota and its association with inflammatory cytokines in endometrial cancer. Int J Cancer 2021;148:1708–1716. doi: 10.1002/ijc.33428 [DOI] [PubMed] [Google Scholar]
- 12.Molina NM Sola-Leyva A Saez-Lara MJ, et al. New opportunities for endometrial health by modifying uterine microbial composition: present or future? Biomolecules 2020;10:593. doi: 10.3390/biom10040593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuźmycz O, Stączek P. Prospects of NSAIDs administration as double-edged agents against endometrial cancer and pathological species of the uterine microbiome. Cancer Biol Ther 2020;21:486–494. doi: 10.1080/15384047.2020.1736483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva-Filho AL Xavier ÉB Cândido EB, et al. Comparative study of different histologic classifications in the degree of differentiation in endometrial adenocarcinoma. Tumori 2016;102:488–495. doi: 10.5301/tj.5000528 [DOI] [PubMed] [Google Scholar]
- 15.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 16.Setiawan VW Yang HP Pike MC, et al. Type I and II endometrial cancers: have they different risk factors? J Clin Oncol 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahid M Dar SA Jawed A, et al. Microbes in gynecologic cancers: causes or consequences and therapeutic potential. Semin Cancer Biol 2021;22:S1044-579X(21)00208-X. doi: 10.1016/j.semcancer.2021.07.013 [DOI] [PubMed] [Google Scholar]
- 18.Borella F Carosso AR Cosma S, et al. Gut microbiota and gynecological cancers: a summary of pathogenetic mechanisms and future directions. ACS Infect Dis 2021;7:987–1009. doi: 10.1021/acsinfecdis.0c00839 [DOI] [PubMed] [Google Scholar]
- 19.Boutriq S González-González A Plaza-Andrades I, et al. Gut and endometrial microbiome dysbiosis: a new emergent risk factor for endometrial cancer. J Pers Med 2021;11:659. doi: 10.3390/jpm11070659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giallourou N, Urbaniak C, Puebla-Barragan S, Vorkas PA, Swann JR, Reid G. Characterizing the breast cancer lipidome and its interaction with the tissue microbiota. Commun Biol 2021;4:1229. doi: 10.1038/s42003-021-02710-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong H Tan Q Xu Y, et al. Convergent alteration of lung tissue microbiota and tumor cells in lung cancer. iScience 2021;25:103638. doi: 10.1016/j.isci.2021.103638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C Song X Wei W, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun 2017;8:875. doi: 10.1038/s41467-017-00901-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winters AD Romero R Gervasi MT, et al. Does the endometrial cavity have a molecular microbial signature? Sci Rep 2019;9:9905. doi: 10.1038/s41598-019-46173-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker JM, Chase DM, Herbst-Kralovetz MM. Uterine microbiota: residents, tourists, or invaders? Front Immunol 2018;9:208. doi: 10.3389/fimmu.2018.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina-Bastidas D, Camacho-Arroyo I, García-Gómez E. Current findings in endometrial microbiome: impact on uterine diseases. Reproduction 2022;163:R81–R96; 1:REP-21-0120. doi: 10.1530/REP-21-0120 [DOI] [PubMed] [Google Scholar]
- 26.Verstraelen H Vilchez-Vargas R Desimpel F, et al. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. PeerJ 2016;4:e1602. doi: 10.7717/peerj.1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno I Cicinelli E Garcia-Grau I, et al. The diagnosis of chronic endometritis in infertile asymptomatic women: a comparative study of histology, microbial cultures, hysteroscopy, and molecular microbiology. Am J Obstet Gynecol 2018;218:602.e1–602.e16. doi: 10.1016/j.ajog.2018.02.012 [DOI] [PubMed] [Google Scholar]
- 28.Liu Y Ko EY Wong KK, et al. Endometrial microbiota in infertile women with and without chronic endometritis as diagnosed using a quantitative and reference range-based method. Fertil Steril 2019;112:707–717.e1. doi: 10.1016/j.fertnstert.2019.05.015 [DOI] [PubMed] [Google Scholar]
- 29.Pelzer ES, Willner D, Buttini M, Huygens F. A role for the endometrial microbiome in dysfunctional menstrual bleeding. Antonie Van Leeuwenhoek 2018;111:933–943. doi: 10.1007/s10482-017-0992-6 [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Grau I Perez-Villaroya D Bau D, et al. Taxonomical and functional assessment of the endometrial microbiota in a context of recurrent reproductive failure: a case report. Pathogens 2019;8:205. doi: 10.3390/pathogens8040205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke MA, Long BJ, Del Mar Morillo A, Arbyn M, Bakkum-Gamez JN, Wentzensen N. Association of endometrial cancer risk with postmenopausal bleeding in women: a systematic review and meta-analysis. JAMA Intern Med 2018;178:1210–1222. doi: 10.1001/jamainternmed.2018.2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh DM Hokenstad AN Chen J, et al. Postmenopause as a key factor in the composition of the endometrial cancer microbiome (ECbiome). Sci Rep 2019;9:19213. doi: 10.1038/s41598-019-55720-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Pol WJ Kumar R Morrow CD, et al. In silico and experimental evaluation of primer sets for species-level resolution of the vaginal microbiota using 16S ribosomal RNA gene sequencing. J Infect Dis 2019;219:305–314. doi: 10.1093/infdis/jiy508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu A Yao B Dong T, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022;185:1356–1372.e26. doi: 10.1016/j.cell.2022.02.027 [DOI] [PubMed] [Google Scholar]
- 35.Crooks TA Madison JD Walsh DM, et al. Porphyromonas somerae invasion of endometrial cancer cells. Front Microbiol 2021;12:674835. doi: 10.3389/fmicb.2021.674835 [DOI] [PMC free article] [PubMed] [Google Scholar]


