ABSTRACT
Mitochondrial DNA (mtDNA) acts as a proinflammatory damage-associated molecular pattern that stimulates innate immune activation via Toll-like receptor 9, similarly to bacterial DNA. A number of clinical studies have measured elevated cell-free mtDNA in the plasma of trauma patients, thought to originate from tissue injury and inflammatory processes; however, the magnitude of this increase, the absolute concentration, and the association with poor outcomes varies considerably across studies. Measurements of cell-free mtDNA in healthy individuals have shown that the majority of “cell-free” mtDNA (>95%) can be centrifuged/filtered from plasma in the size range of 0.45 to 5 μm, suggesting that there are larger forms of mtDNA-containing complexes in the plasma that could be considered cell-free. Whether this is true for trauma patients (and other relevant disease states) and the clinical relevance of the larger forms of mtDNA is unknown. These findings from healthy individuals also suggest that the centrifugation speeds used to generate cell-free plasma (which are rarely consistent among studies) could result in mixed populations of cell-free mtDNA that could confound associations with outcomes. We demonstrate in this study of 25 major trauma patients that the majority of the cell-free mtDNA in trauma patient plasma (>95%) is removed after centrifugation at 16,000g. Despite the larger forms of mtDNA being predominant, they do not correlate with outcomes or expected parameters such as injury/shock severity, multiple organ failure, and markers of inflammation, whereas low-molecular-weight cell-free mtDNA correlates strongly with these variables.
KEYWORDS: Mitochondrial DNA, traumatic injury, inflammation, cell-free DNA, orthopedic trauma
INTRODUCTION
Traumatic injury is a leading cause of death and contributor to global disease burden (1). Although early deaths can occur because of nonsurvivable injuries and exsanguination, improvements in resuscitation practices have led to fewer deaths and a greater number of in-hospital deaths and complications in those who now survive. One of the leading complications of trauma patients in the intensive care unit (ICU) is multiple organ failure (MOF) (2), which is thought to develop, in part, because of a hyperactive systemic inflammatory response characterized by cytokine storms, neutrophil activation, and downstream tissue damage (3,4). It has been shown that postinjury inflammation arises because of the exposure of intracellular contents to the innate immune system. Termed danger-associated molecular patterns or Damage-associated molecular patterns (DAMPs), key components of the intracellular milieu activate pattern recognition receptors on innate immune cells, similarly to bacterial and viral infections, producing a clinical picture not dissimilar to sepsis (5).
Mitochondrial DNA (mtDNA) as a DAMP has been consistently identified as a key driver of postinjury inflammation. Because mitochondria have ancient bacterial origins (6), their DNA contains unmethylated CpG islands that bind to and activate endosomal or cell surface toll-like receptor 9 (TLR-9), leading to upregulation of proinflammatory cytokines and neutrophil activation (7).
The role of mtDNA in systemic inflammatory responses is supported by animal studies demonstrating that intravenous administration of mtDNA to healthy rats leads to increased lung inflammation and plasma interleukin (IL)-6 as well as exacerbating postinjury inflammation and MOF in animal models of trauma and hemorrhagic shock (8). In clinical studies, circulating cell-free mtDNA in trauma patients has been shown to be increased from that of healthy controls and correlate with markers of inflammation, outcomes of mortality, and systemic inflammatory response syndrome, as outlined in a recent systematic review (9).
Recently, two studies of healthy individuals demonstrated that cell-free mtDNA exists in two forms, a low-molecular-weight cell-free mtDNA that cannot be centrifuged/filtered from plasma and a larger form that can be centrifuged/filtered from plasma (10,11). More than 95% of the cell-free mtDNA existed in the larger form. It is unknown whether these two forms of cell-free mtDNA exist in trauma patient plasma and how they differentially contribute to the postinjury inflammatory response. Understanding this is essential in understanding the physical nature of mtDNA as a DAMP and diagnostic/therapeutic target for poor outcomes in trauma patients.
We hypothesize that, like healthy individuals, trauma patients will also exhibit a centrifugable fraction of cell-free mtDNA and a plasma fraction of noncentrifugable cell-free mtDNA. Given the established mechanism by which mtDNA induces inflammation via direct binding to TLR-9, we also hypothesize that mtDNA remaining in plasma after high-speed centrifugation will correlate with inflammatory markers and MOF to a greater degree than mtDNA in larger structures removed by centrifugation.
METHODS
Patient selection and sample collection
This study was carried out at John Hunter Hospital, a level 1 trauma center in Newcastle Australia, and was approved by the Hunter New England Health Human Research Ethics Committee (10/11/17/4.05), with informed consent provided by patients or persons responsible on their behalf. Adult patients presenting with high-energy pelvic, acetabular, femur, and/or tibia fractures requiring open reduction internal fixation had Ethylenediaminetetraacetic acid blood samples collected immediately preoperatively to their orthopedic surgery, 30 minutes postoperatively, and at 1, 3, and 5 days postoperatively. Patients were excluded if pregnant or on immune-modulating medications. Within 1 hour of collection, whole blood was centrifuged at 200g for 10 minutes at room temperature to remove red blood cells and leukocytes. The resultant platelet-rich plasma was centrifuged at 1,200g for 10 minutes at room temperature to remove platelets. The plasma fraction was then centrifuged at 16,000g for 10 minutes at room temperature, and the resultant plasma was collected and stored at −80°C along with the pelleted fraction.
Quantitation of circulating mtDNA
Circulating DNA was quantitated using quantitative polymerase chain reaction. For quantitating mtDNA from plasma, samples were thawed and diluted 1:80 into ddH2O as described previously (12,13). For quantitating mtDNA from pelleted material, DNA was extracted from the pellet using the Qiagen Blood and Tissue DNEasy Kit according to manufacturer's instructions (Qiagen, Hilden, Germany). Quantitative polymerase chain reaction primers were directed at the mitochondrial gene, ND3 as previously described at a final concentration of 500 nM (12). PowerUp SYBR reagent (Thermo-Fisher Scientific, Scoresby, VIC, Australia) was used with concentrations and cycling temperatures as per manufacturer's instructions. Samples were run in triplicate, alongside purified mtDNA standards generated using a Repli-G mtDNA kit (Qiagen, Hilden, Germany) and no template negative controls. For quantitative polymerase chain reaction using 1:80 diluted plasma, mtDNA standards were also spiked with the equivalent amount of healthy control plasma containing negligible amounts of mtDNA, to account for the inhibitory effect of plasma on the PCR reaction. Quantitative polymerase chain reaction was carried out using a QuantStudio Flex 6 (Life Technologies, Foster City, CA), and mtDNA in patient samples was calculated from the standard curve using QuantStudio software. For plasma results, calculated mtDNA concentration was multiplied by 80 to account for the initial dilution while for pellet DNA, calculated mtDNA was corrected to the original concentration by multiplying by the ratio of the elution volume (200 μL) from the extraction column and the initial plasma volume from which the pellet was derived.
Clinical data
Patient data were collected to measure correlations with cell-free mtDNA. Our clinical outcomes recorded were development of postoperative MOF (based on a Denver score >3, 48 hours after injury), ICU length of stay, and mortality. As part of the parent study, a number of clinical variables, immune markers, and standard tests were also measured at the same time points, including demographics, injury and shock severity, transfusions, full blood count, standard coagulation tests, fibrinogen, complement C3 and C4, tryptase, cortisol, and IL-6. All tests were performed by the local pathology service with the exception of IL-6, which was measured from frozen plasma samples using a Human IL-6 uncoated ELISA kit (Invitrogen) according to manufacturer's instructions.
Statistical analysis
Perioperative changes over time and between the plasma and pelleted cell-free mtDNA were compared using a mixed-effects model on log-transformed data. Post hoc multiple comparisons of time points within groups used a Tukey multiple comparisons test and comparison of time points between groups used a Sidak multiple comparison's test. Correlations between cell-free mtDNA and clinical variables were performed using a Spearman test. Differences between cell-free mtDNA according to outcome at each time point were determined using a Mann-Whitney test. Statistical analysis and figure generation used GraphPad Prism version 9.3.
RESULTS
Patient characteristics
Twenty-five major orthopedic trauma patients were included in the study. Patients had an average age of 47 ± 21 years and were predominantly male (68%), with a median Injury Severity Score of 21 (interquartile range, 29) and mean admission base deficit of −0.97 ± 3.27. The median time to surgery was 63 hours (interquartile range, 57 hours), and there were seven patients (28%) admitted to the ICU. Postoperative MOF developed in three patients (12%), with a mean duration of 5.7 days. Single organ failure and organ dysfunction below the Denver MOF score threshold occurred in three patients (12%). No in-hospital deaths occurred.
Natural history of plasma and pelleted cell-free mtDNA
To determine whether trauma patients' plasma contained large and low-molecular-weight forms of mtDNA, high-speed (16,000g) centrifugation was used to generate a pellet of the larger cell-free structures and a remaining cell-free plasma fraction. Concentrations of mtDNA in the plasma and pellet fraction from healthy controls and trauma patients over time are shown in Figure 1. For both healthy controls and trauma patients, the pellet fraction contained significantly more mtDNA than the plasma fraction (58-fold (P = 0.029) and 19-fold (P < 0.0001 preoperative time point, respectively), indicating that the majority of cell-free mtDNA is found in larger structures. At the preoperative time point, trauma patients had significantly higher (5.7-fold) plasma mtDNA than healthy controls (186 [114–339] ng/mL, P < 0.0001). The mtDNA concentration in the pellet fraction of trauma patients (preoperative) was also significantly higher (1.87-fold) than that of healthy controls (P = 0.0013). There were no significant variations in mtDNA across the perioperative time points for both the pellet and plasma fractions (Fig. 2).
Fig. 1.
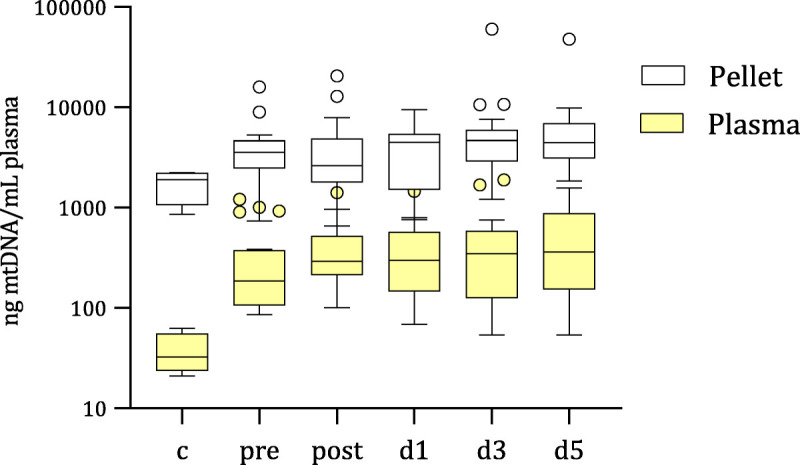
Cell-free MtDNA concentrations in the plasma and pellet fraction after 16,000g centrifugation. After removing cells and platelets from whole blood using sequential 200g and 1,200g centrifugation, plasma was centrifuged at 16,000g for 10 minutes at room temperature, and the resultant pellet and plasma frozen at −80°C. Mitochondria DNA was quantitated from the two fractions with qPCR using primers against the mitochondrial gene, ND3, on DNA extracted from the pellet fraction, and on 1:80-diluted plasma. Ct values were converted to ng/mL using mtDNA standards run on each qPCR plate, accounting for dilution factors. Box and whisker plots with error plotted based on Tukey method are from healthy controls (c) and trauma patients before (pre) and postoperative to (post, d1, d3, d5) major orthopedic surgery. qPCR, quantitative polymerase chain reaction.
Fig. 2.
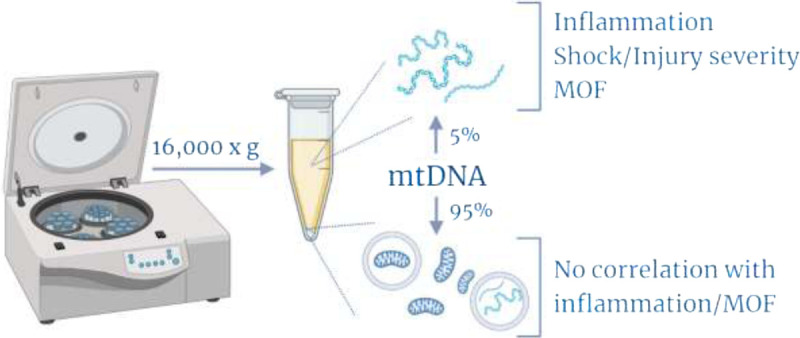
Summary of findings. The majority of cell-free mtDNA in trauma patient plasma and healthy controls exists in the centrifugable fraction, assumed to contain larger mtDNA structures including mitochondria, mitochondria-containing microparticles, or mtDNA-containing microparticles. This fraction does not correlate with expected postinjury inflammatory markers or MOF. Created with BioRender.com.
Correlations with inflammatory markers and outcomes
To determine how the low-molecular-weight plasma mtDNA and the larger mtDNA structures correlated with clinical parameters, plasma and pellet mtDNA concentrations at each time point were used in a Spearman correlation with clinical variables, as shown in Table 1. Overall, plasma mtDNA correlated with injury severity, shock severity, MOF scores, and inflammatory markers, whereas the concentration of mtDNA contained in the pellet did not produce significant correlations for most of these variables.
Table 1.
Spearman correlation with plasma mtDNA, pellet mtDNA, and clinical variables
| Pre | Day 1 | Day 3 | Day 5 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma | Pellet | Plasma | Pellet | Plasma | Pellet | Plasma | Pellet | |||||||||
| Variable | R | P | R | P | R | P | R | P | R | P | R | P | R | P | R | P |
| ISS | 0.45 | 0.024 | 0.22 | 0.360 | 0.14 | 0.526 | 0.02 | 0.931 | 0.19 | 0.394 | 0.31 | 0.211 | 0.03 | 0.900 | −0.25 | 0.312 |
| Admission BD | −0.74 | <0.0001 | −0.49 | 0.034 | −0.18 | 0.405 | −0.10 | 0.678 | −0.33 | 0.144 | −0.30 | 0.234 | −0.13 | 0.567 | 0.26 | 0.278 |
| ICU LOS | 0.64 | 0.001 | 0.33 | 0.154 | 0.39 | 0.064 | −0.01 | 0.966 | 0.41 | 0.061 | −0.03 | 0.902 | 0.23 | 0.325 | 0.06 | 0.822 |
| Vent. days | 0.65 | 0.001 | 0.31 | 0.202 | 0.24 | 0.264 | −0.04 | 0.857 | 0.22 | 0.347 | 0.10 | 0.712 | 0.06 | 0.788 | 0.00 | 0.992 |
| Highest MOF score | 0.46 | 0.021 | 0.13 | 0.577 | 0.20 | 0.351 | 0.08 | 0.723 | 0.33 | 0.136 | −0.09 | 0.729 | −0.01 | 0.973 | −0.11 | 0.644 |
| Length of MOF | 0.48 | 0.014 | 0.37 | 0.104 | 0.18 | 0.414 | −0.13 | 0.567 | 0.29 | 0.188 | −0.01 | 0.959 | −0.13 | 0.563 | −0.22 | 0.367 |
| Denver score | 0.58 | 0.003 | 0.24 | 0.317 | 0.24 | 0.263 | 0.03 | 0.883 | 0.34 | 0.120 | 0.15 | 0.548 | −0.03 | 0.890 | −0.22 | 0.369 |
| PRBC | 0.47 | 0.018 | 0.19 | 0.428 | 0.11 | 0.600 | −0.18 | 0.448 | 0.09 | 0.705 | −0.04 | 0.886 | 0.18 | 0.429 | 0.06 | 0.794 |
| PLT | 0.45 | 0.024 | 0.18 | 0.437 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| FFP | 0.58 | 0.002 | 0.34 | 0.141 | −0.19 | 0.377 | −0.26 | 0.258 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Cryo | 0.55 | 0.005 | 0.41 | 0.076 | −0.27 | 0.210 | 0.03 | 0.887 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Pellet mtDNA | 0.48 | 0.034 | −0.11 | 0.642 | 0.21 | 0.409 | 0.22 | 0.375 | ||||||||
| Plasma mtDNA | 0.48 | 0.034 | −0.11 | 0.642 | 0.21 | 0.409 | 0.22 | 0.375 | ||||||||
| CRP | 0.25 | 0.235 | −0.05 | 0.845 | 0.48 | 0.019 | 0.05 | 0.832 | 0.46 | 0.030 | −0.19 | 0.460 | 0.47 | 0.037 | 0.05 | 0.848 |
| LD | 0.43 | 0.031 | 0.10 | 0.677 | 0.44 | 0.046 | −0.35 | 0.157 | 0.51 | 0.019 | 0.40 | 0.101 | 0.53 | 0.015 | 0.23 | 0.350 |
| Cortisol | 0.32 | 0.118 | 0.03 | 0.900 | 0.44 | 0.042 | −0.12 | 0.632 | 0.24 | 0.300 | −0.19 | 0.453 | 0.16 | 0.532 | −0.14 | 0.605 |
| Fibrinogen | 0.11 | 0.621 | 0.00 | 0.992 | 0.42 | 0.055 | 0.01 | 0.968 | 0.16 | 0.503 | −0.18 | 0.465 | 0.07 | 0.780 | −0.05 | 0.851 |
| C3 | −0.44 | 0.029 | −0.41 | 0.074 | 0.01 | 0.953 | −0.25 | 0.307 | −0.17 | 0.457 | 0.06 | 0.801 | −0.18 | 0.475 | 0.24 | 0.332 |
| C4 | −0.48 | 0.015 | −0.56 | 0.010 | −0.13 | 0.571 | −0.06 | 0.796 | −0.30 | 0.181 | 0.12 | 0.642 | −0.32 | 0.179 | 0.21 | 0.399 |
| IL-6 | 0.25 | 0.230 | 0.19 | 0.427 | 0.45 | 0.027 | −0.06 | 0.788 | 0.58 | 0.005 | 0.15 | 0.548 | 0.14 | 0.560 | −0.04 | 0.864 |
Spearman correlations were performed on outcomes, and clinical variables were measured from the same time point. Spearman correlation coefficient (R) and corresponding P value (P) are shown. Significant findings based on α < 0.05 are shown in bold. Postoperative associations with platelet, FFP, and cryoprecipitate transfusions are shown as n.a. in cases where no transfusions occurred for any patient.
BD, base deficit; CRP, C-reactive protein; FFP, fresh-frozen plasma; ISS, Injury Severity Score; LD, lactate dehydrogenase; LOS, length of stay; n.a., not applicable; PLT, platelet; PRBC, packed red blood cell; Vent., ventilator.
The plasma mtDNA concentration at the preoperative time point had the majority of significant correlations. Plasma mtDNA was associated with both injury severity (via Injury Severity Score) and shock severity (via admission base deficit). Plasma mtDNA was also associated with ICU length of stay, ventilator days, highest MOF score (severity of MOF), duration of MOF, and the daily Denver MOF score. The number of transfusions occurring prior to the preoperative time point also correlated with plasma mtDNA. Postoperative mtDNA up to day 5 did not correlate with any of these clinical parameters. Inflammatory markers, C-reactive protein, lactate dehydrogenase (LD), cortisol, and IL-6 were associated with plasma mtDNA concentration; however, this varied across the perioperative period, with C-reactive protein associating with plasma mtDNA at days 1 to 5, LD at preoperative day 5, cortisol at day 1 only, and IL-6 at days 1 to 3. Negative correlations with plasma mtDNA were observed for C3 and C4 at the preoperative time point only. Fibrinogen did not associate with plasma mtDNA at any time point.
The concentration of mtDNA in the pelleted fraction had limited associations with clinical variables, with significant correlations observed only for admission base deficit, plasma mtDNA, and C4 at the preoperative time point only.
DISCUSSION
Here, we described the nature of mtDNA and its association with postoperative outcomes in orthopedic trauma patients.
This study has demonstrated that, in orthopedic trauma patients, the majority of circulating cell-free mitochondria is in larger structures that can be removed from solution by high-speed centrifugation. Despite only a minority of mtDNA (5%) remaining free in the plasma, we demonstrated that this “low-molecular-weight” cell-free mtDNA is the clinically relevant form, as it associates with relevant outcomes of MOF severity and duration, transfusion rates, and markers of inflammation, whereas higher density mtDNA structures have negligible association with these factors and are unlikely to be proinflammatory. These findings also have implications for any study of cell-free mtDNA in trauma patients and potentially other diseases, indicating that centrifugation speed/s used for plasma preparation can greatly influence the amount of clinically relevant cell-free mtDNA present in the sample.
Similar to our findings, previous studies have shown that the majority of cell-free mtDNA exists in larger structures in the plasma. Chiu et al. (10) demonstrated that, in healthy individuals, >95% of the mtDNA could be removed from acellular plasma with 16,000g centrifugation or filtration between 0.45 and 5 μm. Al Amir Dache et al. (11) found similar results in healthy individuals, removing >78% of the mtDNA using 16,000g, and further demonstrated that the majority of the structures found in this size range were positive for mitochondrial markers, indicating that this form of mtDNA is contained in mitochondrial organelles. Given the estimated size range in the plasma, mitochondria could exist in the form of cell-free mitochondria, platelet microparticles, or other mitochondria-containing extracellular vesicles. In the context of traumatic injury, platelet activation and release of mitochondria-containing microparticles (14), as well as the release of mitochondria and cell fragments resulting from tissue injury, are other potential sources of mtDNA in the high-density fraction (15).
We demonstrated that preoperative plasma mtDNA concentrations depend on the injury and shock severity, as well as the number of blood transfusions. These factors are also known to be independent risk factors for MOF. Previous studies have hypothesized that cell-free mtDNA mediates MOF development through its release from injured cells, coagulation and inflammatory processes, and stimulation of further inflammation via TLR-9, leading to systemic secondary tissue injury (15). In addition, blood components have been shown to contain mtDNA (16), which could explain the association between mtDNA and transfused patients. We found a correlation with plasma mtDNA and postoperative MOF scores, severity, and duration, which has also been shown previously (8,17). Novel to this study was the separation of the larger cell-free mtDNA structures from plasma mtDNA, not previously characterized for clinical relevance, which were found not to be associated with MOF or any other relevant postoperative factors, including ventilator days and ICU length of stay. Based on the findings of Al Amir Dache et al. (11), the higher density mtDNA in healthy individuals is membrane-encapsulated and therefore protected from binding TLR-9. Our findings showing a lack of association between higher density mtDNA structures and inflammatory markers support the idea that mtDNA is membrane encapsulated and, furthermore, unable to stimulate inflammatory responses.
It has been shown previously that mitochondria are proinflammatory. Injection of mitochondrial organelles induces inflammation in rats (18), monocyte-derived mitochondria induce type-1 interferon responses in cultured endothelial cells (19), and mitochondria released by stored platelets promote structural changes in neutrophils (14). However, no clinical data on the ability of whole mitochondria to induce inflammation in trauma patients exist. In our study, the lack of association of the higher density mtDNA with inflammation could be due to there being a mixed population of membrane-encapsulated and nonencapsulated mitochondria, and the possibility remains that, in traumatic injury, mitochondria are simply not directly relevant to inflammation. Future studies of cell-free mitochondria in trauma patients may provide more insight into our current findings.
Previous studies of cell-free mtDNA in trauma patients and other clinical cohorts have used a wide range of sample preparation methods. Our findings support some previous reports that urge mtDNA researchers to carefully consider the preparation of cell-free plasma and which fraction is relevant to the disease in question. Relevant here is the number, g-force, and timing of centrifugation steps. Many studies have used one centrifugation step that would be effective in removing blood cells but leave platelets, microparticles, and their mtDNA in the “cell-free” plasma fraction, which may confound findings. Other studies have used a high-speed centrifugation step after freezing plasma. A recent methodological study demonstrated that high-speed centrifugation before freeze-thawing (as was done here) is essential for correct measurements of mtDNA (20). Freeze-thawing is a common technique for extracting contents from cells, platelets, and vesicles, causing membrane damage, so larger mtDNA-containing structures remaining in plasma upon freezing may be compromised during freeze-thawing, thus contaminating the plasma with mtDNA that is normally in larger structures and possibly irrelevant to inflammation.
Based on the methodology provided in previous reports of mtDNA in trauma patients, many studies are likely to have mtDNA from larger structures contaminating the cell-free plasma compartment, which artificially increases cell-free mtDNA and confounds associations with clinical variables and outcomes. For example, a recent study reported that mtDNA does not correlate with the postinjury inflammatory response (21), which opened up questions about the true role of mtDNA in postinjury inflammation. However, we note that this study froze plasma after three 5,000g centrifugations, which may still retain larger mtDNA-containing structures in the plasma. In fact, the measured mtDNA was markedly higher than that of the present study, being closer to the concentration found in our pelleted fraction, which may explain their finding of a lack of association with inflammation.
In conclusion, despite the majority of mtDNA existing in larger, albeit cell-free structures in the plasma, this is not clinically relevant to postinjury inflammation, with the low-molecular-weight, cell-free mtDNA being robustly correlated with inflammation, MOF severity, and the extent of trauma. To echo the comments of similar studies on healthy individuals, we urge those studying mtDNA in trauma and other inflammatory states where mtDNA is thought to play a role (sepsis (22), myocardial infarction (23), COVID-19 (24)) to consider whether sample preparation methods are preserving clinically relevant cell-free mtDNA.
Footnotes
The authors report no conflict of interests.
Contributor Information
Gabrielle D. Briggs, Email: gabrielle.briggs@newcastle.edu.au.
Scott Gelzinnis, Email: scott.gelzinnis@uon.edu.au.
Simone Meakes, Email: simone.meakes@health.nsw.gov.au.
Kate L. King, Email: kate.king@health.nsw.gov.au.
Zsolt J. Balogh, Email: zsolt.balogh@hnehealth.nsw.gov.au.
REFERENCES
- 1.GBD 2017 DALYs and HALE Collaborators : Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1859–1922, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewar DC, Mackay P, Balogh Z: Epidemiology of post-injury multiple organ failure in an Australian trauma system. ANZ J Surg 79:431–436, 2009. [DOI] [PubMed] [Google Scholar]
- 3.Balogh ZJ, Reumann MK, Gruen RL, Mayer-Kuckuk P, Schuetz MA, Harris IA, Gabbe BJ, Bhandari M: Advances and future directions for management of trauma patients with musculoskeletal injuries. Lancet 380:1109–1119, 2012. [DOI] [PubMed] [Google Scholar]
- 4.Lord JM, Midwinter MJ, Chen YF, Belli A, Brohi K, Kovacs EJ, Koenderman L, Kubes P, Lilford RJ: The systemic immune response to trauma: an overview of pathophysiology and treatment. Lancet 384:1455–1465, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relja B, Mors K, Marzi I: Danger signals in trauma. Eur J Trauma Emerg Surg 44:301–316, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin WF, Garg S, Zimorski V: Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci 370:20140330, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ: Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464:104–107, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aswani A Manson J Itagaki K, et al.: Scavenging circulating mitochondrial DNA as a potential therapeutic option for multiple organ dysfunction in trauma hemorrhage. Front Immunol 9:891, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lubkin DT, Bishawi M, Barbas AS, Brennan TV, Kirk AD: Extracellular mitochondrial DNA and N-formyl peptides in trauma and critical illness: a systematic review. Crit Care Med 46:2018–2028, 2018. [DOI] [PubMed] [Google Scholar]
- 10.Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, Rainer TH, Lo YM: Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem 49:719–726, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Al Amir Dache Z Otandault A Tanos R Pastor B Meddeb R Sanchez C Arena G Lasorsa L Bennett A Grange T, et al.: Blood contains circulating cell-free respiratory competent mitochondria. FASEB J 34:3616–3630, 2020. [DOI] [PubMed] [Google Scholar]
- 12.McIlroy DJ, Minahan K, Keely S, Lott N, Hansbro P, Smith DW, Balogh ZJ: Reduced deoxyribonuclease enzyme activity in response to high postinjury mitochondrial DNA concentration provides a therapeutic target for systemic inflammatory response syndrome. J Trauma Acute Care Surg 85:354–358, 2018. [DOI] [PubMed] [Google Scholar]
- 13.Breitbach S, Tug S, Helmig S, Zahn D, Kubiak T, Michal M, Gori T, Ehlert T, Beiter T, Simon P: Direct quantification of cell-free, circulating DNA from unpurified plasma. PLoS One 9:e87838, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boudreau LH Duchez AC Cloutier N Soulet D Martin N Bollinger J Pare A Rousseau M Naika GS Levesque T, et al.: Platelets release mitochondria serving as substrate for bactericidal group IIA–secreted phospholipase A2 to promote inflammation. Blood 124:2173–2183, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurairajah K, Briggs GD, Balogh ZJ: The source of cell-free mitochondrial DNA in trauma and potential therapeutic strategies. Eur J Trauma Emerg Surg 44:325–334, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons JD, Lee YL, Pastukh VM, Capley G, Muscat CA, Muscat DC, Marshall ML, Brevard SB, Gillespie MN: Potential contribution of mitochondrial DNA damage associated molecular patterns in transfusion products to the development of acute respiratory distress syndrome after multiple transfusions. J Trauma Acute Care Surg 82:1023–1029, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons JD, Lee YL, Mulekar S, Kuck JL, Brevard SB, Gonzalez RP, Gillespie MN, Richards WO: Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann Surg 258:591–596, 2013; discussion 596–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plotnikov EY, Jankauskas SS, Zinovkin RA, Zorova LD, Zorov SD, Pevzner IB, Silachev DN, Zorov DB: A combination of kidney ischemia and injection of isolated mitochondria leads to activation of inflammation and increase in mortality rate in rats. Bull Exp Biol Med 169:213–217, 2020. [DOI] [PubMed] [Google Scholar]
- 19.Puhm F Afonyushkin T Resch U Obermayer G Rohde M Penz T Schuster M Wagner G Rendeiro AF Melki I, et al.: Mitochondria are a subset of extracellular vesicles released by activated monocytes and induce type I IFN and TNF responses in endothelial cells. Circ Res 125:43–52, 2019. [DOI] [PubMed] [Google Scholar]
- 20.Cox DRA, Wong BKL, Yang L, Yoshino O, Testro A, Muralidharan V, Dobrovic A: High speed centrifugation before frozen storage of plasma is critical for quantitative analysis of mitochondrial-derived cell-free DNA. Clin Chem 66:1111–1114, 2020. [DOI] [PubMed] [Google Scholar]
- 21.Stortz JA Hawkins RB Holden DC Raymond SL Wang Z Brakenridge SC Cuschieri J Moore FA Maier RV Moldawer LL, et al.: Cell-free nuclear, but not mitochondrial, DNA concentrations correlate with the early host inflammatory response after severe trauma. Sci Rep 9:13648, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrington JS, Choi AMK, Nakahira K: Mitochondrial DNA in Sepsis. Curr Opin Crit Care 23:284–290, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Haan JJ, Smeets MB, Pasterkamp G, Arslan F: Danger signals in the initiation of the inflammatory response after myocardial infarction. Mediators Inflamm 2013:206039, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valdes-Aguayo JJ Garza-Veloz I Badillo-Almaraz JI Bernal-Silva S Martinez-Vazquez MC Juarez-Alcala V Vargas-Rodriguez JR Gaeta-Velasco ML Gonzalez-Fuentes C Avila-Carrasco L, et al.: Mitochondria and mitochondrial DNA: key elements in the pathogenesis and exacerbation of the inflammatory state caused by COVID-19. Medicina (Kaunas) 57(9):928, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]


