Abstract
The Escherichia coli signal recognition particle (SRP) is a ribonucleoprotein complex that targets nascent inner membrane proteins (IMPs) to transport sites in the inner membrane (IM). Since SRP depletion only partially inhibits IMP insertion under some growth conditions, however, it is not clear why the particle is absolutely essential for viability. Insights into this question emerged from experiments in which we analyzed the physiological consequences of reducing the intracellular concentration of SRP below the wild-type level. We found that even moderate SRP deficiencies that have little effect on cell growth led to the induction of a heat shock response. Genetic manipulations that suppress the heat shock response were lethal in SRP-deficient cells, indicating that the elevated synthesis of heat shock proteins plays an important role in maintaining cell viability. Although it is conceivable that the heat shock response serves to increase the capacity of cells to target IMPs via chaperone-based mechanisms, SRP-deficient cells did not show an increased dependence on either GroEL or DnaK. By contrast, the heat shock-regulated proteases Lon and ClpQ became essential for viability when SRP levels were reduced. These results suggest that the heat shock response protects SRP-deficient cells by increasing their capacity to degrade mislocalized IMPs. Consistent with this notion, a model IMP that was mislocalized in the cytoplasm as the result of SRP depletion appeared to be more stable in a Δlon ΔclpQ strain than in control cells. Taken together, the data provide direct evidence that SRP is essential in E. coli and possibly conserved throughout prokaryotic evolution as well partly because efficient IMP targeting prevents a toxic accumulation of aggregated proteins in the cytoplasm.
The signal recognition particle (SRP) is a soluble ribonucleoprotein complex that was originally identified as an important intermediary in the transport of proteins into the secretory pathway in mammalian cells (reviewed in reference 54). During translation, the 54-kDa polypeptide subunit of SRP (SRP54) binds to hydrophobic targeting signals that are found in both presecretory and integral membrane proteins (29, 31). The targeting signals are generally either amino-terminal signal sequences (3) or, in the case of many membrane proteins that lack discrete signal peptides, the first transmembrane segment (18). Subsequently, SRP targets ribosome-nascent chain complexes to the endoplasmic reticulum (ER), where an interaction between SRP54 and a heterodimeric SRP receptor (SR) catalyzes the release of the nascent polypeptides and their insertion into a translocation channel or “translocon” (19, 37, 38). In the final step of the targeting cycle SRP dissociates from the ER membrane. In mammalian cells, the entry of the vast majority of proteins into the secretory pathway is completely dependent on the SRP targeting pathway.
During the past few years, genes encoding SRP and SR homologs have been identified in every genome that has been sequenced, and components of the SRP pathway have been purified from a variety of eukaryotic, archaeal, and bacterial sources. The genomic and biochemical studies have revealed that the number of SRP subunits varies considerably in different branches of the phylogenetic tree. Whereas mammalian SRP consists of six polypeptides and a 300-nucleotide RNA, the SRP found in mycoplasmas and gram negative bacteria consists of only a single protein, a homolog of SRP54 (Ffh), and an ∼100-nucleotide RNA that corresponds to domain IV of mammalian SRP RNA (4.5S RNA) (43, 46). Likewise, the bacterial SR is a simplified version of its eukaryotic counterpart that consists of a single protein, a homolog of the α-subunit (FtsY) (2, 47). Regardless of size, though, all SRPs appear to have a protein targeting function. SRP has been clearly shown to target presecretory proteins to the ER in Saccharomyces cerevisiae (23) and polytopic inner membrane proteins (IMPs) that lack a signal peptide to the inner membrane (IM) in Escherichia coli (14, 28, 36, 52, 53).
Despite the remarkable conservation of SRP, the particle plays a much smaller role in protein targeting in microbes than in mammalian cells. In yeast, a subset of presecretory proteins can be targeted effectively to the ER in the complete absence of SRP (23). Even proteins that are not translocated efficiently in the absence of SRP, however, can often utilize SRP-independent targeting mechanisms to a significant degree. For example, approximately 50% of the ER luminal protein BiP is still translocated after SRP depletion (23). In E. coli, an even more restricted set of proteins requires SRP for transport out of the cytoplasm. The translocation of most (or all) presecretory proteins is completely unaffected by SRP depletion (14, 43, 46, 52). Although efficient insertion of many IMPs that lack signal peptides requires SRP, the biogenesis of some IMPs is unimpeded by SRP depletion (41, 52). As in yeast, the transport of different SRP substrates shows a variable degree of SRP dependence. At least under relatively slow growth conditions, depletion of SRP blocks IMP insertion by no more than about 50% (13, 14, 41, 52).
In both yeast and bacteria, molecular chaperones play a significant role in protein targeting. Molecular chaperones target proteins to the ER or IM by keeping them in a loosely folded conformation that is required for passage across the membrane (12). Unlike the SRP pathway, chaperone-based targeting pathways appear to function at least in part in a posttranslational mode (34). It has been proposed that posttranslational targeting pathways evolved in rapidly growing organisms to increase the efficiency of secretion by uncoupling translation and translocation (26). Several studies have attributed a protein targeting function to members of the hsp70 family in yeast (11, 16). While the bacterial hsp70 homolog (DnaK) has also been implicated in protein targeting (55), a secretion-specific chaperone called SecB clearly provides the primary targeting pathway for a subset of presecretory proteins (30). Furthermore, there is evidence that the bacterial GroEL-GroES complex may also participate in the targeting of presecretory proteins and/or IMPs (4, 32). It should be emphasized, however, that the transport of some proteins may be partially or entirely independent of both SRP and molecular chaperones because they fold into a transport-incompetent conformation relatively slowly (15).
Since a large fraction of proteins can be targeted to the secretory pathway in an SRP-independent fashion in microorganisms, it might be expected that SRP would not be essential for cell viability. Indeed disruption of SRP genes in S. cerevisiae causes only about a fourfold decrease in growth rate (23). In contrast, the genes that encode Ffh, 4.5S RNA, and FtsY are all absolutely essential for viability in E. coli (7, 35, 42). Mutations that lower the concentration of 4.5S required for viability have been described (6), but these mutations do not bypass the SRP requirement entirely. E. coli requires only very low Ffh concentrations to survive in nutritionally poor media but is inviable when SRP is completely eliminated (H. D. Bernstein, unpublished results). One possible explanation for these observations is that even slight IMP insertion defects severely inhibit a critical cellular process (e.g., cell division). Alternatively, the mislocalization of IMPs or the loss of SRP itself may have unrecognized secondary effects that would prevent cell growth.
In this paper we describe new insights into the function of the SRP pathway that emerged from examining the physiological consequences of introducing modest SRP deficiencies into E. coli. We found that reductions in SRP concentration that did not affect cell growth led to the induction of a heat shock response. This result implied that when SRP is limiting, cells rely on an alternative mechanism to target or process IMPs. Although genes encoding both DnaK and GroEL are in the heat shock regulon (21), neither of these chaperones appeared to provide an obligate default targeting pathway that compensated for the SRP deficiency. In contrast, heat shock regulated proteases that normally are not required for cell viability became essential when SRP levels were reduced. The data strongly suggest that the heat shock response protects cells at least in part by increasing the levels of proteases that degrade mislocalized IMPs. Thus SRP indirectly prevents a potentially toxic aggregation of proteins in the cytoplasm by promoting efficient IMP insertion. Our results may not only help to explain the essentiality of SRP in E. coli, but may also shed light on the striking conservation of SRP throughout the bacterial kingdom.
MATERIALS AND METHODS
Reagents, media, and bacterial manipulations.
Monoclonal antibodies against heat shock proteins (DnaK and GroEL) and a polyclonal antiserum against alkaline phosphatase (AP) were obtained from StressGen and 5 Prime-3 Prime, respectively. Affinity-purified antibodies against Ffh have been described previously (52). Medium preparation and basic bacterial manipulations were performed using standard procedures (39). Unless otherwise noted, all experiments were conducted at 37°C. Selective media contained ampicillin (100 μg/ml) and chloramphenicol (40 μg/ml) as required. The bacterial strains used in this study and their genotypes are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| BB1553 | MC4100 ΔdnaK52 sidB1 | 8 |
| CAG597 | F−rpoH165(Am) zhg::tet lacZ(Am) trp(Am) pho(Am) supC(Ts) mal(Am) rpsL | 1 |
| CAG599 | CAG597 rpoH+ | D. Jin |
| HDB45 | MC4100 ffh::kan-1 pHDB4 | 52 |
| HDB91 | CAG597 ffh::kan-1 pHDB4 | This study |
| HDB90 | CAG599 ffh::kan-1 pHDB4 | This study |
| HDB92 | HDB45 zjd::tet | This study |
| HDB93 | HDB45 groEL44 zjd::tet | 58, this study |
| HDB94 | HDB45 groES619 zjd::tet | 59, this study |
| HDB95 | MC4100 ffh::kan-1 pHDB6 | This study |
| HDB96 | BB1553 ffh::kan-1 pHDB6 | This study |
| HDB97 | SG22622 mal+ | This study |
| HDB98 | SG22623 mal+ | This study |
| HDB99 | HDB97 ffh::kan-1 pHDB4 | This study |
| HDB100 | HDB98 ffh::kan-1 pHDB4 | This study |
| HDB101 | HDB97 ffh::kan-1 ΔclpYQ1172::tet pHDB4 | 27, this study |
| HDB102 | HDB98 ffh::kan-1 ΔclpYQ1172::tet pHDB4 | 27, this study |
| HDB103 | HDB97 ffh::kan-1 zgj203::tet pHDB4 | This study |
| HDB104 | HDB97 ffh::kan-1 ftsH1(Ts) zgj203::tet pHDB4 | 25, this study |
| HDB105 | HDB98 ffh::kan-1 zgj203::tet pHDB4 | This study |
| HDB106 | HDB98 ffh::kan-1 ftsH1(Ts) zgj203::tet pHDB4 | 25, this study |
| HDB107 | HDB98 ΔclpYQ1172::tet | 27, this study |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 thi fib5301 deoC1 ptsF25 rbsR | 10 |
| SG22622 | MC4100 malP::lacI Δara714 cpsB::lacZ | S. Gottesman |
| SG22623 | SG22622 Δlon510 | S. Gottesman |
| Plasmids | ||
| pBAD33-AcrB576 AP | Cmr; Orip15aaraC Para-acrB576-AP | 52, this study |
| pHDB3 | Apr; pBR322 containing a pUC19 polylinker and lacking the HindIII-AvaI fragment | 52 |
| pHBD4 | Cmr; OrislacIqPtrc-ffh | 52 |
| pHDB6 | Apr; OriColE1lacIqPlac-ffh | This study |
| pTRC99-FtsY(G385A) | Apr; OriColE1lacIqPtrc-ftsY(G385A) | 52 |
Plasmid construction.
The isolation of plasmids that produce a Slo phenotype has been described previously (52). Plasmids isolated in the original Slo screen that were used in this study (pH85, pH92, pS134, pS368) are illustrated (see Fig. 2). Plasmid pH92.1 was constructed by first subjecting pH92 to partial digestion with HindIII and complete digestion with NheI. A DNA fragment containing the 3′ end of dnaJ was then generated using PCR and ligated to the prepared vector. A point mutation was introduced into pS368 to produce pS368 DnaK A174T using the QuikChange mutagenesis kit (Strategene) according to the manufacturer's instructions. pS368Δ was generated by removing an EcoRI fragment containing the 3′ end of dnaK and the 5′ end of dnaJ from pS368. To construct pHDB6, a 1.6 kb NheI-NruI fragment from pHDB1 (43, 52) containing the ffh gene was cloned by blunt-end ligation into the EcoRI and HindIII sites of pRB11, a plasmid that contains a lac promoter and a lacIq gene (R. Barber and C. Gross, unpublished). pBAD33-AcrB 576-AP was constructed by inserting a BlpI fragment from plasmid pNU88 (22) containing the AcrB 576-AP fusion into the SmaI site of pBAD33 (45) by blunt-end ligation. The Klenow fragment of E. coli DNA polymerase I was used to create blunt-ended DNA fragments for the above constructs.
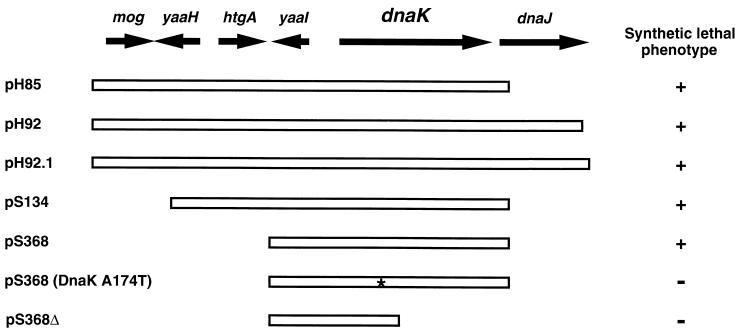
FIG. 2.
Plasmids that drive overexpression of wild-type dnaK are lethal in SRP-deficient cells. As described in reference 52, restriction fragments of E. coli genomic DNA were cloned into pBR322-based multicopy plasmids. HDB45 was transformed with the resulting plasmid libraries and plated onto LB agar containing 10 μM IPTG. Colonies were then replica plated onto LB agar that lacked IPTG. Plasmids that prevented growth on the replica plates were isolated. Such plasmids produced a “Slo” phenotype, that is, they contained genes whose overexpression was synthetically lethal with SRP deficiencies. Four of the plasmids isolated in the screen (pH85, pH92, pS134, and pS368) contained dnaK. The GenBank accession numbers that correspond to the inserts are as follows: pH85, ECAE000111 (bp 8912) to ECAE000112 (bp 3784); pH92, ECAE000111 (bp 8912) to ECAE000112 (bp 4667); pS134, ECAE000111 (bp 10074) to ECAE000112 (bp 3775); pS368, ECAE000112 (bp 787-3775). Derivatives of these plasmids designated pH92.1, pS368 (DnaK A174T), and pS368Δ were constructed as described in Materials and Methods. The pS368 derivatives did not produce a Slo phenotype.
Efficiency of plating assays.
Small (2- to 3-ml) overnight cultures were inoculated with a single colony and grown to saturation in Luria-Bertani (LB) medium containing 10 μM isopropyl-β-l-thiogalactopyranoside (IPTG) (or 200 μM IPTG in the case of strains harboring pHDB6). Cells were then diluted in LB medium and plated in duplicate on LB agar containing 10 μM IPTG or no IPTG (or 200 μM IPTG and 50 μM IPTG in the case of strains harboring pHDB6). The plates were incubated at an appropriate temperature until control (wild-type) colonies were approximately 1.5 mm in diameter. In general, plates that contained 200 to 400 colonies were used to determine plating efficiency. The number of colonies on the duplicate plates was averaged, and a relative plating efficiency was calculated by dividing the number of colonies observed at the lower IPTG concentration by the number of colonies observed at the higher IPTG concentration.
Analysis of the steady-state level of a newly synthesized IMP after inducing expression of a dominant lethal ftsY allele.
Overnight cultures of cells transformed with pTRC or pTRC-FtsY (G385A) (52) and pBAD33-AcrB 576-AP were grown in LB medium containing ampicillin and chloramphenicol. Cells were then washed once and added to 60-ml cultures at an optical density at 550 nm (OD550) of 0.005. When cultures reached an OD550 of 0.05, 2 mM IPTG was added to induce overexpression of the mutant ftsY. After 20 min, 0.2% arabinose was added to all of the cultures to induce synthesis of the AP fusion protein. At various time points portions of each culture were removed and proteins were precipitated with cold 10% trichloroacetic acid (TCA). The levels of AcrB 576-AP were then measured by Western blotting as described below.
Western blotting.
TCA-precipitated proteins were solubilized in buffer A (60 mM Tris [pH 6.8], 2% sodium dodecyl sulfate, 200 mM dithiothreitol, 10% glycerol, 0.001% bromphenol blue). Proteins were then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 8 to 16% acrylamide minigels (Novex) and transferred to nitrocellulose using established methods (24). After incubating filters with a standard blocking buffer and a primary antibody, 125I-goat anti-mouse immunoglobulin G (Amersham) or 35S-protein A (Amersham) was used to detect antibody-antigen complexes. The level of radioactivity in individual bands was quantitated using a Fuji BAS-2500 phosphorimager.
RESULTS
Induction of heat shock in cells that have reduced levels of Ffh.
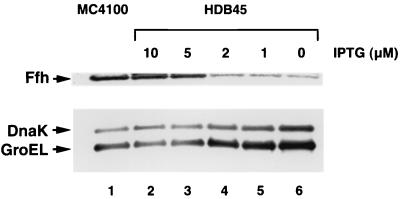
We have previously described a strain (HDB45) in which ffh expression is regulated by the trc promoter (52). In the absence of IPTG, HDB45 cells contain approximately seven- to eightfold less Ffh than the parental strain (MC4100) but show only a very slight growth defect. We hypothesized that the near normal growth rates might mask the induction of a stress response that compensates for the partial loss of SRP. To test this idea, we used Western blotting to measure the levels of two representative heat shock proteins, DnaK and GroEL, in HDB45 that had various levels of SRP. HDB45 grown in LB in the presence of 10 μM IPTG contained approximately the same concentration of Ffh as control MC4100 cells (Fig. 1, lanes 1 and 2). Under these conditions, the levels of DnaK and GroEL were similar in both strains. In the presence of less than 5 μM IPTG, however, the levels of both heat shock proteins were elevated in HDB45, and in the absence of IPTG a severalfold increase in both DnaK and GroEL was observed (Fig. 1, lanes 4 to 6). Thus, the level of heat shock proteins was inversely related to the Ffh concentration. Although these results are consistent with previous data indicating that heat shock is induced after severe depletion of 4.5S RNA (5) or overexpression of a dominant lethal allele of the 4.5S RNA gene (43), they show that even moderate reductions in SRP levels are detected by the stress-sensing machinery of the cell.
FIG. 1.
Inverse relationship between steady-state levels of SRP and heat shock proteins. MC4100 and HDB45 (Ptrc-ffh ffh::kan) were grown overnight in LB containing 10 μM IPTG. Cells were washed and diluted to an OD550 of 0.0002 in LB containing 0, 1, 2, 5, or 10 μM IPTG. At late log phase (OD550, 0.8 to 1.0) samples were removed from each culture and proteins were precipitated with 10% TCA. The concentrations of Ffh, DnaK, and GroEL were measured by Western blotting. Lane 1: MC4100; lanes 2 to 6: HDB45 grown in the presence of the indicated amount of IPTG.
Elevated synthesis of heat shock proteins is required for the viability of SRP-deficient cells.
We previously described a screen for genes whose overexpression is synthetically lethal with reduced expression of ffh (Slo screen) (52). In this screen we isolated multicopy plasmids from a genomic DNA library that allowed growth of HDB45 in the presence of 10 μM IPTG but not in the absence of IPTG. The screen was originally devised to identify genes that encode SRP substrates. We hypothesized that overproduction of a single SRP substrate would have little effect on cell growth when SRP is present in excess (10 μM IPTG) but would inhibit the binding of SRP to other substrates that perform essential cellular functions and lead to a loss of viability when SRP is limiting (no IPTG). Eight of the genes isolated in the screen encode polytopic IMPs. Using a variety of methods, several laboratories have provided strong evidence that these and other IMPs are recognized by SRP and targeted to the IM (14, 28, 36, 52, 53).
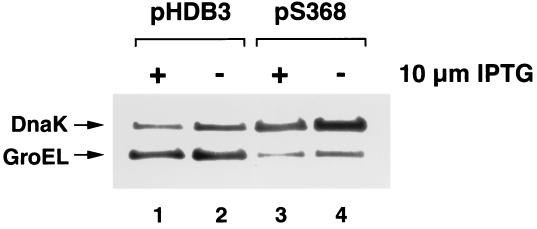
On theoretical grounds we also expected to isolate genes in the Slo screen that fall into at least two other classes. First, we expected to isolate genes encoding proteins that interact with SRP to facilitate progression of the targeting reaction. We surmised that the overproduction of an SR or other binding partner might cause lethality when SRP is limiting by effectively reducing the concentration of free SRP. The isolation of ftsY in the screen was consistent with this prediction (52). Second, we expected to isolate genes whose overexpression interferes with the ability of cells to cope with proteins normally targeted by SRP when the SRP pathway is compromised. In view of this prediction, it is intriguing that several overlapping clones that share only dnaK and the adjacent yaaI gene (pH92, pH85, pS134, and pS368) were isolated in the screen (Fig. 2). Deletion analysis revealed that dnaK was responsible for the synthetic lethality (Fig. 2, plasmid pS368Δ). Since dnaK encodes a protein that is not only a molecular chaperone but also a negative regulator of the heat shock response (50), the gene was most likely isolated because its overexpression inhibits the synthesis of heat shock proteins. Consistent with previous results (51), Western blot analysis of cells containing plasmid pS368 revealed that dnaK overexpression suppressed production of a representative heat shock protein (GroEL) in HDB45 cells both in the presence and absence of 10 μM IPTG (Fig. 3, compare lanes 1 and 2 to lanes 3 and 4). Although it might be postulated that dnaK was isolated in the Slo screen because the overproduction of DnaK inhibits its ability to provide a critical alternative targeting pathway for IMPs, this explanation appears unlikely. In light of previous studies showing that dnaK- and trigger factor (tig)-null alleles are synthetically lethal (17, 49), the observation that tig cells containing plasmid pS368 grow normally (data not shown) strongly suggests that DnaK overproduction is not autoinhibitory. Finally, the finding that cells harboring pS368 contain as much SRP as control cells (H.-Y. Qi and H. D. Bernstein, unpublished results) demonstrates that dnaK overexpression does not interfere with SRP biogenesis.
FIG. 3.
Overexpression of dnaK inhibits the synthesis of heat shock proteins. Overnight cultures of HDB45 transformed with cloning vector pHDB3 or pS368 were grown in LB containing ampicillin and 10 μM IPTG. Cells were washed and diluted to an OD550 of 0.005 in medium containing either 10 μM IPTG or no IPTG. When cells containing pHDB3 reached late log phase (OD550 = 0.8 to 1.0), samples were removed from each culture and proteins were precipitated with 10% TCA. The concentration of DnaK and GroEL was measured by Western blotting. Lanes 1 to 2, cells containing pHDB3; lanes 3 to 4, cells containing pS368. IPTG was added to the cells in lanes 1 and 3.
Experiments performed with a variant of plasmid pS368 provided additional evidence that dnaK was isolated in the Slo screen because its overexpression suppresses heat shock protein synthesis. A recessive mutation (A174T) that abolishes the ability of DnaK to regulate the heat shock response but only partially reduces its molecular chaperone activity (56) was introduced into pS368 by site-directed mutagenesis. Presumably because the presence of the mutant plasmid did not inhibit the synthesis of heat shock proteins, HDB45 cells containing pS368 (A174T) did not exhibit a Slo phenotype (Fig. 2). It is noteworthy that a single missense mutation abolishes the Slo phenotype produced by dnaK but that much more severe mutations (such as truncations) often do not affect the Slo phenotype produced by genes that encode polytopic IMPs (52). The mutagenesis data support the hypothesis that dnaK represents a discrete class of Slo genes in that the phenotype is associated with a specific function of the encoded protein rather than with a distinctive structural feature.
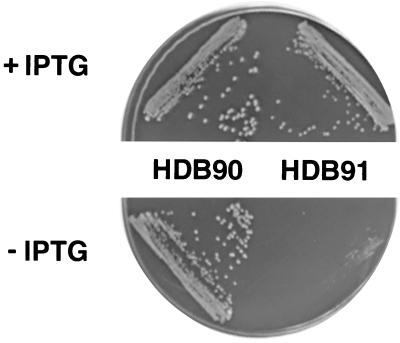
To test directly whether the induction of a heat shock response is required to sustain the viability of SRP-deficient cells, we next examined the effect of reducing the SRP concentration on the growth of a strain that contains a mutant allele of the heat shock sigma factor (ς32) gene (designated rpoH). For these experiments we constructed isogenic strains HDB90 [rpoH+ supC(Ts)] and HDB91 [rpoH165(Am) supC(Ts)] in which the expression of ffh is controlled by the trc promoter. Because SupC is not a completely effective amber suppressor, the expression of heat shock genes is slightly compromised in cells containing the rpoH(Am) allele even at permissive temperatures. We streaked HDB90 and HDB91 cells on LB plates containing either 10 μM IPTG or no IPTG and observed colony formation after incubation at 25°C. As expected, HDB90 grew well on both plates (Fig. 4), but HDB91 grew only on plates containing IPTG. Thus, reduced levels of both SRP and heat shock proteins produce a synthetic lethal effect. Based on these results, we conclude that the elevated synthesis of heat shock proteins observed in SRP-deficient cells is required for their survival.
FIG. 4.
Synthetic lethality of rpoH and SRP deficiencies. Strains HDB90 [Ptrc-ffh ffh::kan supC(Ts) rpoH+] and HDB91 [Ptrc-ffh ffh::kan rpoH(Am) supC(Ts)] were streaked on LB agar containing either 10 μM IPTG (+IPTG) or no IPTG (−IPTG). Plates were incubated at 25°C for 48 h.
Genes encoding heat shock-regulated proteases are essential in SRP-deficient cells.
Although one study has suggested that GroEL-GroES can promote the insertion of LacY into E. coli inverted vesicles (4), the role of molecular chaperones in the targeting of IMPs to the IM has not been extensively investigated. Nevertheless, based largely on the evidence that chaperones play an important role in the targeting of presecretory proteins, we surmised that the heat shock response might serve to increase the level of a chaperone that targets IMPs via a default pathway when the SRP concentration falls below a critical threshold. This hypothesis predicts that cells would not be able to tolerate defects in both the SRP pathway and the alternate targeting pathway simultaneously. To test this possibility, we examined the effect of reducing the SRP concentration in cells that have mutations in molecular chaperone genes.
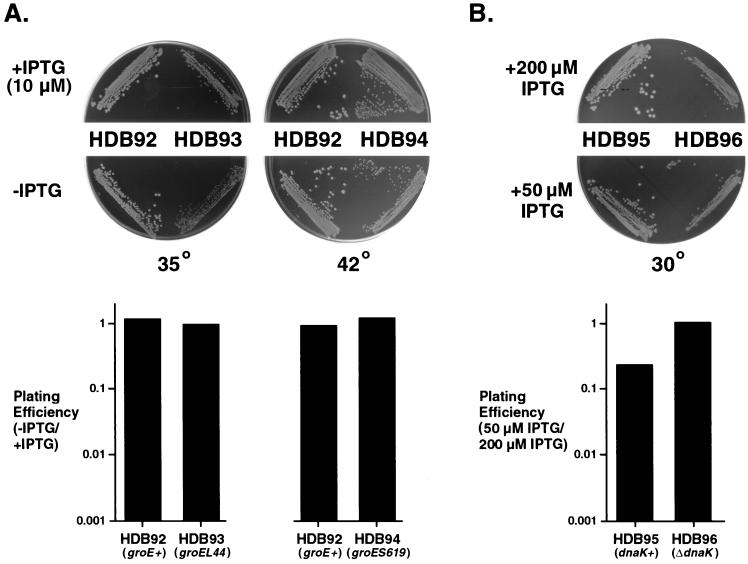
Contrary to our hypothesis, we found that SRP deficiencies did not create an increased dependence on heat shock-regulated chaperones. In one set of experiments we examined the viability of cells that contain both reduced SRP levels and defects in the GroEL-GroES complex. Strains that contain a temperature-sensitive groE mutation and an inducible copy of the ffh gene (Ptrc-ffh) were constructed and streaked on LB plates containing 10 μM IPTG or no IPTG. In order to maximize the magnitude of the GroEL-GroES defects, plates were incubated at the highest temperature cells containing the groE mutation alone could withstand without suffering a loss of plating efficiency. As previously reported, the groES619 allele produced a lethal effect only at temperatures above 42°C (33). When HDB93 (groEL44) and HDB94 (groES619) were incubated along with an isogenic groE+ strain (HDB92) at 35 and 42°C, respectively, both mutant and control strains grew regardless of the IPTG concentration (Fig. 5A, top). Consistent with previous results, the colonies produced by each strain on plates that lacked IPTG were slightly smaller (52). Very similar results were obtained in a quantitative efficiency of plating assay in which the numbers of colonies that formed in the presence and absence of IPTG were compared (see Materials and Methods). Both groE mutant and groE+ strains produced approximately the same number of colonies under the two plating conditions (Fig. 5A, bottom). In a second set of experiments, we assessed the viability of a strain that has a dnaK null allele (HDB96) after reducing the SRP concentration. For practical reasons we constructed this strain using a plasmid in which ffh expression is under the control of a lac rather than a trc promoter. Both HDB96 and a control dnaK+ strain (HDB95) grew well at 30°C when SRP levels were near normal (200 μM IPTG) or moderately reduced (50 μM IPTG) (Fig. 5B, top). The plating efficiency of the dnaK mutant strain at the lower IPTG concentration was actually slightly higher than that of the dnaK+ strain (Fig. 5B, bottom), but this difference may simply reflect the presence of an rpoH mutation in the ΔdnaK strain that is required to suppress cell division defects and genetic instability (8).
FIG. 5.
Mutations in groE and dnaK do not compromise the viability of SRP-deficient cells. (A) Strains HDB92 (Ptrc-ffh ffh::kan), HDB93 (Ptrc-ffh ffh::kan groEL44), and HDB94 (Ptrc-ffh ffh::kan groES619) were streaked on LB agar containing either 10 μM IPTG or no IPTG. Plates were incubated at 35 and 42°C for 20 and 16 h, respectively. Cells were diluted from overnight cultures and incubated at 35 and 42°C on LB agar plates containing either 10 μM IPTG or no IPTG to determine relative plating efficiencies as described in Materials and Methods. (B) Strains HDB95 (Plac-ffh ffh::kan) and HDB96 (Plac-ffh ffh::kan ΔdnaK52 sidB1) were streaked on LB agar containing either 200 μM IPTG or 50 μM IPTG. Cells were diluted from overnight cultures and incubated at 30°C on LB agar plates containing either 200 μM IPTG or 50 μM IPTG to determine relative plating efficiencies. In both panels A and B, plating efficiency was plotted on a logarithmic (log10) scale.
We next considered an alternative explanation for the role of the heat shock response in SRP-deficient cells based on the fact that the genes encoding several key proteases (Lon, ClpP, ClpQ, and FtsH) are all part of the heat shock regulon. We hypothesized that reduction of the SRP concentration below a threshold level might lead to mislocalization of IMPs in the cytoplasm. Since IMPs are extremely hydrophobic and aggregation prone, the heat shock response might be required to increase the proteolytic capacity of the cell so as to ensure efficient degradation of the mislocalized proteins. To test this idea we examined the effect of reducing the SRP concentration on the viability of cells that have mutations in several different protease genes.
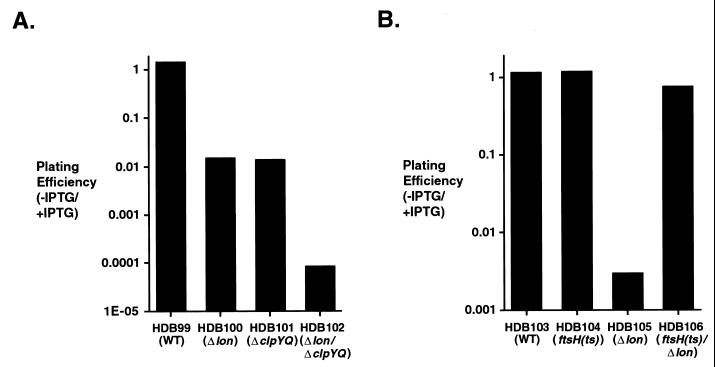
We found that moderate SRP deficiencies were lethal in cells that lack the heat shock regulated proteases Lon and ClpQ. Mutant strains HDB100 (Δlon), HDB101 (ΔclpYQ), and HDB102 (Δlon ΔclpYQ), which contain an inducible copy of the ffh gene (Ptrc-ffh), grew well on LB plates containing 10 μM IPTG. All of the strains showed greatly reduced viability on plates that lacked IPTG, however. Whereas the plating efficiency of a control lon+ clpYQ+ strain (HDB99) was independent of the IPTG concentration, the plating efficiency of HDB100 and HDB101 was approximately 100-fold lower on plates that lacked IPTG than on plates that contained 10 μM IPTG (Fig. 6A). Essentially identical results were obtained with a strain that had a disrupted copy of clpQ, which encodes the proteolytic subunit of ClpYQ, but an intact copy of clpY, which encodes the chaperone subunit (data not shown). Furthermore, the plating efficiency of the double mutant strain HDB102 dropped 4 orders of magnitude in the absence of IPTG. In contrast, disruption of the clpP gene did not reduce the plating efficiency of SRP deficient cells (data not shown). Likewise, a strain that contains an inducible copy of ffh and a temperature-sensitive ftsH allele (HDB104) produced an equal number of colonies in the presence and absence of IPTG upon incubation at a semipermissive temperature (37°C) (Fig. 6B).
FIG. 6.
Synthetic lethality of protease and SRP deficiencies. (A) Overnight cultures of strains HDB99 (Ptrc-ffh ffh::kan), HDB100 (Ptrc-ffh ffh::kan Δlon), HDB101 (Ptrc-ffh ffh::kan ΔclpYQ), and HDB102 (Ptrc-ffh ffh::kan Δlon ΔclpYQ) were diluted, and cells were incubated at 37°C on LB agar plates containing either 10 μM IPTG or no IPTG to determine relative plating efficiencies. (B) Overnight cultures of strains HDB103 [Ptrc-ffh ffh::kan], HDB104 [Ptrc-ffh ffh::kan ftsH(Ts)], HDB105 [Ptrc-ffh ffh::kan Δlon], and HDB106 [Ptrc-ffh ffh::kan ftsH(Ts) Δlon] were diluted, and cells were incubated as described for panel A to determine relative plating efficiencies. In both panels A and B, plating efficiency was plotted on a logarithmic (log10) scale.
Remarkably, the introduction of the ftsH(Ts) allele into lon-negative cells suppressed the lethality brought about by combining SRP and Lon deficiencies. Whereas the Δlon strain HDB105 showed a large reduction in plating efficiency in the absence of IPTG, the isogenic Δlon ftsH(Ts) strain (HDB106) grew as well on plates that lacked IPTG as on plates that contained IPTG (Fig. 6B). This genetic suppression is probably due to the reduced proteolysis of an FtsH substrate. The most likely candidate is the translocon protein SecY, since overproduction of SecYEG complex rescues SRP-deficient Δlon cells as effectively as the ftsH(Ts) allele (data not shown). Moreover, it seems plausible that an increase in the number of translocons would improve the efficiency of IMP insertion and thereby reduce the SRP requirement. It is also conceivable that suppression of the synthetic lethality is due to the hyperstabilization of another FtsH substrate, ς32. An increase in the concentration of ς32 might boost the heat shock response so as to compensate for the loss of Lon. In any case, the genetic suppression data support the argument that the reduced viability of cells containing SRP deficiences and a lon-null allele is due to the simultaneous inhibition of two alternate pathways for handling IMPs and is not simply the result of combining defects in important but unrelated biochemical pathways.
Taken together, our genetic studies strongly suggest that the heat shock response observed in SRP-deficient cells is required to increase expression of lon and clpQ. Introduction of a multicopy plasmid containing lon and clpYQ together with pS368 into HDB45 cells, however, does not suppress the Slo phenotype caused by the overexpression of dnaK (data not shown). Thus, it is likely that the viability of cells that lack sufficient SRP depends on the increased transcription of other heat shock genes as well.
A mislocalized IMP is more stable in Δlon ΔclpYQ cells than in wild-type cells.
The results described above suggest that Lon and ClpQ help sustain the viability of cells that have insufficient SRP by degrading IMPs that accumulate in the cytoplasm. This interpretation of the data predicts that mislocalized IMPs would be more stable in cells that lack Lon and ClpQ than in cells that have a normal complement of proteases. To test this idea, we blocked the SRP pathway in both a wild-type strain (HDB97) and an isogenic Δlon ΔclpYQ strain (HDB107) and assessed the steady-state level of a newly synthesized IMP by Western blotting.
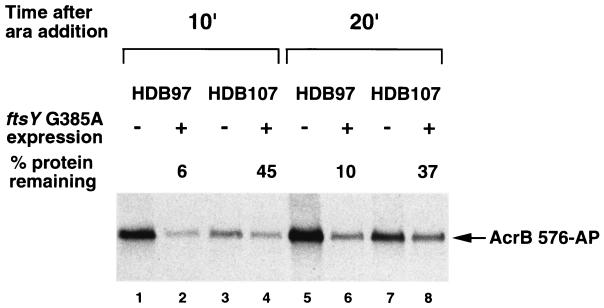
Consistent with our hypothesis, we found that a mislocalized IMP was rapidly degraded in a wild-type strain but only partially degraded in a protease-deficient strain. HDB97 and HDB107 were transformed with either pTRC99 or pTRC99 containing a dominant lethal ftsY allele (ftsY G385A) cloned under the control of the trc promoter and a second plasmid encoding an arabinose-inducible AP fusion to AcrB (AcrB 576-AP). Previous studies have shown that overexpression of ftsY G385A strongly inhibits the SRP pathway (52). Since the araBAD promoter is tightly regulated, only very low levels of fusion protein were detected in the absence of inducer (data not shown). Cultures were grown in LB, and 2 mM IPTG was added at mid-log phase to induce expression of the mutant ftsY allele. After 20 min 0.2% arabinose was added to induce synthesis of AcrB 576-AP and samples were removed after additional incubation periods of 10 and 20 min. At each time point a large amount of fusion protein was detected in HDB97 cells that had a functional SRP pathway (Fig. 7, lanes 1 and 5). Cells in which the insertion of AcrB 576-AP was inhibited by the synthesis of mutant FtsY, however, contained only ∼5 to 10% as much protein (Fig. 7, lanes 2 and 6). Given that mislocalized AcrB 576-AP has been shown to be highly unstable (half-life ≪ 2 min) (45), the simplest interpretation of this result is that almost all of the fusion protein remained in the cytoplasm where it was rapidly proteolyzed. Indeed the steady-state level of several other IMPs has been shown to decline after inhibition of the SRP pathway (48), suggesting that impairment of the insertion process often leads to IMP degradation. The expression of AcrB 576-AP in HDB107 was slightly delayed, presumably because the loss of Lon and/or ClpQ indirectly reduces the rate of arabinose uptake or PBAD activation. Nevertheless, cells that overexpressed ftsY G385A contained ∼35 to 45% as much fusion protein as control cells (Fig. 7, lanes 3 and 4 and 7 and 8). Thus, despite its retention in the cytoplasm, the AcrB 576-AP fusion appeared to be relatively stable. These results provide direct evidence that Lon and ClpQ (as well as other unidentified proteases) are responsible for the destruction of mislocalized IMPs.
FIG. 7.
Increased stability of a mislocalized IMP in Δlon ΔclpYQ cells. Strains HDB97 (lon+ clpYQ+) and HDB107 (Δlon ΔclpYQ) transformed with pTRC or pTRC-ftsY (G385A) were grown in LB containing ampicillin. IPTG (2 mM) was added to induce expression of the mutant ftsY allele and 0.2% arabinose (ara) was added 20 min later to induce expression of the AcrB 576-AP fusion protein. Samples were removed from each culture 10 min (lanes 1 to 4) and 20 min (lanes 5 to 8) after the addition of ara, and proteins were precipitated with 10% TCA. The steady-state level of the AP fusion protein was determined by Western blotting. Lanes 1, 2, 5, and 6, strain HDB97; lanes 3, 4, 7, and 8, strain HDB107. ftsY (G385A) was expressed in lanes 2, 4, 6, and 8. The amount of AcrB 576-AP detected after inhibition of the SRP pathway is expressed as a percentage of the fusion protein detected in control cells.
DISCUSSION
In this report we describe a series of experiments that provide novel insights into the role of SRP in E. coli physiology. Initially we observed that moderate SRP deficiencies that do not cause significant growth defects produce a heat shock response. Genetic experiments then showed that the heat shock response is required to preserve cell viability. These results imply that survival depends on the elevated synthesis of one or more heat shock proteins which compensates for the partial loss of SRP activity. We found that cells containing null alleles of lon and clpQ, two genes that encode heat shock-regulated proteases, are synthetically lethal with moderate SRP deficiencies. The simplest interpretation of these results is that the heat shock response serves at least in part to increase the capacity of the cells to degrade IMPs that are mislocalized as the result of the SRP deficiency. Consistent with this view, we found that a mislocalized IMP that is normally rapidly degraded in the cytoplasm appeared to be partially stabilized in ΔclpYQ Δlon cells. Taken together, our results suggest that by providing an efficient targeting pathway for IMPs, SRP also prevents a potentially toxic accumulation of aggregation-prone proteins in the cytoplasm as a secondary benefit.
In light of evidence that molecular chaperones play a major role in the targeting of presecretory proteins to the IM, it is striking that the heat shock response did not seem to be required to increase the levels of either DnaK or GroEL. The observation that dnaK and groE mutations could be combined with SRP deficiencies without producing synthetic lethality suggests that neither of the major heat shock-regulated chaperone systems provides an obligate default targeting pathway for IMPs in the absence of sufficient SRP. Of course it is conceivable that there is a high degree of redundancy among these and other molecular chaperones. Since DnaK and GroEL have very different roles in protein folding and distinct substrate specificities (9), however, it seems unlikely that they would have overlapping roles in IMP targeting. Another formal possibility is that Lon and ClpQ have unrecognized targeting functions that account for their increased importance in SRP-deficient cells. This interpretation of the data is unlikely for two reasons. First, it would be difficult to explain the observation that an ftsH mutation suppresses the synthetic lethality of lon and SRP defects if Lon were acting as a chaperone instead of a protease. Second, a ΔclpQ mutation that permits continued synthesis of the chaperone subunit of the ClpYQ complex (57) confers as great a sensitivity to SRP defects as a ΔclpYQ mutation.
Indeed, based on the available evidence, SRP may be the only cytoplasmic factor that can target IMPs effectively. The residual insertion of IMPs that is observed after SRP depletion (13, 14, 41, 52) may be due to the fortuitous arrival of insertion-competent nascent IMPs at the translocon rather than to the use of alternative targeting pathways. Inhibition of the SRP pathway produces strong insertion defects under rapid growth conditions (the insertion of AcrB 576-AP is blocked ∼90% in Fig. 7) but smaller defects under slow growth conditions. This finding is consistent with the idea that a specialized targeting system for IMPs is required primarily when biosynthetic rates are high and it is likely that any given nascent chain will attain a sufficient length to fold into an insertion-incompetent conformation before it reaches the IM. By analogy, there is evidence that presecretory proteins can be targeted to the IM without the aid of chaperones, but only if they remain loosely folded during their transit through the cytoplasm (44).
Our analysis of the function of the heat shock response in SRP-deficient cells suggests that SRP is both essential in E. coli and conserved throughout the bacterial kingdom, because the efficient targeting of IMPs solves two distinct problems. Although IMPs play key roles in many fundamental cellular processes, it is not clear that the reduced concentration of properly assembled IMPs that would result from the loss of SRP would in itself be lethal. Our experiments imply that a modest reduction of IMP biogenesis (of a magnitude that leads to the induction of a heat shock response) only subtly affects cell growth. Of course progressively more severe insertion defects would eventually lead to cell death, but the ∼50% reduction of IMP biogenesis observed after depletion of SRP from E. coli grown in minimal medium may not fully account for the dramatic loss of viability. Moreover, the observation that the magnitude of insertion defects correlates with the rate of cell growth suggests that the fraction of IMPs that would be integrated into the IM without SRP might be sufficient to meet the needs of a slow growing organism. Because IMPs are so hydrophobic, however, their retention in the cytoplasm is probably extremely toxic. Thus, SRP may be required not only to optimize the concentration of properly integrated IMPs, but also to prevent the dire consequences of IMP mislocalization. The degree to which bacteria can overproduce proteases to cope with mislocalized IMPs is limited since the proteases themselves are toxic above a threshold concentration (20). Given this constraint, in the complete absence of SRP the aggregation of mislocalized IMPs might simply outpace their degradation by the cytoplasmic proteolytic machinery. In addition, the finding that clpQ and lon are not the only heat shock genes that are required to maintain the viability of SRP-deficient cells raises the possibility that there are as yet undiscovered deleterious effects of eliminating the SRP pathway.
The results presented here also provide insights into the function of ClpYQ. Disruption of clpYQ in wild-type cells produces minor effects on cell growth and a very weak temperature-sensitive phenotype (40). Several studies have shown that the complex participates in the overall proteolysis of prematurely terminated proteins and acts redundantly with other proteases to degrade ς32 and SulA (27, 57). Nevertheless, the function of ClpYQ is still poorly understood. Our results suggest that at least under certain experimental conditions both ClpYQ and Lon can recognize and degrade mislocalized IMPs. Two observations suggest that the two proteases have partially or completely distinct substrate specificities. First, the overexpression of lon does not suppress the synthetic lethal effect of combining clpQ and SRP deficiencies (J. B. Hyndman and H. D. Bernstein, unpublished results). In addition, SRP-deficient cells that contain both lon- and clpQ-null alleles have an ∼100-fold lower plating efficiency than cells that contain only a single protease mutation. Despite these intriguing observations, however, it is not clear that either ClpQ or Lon actually degrades mislocalized IMPs under normal physiological conditions. Because SRP provides a highly efficient targeting pathway, mislocalization of IMPs may be a rare event. The growth of a ΔclpQ Δlon strain is unaffected by significant overproduction of a single IMP (Hyndman and Bernstein, unpublished results), suggesting that E. coli normally contains enough SRP to effectively target an excess load of IMPs. Moreover, the observation that a mislocalized IMP is only partially stabilized by the loss of ClpQ and Lon suggests that there are other as yet unidentified proteases that can also degrade IMPs in the cytoplasm.
Finally, the isolation of dnaK in the Slo screen confirms the prediction that this method can be used to identify genes that reside in parallel pathways as well as in the same pathway. As a corollary, the results formally demonstrate that both underexpression and overexpression of a gene can be equivalent to the introduction of a point mutation and can therefore provide a useful tool to generate a partial loss-of-function phenotype. Interestingly, the observation that heat shock and SRP deficiencies are synthetically lethal led to the identification of parallel pathways that are biochemically distinct. Secondary protein targeting pathways were not identified as a substitute for SRP; instead, degradative pathways were shown to provide an alternate mechanism for processing newly synthesized IMPs. An important implication of these observations is that the function of a gene that resides in a parallel pathway cannot be unequivocably inferred from its isolation in a synthetic lethal screen.
ACKNOWLEDGMENTS
We thank Bernd Bukau, Susan Gottesman, Ding Jin, Takashi Yura, and Jill Zeilstra-Ryalls for providing us with many valuable strains. We also thank Susan Gottesman for helpful discussions during the course of this work and for comments on the manuscript.
REFERENCES
- 1.Baker T A, Grossman A D, Gross C A. A gene regulating the heat shock response in Escherichia coli also affects proteolysis. Proc Natl Acad Sci USA. 1984;81:6779–6783. doi: 10.1073/pnas.81.21.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein H D, Poritz M A, Strub K, Hoben P, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 3.Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkareva E, Seluanov A, Bibi E, Girshovich A. Chaperonin-promoted post-translational membrane insertion of a multispanning membrane protein lactose permease. J Biol Chem. 1996;271:22256–22261. doi: 10.1074/jbc.271.36.22256. [DOI] [PubMed] [Google Scholar]
- 5.Bourgaize D B, Phillips T A, VanBogelen R A, Jones P G, Neidhardt F C, Fournier M J. Loss of 4.5S RNA induces the heat shock response and lambda prophage in Escherichia coli. J Bacteriol. 1990;172:1151–1154. doi: 10.1128/jb.172.2.1151-1154.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S. Mutations in the gene for EF-G reduce the requirement for 4.5S RNA in the growth of E. coli. Cell. 1987;49:825–833. doi: 10.1016/0092-8674(87)90620-9. [DOI] [PubMed] [Google Scholar]
- 7.Brown S, Fournier M J. The 4.5S RNA gene of Escherichia coli is essential for cell growth. J Mol Biol. 1984;178:533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- 8.Bukau B, Walker G C. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 10.Casadaban M J. Transposition and fusion of the lac operon to selected promoters in E. coli using bacteriophage λ and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 11.Chirico W J, Waters M G, Blobel G. 70K heat shock related proteins stimulate protein translocation in microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- 12.Collier D N, Bankaitis V A, Weiss J B, Bassford P J., Jr The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell. 1988;53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 13.deGier J-W, Scotti P A, Sääf A, Valent Q A, Kuhn A, Luirink J, von Heijne G. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:14646–14651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.deGier J-W L, Mansournia P, Valent Q A, Phillips G J, Luirink J, von Heijne G. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 1996;399:307–309. doi: 10.1016/s0014-5793(96)01354-3. [DOI] [PubMed] [Google Scholar]
- 15.Derman A I, Puziss J W, Bassford P J, Jr, Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshaies R J, Koch B D, Werner-Washburne M, Craig E A, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- 17.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in the folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 18.Friedlander M, Blobel G. Bovine opsin has more than one signal sequence. Nature. 1985;318:338–343. doi: 10.1038/318338a0. [DOI] [PubMed] [Google Scholar]
- 19.Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff S A, Goldberg A L. An increased content of protease La, the lon gene product, increases protein degradation and blocks growth in Escherchia coli. J Biol Chem. 1987;262:4508–4515. [PubMed] [Google Scholar]
- 21.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 22.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hann B C, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Using antibodies. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 25.Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D'Ari R, Bouloo P. Cell growth and λ phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci USA. 1993;90:10861–10865. doi: 10.1073/pnas.90.22.10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 1994;13:2686–2696. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanemori M, Nishihara K, Yanagi H, Yura T. Synergistic roles of HslUV and other ATP-dependent proteases in controlling in vivo turnover of ς32 and abnormal proteins in Escherichia coli. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch H G, Hengelage T, Neumann-Haefelin C, MacFarlane J, Hoffschulte H K, Schimz K L, Mechler B, Müller M. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol Biol Cell. 1999;10:2163–2173. doi: 10.1091/mbc.10.7.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krieg U C, Walter P, Johnson A E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumamoto C A, Beckwith J. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurzchalia T V, Wiedmann M, Girshovich A S, Bochkareva E S, Bielka H, Rapoport T A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 32.Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 1989;8:3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landry S J, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Gierasch L M. Characterization of a functionally important mobile domain of GroES. Nature. 1993;364:255–258. doi: 10.1038/364255a0. [DOI] [PubMed] [Google Scholar]
- 34.Lee C A, Beckwith J. Cotranslational and posttranslational protein translocation in prokaryotic systems. Annu Rev Cell Biol. 1986;2:315–336. doi: 10.1146/annurev.cb.02.110186.001531. [DOI] [PubMed] [Google Scholar]
- 35.Luirink J, ten Hagen-Jongman C M, van der Weijden C C, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacFarlane J, Müller M. Functional integration of a polytopic membrane protein of E. coli requires the bacterial signal recognition particle. Eur J Biochem. 1995;223:766–771. doi: 10.1111/j.1432-1033.1995.766_3.x. [DOI] [PubMed] [Google Scholar]
- 37.Meyer D I, Krause E, Dobberstein B. Secretory protein translocation across membranes—the role of the “docking protein”. Nature. 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- 38.Miller J D, Wilhelm H, Gierasch L, Gilmore R, Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- 39.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 40.Missiakis D, Schwager F, Betton J-M, Georgopoulos C, Raina S. Identification and characterization of HslV HslU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 41.Newitt J A, Ulbrandt N D, Bernstein H D. The structure of multiple polypeptide domains determines the signal recognition particle targeting requirement of E. coli inner membrane proteins. J Bacteriol. 1999;181:4561–4567. doi: 10.1128/jb.181.15.4561-4567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips G J, Silhavy T J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 43.Poritz M A, Bernstein H D, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 44.Prinz W A, Spiess C, Ehrmann M, Schierle C, Beckwith J. Targeting of signal sequenceless proteins for export in Escherichia coli with altered protein translocase. EMBO J. 1996;15:5209–5217. [PMC free article] [PubMed] [Google Scholar]
- 45.Qi H-Y, Bernstein H D. SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle. J Biol Chem. 1999;274:8993–8997. doi: 10.1074/jbc.274.13.8993. [DOI] [PubMed] [Google Scholar]
- 46.Ribes V, Römisch K, Giner A, Dobberstein B, Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- 47.Römisch K, Webb J, Herz S, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 48.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 49.Teter S A, Houry W A, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl F U. Polypeptide flux through bacterial hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 50.Tilly K, McKittrick N, Zylicz M, Georgopoulos C. The DnaK protein modulates the heat shock response of Escherichia coli. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- 51.Tomoyasu T, Ogura T, Tatsuta T, Bukau B. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol Microbiol. 1998;30:567–581. doi: 10.1046/j.1365-2958.1998.01090.x. [DOI] [PubMed] [Google Scholar]
- 52.Ulbrandt N D, Newitt J A, Bernstein H D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 53.Valent Q A, Scotti P A, High S, deGier J-W L, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter P, Johnson A E. Signal sequence recognition and protein targeting to the endoplasmic reticulum. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 55.Wild J, Altman A, Yura T, Gross C A. DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 1992;6:1165–1175. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 56.Wild J, Kamath-Loeb A, Ziegelhoffer E, Lonetto M, Kawasaki Y, Gross C A. Partial loss of function mutations in DnaK, the Escherichia coli homologue of the 70-kDa heat shock proteins, affect highly conserved amino acids implicated in ATP binding and hydrolysis. Proc Natl Acad Sci USA. 1992;89:7139–7143. doi: 10.1073/pnas.89.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu W-F, Zhou Y, Gottesman S. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J Bacteriol. 1999;181:3681–3687. doi: 10.1128/jb.181.12.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeilstra-Ryalls J, Fayet O, Baird L, Georgopoulos C. Sequence analysis and phenotypic characterization of groEL mutations that block λ and T4 bacteriophage growth. J Bacteriol. 1993;175:1134–1143. doi: 10.1128/jb.175.4.1134-1143.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeilstra-Ryalls J, Fayet O, Georgopoulos C. Two classes of extragenic suppressor mutations identify functionally distinct regions of the GroEL chaperone of Escherichia coli. J Bacteriol. 1994;176:6558–6565. doi: 10.1128/jb.176.21.6558-6565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]