Abstract
Schizosaccharomyces pombe has an open reading frame, which we named alr1+, encoding a putative protein similar to bacterial alanine racemase. We cloned the alr1+ gene in Escherichia coli and purified the gene product (Alr1p), with an Mr of 41,590, to homogeneity. Alr1p contains pyridoxal 5′-phosphate as a coenzyme and catalyzes the racemization of alanine with apparent Km and Vmax values as follows: for l-alanine, 5.0 mM and 670 μmol/min/mg, respectively, and for d-alanine, 2.4 mM and 350 μmol/min/mg, respectively. The enzyme is almost specific to alanine, but l-serine and l-2-aminobutyrate are racemized slowly at rates 3.7 and 0.37% of that of l-alanine, respectively. S. pombe uses d-alanine as a sole nitrogen source, but deletion of the alr1+ gene resulted in retarded growth on the same medium. This indicates that S. pombe has catabolic pathways for both enantiomers of alanine and that the pathway for l-alanine coupled with racemization plays a major role in the catabolism of d-alanine. Saccharomyces cerevisiae differs markedly from S. pombe: S. cerevisiae uses l-alanine but not d-alanine as a sole nitrogen source. Moreover, d-alanine is toxic to S. cerevisiae. However, heterologous expression of the alr1+ gene enabled S. cerevisiae to grow efficiently on d-alanine as a sole nitrogen source. The recombinant yeast was relieved from the toxicity of d-alanine.
d-Alanine is an essential component of bacterial peptidoglycans and is produced by alanine racemase (EC 5.1.1.1), a pyridoxal 5′-phosphate (PLP)-dependent enzyme (22, 29). However, d-alanine also occurs in various natural compounds produced by other organisms. For example, cyclosporin A contains d-alanine as a component and is produced by a fungus, Tolypocladium niveum (10). Alanine racemase was shown to be involved in the biosynthesis of d-alanine in this fungus and was later purified and characterized. Even though it differs markedly in primary structure from bacterial alanine racemase, it rather resembles yeast threonine aldolase (4, 10).
d-Serine occurs in mammalian brains and serves as an endogenous ligand of the glycine site of the N-methyl-d-aspartate receptor (14). Serine racemase, a PLP-dependent enzyme, was purified from rat brain to homogeneity (35), and a cDNA clone for mouse brain enzyme was obtained (36); this enzyme is distinct from both bacterial and fungal alanine racemases but similar to bacterial threonine dehydratase in primary structure (36). Serine racemase occurs also in vancomycin-resistant Enterococcus gallinarum (1). However, it resembles bacterial alanine racemases but not brain serine racemase. Therefore, alanine racemases and serine racemases are classified into three groups: bacterial alanine racemases plus serine racemase from E. gallinarum, fungal alanine racemase, and serine racemase of mammalian brain. Similar structural divergence has been found between other types of PLP enzymes, as exemplified by ornithine decarboxylases. Bacterial and eucaryotic ornithine decarboxylases differ markedly from each other, and the eucaryotic one shares the same protein fold with bacterial alanine racemase (9).
Contrary to a long-standing belief, various d-amino acids, including d-alanine and d-serine, have been found in yeasts in a peptide-bound form (15). Although no evidence has been obtained for the occurrence of a free form of d-amino acids, they are most probably produced in yeast cells upon proteolytic cleavage of the peptides. Therefore, it is reasonable to assume that amino acid racemases occur in yeasts. In fact, we have found that fission yeast, Schizosaccharomyces pombe, has a gene encoding a putative amino acid racemase similar to bacterial alanine racemases and the serine racemase of E. gallinarum. We have named the gene alr1+ because it shows alanine racemase activity upon cloning and expression in E. coli.
We report here the gene cloning, purification, and characterization of the gene product, alanine racemase ALR1p. Furthermore, we show that the Alr1p protein is involved in the catabolism of d-alanine in S. pombe, which we have confirmed through construction of a deletion mutation of the gene in S. pombe and heterologous expression of the gene in Saccharomyces cerevisiae.
MATERIALS AND METHODS
Materials.
Oligonucleotides were provided by Espec Oligo Service, Tsukuba, Japan; restriction endonucleases were from Takara Shuzo, Kyoto, Japan, or New England Biolabs, Beverley, Mass.; d-amino acid oxidase (EC 1.4.3.3) of hog kidney and l-lactate dehydrogenase (EC 1.1.1.27) of pig heart were from Boehringer GmbH, Mannheim, Germany; and l-alanine dehydrogenase of Bacillus stearothermophilus (EC 1.4.1.1) was from Unitika, Osaka, Japan. All other chemicals were of analytical grade.
Strains and culture conditions.
All S. pombe strains described here were derived from the strain SP1 (h+ leu1-32 ura4-D18) (25). S. cerevisiae ATCC 42752 (MATα ura3) was used for the heterologous expression of the alr1+ gene from S. pombe. S. pombe and S. cerevisiae were routinely grown in a YPD medium (1% yeast extract, 2% peptone, 2% glucose) or an SD medium (0.67% Bacto yeast nitrogen base, 2% glucose). The minimal medium for S. pombe contained 2% glucose, 0.05% KH2PO4, 0.036% CH3COOK, 0.05% MgSO4 · 7H2O, 0.01% NaCl, 0.01% CaCl2 · 2H2O, 0.1% (vol/vol) trace element solution (0.05% H3BO4, 0.004% CuSO4 · 5H2O, 0.01% KI, 0.02% FeCl3 · 6H2O, 0.04% MnSO4 · H2O, 0.02% Na2MoO4 · 2H2O, and 0.04% ZnSO4 · 7H2O), 1% (vol/vol) biotin (10 mg/ml), 1% (vol/vol) vitamin solution (0.1% pantothenic acid calcium salt, 1% nicotinic acid, and 1% myoinositol), and 10 mM ammonium sulfate. Cells were cultivated at 32°C with reciprocal shaking. The utilization of d- or l-alanine by S. pombe was examined as follows. S. pombe cells were harvested at a logarithmic growth phase, washed three times with minimal medium (described above) from which ammonium sulfate was omitted, and then cultured in the minimal medium or the alanine minimal medium, in which ammonium sulfate of the minimal medium described above was replaced by d- or l-alanine as a sole nitrogen source. Each strain was cultivated at 32°C with aeration. Escherichia coli cells were cultured at 37°C in Luria-Bertani broth supplemented with ampicillin (100 μg/ml), if necessary.
Cloning of the alanine racemase gene alr1+.
The alr1+ gene of S. pombe was amplified by PCR with the genomic DNA of S. pombe as a template. The sense primer used for PCR was 5′-CAT TTA AAT TCA TAT GAG AGG TGC AAA GTC CCg-3′ (NdeI site underlined), and the antisense primer was 5′-GGT AGC ATA CCA ATG AAG CTT TGC TC-3′ (HindIII site underlined). The amplified product was digested with NdeI and HindIII, and the resulting fragment was ligated into pET21-a previously digested with the same enzymes to yield the plasmid pETALR1. E. coli BL21(DE3) was used as a host. In order to express the alr1+ gene under the control of the GAL1 promoter in S. cerevisae, another construct of the gene was prepared; BamHI and HindIII sites were introduced at the 5′ and 3′ ends of the alr1+ gene with a sense primer of 5′-GAT TCA TTT AAA GGA TCC ATG AGA GGT GCA AAG-3′ (BamHI site underlined) and an antisense primer of 5′-GGT AGC ATA CCA ATG AAG CTT TGC TC-3′ (HindIII site underlined). The amplified product digested with BamHI and HindIII was introduced between the BamHI and HindIII sites of the p426Gal1 plasmid. S. cerevisiae ATCC 42752 (MATα ura3) was transformed with the resulting plasmid, p426Gal1ALR1. Positive clones were selected on a minimal medium plate lacking uracil.
Assays.
Alanine racemase was assayed in either direction of interconversion between l-alanine and d-alanine. The activity in the direction from d to l was determined with a mixture (1 ml) containing 100 mM CHES [2-(N-cyclohexylamino)ethanesulfonic acid] (pH 9.0), 0.15 U of l-alanine dehydrogenase, 50 mM d-alanine, 2.5 mM NAD+, and enzyme. The increase in absorbance at 340 nm due to NADH formation was monitored at 37°C. The other direction was followed at 37°C in a mixture (1 ml) containing 100 mM CHES (pH 9.0), 0.75 U of d-amino acid oxidase, 5.5 U of l-lactate dehydrogenase, 0.16 mM NADH, 50 mM l-alanine, and enzyme. Decrease in absorbance at 340 nm was monitored. The effect of pH on alanine racemase activity was examined with Bis-Tris propane (pH 6.5 to 9.0), CHES (pH 9.5 to 10.0), or CAPS [3-(cyclohexylamino)-1-propane sulfonic acid] (pH 9.5 to 11.0) replacing CHES (pH 9.0) in the mixtures described above. Activities toward other substrates were determined in a mixture (0.05 ml) containing 100 mM CHES buffer (pH 9.0), 50 mM l-amino acid, and enzyme. The reaction was carried out at 37°C for 30 min. The amount of the corresponding d-amino acid formed was determined by high-performance liquid chromatography after derivatization as described previously (27). Protein was assayed according to the method described by Bradford (2a) with bovine serum albumin as a standard.
Purification of the Alr1p protein.
All procedures were carried out at 4°C. The standard buffer used throughout the purification was a 20 mM potassium phosphate buffer (pH 7.2) containing 0.01% 2-mercaptoethanol and 10 μM PLP. E. coli BL21(DE3) cells harboring pETALR1 (about 50 g, wet weight) were suspended in 200 ml of the standard buffer and sonicated. The lysate was centrifuged at 18,000 × g for 20 min, and protamine sulfate was added to the supernatant solution to a final concentration of 0.4%, followed by centrifugation. After dialysis, the supernatant solution was loaded onto a DEAE-TOYOPEARL column (200 ml) equilibrated with the standard buffer. The column was washed with the standard buffer, and the enzyme was eluted with a linear gradient of 0 to 0.4 M KCl in the standard buffer. The active fractions were combined and brought to 30% saturation with ammonium sulfate. After centrifugation, the supernatant solution was loaded onto a butyl-TOYOPEARL column (50 ml) equilibrated with the standard buffer supplemented with ammonium sulfate (30% saturation). The column was washed with the standard buffer containing a 10% saturation of ammonium sulfate, and then the enzyme was eluted with a linear gradient of 10 to 0% saturation with ammonium sulfate in the standard buffer. The active fractions were combined and dialyzed against the standard buffer. After centrifugation, the supernatant solution was applied to a Q-Sepharose HP column (50 ml) equilibrated with the standard buffer. The column was washed with 0.1 M KCl, and the enzyme was eluted with a linear gradient of 0.1 to 0.3 M KCl in the standard buffer. The pooled active fractions were brought to 30% saturation with ammonium sulfate, and the supernatant solution was loaded onto a butyl-TOYOPEARL column (50 ml) equilibrated with the standard buffer supplemented with 30% saturation of ammonium sulfate. The column was washed with the standard buffer containing 10% saturated ammonium sulfate, and the enzyme was eluted with the standard buffer. The active fractions were combined and dialyzed against a 2 mM potassium phosphate buffer (pH 7.2) containing 0.01% 2-mercaptoethanol and 10 μM PLP. After centrifugation, the supernatant was loaded onto a Gigapite column (30 ml) equilibrated with a 2 mM potassium phosphate buffer (pH 7.2) containing 0.01% 2-mercaptoethanol and 10 μM PLP. The enzyme was eluted with the same buffer, and the active fractions were pooled and dialyzed against the standard buffer. The purity of the enzyme was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Immunoblotting.
The extracts of S. pombe cells were subjected to SDS-PAGE, and proteins were transferred to polyvinylidene difluoride membranes. Western blot analysis was performed with the anti-Alr1p antiserum (raised by standard procedures), and the proteins were detected by chemiluminescence using CDP-Star (Roche Diagnostics, Basel, Switzerland).
Disruption of the alr1+ gene in S. pombe.
For the construction of the alr1::LEU2 yeast strain, a LEU2 deletion cassette for alr1+ was prepared by PCR as follows. Two DNA fragments, one (0.6 kbp) containing the upstream region and the other (0.8 kbp) containing the downstream region of alr1+, were obtained by PCR with the genomic DNA of S. pombe as a template. The primers used were as follows: for the upstream region, 5′-CAG TTT CAT TTC AAA ACG AAT GTT ATA GAA CAT C-3′ (sense primer) and 5′-CGA ATT CCT GCA GCC CGG GGG ATC CAA TTG CTA AAA CAT GAC CGC TTG G-3′ (antisense primer); and for the downstream region, 5′-GCT TAT CGA TAC CGT CGA CCG GAG AGT TCC TTT ACA GTA CAC-3′ (sense primer) and 5′-CAG ACT ACA TAG ATT TAC ACA TCC TTC TTT ACC-3′ (antisense primer). The 3′ end of the upstream region and the 5′ end of the downstream region are complementary to the 5′ and 3′ ends of the LEU2 gene, respectively. The second PCR was carried out with the DNA fragment (2.2 kbp) containing the LEU2 gene as a template and the products of the first PCR as primers. The resulting PCR product (3.6 kbp) containing the LEU2 gene flanked by the upstream and downstream regions of alr1+ was used for the transformation of the haploid yeast strain. Positive clones were selected on a minimal medium not supplemented with l-leucine, and disruption of alr1+ was verified by Southern blotting and PCR analysis. However, the resulting alr1Δ strain (h+ leu1-32 ura4-D18 alr1::LEU2) and the SP1 strain are not suitable for examination of nitrogen sources because even low concentrations of uracil and leucine, which must be added to supplement the defects in mutated ura4 and leu1 genes, respectively, can support the growth without addition of nitrogen sources. Therefore, nitrogen sources for the deletion mutant were examined by introduction of the ura4+ gene on a plasmid, pART2, which we prepared by exchanging the LEU2 marker in the pART1 plasmid with the ura4+ marker derived from the pBSUra4 plasmid at the HindIII and SalI sites. The SP1 strain was transformed with plasmids pART1 and pART2 and then used for examination of the utilization of nitrogen sources.
RESULTS
Cloning of the alanine racemase gene from S. pombe.
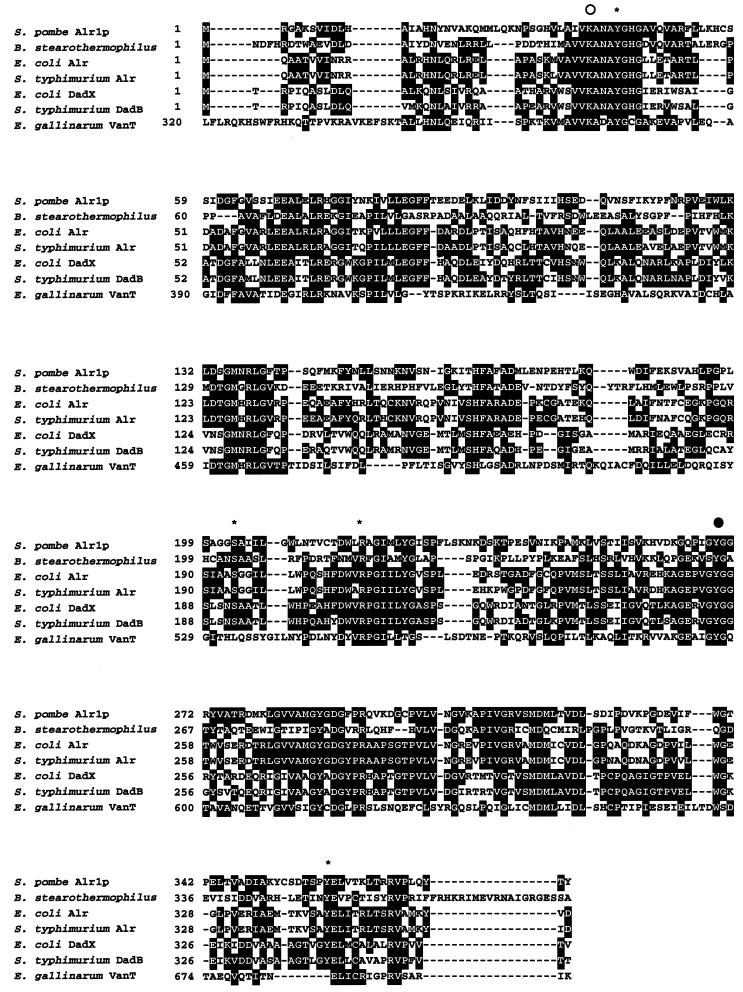
We found by a BLAST search of protein databases that S. pombe contains a gene in its chromosome III (SPCC965.08c) that is homologous to those of bacterial alanine racemases and the serine racemase of E. gallinarum (1) (Fig. 1). Similarities in the amino acid sequence of the putative gene product were 39% to Alr alanine racemase from E. coli and 28% to serine racemase of E. gallinarum. We confirmed by cloning and expression of the gene whether it codes for alanine racemase or serine racemase. We constructed a plasmid, pETALR1, by ligation of the gene amplified by PCR into an expression vector, pET21-a. An E. coli BL21(DE3) clone harboring pETALR1 showed 18 U of alanine racemase activity/mg, which was about 220-fold higher than that of the control clone carrying the empty vector pET21-a. The E. coli BL21/pETALR1 clone also showed racemization activity toward serine, but the activity was much lower than with alanine. Therefore, we named the alanine racemase gene alr1+.
FIG. 1.
Comparison of the amino acid sequences of S. pombe Alr1p (GenBank accession no. AL023590) with the bacterial alanine racemases (plus GenBank accession numbers) of B. stearothermophilus (M19142), E. coli Alr (AE00478), S. enterica serovar Typhimurium Alr (M12847), E. coli DadX (LO2948), S. enterica serovar Typhimurium DadB (KO2119), and E. gallinarum VanT (AF162694). The amino acid sequences were aligned with the MegAlign program of the DNASTAR package (5). Conserved residues are boxed. The open circle indicates the Lys residue that binds PLP and is proposed to abstract α-proton from d-alanine. The closed circle indicates the Tyr residue that is proposed to abstract α-hydrogen from l-alanine. Asterisks show the residues involved in binding with PLP.
The alanine racemase of B. stearothermophilus has been studied extensively by X-ray crystallography and site-directed mutagenesis, and Lys39 and Tyr265 most probably function as catalytic bases abstracting the α-hydrogen of d-alanine and l-alanine, respectively (20, 31, 32). X-ray crystallography of the enzyme also showed that Tyr43, Arg136, Ser204, and Arg219 interact with PLP (20). All these residues are conserved in alanine racemase of S. pombe (Fig. 1).
Purification and characterization of alanine racemase from S. pombe.
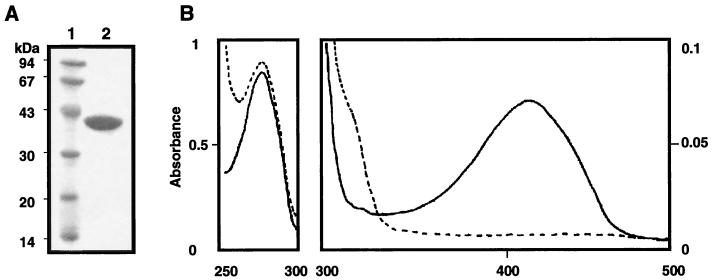
Alanine racemase of S. pombe (Alr1p) was purified to homogeneity through six steps from the cell extract of E. coli BL21(DE3) cells harboring pETALR1 (Fig. 2A). A summary of the purification is shown in Table 1.
FIG. 2.
SDS-PAGE gel (A) and absorption spectra (B) of purified S. pombe Alr1p. (A) Lane 1, marker proteins (the molecular masses are shown in kilodaltons); lane 2, purified S. pombe Alr1p (10 μg). Protein was stained with Coomassie brilliant blue R-250. (B) The spectrum of the enzyme solution (1.0 mg/ml) in a 20 mM potassium phosphate buffer (pH 7.8) containing 0.1% 2-mercaptoethanol is shown as a solid line, while that of the enzyme dialyzed against 500 volumes of a 20 mM potassium phosphate buffer (pH 7.8) containing 0.1% 2-mercaptoethanol and 10 mM sodium borohydride is shown as a dashed line.
TABLE 1.
Purification of ALR1p from E. coli BL21(DE3) cells harboring pETALR1
| Step of purification | Total protein (mg) | Total activity (U) | Sp act (U/mg)a |
|---|---|---|---|
| Crude extracts | 3,200 | 21,000 | 5.5 |
| Protamine sulfate | 1,800 | 19,000 | 11 |
| DEAE-TOYOPEARL | 340 | 14,000 | 42 |
| First Butyl-TOYOPEARL100 | 100 | 12,000 | 120 |
| Q-Sepharose HP | 39 | 8,200 | 210 |
| Second Butyl-TOYOPEARL100 | 25 | 7,500 | 300 |
| Gigapite | 20 | 6,800 | 340 |
Alanine racemase activity was determined as described in Materials and Methods with d-alanine as a substrate at 37°C.
The purified enzyme absorbed at around 420 nm (Fig. 2B). When the enzyme was dialyzed against sodium borohydride, the 420-nm peak disappeared and a new band appeared at around 330 nm (Fig. 2B). The enzyme was completely inactivated by the treatment. The behavior is characteristic of PLP-dependent enzymes, indicating that Alr1p contains PLP as a coenzyme. The sequence around Lys38 is conserved among all bacterial alanine racemases, and X-ray crystallography of the B. stearothermophilus enzyme shows that the corresponding lysine residue (Lys39) of the enzyme forms an aldimine linkage with PLP (20). Therefore, Lys38 of Alr1p is most probably bound to PLP and functions as a catalytic base abstracting α-hydrogen from d-alanine according to the mechanism proposed for the B. stearothermophilus enzyme (31).
X-ray crystallography of the B. stearothermophilus enzyme has shown that it is a dimer of identical subunits (20). Tyr265 of the enzyme functions as the base functioning pairwise with Lys39, as described above, but it comes from another subunit. Tyr265 is conserved (as Tyr269 in Alr1p) as well as many residues functioning as a kind of glue (i.e., residues forming ion pairs, hydrophobic interactions, and hydrogen bonds) at the subunit interface, such as Glu69 (Glu70 in Alr1p), Arg136 (Arg138 in Alr1p), and His166 (His167 in Alr1p) (20). This strongly suggests that Alr1p also occurs as a dimer. However, Alr1p showed an unexpected molecular weight of about 44,000 upon gel filtration through a Superose12 column, suggesting a monomeric structure. Both alanine racemases of Salmonella enterica serovar Typhimurium, DadB and Alr, were thought to be monomers based on the results of gel filtration before X-ray crystallography of the B. stearothermophilus enzyme (6). If all these enzymes, including Alr1p, are dimers, then their unusual behaviors on gel filtration are due to a characteristic nature common to the enzymes, such as sticky surfaces of the protein molecules.
Alr1p is virtually specific toward alanine. Apparent Km and Vmax values were determined: l-alanine, 5.0 mM and 670 μmol/min/mg, respectively; and d-alanine, 2.4 mM and 350 μmol/min/mg, respectively. Vmax/Km values for the conversions from l- to d-alanine and d- to l-alanine were similar to each other: 134 and 146 μmol/min/mg/mM, respectively. The ratio of the values is theoretically unity (3), and the experimental value (0.92) is consistent with the fact that the enzyme catalyzes racemization (3). l-Serine and l-2-aminobutyrate are poor substrates of Alr1p: their rates are 3.7 and 0.37% of that of l-alanine, respectively. The opposite situation can be seen for the VanT serine racemase of E. gallinarum: l-alanine is racemized at a rate that is 14% of that of l-serine (2). No activity of Alr1p was found toward l-valine, l-methionine, l-tryptophan, l-phenylalanine, l-histidine, l-tyrosine, l-threonine, l-asparagine, l-glutamine, l-aspartate, l-glutamate, l-arginine, l-lysine, l-norvaline, l-norleucine, l-homoserine, or l-ornithine.
Alr1p showed the maximum activity toward d- and l-alanine at pHs 9.0 and 9.5, respectively. The alkalophilic nature is common to most bacterial alanine racemases.
Role of d- and l-alanine as a sole nitrogen source in the growth of S. pombe.
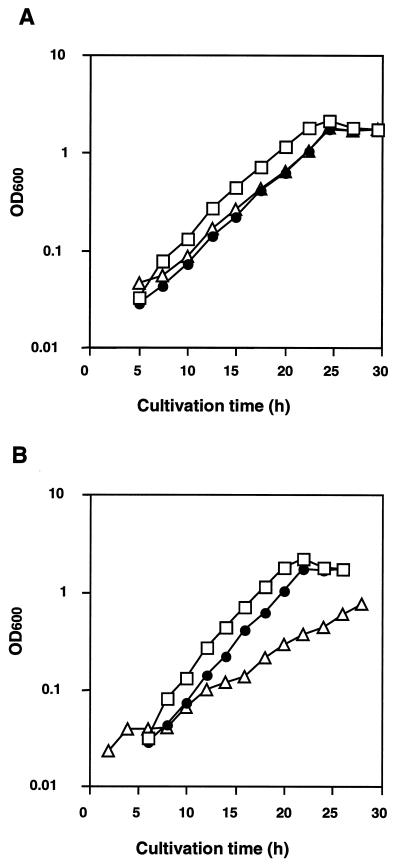
We found that S. pombe grows on either enantiomer of alanine as a sole nitrogen source (Fig. 3A). Since S. pombe has Alr1p, which catalyzes the interconversion between the enantiomers, it is reasonable to assume that the yeast survives on either enantiomer through racemization with only a catabolic pathway for l- or d-alanine. Alternatively, it may have a separate pathway for each enantiomer. Therefore, we constructed a deletion mutant of S. pombe lacking the alr1+ gene (Fig. 3B). The alrlΔ strain grew as well as the original strain on a complete medium such as YPD, indicating that the alr1+ gene is not essential for S. pombe. We found, however, that the deletion mutant grows about 50% less slowly than the original strain on a medium containing d-alanine as a sole nitrogen source (Fig. 3B). The response to the antipode was different: both strains grew equally well on l-alanine as a sole nitrogen source. Moreover, the growth rate on l-alanine was twice that on d-alanine. These results indicate that S. pombe has catabolic pathways for each enantiomer of alanine and that the pathway for l-alanine plays a major role in the catabolism of d-alanine by coupling with racemization. l-Alanine is utilized in Candida maltosa by l-alanine aminotransferase, which is inducibly formed by l-alanine (26). S. pombe also has two putative homologs of this enzyme (GenBank accession no. CAB46671.1 and CAA93786.1). d-Alanine is utilized by d-amino acid oxidase in Rhodotorula gracilis and Candida boidinii (8, 17). We found that S. pombe grown on d-alanine as a sole nitrogen source also shows d-amino acid oxidase activity toward d-alanine as a substrate (data not shown). Thus, these results suggest that alanine racemase participates in the catabolism of d-alanine by providing l-alanine aminotransferase with its substrate l-alanine and thereby supports the growth of the yeast, which would otherwise not be efficient with d-amino acid oxidase alone. We found by Western blotting that Alr1p is produced in a minimal medium containing d-alanine as a sole nitrogen source (Fig. 4). However, no positive bands reactive with the anti-Alr1p antibody were observed either when ammonium sulfate or l-alanine replaced d-alanine or in a complete medium, such as YPD (Fig. 4).
FIG. 3.
Growth characteristics when d- or l-alanine was a sole nitrogen source of wild-type S. pombe (A) and a deletion mutant (alr1Δ) of S. pombe (B). (A) S. pombe SP1 (Leu− Ura−) was made Leu+ Ura+ by introduction of both pART1 and pART2 (see Materials and Methods). (B) alr1::LEU2 strain (Alr1Δ Leu+ Ura−) derived from S. pombe SP1 was transformed with pART2 to become Alr1Δ Leu+ An Ura+. Each recombinant was precultured in the minimal medium described in Materials and Methods and transferred to a culture containing, as a sole nitrogen source, 5 mM ammonium sulfate (squares), 10 mM d-alanine (triangles), or l-alanine (circles). The culture was grown at 32°C with reciprocal shaking.
FIG. 4.
Western blot analysis of Alr1p. S. pombe SP1 cells exponentially growing on YPD medium (lane 1) or a minimum medium containing, as a sole nitrogen source, ammonium sulfate (lane 2), l-alanine (lane 3), or d-alanine (lane 4) were harvested. Cell extracts were subjected to SDS–13.0% PAGE, blotted onto a polyvinylidene difluoride membrane, and analyzed with polyclonal anti-Alr1p antibody. Each lane was loaded with 10 μg of protein.
Effect of heterologous expression of the S. pombe alr1+ gene on S. cerevisiae.
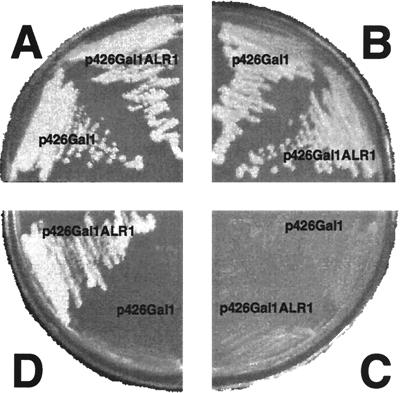
S. cerevisiae contains no genes for homologs of bacterial alanine racemases in its chromosome. Gly1p for threonine aldolase is similar to fungal alanine racemase in primary structure but shows no alanine racemase activity (12). Contrary to S. pombe, d-alanine is toxic to S. cerevisiae (19, 23). When the alr1+ gene of S. pombe was cloned into S. cerevisiae with an expression vector, p426Gal1, controlled by the GAL1 promoter, the recombinant S. cerevisiae carrying the resulting plasmid, p426Gal1ALR1, grew well on a medium containing glucose as a carbon source and either ammonium sulfate or l-alanine as a nitrogen source in the same manner as the control recombinant carrying the empty p426Gal1 vector. However, neither of the two recombinant yeasts grew on d-alanine as a sole nitrogen source (Fig. 5). When the GAL1 promoter was turned on by addition of galactose, only the recombinant carrying the alr1+ gene grew on a medium containing d-alanine as a sole nitrogen source and raffinose as a carbon source (Fig. 5). This indicates that d-alanine is converted to its antipode by Alr1p and is then utilized as a nitrogen source by the recombinant carrying alr1+. Moreover, the growth rate of the recombinant on the d-alanine medium is similar to that on the glucose-ammonium sulfate medium (data not shown). Therefore, d-alanine is not toxic to the recombinant yeast expressing Alr1p. These results indicate that the heterologous Alr1p not only renders S. cerevisiae capable of utilization of d-alanine as a nitrogen source but also relieves the yeast from the toxicity of d-alanine.
FIG. 5.
Heterologous expression of alr1+ gene in S. cerevisiae S. cervisiae cells harboring p426Gal1 or p426Gal1ALR1 were streaked on agar plates containing the minimal medium supplemented with 5 mM ammonium sulfate (see Materials and Methods) (A), 10 mM l-alanine (B), or 10 mM d-alanine (C and D). (D) Glucose in the medium was replaced by a mixture of raffinose (2%) and galactose (0.2%). After 3 days of incubation at 30°C, each plate was photographed.
DISCUSSION
PLP enzymes have been classified into five groups with different fold types (9). Bacterial alanine racemases belong to fold type III (7, 9, 11), while fungal alanine racemase belongs to fold type I (4, 12). This is probably a case of convergent evolution. The enzyme of fold type I has possibly evolved from an ancestor protein common to that of threonine aldolase in the fungus.
The BLAST search in GenBank and EMBL databases showed that, in addition to occurring in eubacteria, the alanine racemase of fold type III occurs only in S. pombe; no homologs could be found in the database of expressed sequence tags or in the genomes of archaea and eucaryotes, including S. cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster. The phylogenetic analysis of alanine racemases indicates that they are classified into three groups (Fig. 6). It is interesting that Alr1p of S. pombe shows high similarity only to alanine racemases of γ-proteobacteria (phylum of gram-negative bacteria) (Fig. 6, group A). Lower similarity was found when Alr1p was compared to other groups: group B, containing mainly high-G+C-content gram-positive bacteria, and group C, containing mainly low-G+C-content gram-positive bacteria. The alr1+ gene of S. pombe was possibly acquired from γ-proteobacteria through some events of horizontal gene transfer, such as conjugation: S. pombe is known to be a recipient of genes from E. coli through direct conjugation (21). Whatever the mechanism of acquisition of the enzyme gene was in the past, it has been kept by S. pombe throughout evolution. The enzyme has probably been required by this yeast in its habitat due to, for example, the availability of d-alanine as a sole nitrogen source.
FIG. 6.
Phylogenetic tree for alanine racemases. Sequences obtained from GenBank were aligned, and a phylogenetic tree was constructed with the MegAlign program by using the CLUSTAL method (5). The scale indicates the percentages of sequence divergence. The accession numbers of alanine racemase genes from various sources are shown.
E. coli and S. enterica serovar Typhimurium contain two isoenzymes of alanine racemases with distinct physiological functions: one is biosynthetic, and the other is catabolic (18). Alr alanine racemases of S. enterica serovar Typhimurium and E. coli are biosynthetic and constitutive, while DadB (S. enterica serovar Typhimurium) and DadX (E. coli) isozymes are catabolic and inducibly formed by l-alanine (13, 30, 34). Alr provides d-alanine for the biosynthesis of peptidoglycans. Both S. enterica serovar Typhimurium and E. coli grow on d-alanine as the sole carbon and energy source because they have d-amino acid dehydrogenase decomposing d-alanine into pyruvate and ammonia. However, they have no direct catabolic pathways for l-alanine and get it racemized with the DadB and DadX racemases (16, 33). S. pombe differs from these bacteria in that it has direct catabolic pathways for each enantiomer of alanine. A methylotrophic yeast, C. boidinii, also grows on d-alanine as a sole nitrogen or carbon source. The utilization of d-alanine is attributed to d-amino acid oxidase (8, 17), but deletion of the enzyme gene showed no significant effect on the growth on d-alanine as a sole nitrogen source (37). This suggests that another degradation pathway for d-alanine occurs in this methylotrophic yeast. Alternatively, it may have alanine racemase and a degradation pathway for l-alanine in the same manner as S. pombe. However, S. cerevisiae is quite different from these yeasts because it cannot utilize d-alanine. Similar examples are known in nitrogen metabolic pathways: a pathway common to S. pombe and Candida is missing in S. cerevisiae, as exemplified by purine degradation through uric acid and allantoin (17, 28). S. cerevisiae lacks uricase and cannot utilize purine as a nitrogen source.
In S. cerevisiae, various d-amino acids are incorporated by the general amino acid permease, which is characterized by its low affinity and high capacity (19, 24). However, some of them, such as d-alanine, d-serine, d-leucine, and d-tyrosine, are toxic to the yeast (23). The toxicity of d-leucine and d-tyrosine is explained by their unfavorable reactions with the corresponding tRNAs by leucyl- and tyrosyl-tRNA synthetases, respectively. d-Tyrosyl-tRNATyr deacylase (DTD1) of S. cerevisiae hydrolyzes resultant d-leucyl-tRNALeu and D-tyrosyl-tRNATyr and thereby reduces their toxicity (23). A similar defense system against the toxicity of d-leucine and d-tyrosine probably occurs in S. pombe because it also has a gene for the DTDI homolog (GenBank accession no. CAB16293.1) although it has not yet been proven biochemically. However, little is known about the mechanism of toxicity of d-alanine and d-serine toward S. cerevisiae. Whatever the mechanism of toxicity, S. cerevisiae recombinant cells expressing alanine racemase are relieved from the toxicity of d-alanine. This may be another role of alanine racemase in S. pombe. A deletion mutant of S. pombe lacking both alr1+ and the d-amino acid oxidase gene is being prepared in order to show the effect of d-alanine and d-serine on this yeast.
ACKNOWLEDGMENTS
We are grateful to H. Suga of the Faculty of Science, Kyoto University, for his helpful discussions.
This work was supported in part by a Grant-in-Aid for Scientific Research, no. 08680683 (to T.Y.), from the Ministry of Education, Science, Sports and Culture of Japan, and a Research Grant from the Japan Society for the Promotion of Science (Research for the Future) to N.E.
REFERENCES
- 1.Arias C A, Martin-Martinez M, Blundell T L, Arthur M, Courvalin P, Reynolds P E. Characterization and modelling of VanT: a novel, membrane-bound, serine racemase from vancomycin-resistant Enterococcus gallinarumBM4174. Mol Microbiol. 1999;31:1653–1664. doi: 10.1046/j.1365-2958.1999.01294.x. [DOI] [PubMed] [Google Scholar]
- 2.Arias C A, Weisner J, Blackburn J M, Reynolds P E. Serine and alanine racemase activities of VanT: a protein necessary for vancomycin resistance in Enterococcus gallinarumBM4174. Microbiology. 2000;146:1727–1734. doi: 10.1099/00221287-146-7-1727. [DOI] [PubMed] [Google Scholar]
- 2a.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Briggs G E, Haldane J B S. Note on the kinetics of enzyme action. Biochem J. 1925;19:338–339. doi: 10.1042/bj0190338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y Q, Walton J D. A eukaryotic alanine racemase gene involved in cyclic peptide biosynthesis. J Biol Chem. 2000;275:4906–4911. doi: 10.1074/jbc.275.7.4906. [DOI] [PubMed] [Google Scholar]
- 5.Clewley J P, Arnold C. MEGALIGN. The multiple alignment module of LASERGENE. Methods Mol Biol. 1997;70:119–129. [PubMed] [Google Scholar]
- 6.Esaki N, Walsh C T. Biosynthetic alanine racemase of Salmonella typhimurium: purification and characterization of the enzyme encoded by the alrgene. Biochemistry. 1986;25:3261–3267. doi: 10.1021/bi00359a027. [DOI] [PubMed] [Google Scholar]
- 7.Fluri R, Kinghorn J R. The all2 gene is required for the induction of the purine deamination pathway in Schizosaccharomyces pombe. J Gen Microbiol. 1985;131:527–532. doi: 10.1099/00221287-131-3-527. [DOI] [PubMed] [Google Scholar]
- 8.Goodman J M, Trapp S B, Hwang H, Veenhuis M. Peroxisomes induced in Candida boidinii by methanol, oleic acid and d-alanine vary in metabolic function but share common integral membrane proteins. J Cell Sci. 1990;97:193–204. doi: 10.1242/jcs.97.1.193. [DOI] [PubMed] [Google Scholar]
- 9.Grishin N V, Phillips M A, Goldsmith E J. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. 1995;4:1291–1304. doi: 10.1002/pro.5560040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann K, Schneider-Scherzer E, Kleinkauf H, Zocher R. Purification and characterization of eucaryotic alanine racemase acting as key enzyme in cyclosporin biosynthesis. J Biol Chem. 1994;269:12710–12714. [PubMed] [Google Scholar]
- 11.Kern A D, Oliveira M A, Coffino P, Hackert M L. Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Struct Fold Des. 1999;7:567–581. doi: 10.1016/s0969-2126(99)80073-2. [DOI] [PubMed] [Google Scholar]
- 12.Liu J Q, Nagata S, Dairi T, Misono H, Shimizu S, Yamada H. The GLY1 gene of Saccharomyces cerevisiae encodes a low-specific l-threonine aldolase that catalyzes cleavage of l-allo-threonine and l-threonine to glycine. Expression of the gene in Escherichia coliand purification and characterization of the enzyme. Eur J Biochem. 1997;245:289–293. doi: 10.1111/j.1432-1033.1997.00289.x. [DOI] [PubMed] [Google Scholar]
- 13.Lobocka M, Hennig J, Wild J, Klopotowski T. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of d-amino acid dehydrogenase and the catabolic alanine racemase. J Bacteriol. 1994;176:1500–1510. doi: 10.1128/jb.176.5.1500-1510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mothet J P, Parent A T, Wolosker H, Brady R O, Jr, Linden D J, Ferris C D, Rogawski M A, Snyder S H. d-serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc Natl Acad Sci USA. 2000;97:4926–4931. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagata Y, Fujiwara T, Kawaguchi-Nagata K, Fukumori Y, Yamanaka T. Occurrence of peptidyl d-amino acids in soluble fractions of several eubacteria, archaea and eukaryotes. Biochim Biophys Acta. 1998;1379:76–82. doi: 10.1016/s0304-4165(97)00084-6. [DOI] [PubMed] [Google Scholar]
- 16.Olsiewski P J, Kaczorowski G J, Walsh C. Purification and properties of d-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur flavoenzyme from Escherichia coliB. J Biol Chem. 1980;255:4487–4494. [PubMed] [Google Scholar]
- 17.Perotti M E, Pollegioni L, Pilone M S. Expression of d-amino acid oxidase in Rhodotorula gracilisunder induction conditions: a biochemical and cytochemical study. Eur J Cell Biol. 1991;55:104–113. [PubMed] [Google Scholar]
- 18.Reitzer L J, Magasanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhart F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Esherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1987. pp. 302–320. [Google Scholar]
- 19.Rytka J. Positive selection of general amino acid permease mutants in Saccharomyces cerevisiae. J Bacteriol. 1975;121:562–570. doi: 10.1128/jb.121.2.562-570.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw J P, Petsko G A, Ringe D. Determination of the structure of alanine racemase from Bacillus stearothermophilusat 1.9-A resolution. Biochemistry. 1997;36:1329–1342. doi: 10.1021/bi961856c. [DOI] [PubMed] [Google Scholar]
- 21.Siroski R S, Michaud W, Levin H L, Boeke J D, Hieter P. Trans-kingdom promiscuity. Nature (London) 1990;345:581–582. doi: 10.1038/345581b0. [DOI] [PubMed] [Google Scholar]
- 22.Soda K, Esaki N. Pyridoxal enzymes acting on d-amino acids. Pure Appl Chem. 1994;66:709–714. [Google Scholar]
- 23.Soutourina J, Plateau P, Blanquet S. Metabolism of d-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiaecells. J Biol Chem. 2000;275:32535–32542. doi: 10.1074/jbc.M005166200. [DOI] [PubMed] [Google Scholar]
- 24.Stanbrough M, Magasanik B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takegawa K, DeWald D B, Emr S D. Schizosaccharomyces pombeVps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J Cell Sci. 1995;108:3745–3756. doi: 10.1242/jcs.108.12.3745. [DOI] [PubMed] [Google Scholar]
- 26.Umemura I, Yanagiya K, Komatsubara S, Sato T, Tosa T. Characteristics of alanine aminotransferase from Candida maltosa. In: Fukui T, Kagamiyama H, Soda K, Wada H, editors. Enzymes dependent on pyridoxal phosphate and other carbonyl compounds as cofactors. Oxford, England: Pergamon Press; 1991. pp. 229–231. [Google Scholar]
- 27.Uo T, Yoshimura T, Shimizu S, Esaki N. Occurrence of pyridoxal 5′-phosphate-dependent serine racemase in silkworm. Bombyx mori. Biochem Biophys Res Commun. 1998;246:31–34. doi: 10.1006/bbrc.1998.8561. [DOI] [PubMed] [Google Scholar]
- 28.Veenhuis M, Hoogkamer-Te Niet M C, Middelhoven W J. Biogenesis and metabolic significance of microbodies in urate-utilizing yeasts. Antonie Leeuwenhoek. 1985;51:33–43. doi: 10.1007/BF00444226. [DOI] [PubMed] [Google Scholar]
- 29.Walsh C T. Enzymes in the d-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem. 1989;264:2393–2396. [PubMed] [Google Scholar]
- 30.Wasserman S A, Walsh C T, Botstein D. Two alanine racemase genes in Salmonella typhimuriumthat differ in structure and function. J Bacteriol. 1983;153:1439–1450. doi: 10.1128/jb.153.3.1439-1450.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe A, Kurokawa Y, Yoshimura T, Kurihara T, Soda K, Esaki N. Role of lysine 39 of alanine racemase from Bacillus stearothermophilusthat binds pyridoxal 5′-phosphate. Chemical rescue studies of Lys39 → Ala mutant. J Biol Chem. 1999;274:4189–4194. doi: 10.1074/jbc.274.7.4189. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe A, Yoshimura T, Mikami B, Esaki N. Tyrosine 265 of alanine racemase serves as a base abstracting alpha-hydrogen from l-alanine: the counterpart residue to lysine 39 specific to d-alanine. J Biochem (Tokyo) 1999;126:781–786. doi: 10.1093/oxfordjournals.jbchem.a022517. [DOI] [PubMed] [Google Scholar]
- 33.Wild J, Klopotowski T. d-Amino acid dehydrogenase of Escherichia coliK12: positive selection of mutants defective in enzyme activity and localization of the structural gene. Mol Gen Genet. 1981;181:373–378. doi: 10.1007/BF00425614. [DOI] [PubMed] [Google Scholar]
- 34.Wild J, Hennig J, Lobocka M, Walczak W, Klopotowski T. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coliK12. Mol Gen Genet. 1985;198:315–322. doi: 10.1007/BF00383013. [DOI] [PubMed] [Google Scholar]
- 35.Wolosker H, Sheth K N, Takahashi M, Mothet J P, Brady R O, Jr, Ferris C D, Snyder S H. Purification of serine racemase: biosynthesis of the neuromodulator d-serine. Proc Natl Acad Sci USA. 1999;96:721–725. doi: 10.1073/pnas.96.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolosker H, Blackshaw S, Snyder S H. Serine racemase: a glial enzyme synthesizing d-serine to regulate glutamate-N-methyl-d-aspartate neurotransmission. Proc Natl Acad Sci USA. 1999;96:13409–13414. doi: 10.1073/pnas.96.23.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yurimoto H, Hasegawa T, Sakai Y, Kato N. Physiological role of the d-amino acid oxidase gene, DAO1, in carbon and nitrogen metabolism in the methylotrophic yeast Candida boidinii. Yeast. 2000;16:1217–1227. doi: 10.1002/1097-0061(20000930)16:13<1217::AID-YEA616>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]