Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein (CRISPR/Cas) system has altered life science research offering enormous options in manipulating, detecting, imaging, and annotating specific DNA or RNA sequences of diverse organisms. This system incorporates fragments of foreign DNA (spacers) into CRISPR cassettes, which are further transcribed into the CRISPR arrays and then processed to make guide RNA (gRNA). The CRISPR arrays are genes that encode Cas proteins. Cas proteins provide the enzymatic machinery required for acquiring new spacers targeting invading elements. Due to programmable sequence specificity, numerous Cas proteins such as Cas9, Cas12, Cas13, and Cas14 have been exploited to develop new tools for genome engineering. Cas variants stimulated genetic research and propelled the CRISPR/Cas tool for manipulating and editing nucleic acid sequences of living cells of diverse organisms. This review aims to provide detail on two classes (class 1 and 2) of the CRISPR/Cas system, and the mechanisms of all Cas proteins, including Cas12, Cas13, and Cas14 discovered so far. In addition, we also discuss the pros and cons and recent applications of various Cas proteins in diverse fields, including those used to detect viruses like severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). This review enables the researcher to gain knowledge on various Cas proteins and their applications, which have the potential to be used in next-generation precise genome engineering.
Keywords: CRISPR, Cas system, Genome engineering, DETECTR, SHERLOCK
Introduction
Genome editing or gene editing is a popular technology used in medicines, therapeutic drugs, infectious studies, and agricultural biotechnology. The genome-editing tools have been employed to study the precise function of a gene by cutting and altering at a programmed locus through insertion, deletion, or replacement of targeted bases. In the beginning, conventional gene-editing techniques like homologous recombination (HR) were utilized for gene inactivation, but the effectiveness of HR was extremely low with the labor-intensive process [1]. Later, targeted gene knock-down utilizing RNA interference (RNAi) technique has provided researchers with rapid and low-cost technology to silence the gene of interest to study its functions. However, it also could not completely knock-down the targeted sequence, which faced unpredictable off-target effects and delivered only temporary or partial inhibition of gene function [2].
The genome-editing technique needs programmable sequence-specific endonucleases to produce the site-specific single-stranded breaks (SSBs) or double-stranded breaks (DSBs) at the targeted site that allow the endogenous repair mechanisms to fill the breaks [3]. These breaks are fixed by either of the two major repair mechanisms, (1) homology-directed repair (HDR) and (2) non-homologous end-joining repair (NHEJ) [4]. To facilitate specific DNA breaks, various genome-editing tools are developed previously, such as meganucleases or homing endonucleases [5], zinc-finger nucleases (ZFNs) [6], and transcription activator-like effector nucleases (TALENs) [7]. But these tools demand laborious efforts for cloning and protein construction to make DSBs, which hinders these tools from routine applications of genome editing. In 2012, the researchers developed clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) system-based genome editing tools [8]. CRISPR/Cas system mediates diverse adaptive immune systems against phages or plasmids. Due to unique features of simplicity in design, cost-effectiveness, and labor intensity, the research community has immediately adopted the CRISPR/Cas system as a user-friendly and robust RNA-guided DNA targeting tool for genome editing in various species [8–11]. Based on the mechanism, the CRISPR system has been divided into two main classes (1 and 2) and six types (I-VI) [12]. In these, types I to III were extensively studied, whereas types IV and VI were recently discovered. Types I, II, and V cut DNA, type VI cleaves RNA, type III cleaves both DNA and RNA and the cleavage activity of type IV has not yet been identified [13].
Cas9 protein has been utilized for diverse applications such as fluorescent imaging, base-editing, and transcriptional activation, apart from targeted cleavage of dsDNA. Like Cas9, the Cas10 protein is also involved in various applications such as fluorescent imaging, base-editing, and RNA tracking. As an alternative to Cas9 and Cas10, the Cas12 protein enhanced genome-editing efficiency by targeting only T-rich motifs without utilizing tracrRNA. So Cas12 system has expanded editing applications such as base-editing and detecting transcriptional variations. Cas13 protein is also used for diverse applications such as imaging, base-editing, and detection of transcriptional variations. Recently, the Cas14 protein has advanced genome-editing efficiency without needing an adjacent protospacer motif (PAM) and performs transcriptional regression and base-editing. This review describes the Cas variants of two classes of CRISPR systems used for genome editing.
History of CRISPR/Cas System
A group of researchers detected CRISPRs by analyzing the alkaline phosphatase gene, which is liable for the isozyme conversion of alkaline phosphatase (iap) in the E. coli K-12 strain [14]. They identified a genomic region that contains a series of 32 nucleotides of distinctive sequences flanked by invariable palindromic repeats on the 3′ end of the iap gene [14]. Later, distinctive parallel sequences were found in the other E. coli strains and Enterobacteria (Shigella dysenteriae and Salmonella enterica) [15]. Similarly, during the study of Mycobacterium tuberculosis strains, researchers identified 36 bp repeats interspaced with unique spacers of 35–41 bp [16].
In subsequent works, the CRISPR array was found in archaea (Haloferax mediterranei and Streptococcus thermopile), and the same was identified in 90% bacterial and 40% archaeal genomes [17]. However, this odd genomic sequence first turned out to be the outline of the CRISPR array. Still, due to the absence of necessary genome sequence data, the biological function of CRISPR remained elusive. Jansen et al. in 2002 described the CRISPR-associated genes [18]. Following this, several CRISPR/Cas genes were identified. In 2005, spacer sequence was identified in many genomes, and unique spacer regions were found within the CRISPR array [19]. These results showed that CRISPR is an adaptive immune system to defend prokaryotic cells against phage infection through the RNA-guided process.
Classification of CRISPR/Cas System
The CRISPR system has been classified into two major classes. In the Class 1 system, the RNA-guided target cleavage needs several effector proteins, but the Class 2 system requires only one RNA-guided endonuclease to cleave the DNA sequences [12, 20]. The class 1 system of CRISPR is divided into three types I, III, and IV, and the Class 2 system is divided into types II, V, and VI [21, 22]. In the type I system, the CRISPR/Cas locus contains the Cas3 signature gene that encodes a large protein with a helicase to unwind DNA-DNA and RNA–DNA duplexes [23]. The type II locus encodes multidomain protein to target and cleave the dsDNA [8]. The type III CRISPR/Cas possesses the Cas10 signature gene, encodes a multidomain protein with palm domain to target, and cleaves ssDNA [24]. Type IV system contains CRISPR-associated splicing factor 1 (Csf1), which encodes a ribonucleic protein, but the detailed function of this system is yet to be identified [21]. Type V locus possesses Cas12 signature gene (known as CRISPR from Prevotella and Francisella 1 (Cpf1), C2c1 or C2c3 protein) encodes RuvC (an E. coli protein involved in DNA repair) domain, which cleaves both dsDNA or ssDNA [25]. Type VI contains Cas13 (C2c2) that encodes higher eukaryotes and prokaryotes' nucleotide-binding domain (HEPN), which cleaves ssRNA [26] (Table 1).
Table 1.
Classification of CRISPR/Cas system with their protein, target molecule, mechanism of spacer acquisition, processing of Pre-CRISPR, self-vs non-self-discrimination and their effectors with the name of isolated organism with references
| Class | Type | Protein | Target | Spacer acquisition strategy | Name of CRISPR/Cas system | Pre-CRISPR processing | Self vs nonself-discrimination | Effectors of CRISPR system | Host organism | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Class1 | I | Cas3 | ssDNA | Cas1/Cas2/ Cas4 | Cas7, Cas5, Cas8, and Cas3 | Cas6 | PAM | Cas3, Cascade, and crRNA | E. coli | [23] |
| III | Cas10 | ssDNA | Cas1/Cas2 | Cas7, Cas5, and Cas1 | Cas6 | CRISPR repeat | Cmr/Csm, crRNA, and Cas10 | S. epidermics | [24] | |
| IV | Csf1 | – | NA | Cas7, Cas5, and Csf1 | – | – | – | – | [21] | |
| Class2 | II | Cas9 | dsDNA | Cas1/Cas2/ Cas4 | Cas9 | RNase III, and tracrRNA | PAM | Cas9, tracrRNA, and crRNA | S. thermophilus and S. pyogenes | [8] |
| V | Cpf1 | ssDNA and dsDNA | Cas1/Cas2/ Cas4 | Cas12 | Cpf1 | PAM | Cpf1, crRNA and tracrRNA | F. novicida | [25] | |
| VI | C2c2 | ssRNA | Cas1/Cas2 | Cas13 | – | – | C2c1, and crRNA | – | [26] |
The CRISPR/Cas system uses three stages to defend against viruses or foreign genetic materials [27] (Fig. 1). In the first stage, protospacers are incorporated into the host CRISPR locus as spacers between crRNA repeats [28]; next, Cas proteins are expressed, a spacer is transcribed into pre-crRNA, and the pre-crRNA is cleaved by Cas proteins and becomes functional as a mature crRNA [28]. In the third stage, Cas protein recognizes the target with the help of crRNA and generates cleavage of the genome [28]. Many CRISPR systems act based on the presence of a sequence-specific PAM, which is adjacent to the crRNA-specific site within the target genome.
Fig. 1.
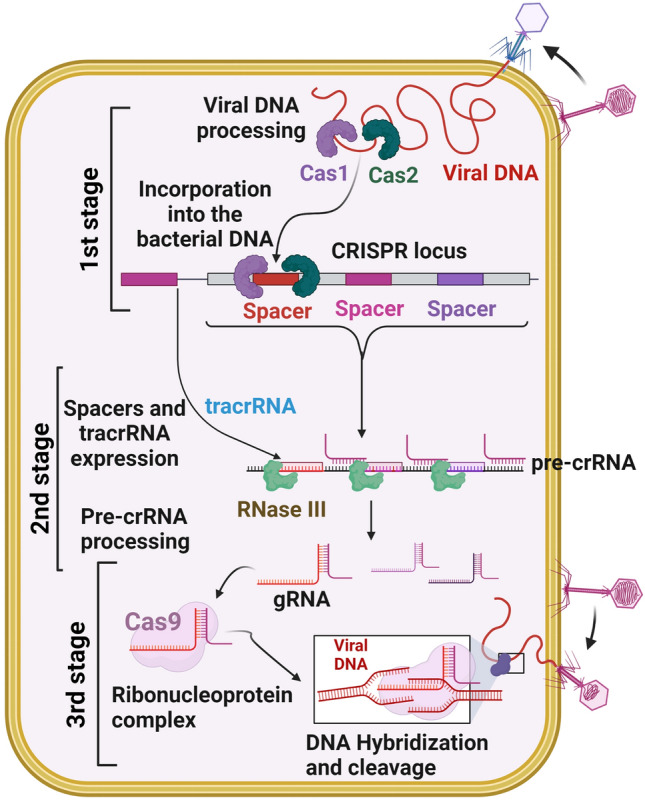
CRISPR/Cas adaptive immunity system. The three stages such as CRISPR adaptation (stage 1), CRISPR RNA biogenesis (stage 2), and CRISPR interference (stage 3) are schematically illustrated. During the adaption stage, the injection of genetic material (virus) into bacterial cells triggers the Cas1 and Cas2 adaption module proteins, which cleaves the invading sequences (spacers) and are then incorporated into the CRISPR array. During CRISPR/RNA biogenesis stage, the CRISPR array is transcribed into a precursor to crRNA molecules (pre-crRNA), which are then cleaved into mature crRNAs. These mature crRNAs form effector complexes with Cas proteins. When foreign genetic material sequences match a CRISPR spacer, the matching crRNA binds to the invading strand and cleaves the invading strand with the help of Cas nuclease (CRISPR interference stage)
CRISPR/Cas System
Scientists have discovered a novel microbial defense system that defends itself from viral and mobile genetic elements. One of the defense mechanisms found in bacterium and archaea is referred to as CRISPR/Cas system. By integrating DNA sequences into their genome identical to previous invaders, bacteria and archaea generate a cellular memory of them [29]. These acquired sequences permit them to spot viruses or mobile genetic invaders resulting in the degradation of the invading sequences and work as an adaptive immune system [29, 30].
CRISPR immunity is denoted by distinct phases. In the adaptation phase, bacterium and archaea gain cellular memory of the invading phages [30]. The phage genome sequences are integrated into the CRISPR locus of the bacterial or archaeal genomes, composed of 24 to 47 base pair (bp) repeats and separated by spacers. The unique loci of the CRISPR system were first discovered in 1987 [14]. Then Bolotin et al. in 2005 identified CRISPR’s function (enzymes encoding Cas genes generate DNA fragmentation) involved in protecting the bacteria and archaea from foreign invaders [31]. Later, scientists revealed that the CRISPR/Cas system works as an adaptive immune system [32] that was eventually adopted as a versatile system for RNA programmable genome editing [8].
Cas Proteins of the CRISPR System
Cas proteins have gained popularity among the research community for broader genome engineering applications and are currently used in diverse fields, including biotechnology, agriculture, and medical research. Recently discovered programmable Cas proteins like Cas 12, Cas 13, and Cas 14 have improved the precision of the CRISPR/Cas-mediated genome editing (Table 2). The mechanism of these different Cas proteins, pros and cons, and their applications are summarized in detail below.
Table 2.
Details on various CRISPR/Cas proteins with their host organism, sgRNA size, PAM sequence with their target molecule and cut site
| Protein name | Host organism | sgRNA sequence size | PAM sequence | Target | Cut site | References |
|---|---|---|---|---|---|---|
| Cas9 | S. pyogenes | 20 | 5ʹ-NGG-3ʹ | dsDNA | 5ʹ of PAM | [33] |
| Cas9 | S. pyogenes | – | 5ʹ-NAC, NTG, NTT, and NCG-3ʹ | DNA | 5ʹ of PAM | [34] |
| Cas9 | F. novicida | 20 | 5ʹ-NGG-3ʹ | DNA | 5ʹ of PAM | – |
| Cas9 | S. aureus | 21 | 5ʹ-NNGRRT-3ʹ | DNA | 5ʹ of PAM | [35] |
| Cas9 | Neisseria meningitidis | 24 | 5ʹ-NNNNGATT-3ʹ | DNA | 5ʹ of PAM | [35] |
| Cas9 | S. thermophilus | 20 | 5ʹ-NNAGAAW 5ʹ | DNA | 5ʹ of PAM | – |
| Cas9 | S. thermophilus | 20 | 5ʹ-NGGNG-3 | DNA | 5ʹ of PAM | [36] |
| Cas9 | Campylobacter jejuna | 22 | NNNNACAC and NNNRYAC | DNA | 5ʹ of PAM | [37] |
| C2c1 | Alicyclobacillus acidoterrestris | 20 | T-rich PAM | DNA | 5ʹ of PAM | [38] |
| Cpf1 | Prevotella and Francisella | 20 | TTTV | DNA | 5ʹ of PAM | [25] |
| Cpf1 | Acidaminococcus sp. | 24 | 5ʹ-TTTN-3ʹ | DNA | 3ʹ of PAM | [39] |
| Cas12a | Acidaminococcus sp. | – | Thymine-rich PAM | DNA | 5ʹ of PAM | [37] |
| Cas13 | Lb | 28 | Non-G nucleotide at the 3ʹ protospacer flanking site (PFS) | ssRNA | – | [26] |
| Cas14 | Uncultivated archaea | – | – | ssDNA | – | [40] |
Cas1 and Cas2 Proteins
Cas1 and Cas2 are generally conserved proteins in the prokaryotic adaptive immune system [28]. CRISPR/Cas system consists of a CRISPR array containing the repeats (∼30 to 40 bp) separated by spacers, an adjacent module comprising Cas1 and Cas2, and the distinctive signature proteins [41].
Mechanism of Cas1 and Cas2 Proteins
Cas1 and Cas2 belong to the Type II CRISPR system found in E. coli [42]. E. coli Cas1-Cas2 complex mediates spacer acquisition in vivo; however, the molecular mechanism behind these proteins throughout immunity is unclear [43]. Cas1 and Cas2 proteins formed an integrase complex consisting of two distal Cas1 dimers bridged by a Cas2 dimer [44]. The Cas1 and Cas2 proteins bind to the pre-spacer like twin-folded DNA. The pre-spacer integrated the proximal leader region of the CRISPR array guided by the leader sequence with a pair of inverted repeats inside the CRISPR repeat [45].
Applications of Cas1 and Cas2 Proteins
Cas1 and Cas2 are the conserved proteins among all CRISPR/Cas systems. The molecular mechanism behind the Cas1 and Cas2 proteins is still unclear. Hence, researchers have not applied these proteins for genome-editing in various fields.
Pros and Cons of Cas1 and Cas2 Proteins
So far, no genome editing studies have been demonstrated via Cas1 and Cas2 proteins; therefore, significant merits and demerits of Cas1 and Cas2 proteins should be evaluated before applying these in diverse fields.
Cas9 Protein
Cas9 is a protein associated with the CRISPR adaptive immune systems of S. pyogenes and is referred to as SpyCas9 protein. The SpyCas9 protein had a large multifunctional domain of 1368 amino acids and acts as a DNA endonuclease in natural and artificial CRISPR/Cas systems.
Mechanism of Cas9 Protein
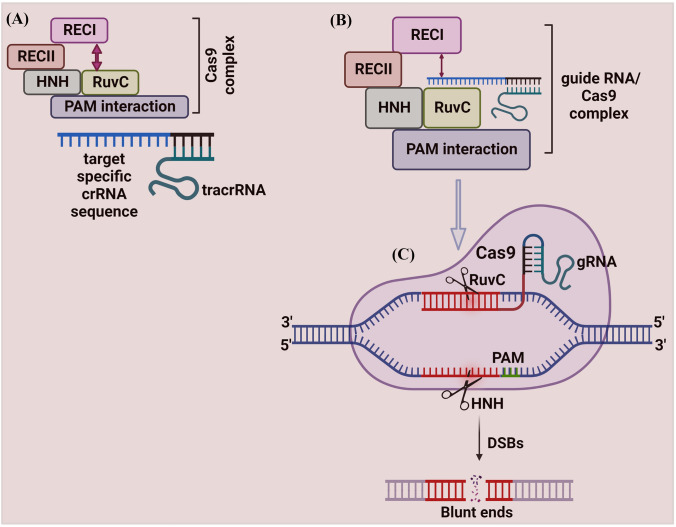
The mechanism of Cas9 protein has been studied extensively. Cas9 protein has six domains (1) Recognition lobe (REC I), (2) REC II, (3) Arginine-rich bridge helix, (4) PAM Interacting, (5) HNH, and (6) RuvC. REC I is the major domain responsible for binding with the gRNA; the REC II function is not studied [46]. The arginine-rich bridge helix initiates cleavage activity upon binding to targeted sequences. The interaction with PAM confers PAM specificity, which is responsible for binding with the target sequence. The HNH and RuvC are nuclease domains to chop the target sequence [46, 47] (Fig. 2).
Fig. 2.
Schematic illustration of CRISPR/Cas9 mechanism. A The Cas9 protein complex contains six domains (Recognition lobe (REC I), REC II, Arginine-rich bridge helix, PAM Interacting, HNH, and RuvC). REC I is the major domain responsible for binding with the gRNA, the REC II function is not studied. The arginine-rich bridge helix initiates cleavage activity upon binding to targeted sequences. The interaction with PAM confers PAM specificity, which is responsible for binding with the target sequence. The HNH and RuvC are nuclease domains to chop the target sequence. The Cas9 protein remains inactive due to the absence of gRNA. B The programmed gRNA binds to the Cas9 and generates changes in the protein, which leads the inactive Cas9 protein into its active form. Once triggered, it searches the target sequence by binding with a sequence that matches the PAM sequence (5′-NGG-3′). Then Cas9 generates DSBs at 3 bp upstream of the PAM using its HNH and RuvC domains
The Cas9 protein remains inactive due to the absence of gRNA. The engineered gRNA forms a T-shape involved in 1 tetra-loop and 3 stem-loops [8, 46]. The gRNA is programmed to own a 5′ end, which is complementary to the target sequence. The programmed gRNA binds to the Cas9 and generates changes in the protein, which leads the inactive Cas9 protein into its active form. Once triggered, it searches the target sequence by binding with a sequence that matches the PAM sequence (5′-NGG-3′). Then Cas9 cuts dsDNA at 3 bp upstream of the PAM using its HNH and RuvC domains. The HNH domain cleaves the DNA strand that is complementary to the 20-nucleotide sequence (gRNA) of crRNA (target strand)) and the RuvC domain cleaves the DNA strand opposite to the complementary strand (non-target DNA strand). The spyCas9 system for cleaving target DNA recognizes a short ‘seed’ sequence with the 5'-NGG-3' di-nucleotide containing PAM [8]. The twin tracrRNA: crRNA was fused into a single guide RNA (sgRNA), making the CRISPR/Cas9 system to cut the targeted dsDNA or ssDNA sequences [8].
Application of Cas9 Protein
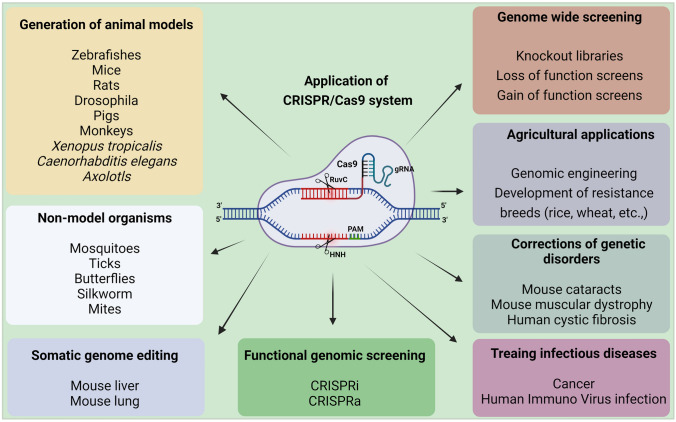
Cas9 protein holds great promise for efficient and targeted genome engineering applications in research, medicine, and biotechnology. The ease of genome modification in many species by simply designing a gRNA sequence enables large-scale genome editing experiments to probe genome function more than traditional gene editing techniques like TALEN and ZFN. Further, the Cas9 protein is also converted into an RNA-guided homing device (dCas9) by inactivating the nuclease domains to slow down the transcription. Using effector fusion (Cas9 proteins or sgRNA) to alter transcription states of specific genomic loci, or rearrange the genome, can significantly expand the genome engineering modification utilizing the CRISPR/Cas9 system. CRISPR/Cas9 system has been widely adopted by researchers and applied in diverse fields, including microbes [48], plants [49–51], animals [52], insects [53, 54], and human cell lines [55] (briefly explained in Fig. 3).
Fig. 3.
Applications of CRISPR/Cas9 system. CRISPR/Cas9 system has revolutionized genome engineering: its accuracy, rapidity, and affordability permit its use in a nearly limitless range of applications. Since its discovery, researchers have been using the CRISPR/Cas9 system to cure diseases, discover new treatments, and for precision medicine. It does not stop there; beyond treating human diseases, CRISPR/Cas9 is also being utilized for studying the model and non-model insects’ biology, somatic genome editing, manufacturing biofuels, and engineering better crops (rice, wheat, etc.), etc. These advances made possible by the invention of the CRISPR/Cas9 system will change the lives of people globally
Pros and Cons of Cas9 Protein
The advantage of the CRISPR/Cas9 system is the design simplicity with more efficiency than existing ZFN and TALEN systems [8]. Multiplexed genome editing is another significant advantage of Cas9, which can be achieved by designing multiple sequence-specific gRNAs simultaneously [8].
Despite numerous advances, CRISPR/Cas9 system faced several disadvantages and also opened numerous queries about risks associated with editing. One example is the gRNA, which guides the Cas9 to cleave dsDNA or ssDNA showed higher on- and off-target mutations in the targeted organisms [9, 55]. Even the finest accessible CRISPR/Cas system that uses HDR also induces undesirable mutations. However, these off-target effects can be reduced by utilizing the modified Cas9 version called null Cas9 (nCas9), which generates a nick in only one strand than the DSBs [55]. However, 100% off-targets cannot be reduced using nCas9, which requires upgraded Cas9 versions in the future.
Cas12 Protein
Cas12 is a versatile protein with more dynamic applications, including epigenome editing. The Cas12 protein belongs to the type V CRISPR system [56]. The Cas12 protein had recently emerged as an effector RNA-guided DNA endonuclease that becomes an alternative to the Cas9 protein for genome editing [25]. The Cas12 protein was isolated from the Acidaminococcus species (AsCas12a) and Lachnospiraceae bacterium (LbCas12a) that fight against invading viruses. Cas9 protein discriminates more accurately against mismatches within the initial ~ 10 bp of the RNA–DNA helix proximal to the PAM sequence [25]. Moreover, Cas12 isn’t a bit like the Cas9 protein; as a result, this protein can process the precursor crRNA by itself, which doesn’t require tracrRNA or RNase III. This process enhanced researchers to use Cas12 protein for multiplex genome editing [25].
Mechanism of Cas12 Protein
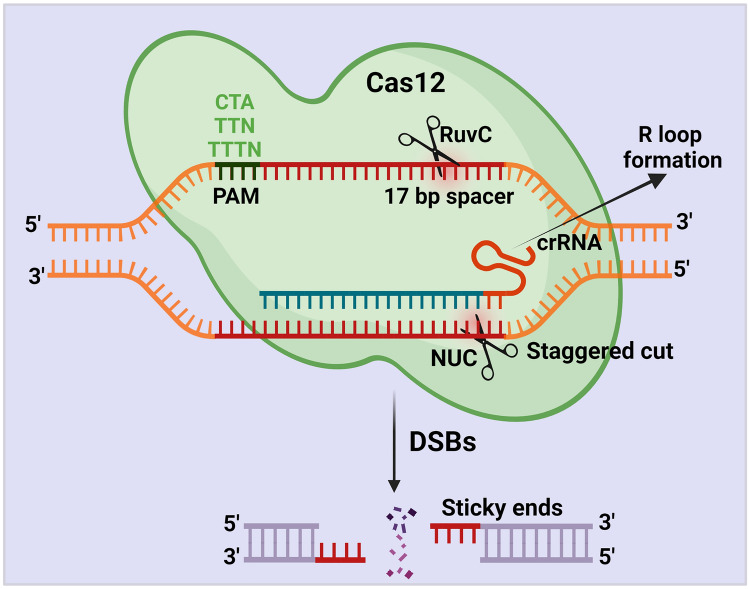
The Cas12 protein requires only the crRNAs to create an efficient cut at ssDNA and dsDNA. Cas12 protein contains the RuvC and nuclease lobe (NUC) domains for cleavage activity [56]. Like Cas9, Cas12 encounters a potential target site beside a PAM sequence. Once Cas12 starts encountering, it initiates R-loop, which forms base-pair hybridization between the crRNA and the target DNA strand. During this step, Cas12 matches the < 17 bp of the target sequence and leads to an R-loop formation [47]. Once R-loop is formed, the Cas12 protein uses its active RuvC domain to cleave the non-target strand with the help of the PAM sequence [56] (Fig. 4). However, the function of the RuvC domain of Cas12 protein in cleaving the targeted DNA strand is not yet clearly studied.
Fig. 4.
Schematic illustration of CRISPR/Cas12 mechanism. The Cas12 protein requires only the crRNAs to generate DSBs. Cas12 protein cleaves the target region beside a PAM sequence (CTA, TTN, TTTN) with the help of the RuvC and nuclease lobe (NUC) domains. Once Cas12 starts encountering, it initiates R-loop, which forms base-pair hybridization between the crRNA and the target DNA strand. During this step, Cas12 matches the < 17 bp of the target sequence and leads to an R-loop formation. Once R-loop is formed, the Cas12 protein uses its active RuvC domain and generates a staggering cut in the non-target strand with the help of the PAM sequence
Application of Cas12 Protein
A more concise system, CRISPR/Cas12 cleaves, dsDNA or ssDNA via the RuvC domain and does not require tracrRNA. Recently, Doudna’s group introduced a novel CRISPR/Cas diagnostic tool termed DNA endonuclease-targeted CRISPR trans-reporter (DETECTR) based on Cas12 protein (Fig. 5b) [57]. This DETECTR technique uses type V enzyme to cleave ssDNA sequence in a three-stage process: (1) Cas12a protein and target crRNA are complemented to the DNA reporter probes; once crRNA recognizes its target sequence through Cas12a protein, the Cas12a protein turns on collateral action and cleaves the target ssDNA or dsDNA. (2) Target DNA probes bind with fluorophores and a quencher molecule. (3) Degradation of the DNA probes releases fluorophore and quencher, producing a robust fluorescent signal for detecting targeted ssDNA or dsDNA cleavage [58]. In addition, this system detected even a single molecule of viral particles (Fig. 5b). For instance, Chen et al. (2018) used the DETECTR technique and detected Human Papilloma Viruses (HPV) [57]. In brief, they combined the non-specific ssDNA of Cas12 with DETECTR and differentiated several HPV16 and HPV18 strains from crude DNA extracts of clinical samples within 1 h. Furthermore, researchers detected African Swine Fever Virus (ASFV) by employing the CRISPR/Cas12 system [59]. They combined a fluorescent-based point of care (POC) system with CRISPR/Cas12 and detected ASFV within 2 h. From this result, they reported that the CRISPR/Cas12 is very specific and can detect even up to a single nucleotide of a targeted virus [59]. Recently, DETECTR is also utilized for detecting novel severe acute respiratory syndrome coronavirus-2 (SARS-Cov-2). Mammoth Biosciences, Inc. targeted two Nucleocapsid (N) and Envelope (E) genes of SARS-Cov-2 and observed faster virus detection within 1 h [60]. In brief, they generated Cas12-gRNAs to specifically target the SARS-CoV-2 and optimized the DETECTR assay for E and N genes. They utilized this upgraded DETECTR assay and detected the SARS-CoV-2 on a lateral flow strip within 1 h. Similar to this, Argentina and CASPR Biotech used a quick and portable SARS-CoV-2 diagnostic method based on CRISPR/Cas12 system [61]. They gathered saliva samples from COVID-19 patients and reported that the naturally occurring proteins in saliva had no inhibitory effects on the CRISPR/Cas12-based paper strip experiment [61]. Additionally, a Chinese institution used the CRISPR/Cas12-based DETECTR system to confirm a clear detection of SARS-CoV-2 [62]. They engineered gRNA that targeted the orf1a, orf1b, N, and E genes of the SARS (Wuhan-Hu-1 strain) and related viruses, in which they detected single nucleotide polymorphisms (SNPs). From these results, researchers stated that CRISPR/Cas12-based detection could be employed for the efficient and rapid diagnosis of COVID-19. Jiang et al. (2021) recently developed a magnetic-pull-down-assisted colorimetric (M-CDC) technique coupled to the CRISPR/Cas12 system to detect SARS-CoV-2. They used gold nanoparticle (AuNP) probes for this technique to detect SARS-CoV-2. Additionally, they screened 41 viral samples and reported that M-CDC is a useful technique for screening SARS-CoV-2 variants without requiring advanced instruments [63]. These applications of Cas12 nuclease enhanced scientists to increase the scope of genome-editing in various fields, including in diagnosing novel viruses [64–66].
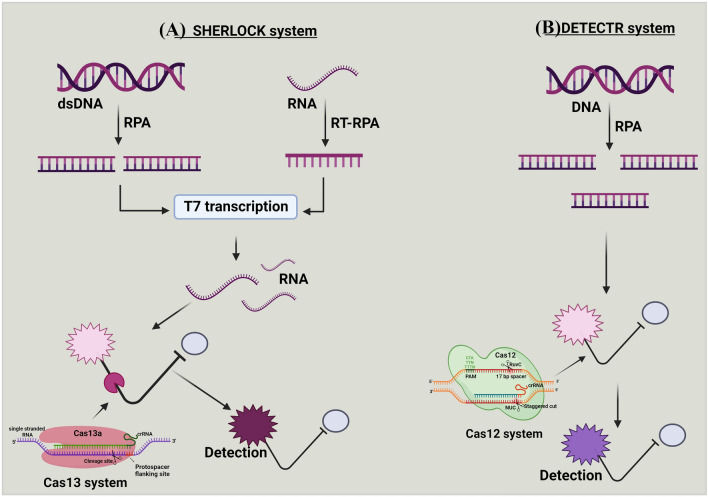
Fig. 5.
a and b Mechanism of SHERLOCK and DETECTR systems. (A) Targeted double-stranded DNA (dsDNA) or RNA is amplified with recombinase polymerase amplification (RPA) or reverse transcription (RT)-RPA. The RPA is coupled with T7 transcription to covert targeted RNA for detection by Cas13 system. This amplification step with the combination of reporter probe, enable specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) to detect the targeted sequence, (B) In DNA endonuclease-targeted CRISPR trans reporter (DETECTR), DNA is amplified with RPA. The Cas12 system pairs with the single-stranded DNA (ssDNA) of interest, and the DNase activity of Cas12 system is initiated. This amplification step, combined with the reporter probe, enables DETECR to detect the targeted sequence
Pros and Cons of Cas12 Protein
Like Cas9, Cas12 protein was also considered as a sole member of the CRISPR family for genome editing. But in most circumstances, Cas12 was deemed superior to Cas9 protein because Cas12 protein generates staggered DSBs and promotes HDR repair mechanism instead of both NHEJ & HDR [67]. Cas12 system also overcomes disadvantages associated with diagnostic strategies. For example, diagnosing SARS-COV-2 utilizing quantitative qRT-PCR demands a longer time to get the results [66]. But the CRISPR/Cas12-based DETECTR detects the SARS-CoV-2 within 1 h [66]. These results proved the advantage of the CRISPR/Cas12 system, which can also be utilized in detecting newly emerging viruses in the future.
The rapid advancement of the CRISPR-Cas12 system for genome editing has proved revolutionary for the life sciences. Despite this technology's wide application areas, the CRISPR/Cas12 system faces several significant flaws. For example, the CRISPR/Cas12 system is dependent on host cell DNA repair machinery with or without the presence of a template [68]. Although this system has been successfully used to acquire accurate DNA insertion into the desired genomic loci, its effectiveness varies depending on the cell type. DNA repair through HDR is also associated with active cell division, making these tools ineffective in cell division (for example, neurons) [69]. However, significant continuing research aims to tailor the Cas12 system further to ensure accurate DNA insertion into the targeted genome. Apart from this drawback, this system has a wide range of applicability, and ongoing endeavors are striving to generate enhanced CRISPR/Cas12 for robust genome engineering.
Cas13 Protein
The most recently discovered Cas protein is Cas13. CRISPR/Cas13 system functions as an ‘adaptive’ immune system in archaea and bacteria to defend against the invading RNA elements [70]. The Cas13 protein family contains two subtypes: (1) Cas13a protein from Leptotrichia shahii bacterium (LshCas13a), which is formally known as C2c2 and belongs to type VI, and (2) Cas13b from Prevotella sp. (PspCas13b) belongs to the type III CRISPR/Cas system. This system targets and cleaves only the ssRNA, not the ssDNA or dsDNA [71].
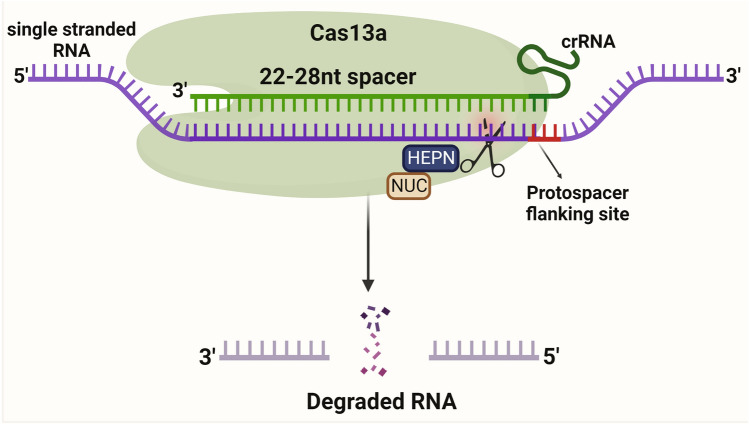
Mechanism of Cas13a Protein
Cas13a protein is activated through a single crRNA, like Cas12 protein from pre-crRNA processing. Cas13a protein comprises crRNA, NUC lobes, and two nucleotide-binding (HEPN) RNase domains for targeting RNA (Fig. 6). The LshCas13a cleaves ssRNA, upon recognizing the target sequence (22–28 nt) complementary to the crRNA spacer [72, 73]. The target sequence is flanked by a protospacer-flanking site (PFS) at the 3′-end, which has a bias to adenosine (A), uracil (U), and cytosine(C). LshCas13a and crRNA bind together and cleave the target region of ssRNA without the tracrRNA.
Fig. 6.
Schematic illustration of CRISPR/Cas13a mechanism. Cas13a protein is activated through a single crRNA. Cas13a protein comprises crRNA, NUC lobes, and two nucleotide-binding (HEPN) RNase domains for targeting RNA. The Cas13a cleaves ssRNA, upon recognizing the target sequence (22–28 nt) complementary to the crRNA spacer. The target sequence is flanked by a protospacer-flanking site (PFS) at the 3′-end and crRNA binds together and cleaves the target region of ssRNA without the tracrRNA
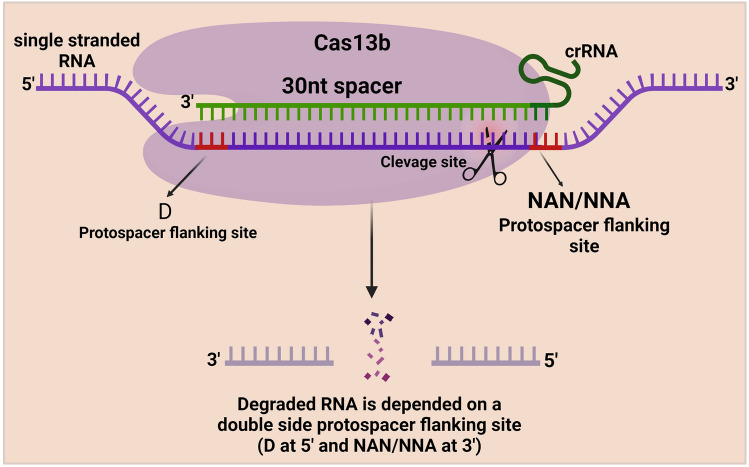
Mechanism of Cas13b Protein
The Cas13b protein is more precise than the Cas13a protein since a PFS flanks RNA targeting with A, U, or G at the 5′ end and PAM (NAN/NNA) at the 3′ end [74]. Cas13b protein is associated with the mature crRNA. ÇRISPR/Cas13b complex searches for the target ssRNA and induces conformational changes at the ssRNA target, resulting in nonspecific RNA cleavage [75]. However, the mechanism behind the Cas13b protein is not fully revealed, but scientists tested the ability of the cas13b protein for RNA editing (Fig. 7).
Fig. 7.
Schematic illustration of CRISPR/Cas13b mechanism. Cas13b protein is associated with the mature crRNA. This ÇRISPR/Cas13b complex searches for the target ssRNA and induces precise conformational changes at the ssRNA target with the help of the Protospacer flanking site (PFS), which flanks RNA targeting at the 5′ end and PAM sequence (NAN/NNA) at the 3′ end, resulting in nonspecific RNA cleavage
Application of Cas13 Protein
Cas13 proteins had wider application in various genome-editing and diagnostic fields. Beyond other applications like base editing, the Cas13a is also utilized for single-nucleotide detection at any site on the target sequence [25]. Like the DETECTR system, which depends on the activity of Cas12 protein, the specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) technique depends on Cas13 protein (Fig. 5a) [76, 77]. The SHERLOCK system works by integrating the targeted RNA fragments with Cas13 crRNA and fluorescent RNA probes. If the corresponding target sequence exists in the sample, Cas13 recognizes it via crRNA and cleaves fluorescent DNA probes, influencing fluorophore and quencher. This results in detecting the target via a fluorescent signal [73]. In addition, while SHERLOCK combined with reverse transcription-recombinase polymerase amplification (RT-RPA) or isothermal-RPA, the crRNA-Cas13a complex binds and cleaves a specific target sequence with high specificity [72]. SHERLOCK is used for various functions, including RNA detection, sensitive detection of nucleic acid contamination, and general RNA/DNA quantitation of specific qPCR assays [77].
Additionally, Cas13 nuclease could potentially track the allele-specific expression of transcripts or disease-associated mutations in cells. These attractive features opened new avenues to discriminate single nucleotide changes with high sensitivity in many organisms, including viruses (Fig. 5a). For instance, Gootenberg et al. (2018) diagnosed E. coli and Pseudomonas aeruginosa with the DNA isolated from these strains using the SHERLOCK system. They have also distinguished the isolates of Klebsiella pneumonia with two types of resistance genes, such as K. pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase 1 [70]. Furthermore, SHERLOCK differentiated African and American strains of the zika virus, the dengue virus (DENV) serotype, and cancer-related DNA targets. In addition, SHERLOCK detected Zika virus and DENV with higher sensitivity within 30 min. Researchers also developed a unique technique for detecting viral infection by combining SHERLOCK with heating unextracted diagnostic samples to obliterate nucleases (HUDSON) [75]. Within 2 hours, updated SHERLOCK identified DENV in a patient's saliva, blood, and serum. From these existing results, SHERLOCK Biosciences, Inc. and Mammoth Biosciences, Inc. used the SHERLOCK technique for the detection of SARS-CoV-2. They targeted S and Orf1ab genes and detected the SARS-Cov-2 RNA sequence within an hour. Metsky et al. (2020) also detected the SARS-CoV-2 RNA using the Cas13a proteins. They combined synthetic RNA targets with fluorescent visualizers and detected SARS-CoV-2-RNA [76]. To test the effectiveness of detecting SARS-CoV-2, Rauch et al. (2020) recently utilized the Cas13 based-Ragged-Equitable-Scalable Testing (CREST) method [77]. This technology uses portable and lightweight LED visualizers built with plastic filters. Researchers also used lateral flow immunochromatography strips to detect target RNAs, but these strips are expensive. Therefore, they created a P51 cardboard fluorescent visualizer, which was less expensive and could detect 10 copies of target RNA per millilitre. Additionally, they utilised the smartphone camera to capture data, which they then uploaded to the cloud to enable POC testing [77]. It improves nucleic acid detection based on CRISPR and demonstrates its power to change diagnostic and surveillance efforts by direct screening even on a vast population. Later, the CRISPR/Cas13a test based on single-molecule RNA diagnosis was developed by the Tian et al. (2020) group, which eliminated reverse transcriptase and nucleic acid extension procedures [78]. They stated that the SARS-CoV-2 would be easily detectable using the CRISPR/Cas13a test [78, 79]. These advantages of Cas13 protein enabled researchers to target any non-targetable sequences in diverse organisms [80–82].
Pros and Cons of Cas13 Protein
Like Cas9 and Cas12, CRISPR/Cas13 is also a robust, precise, and versatile RNA-targeting system, which opens novel research horizons in diverse fields. Compared with earlier RNA manipulation systems, CRISPR/Cas13 offers numerous advantages. For example, its modular construction, which consists of a single protein effector module and an RNA guide module, allows for significant scalability by enabling the production of whole libraries of various guide RNAs in addition to easy and quick design [83]. The recently discovered Cas13 mutant versions (dCas13, Cas13x), which function as programmable RNA-binding proteins, efficiently target different effectors to specific RNAs to induce specific mutations [83]. Due to the inherent crRNA biogenesis, multiple RNAs can be targeted using Cas13 precisely. Compared with RNAi, CRISPR/Cas13 mediated genome modifications are not limited to targeting cytoplasmic transcripts. In addition, Cas13 enables faster downregulation of gene expression by directly knocking out the cytoplasmic mRNA transcripts [83]. Recently, CRISPR/Cas13-based SHERLOCK and SHERLOCKv2 also played a vital role in developing novel molecular diagnostic tools to detect viruses, including SARS-CoV-2. Apart from these major advantages, CRISPR/Cas13 faced off-target mutations [55], which is the major drawback of this system. However, future research would help overcome this obstacle and aid in developing novel RNA knockdown approaches with more specificity and efficiency.
Cas14 Protein
Doudna’s group investigated other and similar types (Cas9, Cas12, and Cas13) of Cas systems that were available in nature [84]. They explored by creating a metagenomic database of the bacterial genome to search for uncharacterized Cas genes [84]. Surprisingly, they found Cas14 protein, which codes for smaller Cas protein with MW 40–70 kd. This Cas14 protein is extremely smaller (400–700 amino acids) than the other characterized Cas proteins [84]. Due to their small size, Doudna’s lab reported that Cas14 protein can target ssDNA without a PAM [84].
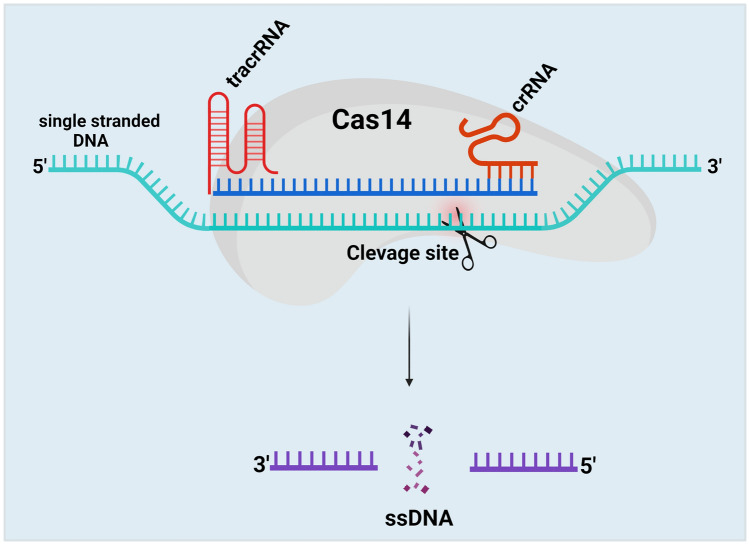
Mechanism of Cas14 Protein
This Cas14 cleaves ssDNA and confers immunity against viruses with ssDNA genomes or mobile genetic elements (MGEs) [84]. Cas14 protein recognizes the ssDNA, mediates seed sequence interaction with the target ssDNA, and cleaves the ssDNA, not dsDNA or ssRNA. Like Cas9, the Cas14 protein requires both tracrRNA and crRNA to target the ssDNA (Fig. 8). The cleavage efficiency of the Cas14 protein is more specific than Cas9, Cas12, and Cas 13 proteins without the presence of the PAM region [40]. Thus, this system meets all the criteria for high-fidelity genome editing.
Fig. 8.
Schematic illustration of CRISPR/Cas14 mechanism. The Cas14 protein comprises both tracrRNA and crRNA to target ssDNA. Cas14 protein recognizes the ssDNA with the help of tracrRNA and crRNA, mediates seed sequence interaction with the target ssDNA, and cleaves the ssDNA, not dsDNA or ssRNA. The cleavage efficiency of the Cas14 protein is more specific than Cas9, Cas12, and Cas 13 proteins without the presence of the PAM region
Application of Cas14 Protein
CRISPR/Cas14 system is now harnessed as superior to the Cas13 system. Researchers combined the CRISPR/Cas14 system with the DETECTR as a diagnostic approach for high-fidelity detection of ssDNA. Harrington et al. (2018) first used CRISPR/Cas14 system with the DETECTR technique. They designed gRNA targeting the HECT and RLD Domain Containing E3 Ubiquitin Protein Ligase 2 (HERC2) gene of human saliva samples from blue-eyed single nucleotide polymorphisms (SNP) individuals. They exhibited strong activation of recognition of the blue-eyed SNP by Cas14, while the Cas12 system failed to detect the blue-eyed SNP [40]. This result represents a cost-effective method for screening pathogenic mutations and provides a great opportunity for mapping the candidate genes associated with different pathogens. Another group proposes the Cas14 for viral diagnostic in combination with the simplified nucleic acid extraction, which does not involve complicated sample extraction like heating unextracted diagnostic samples to obliterate nucleases (HUDSON). The results reported that Cas14 could provide an advanced platform in the future for viral screening [72, 79]. This novel CRISPR/Cas14 system crosses criteria with user-friendly, sensitive, and more specific applications for future genome engineering. It can be harnessed for other purposes, including diagnostic applications like phylogenetic association (new viruses), epidemiology association with other pathogens, and taxonomic analysis [85].
Pros and Cons of Cas14 Protein
The Cas14 protein, which edits dsDNA, ssDNA, and RNA, holds various advantages over traditional Cas9 protein. For example, the Cas14 protein that has around 500 amino acid is very small, so this protein could be easily delivered to target any tissue than Cas9 protein [86]. Additionally, the Cas14 protein's selectivity improves the fidelity of single nucleotide polymorphism (SNP) [83]. Finally, the Cas14 protein has less limiting PAM requirements (uses only T-rich sequences) [83], allowing it to edit more targeted genomic sequences than the Cas9 protein. However, only a few studies have been demonstrated, which requires extensive studies to assist Cas14 in several fields of research in the future.
Conclusion and Future Perspectives
The exploitation of CRISPR/Cas tools made a breakthrough in genome engineering in the last few years. The discovery of Cas proteins has revolutionized natural science research and enabled advances in basic research, therapeutics, and diagnostics. The ease to use, the requirement of very little equipment, and the low cost have made many laboratories utilize these CRISPR/Cas-based systems to study the function of genes in various organisms. The initial discovery of the Cas9 protein, which shows its spotlight in the CRISPR system, has enhanced researchers to find different Cas variants like Cas12, Cas13, and Cas14 proteins. This research improvement in Cas proteins has provided exciting platforms in genome engineering, including detecting RNA viruses from plants, animals, and humans and treating infections. Several studies are currently underway to find novel Cas variants from nature. However, numerous aspects of the mechanism of Cas proteins still lack sufficient understanding. Therefore, understanding the molecular mechanism behind the Cas proteins, identification of PAM-less Cas proteins, and accurate targeting specificity to minimize off-target effects will increase the potential of utilizing Cas proteins for precise applications of genome engineering. In addition, identifying the uncharacterized advance Cas proteins that may still exist in many bacteria or archaea will revolutionize multiple fields, including diagnosing novel viruses, therapeutics, agriculture, breeding, etc. If these proteins are fully addressed with improved genome-editing abilities, it might kick-start new CRISPR “fever” offering influential and groundbreaking CRISPR technologies into mainstream use in the near future.
Author Contributions
All authors contributed to the study's conception and design. EHV and SAC wrote the article. EHV and SAC drew the images.
Funding
No funding was received for this article.
Declarations
Competing interest
The authors declare that there are no competing interests associated with the manuscript.
Ethical Approval
No ethics approval is needed for this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Capecchi MR. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nature Reviews Genetics. 2005;6(6):507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 2.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nature Reviews Genetics. 2002;3(10):737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 3.Rudin N, Sugarman E, Haber JE. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122(3):519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Letters. 2010;584(17):3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoddard BL. Homing endonuclease structure and function. Quarterly Reviews of Biophysics. 2005;38(1):49. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 6.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Qi Y. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nature Methods. 2011;8(1):67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nature Biotechnology. 2012;30(5):460. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceasar SA, Rajan V, Prykhozhij SV, Berman JN, Ignacimuthu S. Insert, remove or replace: A highly advanced genome editing system using CRISPR/Cas9. Biochimica et Biophysica Acta - Molecular Cell Research. 2016;1863(9):2333–2344. doi: 10.1016/j.bbamcr.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E. The biology of CRISPR-Cas: Backward and forward. Cell. 2018;172(6):1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Li Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Computational and Structural Biotechnology Journal. 2020;18:2401–2415. doi: 10.1016/j.csbj.2020.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Yakunin AF. Evolution and classification of the CRISPR–Cas systems. Nature Reviews Microbiology. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Wang L, Zou X, Duan S, Li Z, Deng Z, Chen S. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnology Advances. 2019;37(5):708–729. doi: 10.1016/j.biotechadv.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology. 1987;169(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakata A, Amemura M, Makino K. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. Journal of Bacteriology. 1989;171(6):3553–3556. doi: 10.1128/jb.171.6.3553-3556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermans PW, Van Soolingen D, Bik EM, De Haas PE, Dale JW, Van Embden JD. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infection and Immunity. 1991;59(8):2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojica FJM, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria. Molecular Microbiology. 2000;36(1):244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 18.Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 19.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Computational Biology. 2005;1(6):e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Current Opinion in Microbiology. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin EV, Makarova KS. Mobile genetic elements and evolution of CRISPR-Cas systems: All the way there and back. Genome Biology and Evolution. 2017;9(10):2812–2825. doi: 10.1093/gbe/evx192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends in Ecology & Evolution. 2008;23(10):578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Van Der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Regev A. RNA targeting with CRISPR–Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 29.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 30.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proceedings of the National Academy of Sciences. 2011;108(25):10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151(8):2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 32.Koonin EV, Wolf YI. Is evolution Darwinian or/and Lamarckian? Biology Direct. 2009;4(1):1–14. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of Molecular Evolution. 2005;60(2):174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 34.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nature Methods. 2013;10(11):1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. Journal of Bacteriology. 2008;190(4):1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim E, Koo T, Park SW, Kim D, Kim K, Cho H-Y, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim J-S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nature Communications. 2017;8(1):14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167(7):1814–1828.e12. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, Ishitani R, Zhang F, Nureki O. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165(4):949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Doudna JA. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17(6):904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 42.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nature Communications. 2012;3(1):1–7. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 43.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Binkowski A. A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Molecular Microbiology. 2011;79(2):484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Research. 2012;40(12):5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright AV, Doudna JA. Protecting genome integrity during CRISPR immune adaptation. Nature Structural & Molecular Biology. 2016;23(10):876. doi: 10.1038/nsmb.3289. [DOI] [PubMed] [Google Scholar]
- 46.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu R, Chen L, Jiang Y, Zhou Z, Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discovery. 2015;1(1):1–11. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Massel K, Godwin ID, Gao C. Applications and potential of genome editing in crop improvement. Genome Biology. 2018;19(1):1–11. doi: 10.1186/s13059-018-1586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hillary VE, Ceasar SA. Application of CRISPR/Cas9 genome editing system in cereal crops. The Open Biotechnology Journal. 2019;13(1):173–179. doi: 10.2174/1874070701913010173. [DOI] [Google Scholar]
- 51.Ceasar SA, Maharajan T, Hillary E, Krishna TPA. Insights to improve the plant nutrient transport by CRISPR/Cas system. Biotechnology Advances. 2022;59:107963. doi: 10.1016/j.biotechadv.2022.107963. [DOI] [PubMed] [Google Scholar]
- 52.Lee H, Yoon DE, Kim K. Genome editing methods in animal models. Animal Cells and Systems. 2020;24(1):8–16. doi: 10.1080/19768354.2020.1726462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hillary VE, Ceasar SA, Ignacimuthu S. Genome engineering via CRISPR-Cas9 system. Elsevier; 2020. Genome engineering in insects: focus on the CRISPR/Cas9 system; pp. 219–249. [Google Scholar]
- 54.Hillary VE, Ceasar SA. Genome engineering in insects for the control of vector borne diseases. Progress in Molecular Biology and Translational Science. 2021;179:197–223. doi: 10.1016/bs.pmbts.2020.12.017. [DOI] [PubMed] [Google Scholar]
- 55.Rodríguez-Rodríguez DR, Ramírez-Solís R, Garza-Elizondo MA, Garza-Rodríguez MDL, Barrera-Saldaña HA. Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases. International Journal of Molecular Medicine. 2019;43(4):1559–1574. doi: 10.3892/ijmm.2019.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532(7600):517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 57.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Horvath P. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nature Reviews Microbiology. 2020;18(2):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Q, Yu D, Bao M, Korensky G, Chen J, Shin M, Du K. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosensors and Bioelectronics. 2020;154:112068. doi: 10.1016/j.bios.2020.112068. [DOI] [PubMed] [Google Scholar]
- 60.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Sotomayor-Gonzalez A. CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Satyanarayana, M. (2020). A COVID-19 diagnostic that uses CRISPR gets a nod from the FDA chemical & engineering news.
- 62.Feng M, Li X. Land cover mapping toward finer scales. Science Bulletin. 2020;65(19):1604–1606. doi: 10.1016/j.scib.2020.06.014. [DOI] [PubMed] [Google Scholar]
- 63.Jiang Y, Hu M, Liu A-A, Lin Y, Liu L, Yu B, Pang DW. Detection of SARS-CoV-2 by CRISPR/Cas12a-enhanced colorimetry. ACS Sensors. 2021;6(3):1086–1093. doi: 10.1021/acssensors.0c02365. [DOI] [PubMed] [Google Scholar]
- 64.Bandyopadhyay A, Kancharla N, Javalkote VS, Dasgupta S, Brutnell TP. CRISPR-Cas12a (Cpf1): A versatile tool in the plant genome editing tool box for agricultural advancement. Frontiers in Plant Science. 2020;11:1589. doi: 10.3389/fpls.2020.584151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsukamoto T, Sakai E, Iizuka S, Taracena-Gándara M, Sakurai F, Mizuguchi H. Generation of the adenovirus vector-mediated CRISPR/Cpf1 system and the application for primary human hepatocytes prepared from humanized mice with chimeric liver. Biological and Pharmaceutical Bulletin. 2018;41(7):1089–1095. doi: 10.1248/bpb.b18-00222. [DOI] [PubMed] [Google Scholar]
- 66.Teng F, Li J, Cui T, Xu K, Guo L, Gao Q, Zhou Q. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biology. 2019;20(1):1–6. doi: 10.1186/s13059-019-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hillary VE, Ceasar SA. Prime editing in plants and mammalian cells: Mechanism, achievements, limitations, and future prospects. BioEssays. 2022;44:2200032. doi: 10.1002/bies.202200032. [DOI] [PubMed] [Google Scholar]
- 68.Hillary VE, Ignacimuthu S, Ceasar SA. Potential of CRISPR/Cas system in the diagnosis of COVID-19 infection. Expert Review of Molecular Diagnostics. 2021;21(11):1179–1189. doi: 10.1080/14737159.2021.1970535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Shayeb B, Sachdeva R, Chen LX, Ward F, Munk P, Devoto A, Amano Y. Clades of huge phages from across Earth’s ecosystems. Nature. 2020;578(7795):425–431. doi: 10.1038/s41586-020-2007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, Doudna JA. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu L, Li X, Wang J, Wang M, Chen P, Yin M, Wang Y. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168(1–2):121–134. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- 72.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Freije CA. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, Makarova KS. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Molecular Cell. 2017;65(4):618–630. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nature Protocols. 2020;15(3):1311. doi: 10.1038/s41596-020-0302-z. [DOI] [PubMed] [Google Scholar]
- 77.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nature Protocols. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian T, Shu B, Jiang Y, Ye M, Liu L, Guo Z, Zhou X. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano. 2020;15:1167. doi: 10.1021/acsnano.0c08165. [DOI] [PubMed] [Google Scholar]
- 79.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Parham LA. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Metsky HC, Freije CA, Kosoko-Thoroddsen TSF, Sabeti PC, Myhrvold C. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. BioRxiv. 2020 doi: 10.1101/2020.02.26.967026. [DOI] [Google Scholar]
- 81.Rauch S, Roth N, Schwendt K, Fotin-Mleczek M, Mueller SO, Petsch B. mRNA based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus neutralizing antibodies and mediates protection in rodents. bioRxiv. 2020;11:12. doi: 10.1038/s41541-021-00311-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Palaz F, Kalkan AK, Can O, Demir AN, Tozluyurt A, Özcan A, Ozsoz M. CRISPR-Cas13 system as a promising and versatile tool for cancer diagnosis, therapy, and research. ACS Synthetic Biology. 2021;10(6):1245–1267. doi: 10.1021/acssynbio.1c00107. [DOI] [PubMed] [Google Scholar]
- 83.Lotfi M, Rezaei N. CRISPR/Cas13: A potential therapeutic option of COVID-19. Biomedicine & Pharmacotherapy. 2020;131:110738. doi: 10.1016/j.biopha.2020.110738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Banfield JF. New CRISPR–Cas systems from uncultivated microbes. Nature. 2017;542(7640):237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nayak S, Blumenfeld NR, Laksanasopin T, Sia SK. Point-of-care diagnostics: Recent developments in a connected age. Analytical Chemistry. 2017;89(1):102–123. doi: 10.1021/acs.analchem.6b04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mahas A, Stewart CN, Jr, Mahfouz MM. Harnessing CRISPR/Cas systems for programmable transcriptional and post-transcriptional regulation. Biotechnology Advances. 2018;36(1):295–310. doi: 10.1016/j.biotechadv.2017.11.008. [DOI] [PubMed] [Google Scholar]