Abstract
Purpose
To inhibit the transmission of SARS-CoV-2, we developed engineered exosomes that were conjugated with anti-spike nanobodies and type I interferon β (IFN-β). We evaluated the efficacy and potency of nanobody-IFN-β conjugated exosomes to treatment of SARS-CoV-2 infection.
Methods
Milk fat globule epidermal growth factor 8 (MFG-E8) is a glycoprotein that binds to phosphatidylserine (PS) exposed on the exosomes. We generated nanobody-IFN-β conjugated exosomes by fusing an anti-spike nanobody and IFN-β with MFG-E8. We used the SARS-CoV-2 pseudovirus with the spike of the D614G mutant that encodes ZsGreen to mimic the infection process of the SARS-CoV-2. The SARS-CoV-2 pseudovirus was infected with angiotensin-converting enzyme-2 (ACE2) expressing adenocarcinomic human alveolar basal epithelial cells (A549) or ACE2 expressing HEK-blue IFNα/β cells in the presence of nanobody-IFN-β conjugated exosomes. By assessing the expression of ZsGreen in target cells and the upregulation of interferon-stimulated genes (ISGs) in infected cells, we evaluated the anti-viral effects of nanobody-IFN-β conjugated exosomes.
Results
We confirmed the anti-spike nanobody and IFN-β expressions on the exosomes. Exosomes conjugated with nanobody-hIFN-β inhibited the interaction between the spike protein and ACE2, thereby inhibiting the infection of host cells with SARS-CoV-2 pseudovirus. At the same time, IFN-β was selectively delivered to SARS-CoV-2 infected cells, resulting in the upregulation of ISGs expression.
Conclusion
Exosomes conjugated with nanobody-IFN-β may provide potential benefits in the treatment of COVID-19 because of the cooperative anti-viral effects of the anti-spike nanobody and the IFN-β.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11095-022-03400-0.
Keywords: anti-spike nanobody, engineered exosome, SARS-CoV-2, type I IFN
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a single-stranded RNA coronavirus that has threatened global public health since December 2019. The first step of SARS-CoV-2 infection stems from the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein (S-glycoprotein), which binds to its receptor, angiotensin-converting enzyme-2 (ACE2), on host cells [1]. Therefore, blocking the spike-ACE2 interaction is a crucial strategy for preventing SARS-CoV-2 infection, and anti-spike neutralizing antibodies serve as crucial agents for Covid-19 [2–4]. For instance, bamlanivimab and etesevimab are neutralizing anti-spike antibodies developed by AbCellera Biologics and Eli Lilly and have been used to treat COVID-19 [5]. In addition, FDA-approved COVID-19 vaccines produced by Pfizer-BioNTech and Moderna have been applied to individuals worldwide to produce neutralizing antibodies to prevent SARS-CoV-2 infection [6, 7].
In addition to conventional antibodies, a nanobody is a small (12-15 kDa) single domain antibody fragment that exposes a convex paratope that forms a prolate shape, allowing it to efficiently blocks the essential epitopes on pathogens that are inaccessible to conventional antibodies [8, 9]. In contrast, RNA viruses, such as SARS-CoV-2, typically have higher mutation rates that allow escape from neutralizing antibodies or nanobodies [10]. The current SARS-CoV-2 variants, such as the Delta variant and the Omicron variant, mutations occur on their spike protein and are more resistant to neutralization antibodies [11, 12]. Hence the anti-spike nanobodies or antibodies are insufficient to prevent the infection of SARS-CoV-2 variants as a monotherapy.
We hypothesized that the combining an innate anti-viral immune response with a neutralizing nanobody would enhance anti-SARS-CoV-2 immunity efficiently. Following viral infection, type I interferons (IFNs) are the first cytokines that produced by viral-infected cells. The secreted type I IFNs activate neighboring cells, leading to the induction of hundreds of IFN-stimulatory genes (ISGs), which limit viral replication [13]. Type I IFNs include IFN-α, IFN-β, IFN-ω, etc. Especially, IFN-β inhibits coronavirus replication more efficiently than IFN-α [14]. Furthermore, patients with COVID-19 treated with IFN-β were found in a study to have recovered rapidly [15].
In recent years, the use of newly developed nanomaterials for drug delivery has become widespread [16, 17]. Among nanoparticle materials, exosomes have recently been well engineered because of their low immunogenicity, high stability, and wide distribution throughout biological fluids [18–23]. For example, a recent study showed that extracellular vesicles expressing ACE2 blocked SARS-CoV-2 receptor-binding domain (RBD) more efficiently than soluble recombinant ACE2 and may be used as potential therapeutics for SARS-CoV-2 [24].
Here, we developed nanobody-IFN-β conjugated exosomes to inhibiting SARS-CoV-2 infection by combining the nanobody-mediated blocking effects with IFN-β-induced anti-viral immune response. In addition to inhibiting SARS-CoV-2 infection, nanobody-IFN-β conjugated exosomes also upregulated ISGs expression in infected cells, providing a superior anti-SARS-CoV-2 effects.
Materials and Methods
Cell Lines
Human embryonic kidney cells (HEK293T, Riken BRC), HEK-blue IFNα/β cells (InvivoGen, hkb-ifnab), and adenocarcinomic human alveolar basal epithelial cells (A549, ATCC, CCL-185) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco), supplemented with 10% heat-inactivated fetal calf serum (FCS, Gibco), 100U/ml penicillin, and 100 U/ml streptomycin (FUJIFILM Wako). The cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. To prepare exosome-free FCS, FCS was mixed with polyethylene glycol (PEG) 10,000 (Sigma-Aldrich) at a ratio of 5:1 and rotated at 4°C for 3 h. PEG was removed by centrifuging at 2,000 g for 20 min. The supernatant was filtered through a 0.22 µm filter and used as exosome-free FCS. The exosome-producing cell lines were cultured in advanced DMEM (Gibco) supplemented with 2% exosome-free FCS, 1nmol/L of sodium pyruvate (Nacalai Tesque), 100U/ml penicillin, and 100 U/ml streptomycin (FUJIFILM Wako).
Preparation of Plasmids
The nanobody-MFG-E8-IFN-β encoding pCAG vector was prepared using the Gibson assembly reaction. In particular, the FLAG sequence (DYKDDDDK) was inserted after the signal sequence, followed by an anti-spike nanobody (clone N112) fused to the N-terminus of mutated human MFG-E8 (D48E). IFN-β was fused to the C-terminus of human MFG-E8. A flexible linker (GGGGS) was used to connect among nanobody, MFG-E8, and IFN-β. The DNA sequences of the soluble anti-spike nanobody-IFN-β recombinant proteins were synthesized by Eurofins Genomics and inserted into the pCAG vector.
Preparation of Engineered Exosomes
4×106 293T cells were plated in a 10-cm-dish a day before transfection. Cells were transfected with 10 µg of the plasmid using polyethylenimine (PEI; Polysciences). The cell culture media was replaced with exosome-free medium 6 h after transfection, and cells were cultured in exosome-free medium for another 2 days to produce exosomes. To harvest the exosomes, the supernatant was collected and centrifuged at 300 g for 5 min to remove cell debris, followed by centrifugation at 2,000 g for 20 min to remove the cell debris and apoptotic bodies. The supernatant was centrifuged at 10,000 g for 30 min to remove apoptotic bodies and large extracellular vesicles. The supernatant was centrifuged at 100,000 g for 4 hours, and the pellet was washed with PBS and centrifuged at 100,000 g for 4 hours. The pellet was resuspended with PBS and used as engineered exosomes. The numbers and characteristics of engineered exosomes were quantified using NanoSight LM10 (Malvern Panalytical, Malvern, United Kingdom), and used for further experiments.
Preparation of the Soluble Anti-spike Nanobody-IFN-β Recombinant Proteins
The soluble anti-spike nanobody-IFN-β recombinant proteins were produced in 293T cells using transient transfection and purified using anti-DYKDDDDK tag antibody beads (FUJIFILM WAKO) as previously described [25]. Briefly, The soluble anti-spike nanobody-IFN-β recombinant proteins were captured with anti-FLAG tag antibody beads from the supernatant, washed, and competitively eluted using 150ug/ml FLAG peptide. The eluates were concentrated with Amicon Ultra centrifugal filter 3K (Millipore, Germany) to remove the residual FLAG peptide. LILRB3-FLAG was used for the control recombinant protein.
Bioassay of IFN-β
HEK-blue IFNα/β reporter cells were seeded in a 96-well plate at 5×104 cells/well density. Serial dilutions of recombinant human IFN-β (Abcam, ab71475) were added to create a standard curve. The cells were cultured with recombinant human IFN-β or the engineered exosomes or the soluble recombinant proteins for 24 h. The QUANTI-Blue solution was prepared according to the manufacturer’s instruction. The QUANTI-Blue solution was mixed with the supernatant and incubated at 37°C for 30min. The IFN-induced secreted embryonic alkaline phosphatase (SEAP) level was determined using spectrophotometer at 650 nm. All samples were performed in triplicates.
Flow Cytometry
APC-conjugated monoclonal antibodies (mAbs) to FLAG (clone: L5) and phycoerythrin-Cy7 (PE-Cy7)-conjugated mAbs to CD81 (clone: 5A6) were purchased from BioLegend. Alexa Fluor 488-conjugated mAbs to human ACE2 (clone: 535919) and APC-conjugated monoclonal antibodies (mAbs) to MFG-E8 (clone:278918) were purchased from R&D Systems. Staining for surface markers was performed using a PS Capture Exosome Flow Cytometry Kit (FUJIFILM Wako) according to the manufacturer’s instruction. Flow cytometric analysis was performed using a BD FACSCANTO II. Flowjo10 (Treestar) software was used for the analysis.
Generation of ACE2-expressing A549 Cells and HEK-blue IFNα/β Cells
The human ACE2 full-length sequence was synthesized by gene synthesis (Eurofins) and inserted into a pLJM1 lentiviral vector. The virus supernatant was prepared by transfecting 293T cells. 5×105 A549 cells were plated in a 6-well-plate one day before infection, and the lentivirus with 10 µg/ml polybrene was used to infect A549 cells or HEK-blue IFNα/β cells. The A549 cells or HEK-blue IFNα/β cells with high ACE2 expression were sorted using BD FACSMelody.
SARS-CoV-2 Infection Assay
The SARS-CoV-2 pseudovirus (Takara, 632673) encoding ZsGreen was prepared according to the manufacturer’s instruction. 4×104 ACE2-expressing A549 cells or 4×104 ACE2-expressing HEK-blue IFNα/β cells were plated in a 96-well-plate 24 h before infection. The SARS-CoV-2 pseudovirus were added to the well, then the engineered exosomes or the soluble recombinant proteins were mixed into the well. The applied amount of hIFN-β or mIFN-β on engineered exosomes or the soluble recombinant proteins was quantified by human IFN-β ELISA kit (Abcam) or LEGEND MAX mouse IFN-β ELISA kit (Biolegend). 2×109; 4×108; 8×107 particles of anti-spike nanobody-hIFN-β-conjugated exosomes contain 4.5ng, 1.1ng and 0.28ng of hIFN-β, respectively. The same amounts of IFN-β in the soluble recombinant proteins were used for the experiments. Two days after infection, the infection rate of SARS-CoV-2 pseudovirus was evaluated by the expression level of ZsGreen using flow cytometry.
Quantitative PCR
3,000 infected ACE2-A549 cells (ZsGreen+) and 3,000 non-infected cells (ZsGreen-) were sorted using a BD FACSMelody. mRNA was isolated using a NucleoSpin RNA PLUS XS kit (Takara, 740990.50). qPCR was performed using QuantStudio 3 (ThermoFisher) with the One-Step TB Green PrimeScript RT-PCR Kit II (Takara, RR086A). Primers used for qPCR were described elsewhere [26].
Results
Generation of Anti-spike Nanobody-IFN-β Conjugated Exosomes
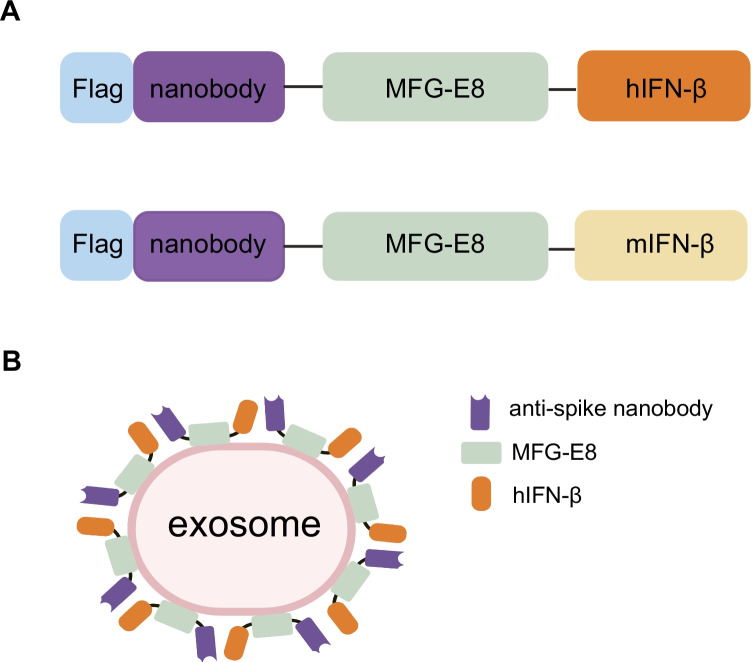
MFG-E8 is a secreted protein that binds phosphatidylserine (PS) on the surface of exosomes via its tandem discoidin-like domains (C1 and C2 domains) [27]. MFG-E8 contains an Arg-Gly-Asp (RGD) motif, that binds to αvβ3 integrin and promotes phagocytosis [28]. Since an RGD motif may shorten the half-life of MFG-E8 conjugated exosomes in vivo, we utilized the MFG-E8 (D48E) mutant [29], which does not bind to integrin. To create engineered exosomes that conjugated anti-spike nanobodies and human IFN-β (hIFN-β), we developed a fusion protein with MFG-E8 (Fig. 1). We selected a clone of anti-spike nanobody, N112, which was screened in a previous study as a lead anti-spike nanobody candidate with a high binding affinity [30]. FLAG sequence was fused to the N-terminal of the nanobody (Supplementary Fig. 1A, B). To evaluate the function of hIFN-β, we prepared an anti-spike nanobody-MFG-E8- mouse-IFN-β fusion protein as a control since mouse-IFN-β (mIFN-β) does not cross-react with the human IFN receptor. We transfected pCAG vectors that encoded fusion proteins into 293T cells and isolated engineered exosomes from the culture supernatant by serial centrifugation. As a control, we also purified the soluble anti-spike nanobody-IFN-β recombinant proteins (Supplementary Fig. 1C, D).
Fig. 1.
Schematic diagram of the engineered exosomes. (A) Schematic diagram of the fusion proteins. The anti-spike nanobody and either human IFN-β (hIFN-β) or mouse IFN-β (mIFN-β) were fused with MFG-E8. A FLAG sequence was inserted before the anti-spike nanobody. (B) Schematic diagram of the engineered exosomes. The anti-spike nanobody-hIFN-β fusion protein was conjugated with the exosomes via the C1C2 domains of MFG-E8.
Characteristics of Engineered Exosomes
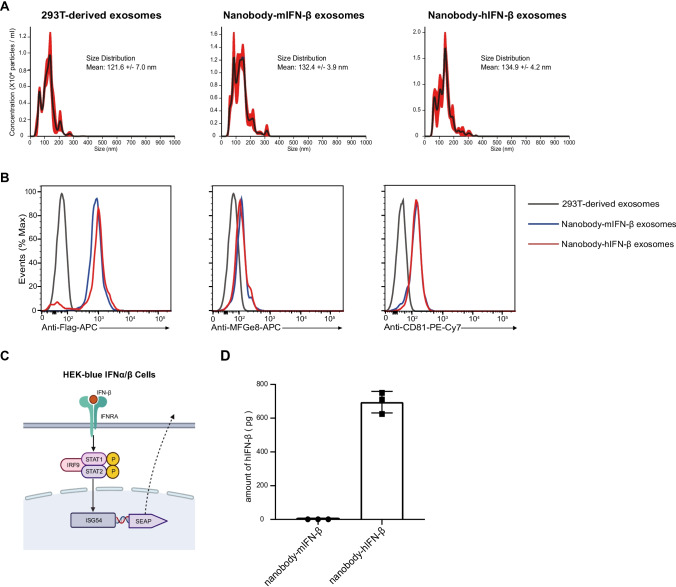
First, we performed a nanoparticle tracking analysis (NTA) to evaluate the size of engineered exosomes. We found that the engineered exosomes had a similar size distribution compared to the exosomes from 293T cells (Fig. 2A). To confirm the expressions of nanobody and IFN-β on the exosomes, we performed fluorescence-activated cell sorting (FACS) analysis using the PS Capture Exosome Flow Cytometry Kit. The expression of CD81 reflected the number of exosomes, allowing us to compare the expression levels of the nanobody-hIFN-β fusion protein and the nanobody-mIFN-β fusion protein on the exosomes. We observed the comparable expression of nanobody-hIFN-β and nanobody-mIFN-β on the exosomes (Fig. 2B). Next, we verified the function of hIFN-β on the exosomes. We performed a bioassay of hIFN-β on exosomes using HEK-blue type I interferon reporter cells. Reporter cells produce SEAP under the control of the IFN-α/β inducible ISG54 promoter (Fig. 2C). We stimulated the reporter cells with nanobody-hIFN-β or nanobody-mIFN-β exosomes for one day and evaluated the level of SEAP in the cell culture supernatant. The results showed that hIFN-β on exosomes stimulated the reporter cells (Fig. 2D). We also observed that the nanobody-mIFN-β exosomes did not stimulate reporter cells, indicating that mIFN-β did not cross-react with the human type I IFN receptor (Fig. 2D). The soluble anti-spike nanobody-hIFN-β recombinant protein but not anti-spike nanobody-mIFN-β recombinant protein also stimulated reporter cells (Supplementary Fig. 2).
Fig. 2.
Evaluation of the engineered exosomes. (A) The size distribution of isolated exosomes was analyzed by Nano Tracking Analysis (NTA) using NanoSight LM10. The red area surrounding the lines represents the standard error of the mean. (B) The fusion protein expression on the exosomes was analyzed with flow cytometry using the PS Capture Exosome Flow Cytometry Kit. Histograms show expressions of FLAG, MFG-E8 and CD81 on the exosomes. (C) Schematic diagram of the HEK-blue IFNα/β reporter cell assay. (D) The activity of hIFN-β on the engineered exosomes. 1×109 particles of engineered exosomes were applied to the HEK-blue IFNα/β cells. After 24 h, the IFN-induced secreted embryonic alkaline phosphatase (SEAP) level was determined using a spectrophotometer at 650 nm.
Anti-spike Nanobody-IFN-β Conjugated Exosomes Exerted a Potent Anti-viral Effect
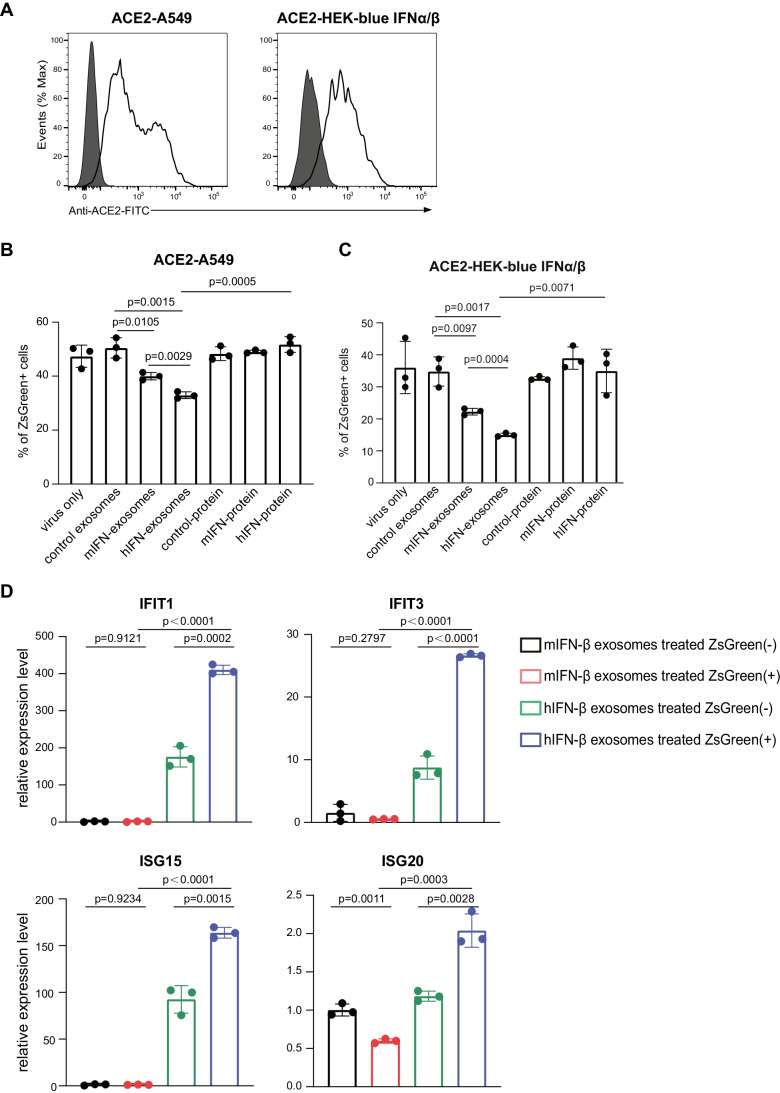
To evaluate whether anti-spike nanobody-hIFN-β-conjugated exosomes inhibit the infection of SARS-CoV-2, we used the SARS-CoV-2 pseudovirus with the spike of D614G mutant to mimic the infection process of SARS-CoV-2. We generated A549 cells and HEK-blue IFNα/β cells stably expressing ACE2 as target cells (Fig. 3A). We added the SARS-CoV-2 pseudovirus that encodes ZsGreen to the target cells, and then we mixed either nanobody-mIFN-β exosomes, nanobody-hIFN-β exosomes, or soluble anti-spike nanobody-IFN-β recombinant proteins. We observed that nanobody-mIFN-β exosomes partially blocked SARS-CoV-2 pseudovirus infection. In contrast, nanobody-hIFN-β conjugated exosomes showed a superior ability to inhibit viral infection (Fig. 3B, C and Supplementary Fig. 3). Note that anti-spike nanobody-IFN-β conjugated exosomes are much more effective in blocking SARS-CoV-2 pseudovirus infection than soluble anti-spike nanobody-hIFN-β recombinant protein. To further confirm whether hIFN-β was selectively delivered to SARS-CoV-2-infected cells, we sorted SARS-CoV-2 pseudovirus infected cells (ZsGreen+) and non-infected cells (ZsGreen-) and evaluated the expression of interferon stimulated genes (ISGs) family by quantitative PCR (qPCR). The qPCR results showed upregulation of ISGs mRNA in ZsGreen+ A549 cells supplemented with nanobody-hIFN-β conjugated exosomes (Fig. 3D). Upregulation of ISG mRNA was not observed in ZsGreen+ A549 cells that were supplemented with nanobody-mIFN-β conjugated exosomes. These results indicated that nanobody-hIFN-β conjugated exosomes blocked the entry of SARS-CoV-2 pseudovirus into the target cells, and additionally, hIFN-β signals enhanced the expression of ISGs to mount an anti-viral state (Fig. 4).
Fig. 3.
Anti-viral efficacy of engineered exosomes. (A) ACE2 was lentivirally infected to the A549 cells or HEK-blue IFNα/β cells.. The expression of ACE2 on A549 cells or HEK-blue IFNα/β cells were analyzed using flow cytometry. (B) Engineered exosomes or the soluble nanobody-IFN-β recombinant proteins containing 0.28ng of IFN-β were used to block the infection. The expression of ZsGreen in ACE2-A549 cells represents the infection of the SARS-CoV-2 pseudovirus. The percentage of ZsGreen positive cells was analyzed by flow cytometry. (C) The percentage of ZsGreen positive cells in ACE2- HEK-blue IFNα/β cells was analyzed by flow cytometry. (D) The ZsGreen positive cells and negative cells were sorted. The expressions of ISGs were analyzed using qPCR. The Mean ± SEM of triplicates experiments was calculated.
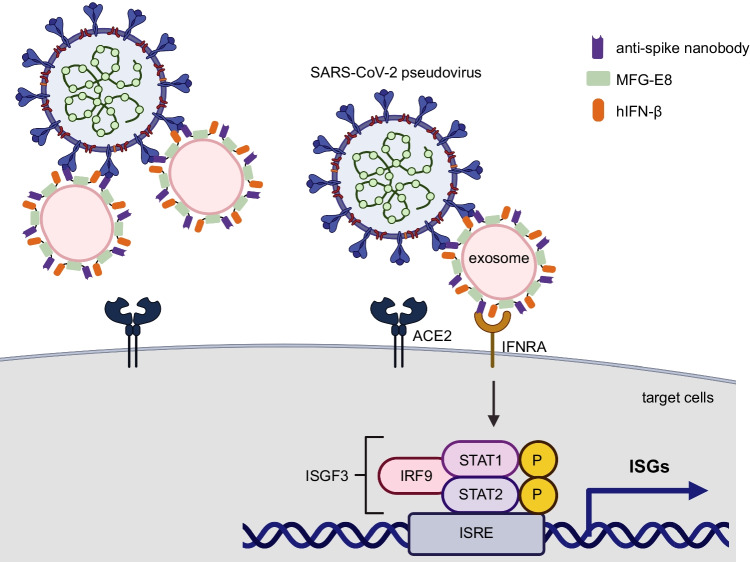
Fig. 4.
An illustration of how engineered exosomes play a dual role in protecting against viral infection. Exosomes conjugated with the anti-spike nanobody and IFN-β blocked the entry of the SARS-CoV-2 pseudovirus into the host cells. At the same time, IFN-β binds to the SARS-CoV-2 pseudovirus. Therefore, the host cells infected with the SARS-CoV-2 pseudovirus received IFN-β and upregulated ISGs, creating an anti-viral state.
Discussion
In this study, we developed the engineered exosomes that conjugated with anti-spike nanobodies and IFN-β. The engineered exosomes selectively delivered IFN-β to SARS-CoV-2 pseudovirus infected cells and upregulated ISGs, which in turn induced an anti-viral state.
It has been demonstrated that the non-structural protein (NSP) of SARS-CoV-2, such as NSP1, NSP3, ORF3, and ORF6, suppresses the transcription of ISGs, inhibiting the host’s type I interferon response [31–33]; however, an additional IFN-β treatment induces ISGs in the infected host cells. In addition, pre-exposure to IFN-β can inhibit SARS-CoV-2 replication [34], indicating that the timing of exposure to IFN-β is key to control SARS-CoV-2 replication. However, the systemic administration of IFN-β may cause a series of side effects, including flu-like symptoms, menstrual disorders, fatigue, or worsened depression or headache [35, 36]. Considering that our engineered exosomes selectively delivered IFN-β to the infected cells, it may be possible to control the replication of SARS-CoV-2 with less side effects.
It is well known that proteins of interest (POI) fused with the C1 and C2 domains of MFG-E8 can be expressed on the exosomes [37]. To selectively deliver IFN-β to target cells, we prepared the nanobody-MFG-E8-hIFN-β conjugated engineered exosomes and the soluble nanobody-hIFN-β recombinant protein. We found that engineered exosomes more efficiently blocked SARS-CoV-2 pseudovirus infection than the soluble nanobody-IFN-β recombinant proteins. The higher blocking efficacy of engineered exosomes is probably because several dozen nanobody-MFG-E8-hIFN-β are conjugated on a single exosome [37], so the engineered exosomes delivered a higher concentration of nanobody and IFN-β to the target cells. On top of that, the advantage of engineered exosomes over recombinant proteins is their convertibility [37]. For example, integrins expression has been reported to determine the orientation of the exosomes in specific organs [38]. Adding specific integrins to the engineered exosomes would enable delivery to the infection site. Furthermore, ribonucleases could be incorporated into the engineered exosomes so that the viral genome could be digested, or chemicals that inhibit the replication of the virus could be included. Although further studies are needed to evaluate the pharmacokinetics of nanobody-MFG-E8-hIFN-β conjugated exosomes in-vivo [18–22], it is possible that our engineered exosomes that block virus entry and selectively deliver IFN-β may be applicable to other viruses as well as variants of SARS-CoV-2.
Conclusion
Our engineered exosomes conjugated with multiple anti-spike nanobody-IFN-β fusion proteins induced dual anti-virus effects. The anti-spike nanobody on the exosomes blocked the entry of SARS-CoV-2 into the host cells. IFN-β signaling upregulates the expression of ISGs, which induces an anti-viral state. Since anti-spike monoclonal antibodies are insufficient to prevent the highly transmissible SARS-CoV-2 variants, our nanobody-hIFN-β conjugated exosomes may be a promising treatment for SARS-CoV-2 treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments and Disclosures
We thank M. Ueda for the technical assistance. We also thank T. Yoshida and K. Hirayasu for their helpful discussions. The authors declare no conflict of interest.
Author Contributions
X.L., T.Y., and S.I. performed the experiments. X.L., T.Y., and R.H. designed the experiments, analyzed the results, and wrote the paper.
Funding
This work was supported by the Japan Science and Technology Agency (JST) Precursory Research for Embryonic Science and Technology (PRESTO) (No. JPMJPR19HA to TY) and Core Research for Evolutional Science and Technology (CREST) from JST (No. JPMJCR18H4 to RH), and Science and Technology Platform for Advanced Biological Medicine from the Japan Agency for Medical Research and Development (No. 22am0401019h0004 to RH).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tomoyoshi Yamano, Email: tomoyoshi.yamano@med.kanazawa-u.ac.jp.
Rikinari Hanayama, Email: hanayama@med.kanazawa-u.ac.jp.
References
- 1.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, Robbiani DF, Nussenzweig MC, West AP, Jr, Bjorkman PJ. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, Cordon-Cardo C. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370(6521):1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo YM, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe JE., Jr Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584(7821):443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dougan M, Nirula A, Azizad M, Mocherla B, Gottlieb RL, Chen P, Hebert C, Perry R, Boscia J, Heller B, Morris J, Crystal C, Igbinadolor A, Huhn G, Cardona J, Shawa I, Kumar P, Adams AC, Van Naarden J, Custer KL, Durante M, Oakley G, Schade AE, Holzer TR, Ebert PJ, Higgs RE, Kallewaard NL, Sabo J, Patel DR, Dabora MC, Klekotka P, Shen L, Skovronsky DM. Bamlanivimab plus Etesevimab in mild or moderate covid-19. N Engl J Med. 2021;385(15):1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Jr, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC. Safety and Efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 9.Steeland S, Vandenbroucke RE, Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today. 2016;21(7):1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann M, Krüger N, Schulz S, Cossmann A, Rocha C, Kempf A, Nehlmeier I, Graichen L, Moldenhauer AS, Winkler MS, Lier M, Dopfer-Jablonka A, Jäck HM, Behrens GMN, Pöhlmann S. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185(3):447–456.e411. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1(6):519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monk PD, Marsden RJ, Tear VJ, Brookes J, Batten TN, Mankowski M, Gabbay FJ, Davies DE, Holgate ST, Ho LP, Clark T, Djukanovic R, Wilkinson TMA. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med. 2021;9(2):196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, Acosta-Torres LS, Diaz-Torres LA, Grillo R, Swamy MK, Sharma S, Habtemariam S, Shin HS. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnology. 2018;16(1):71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20(2):101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol Pharm Bull. 2008;31(6):1059–1062. doi: 10.1248/bpb.31.1059. [DOI] [PubMed] [Google Scholar]
- 19.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 20.Vella LJ, Greenwood DL, Cappai R, Scheerlinck JP, Hill AF. Enrichment of prion protein in exosomes derived from ovine cerebral spinal fluid. Vet Immunol Immunopathol. 2008;124(3–4):385–393. doi: 10.1016/j.vetimm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 22.Jafari D, Shajari S, Jafari R, Mardi N, Gomari H, Ganji F, ForouzandehMoghadam M, Samadikuchaksaraei A. Designer exosomes: a new platform for biotechnology therapeutics. BioDrugs. 2020;34(5):567–586. doi: 10.1007/s40259-020-00434-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunggulawa EJ, Wang W, Yin T, Wang N, Durkan C, Wang Y, Wang G. Recent advancements in the use of exosomes as drug delivery systems. J Nanobiotechnol. 2018;16(1):81. doi: 10.1186/s12951-018-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Shennawy L, Hoffmann AD, Dashzeveg NK, McAndrews KM, Mehl PJ, Cornish D, Yu Z, Tokars VL, Nicolaescu V, Tomatsidou A, Mao C, Felicelli CJ, Tsai CF, Ostiguin C, Jia Y, Li L, Furlong K, Wysocki J, Luo X, Ruivo CF, Batlle D, Hope TJ, Shen Y, Chae YK, Zhang H, LeBleu VS, Shi T, Swaminathan S, Luo Y, Missiakas D, Randall GC, Demonbreun AR, Ison MG, Kalluri R, Fang D, Liu H. Circulating ACE2-expressing extracellular vesicles block broad strains of SARS-CoV-2. Nat Commun. 2022;13(1):405. doi: 10.1038/s41467-021-27893-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahrini I, Hanayama R. Development of a method that delivers drugs to enveloped viruses. Biol Pharm Bull. 2019;42(6):977–981. doi: 10.1248/bpb.b18-01000. [DOI] [PubMed] [Google Scholar]
- 26.Jiao P, Fan W, Cao Y, Zhang H, Tian L, Sun L, Luo T, Liu W, Li J. Robust induction of interferon and interferon-stimulated gene expression by influenza B/Yamagata lineage virus infection of A549 cells. PLoS One. 2020;15(4):e0231039. doi: 10.1371/journal.pone.0231039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen MH, Graversen H, Fedosov SN, Petersen TE, Rasmussen JT. Functional analyses of two cellular binding domains of bovine lactadherin. Biochemistry. 2000;39(20):6200–6206. doi: 10.1021/bi992221r. [DOI] [PubMed] [Google Scholar]
- 28.Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417(6885):182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 29.Asano K, Miwa M, Miwa K, Hanayama R, Nagase H, Nagata S, Tanaka M. Masking of phosphatidylserine inhibits apoptotic cell engulfment and induces autoantibody production in mice. J Exp Med. 2004;200(4):459–467. doi: 10.1084/jem.20040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esparza TJ, Martin NP, Anderson GP, Goldman ER, Brody DL. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci Rep. 2020;10(1):22370. doi: 10.1038/s41598-020-79036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shemesh M, Aktepe TE, Deerain JM, McAuley JL, Audsley MD, David CT, Purcell DFJ, Urin V, Hartmann R, Moseley GW, Mackenzie JM, Schreiber G, Harari D. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoS Pathog. 2021;17(8):e1009800. doi: 10.1371/journal.ppat.1009800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, Guo F, Zhao Z, Zhou Z, Xiang Z, Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11(1):3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Ishida R, Strilets T, Cole J, Lopez-Orozco J, Fayad N, Felix-Lopez A, Elaish M, Evseev D, Magor KE, Mahal LK, Nagata LP, Evans DH, Hobman TC. SARS-CoV-2 nonstructural protein 1 inhibits the interferon response by causing depletion of key host signaling factors. J Virol. 2021;95(13):e0026621. doi: 10.1128/JVI.00266-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebendenne A, Valadão ALC, Tauziet M, Maarifi G, Bonaventure B, McKellar J, Planès R, Nisole S, Arnaud-Arnould M, Moncorgé O, Goujon C. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J Virol. 2021;95(8). [DOI] [PMC free article] [PubMed]
- 35.Walther EU, Hohlfeld R. Multiple sclerosis: side effects of interferon beta therapy and their management. Neurology. 1999;53(8):1622–1627. doi: 10.1212/WNL.53.8.1622. [DOI] [PubMed] [Google Scholar]
- 36.Neilley LK, Goodin DS, Goodkin DE, Hauser SL. Side effect profile of interferon beta-1b in MS: results of an open label trial. Neurology. 1996;46(2):552–554. doi: 10.1212/WNL.46.2.552. [DOI] [PubMed] [Google Scholar]
- 37.Corso G, Heusermann W, Trojer D, Görgens A, Steib E, Voshol J, Graff A, Genoud C, Lee Y, Hean J, Nordin JZ, Wiklander OPB, El Andaloussi S, Meisner-Kober N. Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule - single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J Extracell Vesicles. 2019;8(1):1663043. doi: 10.1080/20013078.2019.1663043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.