Abstract
Elongation factor 3 (EF3) is considered a promising drug target for the control of fungal diseases because of its requirement for protein synthesis and survival of fungi and a lack of EF3 in the mammalian host. However, EF3 has been characterized only in ascomycete yeast. In order to understand the role of EF3 in a basidiomycete yeast, we cloned the gene encoding EF3 from Cryptococcus neoformans (CnEF3), an important fungal pathogen in immunocompromised patients, including those infected with human immunodeficiency virus. CnEF3 was found to encode a 1,055-amino-acid protein and has 44% identity with EF3 from Saccharomyces cerevisiae (YEF3). Expressed CnEF3 exhibited ATPase activity that was only modestly stimulated by ribosomes from S. cerevisiae. In contrast, CnEF3 showed tight binding to cryptococcal ribosomes, as shown by an inability to be removed under conditions which successfully remove Saccharomyces EF3 from ribosomes (0.5 M KCl or 2 M LiCl). CnEF3 also poorly complemented a YEF3 defect in a diploid null mutant and two temperature-sensitive mutants which have been shown previously to be complemented well by EF3 from other ascomycetes, such as Candida albicans. These data clearly identify the presence of a functioning EF3 in the basidiomycete yeast C. neoformans, which demonstrates an evolutionary divergence from EF3 of ascomycete yeast.
Cryptococcus neoformans is an important fungal pathogen which causes a lethal meningoencephalitis in a significant number of persons with AIDS and afflicts an increasing number of immunocompromised patients on steroids, chemotherapy, or posttransplant immunosuppressives (18). Therapy of cryptococcosis is limited by toxicity of such agents as amphotericin B (1); newer agents such as the azole inhibitors are less toxic, but increasing reports of resistance may limit their eventual usefulness (3, 15, 17). Echinocandins and pneumocandins are important new antifungal agents which are inhibitors of 1,3-β-glucan synthetases and show excellent activity against ascomycete pathogens such as Candida albicans and Aspergillus fumigatus, but alterations in this enzyme from basidiomycetes make this important class of agents ineffective against C. neoformans (27). The latter example shows the potential gap in antifungal coverage which may occur when inhibitors are chosen without consideration of possible evolutionary differences in drug targets within various fungal pathogens.
Elongation factor 3 (EF3) has been shown to be a required translation cofactor in the ascomycete Saccharomyces cerevisiae (7). The factor is also present in a variety of pathogenic ascomycete fungi, including Candida albicans (8) and the pathogen Pneumocystis carinii (34), which has been shown to be closely related to ascomycete yeasts based on analysis of its rRNA gene as well as genes encoding dihydrofolate reductase, thymidylate synthetase, β-tubulin, and ATP (for a review, see reference 31). While an anti-EF3 antibody has been shown to react with basidiomycete yeasts (4), EF3 has not been characterized from this class of fungi. When present, EF3 is a required translational cofactor essential for growth of the organism. EF3 is believed to modulate the inverse relationship between protein translation rate and amino acid fidelity by altering the binding affinities of the ternary complex to the ribosomal A site and that of the deacylated tRNA to the E site (29). It is unique among the translational factors in that it is not present or required in mammalian translational systems (4). This makes EF3 a putative drug target for a wide variety of fungal pathogens while offering the possibility of a favorable side effect profile in the mammalian host. Many valuable antibacterial agents such as the macrolides and aminoglycosides are inhibitors of the prokaryotic translational apparatus. These antibiotics have had a profound and long-lasting impact on the outcome of bacterial infections and show the extensive precedent for the role of translational inhibitors in the chemotherapy of infectious agents.
While studies in model yeasts such as the ascomycete Saccharomyces may yield important information about essential biological systems, recent advances in the molecular biology of C. neoformans allow the study of drug targets such as EF3 in the pathogen itself, allowing direct application of findings to the rational design of antifungal agents. The present study seeks to identify and characterize EF3 from C. neoformans in order to extend the role for this factor to basidiomycete fungi. This will allow further study of its properties in protein translation and may enable the design of antifungal compounds directed against recombinant cryptococcal EF3.
MATERIALS AND METHODS
Strains.
C. neoformans ATCC 34873 was a generous gift of K. J. Kwon-Chung. S. cerevisiae strain BJ3505/G was from Eastman Kodak (New Haven, Conn.). Escherichia coli SURE (Stratagene, La Jolla, Calif.) was the host strain used for screening the cDNA library after mass excision of the Uni-Zap cDNA library. E. coli XL1-Blue (Stratagene) was the recipient strain of the Bluescript phagemid following in vivo excision for the Uni-Zap XR vector (Stratagene)-containing cDNA clones. E. coli DH10B (Life Technologies, Bethesda, Md.) was the host strain for recovery of ligated plasmids.
Enzyme assay.
The pyruvate kinase/lactate dehydrogenase-coupled ADP assay for EF3 ATPase activity was performed according to the method of Sarthy et al.(25). Activity was expressed in nanomoles of ADP produced per minute at 30°C.
Nested PCR amplification and screening a cryptococcal cDNA library.
A stationary-phase cryptococcal cDNA library in Uni-Zap described previously (32) was mass excised and inserted into E. coli SURE according to the manufacturer's directions (Stratagene). Library plasmid was prepared from cells (Qiagen, Valencia, Calif.) and subjected to endonuclease digestion with NotI. Linearized library plasmids were used as the template (50 ng) and subjected to 25 cycles of amplification by PCR using Taq polymerase (Life Technologies, Bethesda, Md.), an annealing temperature of 40°C, and degenerate primers constructed from amino acid sequence contained in two of the ATP-binding regions of S. cerevisiae EF3 (20) (primer 1360S, CCNAAYGGNTGYGGNAAA, and primer 2760A, RTARTTNGTNGGYTCRTC). Products of 1,200 to 1,800 bp were gel purified and subjected to a second round of 25 cycles of PCR amplification using an annealing temperature of 48°C and internal degenerate primers from S. cerevisiae EF3 (primer 1690S, GAYCCNACNAAYCAT, and primer 2100A, YTTNCCNGCNCCRTTNGG). A 420-bp single band was gel purified, labeled with [α-32P]ATP, and used to screen the cryptococcal stationary-phase cDNA library by standard techniques (23). Clone p5a was selected based on the size of its insert and its ability to bind the PCR probe described above, and both strands were sequenced by automated methods (CRC-DNA Sequencing Facility, University of Chicago).
Northern and Southern blot analysis of CnEF3.
Cryptococcal cells were grown to mid-log phase (A600 < 1.0), and total RNA was obtained as described (32). DNA was obtained from cryptococcal cells as described (30). Northern blots were performed by standard methods (23) using CnEF3 from plasmid p5a.
Construction of expression plasmid pmyep.NS-CnEF3.
A double-stranded oligonucleotide (sense, TCGACCACCACCACCACCACCACTAGGCTAGC; antisense, TAGCCTAGTGGTGGTGGTGGTGGTGG) was inserted into S. cerevisiae expression plasmid YEpFLAG-1 (IBI/Kodak, Rochester, N.Y.) between restriction sites ApaI and SacII to produce the plasmid pmyep (pmyep was kindly obtained from K. Williamson). The Flag affinity tag and hydrophobic leader sequence were removed from pmyep by divergent PCR using primers YEP-1496S (GCCGCCGAATTCCTCGAGCCCGGG) and YEP-1217A (GCCGCCGAATTCTGAAGGAAATCTCATCGC) to create plasmid myep.NS. Plasmid myep.NS was selected for its correct size, ability to transform S. cerevisiae strain BJ3505/G by a polyethylene glycol-lithium acetate (PEG-LiAc) protocol (Stratagene), and the presence of an a EcoRI site. The CnEF3 open reading frame (ORF) was PCR amplified using Pfu polymerase (Stratagene, La Jolla, Calif.) using primers CnEF-5Ba (GCCGCCGGATCCCTCCTGCTGCTACCGCTGCTG) and CnEF3Apa1 (GCCGCCGGGCCCAAGCTCTTCATCACTGAAGAC). The amplified product was restriction endonuclease digested with BamHI and ApaI and inserted into the respective sites of myep.NS to produce plasmid myns.CnEF3.1. Use of the multiple cloning site of myep.NS resulted in a 43-nucleotide addition to the 5′ region of the CnEF3 ORF, resulting in a putative polypeptide having 14 additional amino acids at the N terminus with the sequence MRFPSEFREIYRWIP. Integrity of sequence was verified by automated sequencing of the CnEF3 ORF (CRC-DNA Sequencing Facility).
Expression and purification of recombinant CnEF3.
S. cerevisiae strain BJ 3505/G was transformed with myns.CnEF3.1 by means of a PEG-LiAc protocol (Stratagene). Transformants containing the myns.CnEF3.1 plasmid were selected by Southern blot and for ability to overexpress a 116-kDa protein band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (approximately 20-fold that of untransformed BJ3505/G). To induce recombinant protein, a selected transformant was grown in yeast expression medium (3% glycerol, 1% glucose, 20 mM CaCl2, 1% yeast extract, 8% peptone) according to the manufacturer's instructions (Invitrogen). Cells were isolated by centrifugation and subjected to glass bead (0.45 μm) breakage in a Braun homogenizer (Germany) for 1.5 min at 4°C in buffer A containing 10 mM Tris-HCl(pH 7.5), 10 mM MgSO4, 0.2 mM EDTA, and 10% glycerol. Cell debris was removed by centrifugation, and the supernatant was adsorbed on a 20-ml column of carboxymethyl (CM)-Sepharose equilibrated in buffer A, washed, and eluted with buffer A containing 100 mM KCl. Fractions were assayed by SDS-PAGE, and fractions containing the predominant 116-kDa band were pooled, dialyzed in buffer B (20 mM Tris-HCl pH[7.0], 50 mM KCl, 10 mM magnesium acetate, 0.2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], 10% [vol/vol] PMSF, glycerol) and adsorbed on a 1-ml ATP-agarose column (Sigma) equilibrated in buffer C, and then washed and eluted with buffer C containing 2 mM ATP. Fractions were again assayed on SDS-PAGE, and suitable fractions were pooled, tested for ATPase activity, and subjected to N-terminal sequencing.
Cryptococcal ribosome isolation.
The method used was based on that by Otaka and Kobata (16). Briefly, cryptococcal cells were grown in 2 liters of YPD (2% glucose, 1% yeast extract, 2% Bactopeptone) to an A600 of 0.8, followed by homogenization in a Braun homogenizer using 0.46-μm glass beads for 2 min at 4°C. Two milliliters of cold buffer A (50 mM Tris-HCl [pH 7.0], 50 mM NH4Cl, 10 mM magnesium acetate, 5 mM DTT, 0.1 M EDTA, 0.2 mM PMSF, 10% glycerol) was added, and the mixture was centrifuged at 5,000 × g for 10 min. The supernatant was clarified by centrifuging twice at 35,000 × g for 15 min, and the supernatant was then centrifuged for 3 h at 4°C at 150,000 × g. The pellet from the 150,000 × g centrifugation was resuspended in buffer B (buffer A plus 0.5 M KCl) and centrifuged at 10,000 × g for 10 min to remove aggregates, and the supernatant was overlaid on a cushion of buffer C (buffer B plus 25% glycerol) and centrifuged at 150,000 × g at 4°C for 3 h, and the pellet was recovered. These two sets of low-speed and high-speed centrifugations were repeated once again in the same way, and the pellet was resuspended in buffer A containing 25% glycerol. Cryptococcal ribosomes were also prepared in the presence of buffers excluding 0.5 M KCl. Cryptococcal ribosomes were assayed by ultracentrifugation using a 5 to 30% sucrose gradient (110,000 × g for 10 h) and showed a protein profile similar to that of S. cerevisiae ribosomes prepared as described (16).
Antibody production to recombinant CnEF3.
Purified recombinant cryptococcal EF3 was subjected to SDS-PAGE, and the 116-kDa band was excised, homogenized in the presence of Freund's adjuvant, and administered to 6-week-old CAF1/J mice (20 μg of protein/mouse) by intraperitoneal injection on days 0, 21, 49 and 74. Serum was obtained from the mice prior to the first injection and 1 week after the fourth injection. Western blots and dot blots were performed using the indicated amount of protein and primary antibody and a 1:1,000 dilution of horseradish peroxidase (HRP)-labeled anti-mouse immunoglobulin (Ig) antibody (Accurate Antibodies, Westbury, N.Y.).
ELISA.
For the enzyme-linked immunosorbant assay (ELISA), 96-well plates were incubated with antigen (10 mg/mlμ) overnight at 4°C, washed three times with phosphate-buffered saline (PBS), and blocked with 5% milk for 1 h. The serum was then diluted as indicated with PBS and added to the wells. Following an overnight incubation at 4°C, the wells were washed PBS, and then alkaline phosphatase-labeled goat anti-mouse Ig antibody (1:2,000 dilution) was added. After incubation at room temperature for 1 h, the wells were washed with PBS, 3 mM p-nitrophenyl phosphate in 0.05 M NaCO3–0.05 mM MgCl2 was added, and the optical density of 405 nm of the wells was read on a microtiter plate reader (Titertek, Huntsville, Ala.).
Construction of complementation plasmid Y24EF-CnEF3 7.
Divergent PCR in the direction away from the plasmid insert was used to remove the S. cerevisiae EF3 ORF and insert a NotI site into plasmid YEpEF3 using primers YE24-EF5 (GCCGCCGCGGCCGCATCTTTTAATGTTATCGATGGATT) and YE24-EF3 (GCCGCCGCGGCCGCATCACTGCTTTCACAGTTTTCTT). The modified YepEF3 plasmid retained sufficient 5′ untranslated region and 3′ termination sequence necessary for complementation and expression in S. cerevisiae (24). Plasmid YEpEF3φ-4 was selected for its correct size, ability to transform S. cerevisiae strain BJ3505/G by a PEG–LiAc protocol (Stratagene), and presence of a NotI site. The CnEF3 ORF was PCR amplified in from NotI endonuclease-treated p5a using Pfu polymerase (Stratagene) and primers Crypto 5 (GCCGCCGCGGCCGCCTGCTACCGCTGCTGCCTC) and Crypto 3 (GCCGCCGCGGCCGCTTAAAGCTCTTCATCACTGAAGA). The amplified fragment was ligated to NotI digested plasmid YEpEF3φ-4. Insertion of the NotI site resulted in a nine-nucleotide substitution after the ATG codon of the ORF which would be expected to lead to a two-amino-acid alteration of the CnEF3 ORF at the N terminus, resulting in an expected N-terminal sequence of MRPPATA in the recombinant protein. Suitable clones were screened for the presence of a suitably sized insert in the correct orientation by means of PCR using appropriate primers. Clone Y24EF-CnEF3#7 was further characterized by automated sequencing (CRC-DNA Sequencing Facility) to establish fidelity of sequence.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been submitted to GenBank and assigned accession number AF316889.
RESULTS
Isolation and sequence of C. neoformans EF3 gene.
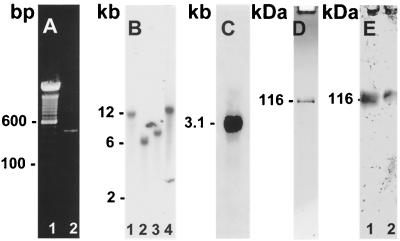
Initial unsuccessful cloning attempts using full-length S. cerevisiae EF3 cDNA clones suggested poor conservation of nucleotide sequence of this factor between ascomycetes and basidiomycetes. Thus, a nested PCR approach was used to produce a suitable probe for cloning using fully degenerate oligonucleotide primers derived from conserved S. cerevisiae sequence within the ATP-binding cassettes and use of a cryptococcal cDNA template from a mass-excised stationary-phase library of C. neoformans to avoid untranscribed sequences. This approach yielded a single 420-bp amplified fragment of the expected size (Fig. 1A), which was then radiolabeled and used to screen a C. neoformans cDNA library (a gift from J. Edman). Two cDNA clones of appropriate size were obtained; p5a was selected, and both strands were sequenced. Southern blots of cryptococcal DNA showed the presence of a single-copy gene (Fig. 1B) using restriction enzymes predicted to cut outside the CnEF3 ORF according to sequence generated from clone p5a. Northern blot analysis of CnEF3 showed the presence of a 3.1-kb transcript which was present in cells obtained in log-phase growth (Fig. 1C).
FIG. 1.
Cloning and characterization of cryptococcal EF3. (A) Lane 1, 100-bp ladder; lane 2, PCR-amplified product from cryptococcal cDNA library. (B): Southern blot of CnEF3. Digestions: lane 1, SpeI and BamHI; lane 2, SpeI and ApaI; lane 3, SpeI and XbaI; lane 4, SpeI alone. (C) Northern blot of CnEF3. (D) SDS-PAGE of 1 μg of recombinant cryptococcal EF3. (E) Western blot of whole-cell extract of C. neoformans (lane 1) and 1 μg of purified recombinant CnEF3 (lane 2).
Analysis of CnEF3 suggests evolutionary sequence divergence from other fungi.
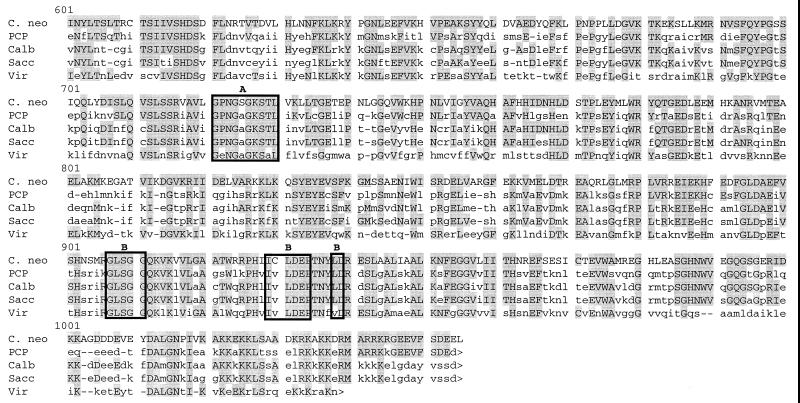
Clone p5a was found to contain a 3,447 bp insert containing a 3,165-bp. ORF which encoded a putative 1,055 amino acid polypeptide having a calculated molecular mass of 116.4 kDa (Fig. 2). Amino acid sequence was used to query the nonredundant combined nucleotide databases of GenBank, EMBL, DDBI, and PDB using MacVector software and the Blast search engine (1). Comparison of amino acid sequence showed significant amino acid homology to EF3 of S. cerevisiae, especially within the two ATP-binding “cassettes” (motifs A and B). Overall conservation of amino acid sequence between C. neoformans and S. cerevisiae (44% identity) is lower than that reported between S. cerevisiae and Candida (78% identity) (8) and between S. cerevisiae and Pneumocystis (57% identity) (34). In addition, there is no serine at position 277, a proposed phosphorylation site for EF3 in S. cerevisiae (20). However, there were three putative phosphorylation sites that were conserved between ascomycetes and C. neoformans, T244, T680, and T772. There were also observed regions of homology between ribosome-binding sites within the N terminus and C terminus of the CnEF3 protein.
FIG. 2.
Predicted amino acid sequence of CnEF3 product compared to EF3 from other organisms. Shaded boxes indicate regions of identity, and the arrow indicates the previously proposed serine phosphorylation site. amino acid sequences encoding ABC cassettes A and B and associated hydrophobic elements are indicated by boxes. Putative phosphorylation sites proposed by Qin et al. (25) are shown by arrows. Sequences shown are form C. neoformans (C. neo) this study), P. carinii (PCP) (34), C. albicans (Calb) (8), S. carevisiae (Sacc) (19), and Chlorella virus CVK2 (Vir) (33).
Production and analysis of recombinant cryptococcal EF3.
To assess the ability of cryptococcal EF3 to function with S. cerevisiae ribosomes, recombinant CnEF3 was heterologously expressed in S. cerevisiae using a new expression plasmid, pmyep.NS, designed for intracellular expression of proteins using the ADH2-promoter. Plasmid pmyep was constructed from the yeast expression plasmid FLAG1 by addition of a terminal histidine affinity tag, divergent PCR was used to remove the pro-α-leader sequence, and Flag affinity sequence was used to construct plasmid pmyep.NS. The CnEF3 ORF was inserted into pmyep.NS and used to transform S. cerevisiae. Transformed yeast cells expressed 20-fold-higher levels of CnEF3 over background YEF3. Recombinant CnEF3 was successfully purified from native YEF3 because CnEF3 did not bind to negatively charged affinity matrices and showed only minimal binding to positively charged matrices such as DEAE-Sepharose (eluted with 0.1 M KCl) in contrast to YEF3, which binds more strongly to both (25). As shown in Fig. 1D, recombinant cryptococcal EF3 purified as a 116 kDa protein, consistent with its predicted size based on cDNA sequence. Western blot analysis of cryptococcal cell extract using antibody prepared from recombinant CnEF3 showed a single band at 116 kDa (Fig. 1E) which showed no reactivity against Saccharomyces cell extracts, ribosomes, or EF3 (data not shown). N-terminal sequencing of the purified factor showed exclusively the expected CnEF3 fusion protein sequence MRFPS. Steady-state kinetics of ATP hydrolysis by CnEF3 using heterologous S. cerevisiae ribosomes showed a ribosome-dependent alteration in Km (709 μM with versus 318 μM without ribosome), whereas there was no significant ribosome-dependent alteration in Vmax (11 versus 13 nM/min). This is in contrast to previous work showing a 100-fold increase in Vmax of the S. cerevisiae factor with the same species ribosomes (9). The stimulation in Km observed in the present case was also not as marked as that reported previously for Saccharomyces EF3 and may be due to a suboptimal interaction between CnEF3 and ribosomes from S. cerevisiae. It is possible that the suboptimal interaction between recombinant CnEF3 and ribosomes could have been due to the additional 14 amino acids at the N terminus of the expressed protein construct, although a lack of significant homology between the elongation factors within the first 100 amino acids makes this less likely to be a region of critical importance. Stimulation of CnEF3 by cryptococcal ribosomes could not be assessed because of an inability to remove ATPase activity from cryptococcal ribosomes, as described in the next section. Western blot of whole cryptococcal extract using an anti-CnEF3 antibody showed a single 116-kDa band, consistent with the size of the recombinant protein and the expected size based on the cDNA sequence (Fig. 1E). Anti-CnEF3 antibody did not cross-react with either cell wall extract or ribosomes from S. cerevisiae (data not shown).
Analysis of CnEF3 binding to cryptococcal ribosomes.
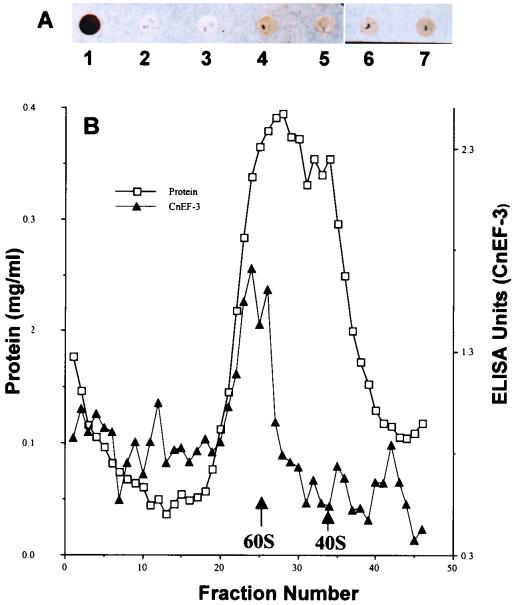
Typically, extraction with 0.5 M KCl removes essentially all ATPase activity from S. cerevisiae ribosomes and allows the ready separation of elongation factors from ribosomal proteins in S. cerevisiae systems. In contrast, we found that extraction of C. neoformans ribosomes yielded a preparation with a large amount of residual ATPase activity which prevented assay of ribosome-dependent ATPase activity of the cryptococcal EF3. For example, ribosomes prepared in the absence of 0.5 M KCl exhibited ATPase activity of 78 nmol min−1 A260-rib−1 which, after washing three times with 0.5 M KCl, continued to show an ATPase activity of 200 nmol min−1 A260-rib−1. (An increase in ATPase activity with washing may have been due to the removal of ATPase inhibitors.) Since one source of ATPase activity could be cryptococcal EF3, we investigated the binding properties of the cryptococcal factor for its homologous ribosome. Dot-blot analysis of nitrocellulose-absorbed cryptococcal ribosome preparations showed that extraction of ribosomes with 0.5 M KCl, 1 M LiCl, or 2 M LiCl did not remove significant amounts of immunoreactive EF3 from the ribosome preparation (Fig. 3A). Western blot of 0.5 M KCl-washed ribosomes using anti-CnEF3 antibody showed an immunoreactive band at 116 kDa, consistent with the presence of CnEF3 (data not shown). These conditions have been shown previously to remove S. cerevisiae but not mammalian ATPase activity from ribosomes (13).
FIG. 3.
Extraction of cryptococcal ribosomes with KCl and LiCl. (A) Dot blots of ribosomes from S. cerevisiae and C. neoformans. Lane 1, 0.6 μg of recombinant CnEF-2; lane 2, 0.5 μg of unwashed S. cerevisiae ribosomes; lane 3, 0.5 μg of S. cerevisiae ribosomes washed with 0.5 M KCl; lane 4, 0.5 μg of unwashed ribosomes from C. neoformans; lanes 5 to 7, 0.5 μg of C. neoformans ribosomes washed with 0.5 M KCl (lane 5), 1 M LiCl (lane 6), or 2 M LiCl (lane 7). Blots were treated with anti-CnEF3 antibody (1:1,000) and HRP-conjugated anti-mouse Ig as described in Materials and Methods. (B): Cryptococcal ribosomes were washed three times with 0.5 M KCl and subjected to sucrose density centrifugation as described in Material and Methods. Fractions were assayed for protein and CnEF3 protein by ELISA. 40S and 60S refer to expected positions of cryptococcal ribosomes based on centrifugation of purified S. cerevisiae ribosome subunits.
Since aggregation of CnEF3 under these extraction conditions could also result in coprecipitation of CnEF3 with ribosomes, additional experiments were performed to assess the molecular mass of the precipitated CnEF3-containing ribosomes after extraction with salt. Purified cryptococcal ribosomes were extracted with 0.5 M KCl, subjected to 5 to 30% sucrose gradient ultracentrifugation in buffer containing 5 mM DTT and 0.1 M EDTA, and assayed for fractions containing CnEF3 by ELISA. As shown in Fig. 3B, CnEF3 comigrates with ribosomal subunits (fractions 20 to 30) even after extraction with 0.5 M KCl, whereas fractions either at the top of the gradient (fraction 47) or in the pellet (fraction 1) did not contain significant amounts of CnEF3 reactivity. The magnesium-free conditions of the gradient were severe enough to dissociate the cryptococcal ribosome, as has been reported previously for S. cerevisiae (16), but still did not dissociate CnEF3 from cryptococcal ribosomes, as shown by lack of soluble CnEF3 at the top of the gradent (fraction 47), and did not cause aggregation, as shown by lack of CnEF3 reactivity in the pellet (fraction 1). These data suggest that cryptococcal EF3 remains associated with the ribosome even after 0.5 M KCl extraction.
Complementation between YEF3 of S. cerevisiae and CnEF3 of C. neoformans.
Diploid strain KC44 of S. cerevisiae, bearing one wild-type and one disrupted YEF3 gene (MATa/ MATα Δyef3::LEU2/leu2-3,112/leu2-3,112 lys2/+ met2-1/+trp1-7/+ura3-52/ura3-52) (24) was transformed with plasmid-borne CnEF3 driven by the YEF3 promoter in a 2μm, Y24 vector (y24-CnEF3.7). The diploid yeasts were sporulated and dissected by standard protocols (26). The viable spores showed a segregation pattern of 2:2, 3:1, and 4:0. In all dissected spores, two of the spores grew equally to the wild-type strain, while the rest grew at a much slower rate (data not shown). Complementation of two strains of haploid S. cerevisiae bearing a temperature-sensitive mutation in the YEF3 gene (strains ts22 and ts27) revealed the appearance of significantly slower-growing colonies at the nonpermissive temperature of 34°C. Similar complementation with wild-type YEF3 genes from S. cerevisiae and C. albicans gave normal-sized colonies, as reported previously (6, 8). From these results, we conclude that CnEF3 complements the function of YEF3 poorly, although it does show that CnEF3 has functional capability. To verify that the cryptococcal factor was expressed from y24-CnEF3.7 under the YEF3 promoter, CnEF3 protein was purified from ts22 complemented with y24-CnEF3.7 using the purification procedure which resolves CnEF3 from YEF3 (see Materials and Methods). The protein was readily expressed at the permissive temperature as a protein having the predicted 116-kDa size on SDS-PAGE, intact ATPase activity (Vmax = 90 mol/min/mg), and the expected unique N-terminal sequence of the cryptococcal fusion protein from Y24EF-CnEF3#7, MRPPA. It is possible that poor complementation was due to the two-amino-acid substitution in the N terminus of the cryptococcal fusion protein, although this is unlikely, as very little homology exists between the cryptococcal factor and the Saccharomyces factor in the first 100 amino acids of the N terminus.
DISCUSSION
While EF1 and EF2 are required for efficient protein translation for all eukaryotic organisms, EF3 is unique in its identification and requirement only within the kingdom Fungi. Since inhibition of protein synthesis by targeting EF3 might represent an effective drug strategy for the control of fungal diseases, identification and characterization of this factor among diverse fungi of medical importance are essential to allow design of pharmacological inhibitors having the broadest application. Thus far, EF3 has only been cloned and characterized from yeasts of the order Ascomycota as well as the fungus-like organism Pneumocystis carinii (6). Basidiomycete yeasts are distinct from ascomycete yeasts in both morphology (2) and 18S ribosomal DNA sequence criteria (21). Of the more than 20,000 species described (11), C. neoformans represents the most important human pathogen within this phylum. Various properties of this fungus, such as less efficient homologous recombination, complex transcriptional processes, and a promiscuous telomerase (2), suggest that essential cellular machinery may be distinct from that in the more highly studied ascomycetes such as S. cerevisiae and Neurospora crassa.
In order to compare the functional properties of EF3 among fungi, we undertook the characterization of EF3 from the basidiomycete C. neoformans. Consistent with its more distant relationship to the ascomycetes, CnEF3 showed less homology to EF3 from S. cerevisiae (46% identity) than to EF3 from ascomycetes such as C. albicans (78% identity to S. cerevisiae) and the ascomycete-like organism P. carinii (57% identity to S. cerevisiae) (5). An EF3-like gene cloned from the Chlorella virus shows a similar low identity (36%), but the functional significance of this latter gene has not been established (33). In contrast, the homology of EF1 between the two classes is much higher (84.7% identity) (28) and suggests a greater evolutionary drift of the third translational cofactor within phyla of Fungi. Low identity between CnEF3 and ascomycete fungi may also help in determining the functional significance of amino acids within the protein. For example, lack of a conserved serine at position 277, corresponding to a proposed phosphorylation site in YEF3 (19), makes this amino acid unlikely to have a role in regulation by phosphorylation in C. neoformans. In contrast, important ATP-binding regions A and B that form the ATP-binding cassette are conserved in CnEF3, consistent with their role in ATP hydrolysis and protein translation in Saccharomyces (4).
Ribosome binding is believed to be an important function of EF3 and to be essential for the factor's translation-modulating effects (29). Analysis of peptide regions of EF3 believed to be involved in ribosome binding may shed light on possible structural differences in EF3-ribosome binding interactions between ascomycetes and basidiomycetes. Previous studies have shown that an acidic N-terminal region of S. cerevisiae (amino acids 98 to 388) (10) and a basic C-terminal region (775 to 1044) (12) of EF3 are both involved in ribosome binding. Comparison of the amino acid sequence of the homologous regions of CnEF3 (N-terminal residues 108 to 404 and C-terminal residues 796 to 1055) show that the cryptococcal N-terminal region is less acidic (12 versus 17 D and 16 versus 23 E) with a calculated pI of the cyptococal fragment of 8.98 versus 4.85 for the S. cerevisiae fragment. In addition, the cyptococal C-terminal fragment is much less basic (26 versus 31 K), yielding a calculated pI of 6.73, versus 8.82 for the S. cerevisiae fragment. Indeed, differences in surface charge of the cryptococcal factor were also suggested by its lack of binding to CM-cellulose and its relatively poor binding to DEAE-cellulose, in contrast to the stronger binding of the S. cerevisiae factor to both matrices. Such differences in amino acid sequence and CnEF3 binding characteristics suggest an evolutionary drift in the nature of CnEF3-ribosome binding from that of ascomycetes.
Recombinant CnEF3 showed significant ATPase activity, as has been shown for YEF3 (4). Since ribosome-dependent ATPase activity is a key characteristic of EF3, the ability of S. cerevisiae ribosomes to stimulate CnEF3 ATPase activity was assessed. While previous reports have shown a 50- to 100-fold increase in Vmax of YEF3 ATPase activity in the presence of homologous ribosomes, we found no significant increase in ATPase Vmax of the cryptococcal EF3 in the presence of S. cerevisiae ribosomes and only a small decrease in Km. This suggests that evolutionary divergence from ascomycetes exhibited by the CnEF3 sequence results in alterations in properties leading to poor EF3-S. cerevisiae ribosome interactions.
In order to study the nature of the interaction of CnEF3 with cryptococcal ribosomes, attempts were made to remove CnEF3 from a ribosome preparation with 0.5 M KCl, which typically removes all S. cerevisiae elongation factors. Even after extraction with potassium (0.5 M) or lithium (2 M) salts, significant ribosome-ATPase activity remained, preventing the measurement of CnEF3-cryptococcal ribosome-dependent ATPase activity. Both dot-blot and sucrose sedimentation experiments showed that CnEF3 exhibited an unusually strong binding to ribosomes, which may account for at least part of the observed residual ATPase activity of salt-extracted cryptococcal ribosomes. It is interesting that in higher eukaryotes such as mammals, 80S ribosomes are able to hydrolyze ATP without the addition of soluble protein factors. In addition, mammalian ribosomal ATPase activity cannot be removed with 0.5 M KCl and is only poorly removed with 2 M LiCl (14, 22). However, there has been no evidence of a third elongation factor in mammalian ribosomes, and the identity of the mammalian ribosomal ATPase activity remains unknown.
In order to further characterize the functional significance of EF3 amino acid differences between basidiomycetes and ascomycetes, CnEF3 was tested for its ability to replace the function of YEF3 in vivo. It was found that CnEF3 only poorly complemented either of two temperature-sensitive mutants of S. cerevisiae (ts22 and ts27). In addition, the sizes of colonies of a KC44 Δyef3::LEU2 null mutant rescued by CnEF3 were significantly smaller than those rescued by YEF3. These results are in contrast to those of Cholthurst et al., who showed good complementation and normal colony size after complementation of these same strains using EF3 from the ascomycete C. albicans (6). The poor complementation by CnEF3 in vivo may again be a reflection of structural or functional differences between the elongation factors of ascomycetes and basidiomycetes.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI38258-04 and a grant from the American Lung Association.
We appreciate helpful comments from K. J. Kwon-Chung.
REFERENCES
- 1.Andriole V T, Kravetz H M. The use of amphotericin B in man. JAMA. 1962;180:269–272. doi: 10.1001/jama.1962.03050170001001. [DOI] [PubMed] [Google Scholar]
- 2.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 3.Casadevall A, Spitzer E D, Webb D, Rinaldi M G. Susceptibilities of serial Cryptococcus neoformans isolates from patients with recurrent cryptococcal meningitis to amphotericin B and fluconazole. Antimicrob Agents Chemother. 1993;37:1383–1386. doi: 10.1128/aac.37.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraburtty K, Triana-Alonso F J. Yeast elongation factor 3: structure and function. Biol Chem. 1998;379:831–840. doi: 10.1515/bchm.1998.379.7.831. [DOI] [PubMed] [Google Scholar]
- 5.Colthurst D R, Santos M, Grant C M, Tuite M F. Candida albicans and three other Candida species contain an elongation factor structurally and functionally analogous to elongation factor 3. FEMS Microbiol Lett. 1991;64:45–49. doi: 10.1016/0378-1097(91)90207-q. [DOI] [PubMed] [Google Scholar]
- 6.Colthurst D R, Schauder B S, Hayes M V, Tuite M F. Elongation factor 3 (EF3) from Candida albicans shows both structural and functional similarity to EF3 from Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1025–1033. doi: 10.1111/j.1365-2958.1992.tb02168.x. [DOI] [PubMed] [Google Scholar]
- 7.Dasmahapatra B, Chakraburtty K. Protein synthesis in yeast. I. Purification and properties of elongation factor 3 from Saccharomyces cerevisiae. J Biol Chem. 1981;256:9999–10004. [PubMed] [Google Scholar]
- 8.Di Domenico B J, Lupisella J, Sandbaken M, Chakraburtty K. Isolation and sequence analysis of the gene encoding translation elongation factor 3 from Candida albicans. Yeast. 1992;8:337–352. doi: 10.1002/yea.320080502. [DOI] [PubMed] [Google Scholar]
- 9.Goldman D L, Fries B C, Franzot S P, Montella L, Casadevall A. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci USA. 1998;95:14967–14972. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gontarek R R, Li H, Nurse K, Prescott C D. The N terminus of eukaryotic translation elongation factor 3 interacts with 18 S rRNA and 80 S ribosomes. J Biol Chem. 1998;273:10249–10252. doi: 10.1074/jbc.273.17.10249. [DOI] [PubMed] [Google Scholar]
- 11.Hawksworth D L, Ainsworth G C. Ainsworth and Bisby's dictionary of the fungi. Walingford, Oxon, United Kingdom: CAB International; 1995. [Google Scholar]
- 12.Kambampati R, Chakraburtty K. Functional subdomains of yeast elongation factor 3: localization of ribosome-binding domain. J Biol Chem. 1997;272:6377–6381. doi: 10.1074/jbc.272.10.6377. [DOI] [PubMed] [Google Scholar]
- 13.Kovalchuke O, Ziehler J, Chakraburrty K. Comparative analysis of ATPase of yeast elongation factor 3 and ATPase associated with Tetrahymena ribosomes. Biochimie. 1995;77:713–718. doi: 10.1016/0300-9084(96)88187-9. [DOI] [PubMed] [Google Scholar]
- 14.Kovalchuke O, Chakraburrty K. Comparative analysis of ribosome-associated adenosinetriphosphatase (ATPase) from pig liver and the ATPase of elongation factor 3 from Saccharomyces cerevisiae. Eur J Biochem. 1994;226:133–140. doi: 10.1111/j.1432-1033.1994.tb20034.x. [DOI] [PubMed] [Google Scholar]
- 15.Lamb D C, Corran A, Baldwin B C, Kwon-Chung J, Kelly S L. Resistant P45051A1 activity in azole antifungal tolerant Cryptococcus neoformans from AIDS patients. FEBS Lett. 1995;368:326–330. doi: 10.1016/0014-5793(95)00684-2. [DOI] [PubMed] [Google Scholar]
- 16.Otaka E, Kobata K. Yeast ribosomal proteins. I. Characterization of cytoplasmic ribosomal proteins by two-dimensional gel electrophoresis. Mol Gen Genet. 1978;162:259–268. doi: 10.1007/BF00268851. [DOI] [PubMed] [Google Scholar]
- 17.Paugam A, Dupouy-Camet J, Blanche P, Gangneux J P, Tourte-Schaefer C, Sicard D. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin Infect Dis. 1994;19:975–976. doi: 10.1093/clinids/19.5.975-a. [DOI] [PubMed] [Google Scholar]
- 18.Pinner R W, Hajjeh R A, Powderly W G. Prospects for preventing cryptococcosis in persons infected with human immunodeficiency virus. Clin Infect Dis. 1995;21(Suppl. 1):S103–S107. doi: 10.1093/clinids/21.supplement_1.s103. [DOI] [PubMed] [Google Scholar]
- 19.Qin S L, Moldave K, McLaughlin C S. Isolation of the yeast gene encoding elongation factor 3 for protein synthesis. J Biol Chem. 1987;262:7802–7807. [PubMed] [Google Scholar]
- 20.Qin S L, Xie A G, Bonato M C, McLaughlin C S. Sequence analysis of the translational elongation factor 3 from Saccharomyces cerevisiae. J Biol Chem. 1990;265:1903–1912. [PubMed] [Google Scholar]
- 21.Restrepo B I, Barbour A G. Cloning of 18S and 25S rDNAs from the pathogenic fungus Cryptococcus neoformans. J Bacteriol. 1989;171:5596–5600. doi: 10.1128/jb.171.10.5596-5600.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodnina M V, Serebryanik A I, Ovcharenko G V, El'Skaya A V. ATPase strongly bound to higher eukaryotic ribosomes. Eur J Biochem. 1994;225:305–310. doi: 10.1111/j.1432-1033.1994.00305.x. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Sandbaken M G, et al. Protein synthesis in yeast: structural and functional analysis of the gene encoding elongation factor 3. J Biol Chem. 1990;265:15838–15844. [PubMed] [Google Scholar]
- 25.Sarthy A V, McGonigal T, Capobianco J O, Holzman T H, Walter K A, Egan D A, Goldman R C. High-level overexpression of yeast elongation factor 3 and detailed kinetic analysis using a coupled spectrophotometric assay. Anal Biochem. 1997;254:288–290. doi: 10.1006/abio.1997.2456. [DOI] [PubMed] [Google Scholar]
- 26.Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J R, Douglas C M, Li W, Jue C K, Pramanik B, Yuan X, Rude T H, Tofaletti D L, Perfect J R, Kurtz M. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J Bacteriol. 1999;181:444–453. doi: 10.1128/jb.181.2.444-453.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thornewell S J, Peery R B, Skatrud P L. Cloning and molecular characterization of CnTEF1 which encodes translation elongation factor 1 alpha in Cryptococcus neoformans. Fungal Genet Biol. 1997;22:84–91. doi: 10.1006/fgbi.1997.1002. [DOI] [PubMed] [Google Scholar]
- 29.Triana-Alonso F J. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 30.Varma A, Kwon-Chung K J. Rapid method to extract DNA from Cryptococcus neoformans. J Clin Microbiol. 1991;29:810–812. doi: 10.1128/jcm.29.4.810-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walzer P D. Pneumocystis carinii. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. Philadelphia, Pa: Churchill Livingstone; 2000. pp. 2781–2795. [Google Scholar]
- 32.Williamson P R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada T, Fukuda T, Tamura K, Furukawa S, Songsri P. Expression of the gene encoding a translational elongation factor 3 homolog of Chlorella virus CVK2. Virology. 1993;197:742–750. doi: 10.1006/viro.1993.1650. [DOI] [PubMed] [Google Scholar]
- 34.Ypma-Wong M F, Fonzi W A, Sypherd P S. Fungus-specific translation elongation factor 3 gene present in Pneumocystis carinii. Infect Immun. 1992;60:4140–4145. doi: 10.1128/iai.60.10.4140-4145.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]