Abstract
Mitomycin C (MMC), a DNA-damaging agent, is a potent inducer of the bacterial SOS response; surprisingly, it has not been used to select resistant mutants from wild-type Escherichia coli. MMC resistance is caused by the presence of any of four distinct E. coli genes (mdfA, gyrl, rob, and sdiA) on high-copy-number vectors. mdfA encodes a membrane efflux pump whose overexpression results in broad-spectrum chemical resistance. The gyrI (also called sbmC) gene product inhibits DNA gyrase activity in vitro, while the rob protein appears to function in transcriptional activation of efflux pumps. SdiA is a transcriptional activator of ftsQAZ genes involved in cell division.
Mitomycin C (MMC), an antitumor agent isolated from Streptomyces cultures, is used in chemotherapy (30). It interacts with DNA by intercalation and adduct formation (37). These actions trigger the SOS response, the concerted induction of several DNA repair, and recombination activities controlled by the lexA-specified repressor in Escherichia coli (42). This interaction has been studied by in vitro and in vivo methods. The breadth of the SOS regulatory circuit has been approximated by screening a collection of Escherichia coli promoter-lacZYA gene fusions for those which displayed increased β-galactosidase activity in the presence of low levels of MMC (19). Similarly, two-dimensional electrophoretic separation of E. coli polypeptides induced by a DNA cleaving treatment has been catalogued (38). The expression of at least 29 genes is induced by DNA damage; 18 of these are regulated by the SOS response, while 11 are lexA independent (42).
Global regulatory circuits do not act in isolation (29). Rather, a stress treatment may induce many regulons, as has been observed by both two-dimensional protein separation methodology in studies of Salmonella enterica serovar Typhimurium (13) and gene fusion-based analyses of E. coli (7) after hydrogen peroxide treatment. The concerted action of all such induced regulons describes the responses to the stress caused by an individual chemical treatment. Such interactions may also be suggested by the analysis of pleiotropic mutants (20) resistant to DNA-damaging agents.
Despite the clinical, molecular biological, and historical importance of MMC-DNA interactions, the selection of MMC-resistant mutants has been limited to a single pseudoreversion study (24, 26). Further genetic studies might enhance our understanding of the cellular interactions with MMC. Demanding overexpression of a gene product is a classic means of overcoming chemical toxicity (20). Such selections were used to define the regulatory circuit controlling his operon expression in Salmonella serovar Typhimurium (32). This selection system was later exploited to select gene amplification events in bacteria (4). Similar selections with PALA, N-(phosphonacetyl)-l-aspartate, and methotrexate led to amplification of specific genes encoding enzymes targeted by the inhibitors in mammalian cells (34, 41). With the construction of yeast genomic DNA libraries in high-copy-number plasmids, the selection of inhibitor-resistant lines in yeast accelerated. Targets of inhibitor action were verified by using these methods (31), while the ability to define both target-specifying and unexpected resistance genes was uncovered (17). More recently, these methodologies were extended to a tumoricidal agent with an ill-defined mode of action using an E. coli-based multicopy plasmid library and a selection scheme (12). Such techniques have also been used to define the action of amino acid biosynthetic inhibitors (16; Z. Xue, D. R. Smulski, D. Delduco, S.-Y. Choi, M. H. Jia, and R. A. LaRossa, unpublished results; D. R. Smulski, L. X. Huang, T. K. Van Dyk, and R. A. LaRossa, unpublished results).
In this study, the response to an MMC challenge was illuminated by the isolation of inhibitor-resistant mutants due to the presence of E. coli genomic fragments in multicopy plasmids. The sequencing of insert-vector junctions defines genes that confer inhibitor resistance when present in high copy. The application of such technology to MMC action is described here.
MATERIALS AND METHODS
Strains and media.
The E. coli strains used in this study are all K-12 derivatives (Table 1). The strains were grown in Luria-Bertani (LB) medium. Ampicillin (at 100 or 150 μg/ml) or kanamycin (at 25 μg/ml) was added to the medium when necessary. The standard growth temperature was 37°C. Liquid cultures were aerated by rotary shaking at 250 rpm.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli | ||
| MG1655 | F− λ−rph-1 | 5 |
| W3110 | F− λ−rph-1 IN(rrnD-rrnE) | 5 |
| RFM443 | rpsL200 galK2 lacΔ74 | 22 |
| WX2 | Δlac-pro met pro zzz::Tn10 sdiA::Kan thy supD (rK− mK−) | 43 |
| DM800 | F−metA28 lacY1 or lacZ4 thi-1 xyl-5 or xyl-7 galK2 tsx-6 | 25 |
| DM803 | Same as DM800 but lexA1a | 25 |
| DPD2272 | F− λ−rph-1 sdiA::Kan | This studyb |
| DPD2668 | RFM443(pUC19) | This study |
| DPD2669 | RFM443(pDEW140) | This study |
| Plasmids | ||
| pBR322 | Cloning vector | 33 |
| pUC18 | Cloning vector | 33 |
| pUC19 | Cloning vector | 33 |
| pDEW133 | pUC18 plus gyrI | This study |
| pDEW140 | pUC19 plus sdiA (EcoRI) | This study |
| pDEW141 | pUC19 plus rob (EcoRI) | This study |
lexA1 [also lexA(Ind)] is a noninducible allele of lexA.
P1(WX2) × MG1655 → Kanr.
Strain construction.
DPD2272 was constructed by P1vir phage-mediated transduction with the donor strain, WX2, and the recipient strain, MG1655. Recombinants were selected using kanamycin (23). An electrotransformation method (33) was used to introduce plasmids into bacterial strains.
MIC of MMC.
LB agar plates containing different MMC concentrations (10, 8, 5, 3, 1, 0.5, 0.1, 0.05, 0.01, and 0 μg/ml) were prepared. Strain RFM443 was streaked onto each plate, followed by incubation overnight at 37°C. The growth of RFM443 was checked by scoring for colony formation. The lowest concentration of MMC that inhibited colony formation of RFM443 was defined as the MIC of MMC. Strain RFM443 grew on all the plates with an MMC concentration of ≤1 μg/ml; growth was prevented by a concentration of ≥3 μg/ml. The MIC of MMC on plates for RFM443 was thus defined to be 3 μg/ml.
Identification of MMC-resistant clones.
Libraries were previously prepared from E. coli strain W3110 genomic DNA partially digested with Sau3AI to ∼4-kbp fragments (16). For each round of multicopy titration, 0.2 ng of the pBR322-based or 0.3 ng of the pUC18-based library was electrotransformed into RFM443. The colonies underwent single colony purification on the same medium. Plasmids were isolated from 1.5-ml overnight cultures of the single colonies in LB medium supplemented with ampicillin (150 μg/ml) using the Qiagen 96-well Turbo Plasmid Prep kit (Qiagen, Inc., Valencia, Calif.). DNA sequence data from both ends of each insert were obtained. The M13/pUC sequencing primer (−40) and the M13/pUC reverse sequencing primer (−48) were used in sequencing the pUC18-based inserts (33). The primers used in sequencing the pBR322-based insertions were 5′-GCC ACT ATC GAC TAC GCG-3′ and 5′-CGA TAT AGG CGC CAG CAA C-3′. BLASTn (3) searches identified the chromosomal segments harbored on each plasmid.
Subcloning of gyrI, sdiA, and rob.
Primers were designed for PCR amplification of gyrI, sdiA, and rob, each with the flanking intergenic regions containing the corresponding promoter (Table 2). PCR amplification was carried out using the PCR AmpliTaq kit (Roche, Palo Alto, Calif.). The EcoRI-BamHI double-digested PCR product of the gyrI region was subcloned into the EcoRI-BamHI site of pUC18, and the resultant plasmid was designated pDEW133. The PCR products of the sdiA region and the rob region were each inserted into the polylinker site of pCR2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen Co., Carlsbad, Calif.), and both inserts were subsequently placed into the EcoRI site of pUC19. The resultant plasmids were named pDEW140 and pDEW141, respectively. Their structures were confirmed by sequencing the insert junctions using the M13/pUC sequencing primer (−40) and the M13/pUC reverse sequencing primer (−48). pDEW133, pDEW140, and pDEW141 were electrotransformed into strains RFM443, DM800, and DM803 for further characterization.
TABLE 2.
Primers used in genetic constructions
| Primer | Sequence (5′-3′) |
|---|---|
| gyrI Bam-left | CGGGATCCCGCCGCGCACCAGACTAACAT |
| gyrI Eco-right | GGAATTCCGTCGGAACGGAACGCATCTGGTA |
| sdiAf | TGGCACGCAGGACAGAA |
| sdiAd | TAACAAATCAGCATAACTCAT |
| rob1 | ACGACGGATCGGAATCAGCA |
| rob2 | TAGTTCGTCACGGTAA |
Zone of inhibition assays.
These were performed using the method modified (21) from that of Stephens et al. (36). Briefly, the test strains were grown overnight in LB medium supplemented with 150 μg of ampicillin per ml. Then, 0.1-ml portions of each culture and 2.5 ml of melted LB soft agar with 150 μg of of ampicillin per ml were mixed and poured over a 30-ml LB agar plate appended with 150 μg of ampicillin per ml. After the top layer was solidified, a sterile filter disc (7 mm in diameter) containing the desired amount of a chemical was placed at the center of the plate. For MMC, 15 or 30 μg was used; for nalidixic acid, 75 μg was used. The diameters of the inhibition zones were measured after overnight incubation at 37°C.
Microscopic examination of cultures.
Both overnight and early-exponential-phase aliquots of a control strain, DPD2668, and one having an sdiA multicopy plasmid, DPD2669, were examined by confocal laser scanning microscopy. Portions (1 ml) of cultures, grown at 37°C in LB medium, were stained with 1 μl of SYTO13, a cell-permeant green fluorescent nucleic acid stain (5 mM solution in dimethyl sulfoxide, Molecular Probes, Eugene, Oreg.). Samples (ca. 1 μl) were spotted onto microscope slides and dried for 20 min by placement on a surface heated to 60°C. They were rehydrated in a drop of a glycerol-based mounting medium, Citifluor (Ted Pella, Inc., Redding, Calif.), designed to reduce photobleaching, and a coverslip was placed over the sample prior to examination by confocal laser scanning microscopy. Images were archived on a personal computer.
RESULTS
sdiA, gyrI, rob, and mdfA in multicopy conferred resistance to MMC upon E. coli.
MMC-resistant clones were selected on LB agar plates with 6 μg of MMC per ml (twice the MIC determined in this study) and 150 μg of ampicillin per ml. Resistant colonies appeared after 1 day of incubation at 37°C. E. coli genomic DNA libraries in pUC18 and pBR322 were separately transformed into strain W3110. Thirty MMC-resistant isolates were found among approximately 109 ampicillin-resistant colonies obtained from the pUC18 library. The plating of approximately 108 ampicillin-resistant colonies obtained from the pBR322 library yielded 16 MMC-resistant clones. Plasmids were isolated from each of these 46 lines and used to retransform strain RFM443 selecting for ampicillin resistance. In each backcross, MMC resistance was coinherited with ampicillin resistance, indicating that the MMC resistance determinants were plasmid-borne. The ends of these 46 inserts conferring MMC resistance were sequenced to identify the regions of the E. coli chromosome harbored within each multicopy plasmid. Inserts were derived from four distinct chromosomal loci.
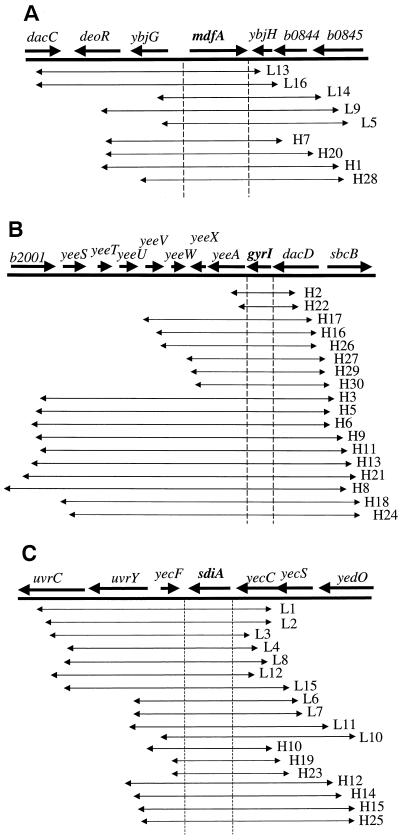
Four pUC18-derived plasmids and five pBR322-derived plasmids contained a region of the genome that mapped to min 19 (Fig. 1A). The only gene present in all nine plasmids was mdfA, suggesting that mdfA in multiple copies conferred MMC resistance. Eighteen other pUC18-derived plasmids clustered at a second locus, the gyrI (also known as sbmC) region of the chromosome at min 44 (Fig. 1B). gyrI was the single gene in common among the 18 inserts. Another cluster of 7 pUC18-derived plasmids and 11 pBR322-derived plasmids shared a single common gene, sdiA from min 43 (Fig. 1C). The final pUC18-derived plasmid conferring MMC resistance contained three intact E. coli genes, i.e., rob, creA, and creB (map not shown), in the vicinity of min 100.
FIG. 1.
Physical maps of E. coli genomic inserts conferring mitomycin resistance. “L” and “H” are plasmid names (e.g., L13 and H7), with “L” referring to pBR322-based plasmids and “H” referring to pUC18-based plasmids. (A) Region from min 9 in E. coli, where nine MMC-resistant clones were clustered (GenBank accession no. AE000186). (B) Region from min 44, where 18 MMC-resistant clones were clustered (GenBank accession no. AE000292). (C) Region from min 43, where 18 MMC-resistant clones were clustered (GenBank accession no. AE000284).
To confirm these assignments of resistance determinants, the gyrI, sdiA, and rob genes with just their flanking intergenic regions were individually inserted into either pUC18 or pUC19 (see Materials and Methods). The resultant plasmids, when introduced by transformation into strain RFM443, were tested for their abilities to alter the strains' responses to MMC and nalidixic acid, an inhibitor of DNA gyrase that causes strand scissions through interference with the gyrase ligation reaction (42). The results (Table 3) demonstrated that each of these three genes in multicopy conferred resistance to both MMC and nalidixic acid, since strains bearing these genes in high copy displayed smaller zones of inhibition than strains containing control plasmids.
TABLE 3.
Zone of inhibition assaysa
| Plasmid | Inhibition zone diam (mm) with:
|
|
|---|---|---|
| MMC (15 μg) | Nalidixic acid (75 μg) | |
| pUC19 | 22 | 23 |
| pDEW140 (sdiA) | 16 | 16 |
| pDEW141 (rob) | 19 | 18 |
| pUC18 | 19 | 19 |
| pDEW133 (gyrI) | 12 | <7 |
RFM443, harboring various plasmids, was challenged with either 15 μg of MMC or 75 μg of nalidixic acid. The diameter of each resultant inhibition zone is presented.
An sdiA null mutant did not show hypersensitivity to MMC.
Inhibition zone assays were performed on the isogenic sdiA+ and sdiA strains MG1655 and DPD2272. Obvious differences in the zones of inhibition caused by exposure to MMC or nalidixic acid were not observed. Both strains showed 19-mm-diameter zones of inhibition with 15 μg of MMC or 75 μg of nalidixic acid.
Modulation of multicopy resistance to DNA-damaging agents by lexA (Ind).
Strain DM800 (lexA+) and strain DM803 [lexA (Ind)], each individually transformed with a set of plasmids (pUC18, pUC19, pDEW133 [gyrI], pDEW140 [sdiA], and pDEW141 [rob]), were tested for MMC and nalidixic acid sensitivities (Tables 4 and 5). The lexA (Ind) product is resistant to proteolysis by activated RecA and thus prevents induction of the SOS response (42). As expected, the lexA (Ind) mutants displayed larger zones of inhibition than those of the isogenic lexA+ strain. A multicopy gyrI plasmid (pDEW133) in the lexA(Ind) background did not confer significant resistance to either MMC or nalidixic acid. Thus, the SOS response was needed for the gyrI-associated phenotype. In contrast, the presence of sdiA in high copy (pDEW140) conferred resistance in both the lexA+ and lexA(Ind) backgrounds. Thus, the phenotype conferred by sdiA amplification did not rely upon the SOS response. rob in high copy (pDEW141) did not confer significant resistance in the DM800/DM803 background used for testing lexA(Ind) dependence (data not shown).
TABLE 4.
gyrI-mediated multicopy resistance is lexA dependent
| Strain | Inhibition zone diam (mm) with:
|
|
|---|---|---|
| MMC (30 μg) | Nalidixic acid (75 μg) | |
| DM800/pUC18 | 17 | 19 |
| DM803/pUC18 | 22 | 22 |
| DM800/pDEW133 (gyrI) | 12 | 19 |
| DM803/pDEW133 (gyrI) | 22 | 22 |
TABLE 5.
Modulation of sdiA multicopy resistance
| Strain | Inhibition zone diam (mm) with:
|
|
|---|---|---|
| MMC (15 μg) | Nalidixic acid (75 μg) | |
| DM800/pUC19 | 20 | 20 |
| DM803/pUC19 | 27 | 25 |
| DM800/pDEW140 (sdiA) | <7 | 8 |
| DM803/pDEW140 (sdiA) | 16 | 13 |
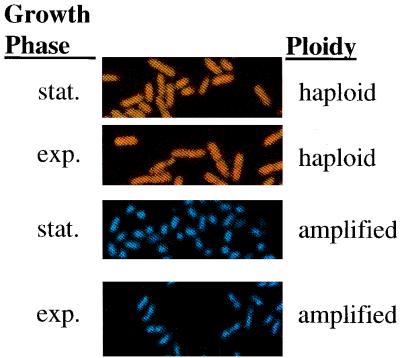
Microscopic examination of cultures harboring an sdiA plasmid.
In the early exponential phase the DPD2668 (control) culture displayed a typical rod-shaped morphology, and in stationary phase the culture was composed of rods that might be slightly shorter than cells in the exponential phase of growth. In contrast, the stationary-phase culture of DPD2669 (harboring sdiA in high copy) was dominated by rounded cells that transformed into rods upon reaching logarithmic growth after subculturing. These rods were significantly shorter than those observed in the early-exponential-phase control culture. Representative fields of equal magnification are shown in Fig. 2.
FIG. 2.
Micrographs of E. coli cells. Each frame is enlarged to the same extent. The top two panels capture images of cells with a haploid sdiA (DPD2668) content, while the bottom two panels are of an isogenic strain containing multiple copies of sdiA (DPD2669). The first and third frames from the top are pictures of stationary (stat.)-phase cultures, while the second and fourth panels from the top are images of exponential (exp.)-phase cultures. Color differences in the images of the two strains are a result of computer processing.
In contrast, exponential- and stationary-phase E. coli cultures harboring either mdfA or rob on a multicopy plasmid displayed a morphology indistinguishable from that of the control cultures of DPD2668. The gyrI-containing plasmid, however, was associated with an intermediate phenotype having a rounded morphology in the stationary phase but a normal shape in the exponential phase.
DISCUSSION
Random E. coli genomic DNA fragments on a medium-copy-number vector, pBR322, and a high-copy-number vector, pUC18, were screened in an E. coli K-12 strain for variants resistant to MMC, a DNA-damaging agent. The clones obtained were clustered at four chromosomal regions. Only one gene in each region was responsible for the MMC resistance phenotype; these genes are mdfA, sdiA, rob, and gyrI.
Since mdfA is a multidrug resistance locus (15), the demonstration that resistance was conferred by minimal plasmids expressing only one E. coli gene was restricted to the analysis of rob, sdiA, and gyrI. One chromosomal region, defined by a single plasmid, contained rob and creBC. Since rob is involved in drug resistance (45), while creBC functions in carbon metabolism (10), rob was subcloned and shown to confer resistance to MMC when present at a high copy number.
The finding that these four genes confer resistance, though unanticipated, can be incorporated into a plausible model. The gyrI product inhibits DNA gyrase activity in vitro (28), while the sdiA product activates the transcription of ftsQAZ, genes involved in septum formation at an early stage of cell division (43). SdiA also activates the expression of several other genes (1, 44), including uvrY and uvrC, which are involved in the protection of the cells from UV irradiation, and the acrA, acrB, acrD, acrE, and acrF genes, whose products are responsible for acridine efflux. The rob product also appears to function in transcriptional activation of efflux pump genes, including the acrAB operon (9, 27, 45). AcrAB connects to form an export channel with the tolC-encoded outer membrane porin (2). Since tolC mutants are hypersensitive to MMC (14), we propose that MMC is exported from cells by the action of the efflux pumps, while DNA gyrase activity facilitates the intercalation of MMC into the chromosome. Thus, amplification of gyrI and mdfA may prevent MMC from interacting with DNA, its macromolecular target. Amplification of gyrI has previously been shown to protect cells from the action of microcin B17, a DNA-cleaving agent (6). This compound causes double-stranded DNA breaks in vivo and in vitro only in concert with DNA gyrase (39). Perhaps, rob overexpression also acts to enhance efflux, while elevated levels of the sdiA product might overcome cell division arrest imposed by DNA damage. Thus, exhaustive selection of multicopy resistance, in conjunction with previous knowledge of MMC action, has allowed us to define the integrated response to this chemical insult.
Both MMC and nalidixic acid are known to induce the SOS response in E. coli (38, 42). While the lexA+ strain, DM800, showed sensitivity to MMC and nalidixic acid, strain DM803 [lexA(Ind)], which is incapable of mounting the SOS response due to a noncleavable form of the LexA repressor, showed an increased sensitivity to both chemicals. In the lexA(Ind) background, the strains harboring pUC18 or the gyrI-containing pDEW133 both displayed inhibition zones of the same size when challenged with MMC. This indicates that gyrI in multicopy did not confer resistance in DM803. Thus, gyrI multicopy-mediated resistance to MMC was dependent upon lexA function; this gyrI function was thus defined to be a part of the SOS response; a result congruent with other studies of this gene (6). In contrast, multicopy sdiA conferred resistance to MMC or nalidixic acid in both lexA+ and lexA(Ind) backgrounds, suggesting that this phenotype was at least not directly related to the lexA and recA circuitry which defines the SOS response. It was not clear to us why gyrI amplification in strain DM800 did not confer resistance to nalidixic acid as it did to MMC. This, at least, reflects differences in the action of the two chemicals. That rob amplification did not confer resistance in a DM800 or DM803 strain background reinforces that MMC resistance is a complex trait influenced by several genetic factors.
While sdiA conferred resistance in multicopy, a null mutation in sdiA did not result in hypersensitivity to MMC. It has been observed that the strain with sdiA in high copy, DPD2669, forms rounder and shorter cells than control cells in both the exponential and the stationary growth phases, and the same strain forms slightly shorter cells in the stationary phase than those in the exponential phase. These observations agree with the knowledge that sdiA overexpression speeds up cell division (43), and the expression of sdiA is decreased 50 to 80% in mid- to late-exponential growth phase with the appearance of an extracellular factor in the growth medium that specifically downregulates sdiA expression (18). It is also known that sdiA null mutants did not have obviously different phenotypes in cell division or growth (18). This may be because sdiA encodes a transcriptional activator only partially responsible for expression of the cell division genes ftsQAZ. Expression of ftsQAZ is controlled by at least two regulators, RpoS and SdiA (35). Eliminating SdiA function does not prevent ftsQAZ expression, while amplification of sdiA results in overproduction of ftsQAZ transcripts (44). In contrast, amplification of gyrI encoding a protein that inhibits DNA supercoiling, increased resistance to MMC (this work), while a null mutation in gyrI resulted in a strain that is twofold more sensitive to MMC and another DNA-damaging agent, microcin B17 (6).
Biosensors that can detect genotoxic agents have been developed (8, 40), providing one means of categorizing different DNA-damaging agents. The genetic titration of inhibitor action, coupled with the completed E. coli genomic sequence (11) and the availability of high-throughput, automated sequencing facilities, has proven to be a very powerful technique for the characterization of one DNA-damaging agent, MMC. The broad application of this methodology to several DNA-damaging agents may be quite informative.
ACKNOWLEDGMENT
Tina Van Dyk's provision of the pUC18-based and pBR322-based E. coli genomic DNA libraries was an instrumental starting point for this work. Prior work of Dana Smulski and David Elsemore on genetic titration of amino acid biosynthetic antagonists provided an experimental path for this study. We thank them also for reagents and methods. We thank Tim Bouret and Rick Howard (Nutrition and Health, DuPont) for sharing their expertise in microscopy and their insightful comments.
REFERENCES
- 1.Ahmer B M M, van Reeuwijk J, Timmers C D, Valentine P J, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun M N, Levy S B. Bacterial drug resistance: response to survival threats. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 323–366. [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Anderson R P, Roth J R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 6.Baquero M R, Bouzon M, Varea J, Moren F. sbmC, a stationary-phase induced SOS Escherichia coli gene, whose product protects cells from the DNA replication inhibitor microcin B17. Mol Microbiol. 1995;18:301–311. doi: 10.1111/j.1365-2958.1995.mmi_18020301.x. [DOI] [PubMed] [Google Scholar]
- 7.Belkin S, LaRossa R A. Biotechnological applications of microbial stress responses: new trends in environmental monitoring. In: Martins M T, Sato M I Z, Tiedje J M, Hagler L C N, Döbereiner J, Sanchez P S, editors. Progress in microbial ecology. Sao Paulo, Brazil: Brazilian Society for Microbiology; 1997. pp. 565–570. [Google Scholar]
- 8.Belkin S, Vollmer A C, Van Dyk T K, Smulski D R, Reed T R, LaRossa R A. Oxidative and DNA damaging agents induce luminescence in E. coli harboring lux fusions to stress promoters. In: Cambell A K, Kricka L J, Stanley P E, editors. Bioluminescence and chemiluminescence: fundamentals and applied aspects. Chichester, England: John Wiley & Sons, Ltd.; 1994. pp. 509–512. [Google Scholar]
- 9.Bennik M H, Pomposiello P J, Thorne D F, Demple B. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J Bacteriol. 2000;182:3794–3801. doi: 10.1128/jb.182.13.3794-3801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berlyn M K, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 11.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee P K, Sternberg N L. A general genetic approach in Escherichia coli for determining the mechanism(s) of action of tumoricidal agents: application to DMP840, a tumoricidal agent. Proc Natl Acad Sci USA. 1995;92:8950–8954. doi: 10.1073/pnas.92.19.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:735–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 14.Davidov Y, Rozen R, Smulski D R, Van Dyk T K, Vollmer A C, Elsemore D A, LaRossa R A, Belkin S. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat Res. 2000;46:97–107. doi: 10.1016/s1383-5718(99)00233-8. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsemore, D. A., R. A. LaRossa, and T. K. Van Dyk. September 1998. A method for identifying the site of action of xenobiotic chemicals. U.S. patent WO98/38336, patent PCT/US98/03684.
- 17.Falco S C, Dumas K S. Genetic analysis of mutants of Saccharomyces cerevisiae resistant to the herbicide sulfometuron methyl. Genetics. 1985;109:21–35. doi: 10.1093/genetics/109.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Lara J, Shang L H, Rothfield L I. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J Bacteriol. 1996;178:2742–2748. doi: 10.1128/jb.178.10.2742-2748.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenyon C J, Walker G C. DNA damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaRossa R A. Mutant selections linking physiology, inhibitors, and genotypes. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2527–2587. [Google Scholar]
- 21.LaRossa R A, Schloss J V. The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J Biol Chem. 1984;259:8753–8757. [PubMed] [Google Scholar]
- 22.Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for β-galactosidase activity. Anal Biochem. 1989;181:40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 24.Mount D W. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc Natl Acad Sci USA. 1977;74:300–304. doi: 10.1073/pnas.74.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mount D W, Low K B, Edmiston S J. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light-induced mutation. J Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mount D W, Walker A C, Kosel C. Suppression of lex mutations affecting deoxyribonucleic acid repair in Escherichia coli K-12 by closely linked thermosensitive mutations. J Bacteriol. 1973;116:950–956. doi: 10.1128/jb.116.2.950-956.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima H, Kobayashi K, Kobayashi M, Asako H, Aono R. Overexpression of the rob gene increases organic solvent tolerance and multiple antibiotic and heavy metal ion resistance in Escherichia coli. Appl Environ Microbiol. 1995;61:2302–2307. doi: 10.1128/aem.61.6.2302-2307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakanishi A, Oshida T, Matsushita T, Imajoh-Ohmi S, Ohnuki T. Identification of DNA gyrase inhibitor (GyrI) in Escherichia coli. J Biol Chem. 1998;273:1933–1938. doi: 10.1074/jbc.273.4.1933. [DOI] [PubMed] [Google Scholar]
- 29.Neidhardt F C, Savageau M F. Regulation beyond the operon. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1310–1324. [Google Scholar]
- 30.Paz M M, Das T A, Tomasz M. Mitomycin C linked to DNA minor groove binding agents: synthesis, reductive activation, DNA binding and cross-linking properties and in vitro antitumor activity. Bioorg Med Chem. 1999;7:2723–2726. doi: 10.1016/s0968-0896(99)00223-0. [DOI] [PubMed] [Google Scholar]
- 31.Rine J, Hansen W, Hardeman E, Davis R W. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci USA. 1983;80:6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth J R, Anton D N, Hartman P E. Histidine regulatory mutants in Salmonella typhimurium. I. Isolation and general properties. J Mol Biol. 1966;22:305–323. doi: 10.1016/0022-2836(66)90134-3. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schimke R T, Kaufman R J, Alt F W, Kellems R F. Gene amplification and drug resistance in cultured murine cells. Science. 1978;202:1051–1055. doi: 10.1126/science.715457. [DOI] [PubMed] [Google Scholar]
- 35.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens J C, Artz S W, Ames B N. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino acid deficiency. Proc Natl Acad Sci USA. 1975;72:4389–4393. doi: 10.1073/pnas.72.11.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomasz M, Palom Y. The mitomycin bioreductive antitumor agents: cross-linking and alkylation as the molecular basis of their activity. Pharmacol Ther. 1997;76:73–87. doi: 10.1016/s0163-7258(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 38.VanBogelen R A, Abshire K Z, Pertsemlidis A, Clark R L, Neidhardt F C. Gene-protein database of Escherichia coli K-12, edition 6. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2067–2117. [Google Scholar]
- 39.Vizan J L, Hernandez-Chico C, del Castillo I, Moreno F. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 1991;10:467–476. doi: 10.1002/j.1460-2075.1991.tb07969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vollmer A C, Belkin S, Smulski D R, Van Dyk T K, LaRossa R A. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahl G M, Padgett R A, Stark G R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-l-aspartate-resistant hamster cells. J Biol Chem. 1979;254:8679–8689. [PubMed] [Google Scholar]
- 42.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 43.Wang X D, de Boer P A, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei Y, Lee J-M, Smulski D R, LaRossa R A. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White D G, Goldman J D, Demple B, Levy S. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]