Abstract
In Escherichia coli the amplification of sdiA, a positive activator of ftsQAZ, genes that are essential for septation, results in mitomycin C resistance. To help us understand this resistance phenotype, genes whose expression was altered by increased sdiA dosage were identified using a DNA microarray-based, comprehensive transcript profiling method. The expression of 62 genes was reduced by more than threefold; of these, 41 are involved in motility and chemotaxis. Moreover, the expression of 75 genes, 36 of which had been previously characterized, was elevated at least threefold. As expected, increased sdiA dosage led to significantly elevated sdiA and ′ddlB-ftsQAZ-lpxC operon expression. Transcription of two genes, uvrY and uvrC, located downstream of sdiA and oriented in the same direction, was elevated about 10-fold, although the intervening gene, yecF, of opposite polarity was unaffected by increased sdiA dosage. Three genes (mioC and gidAB) flanking the replication origin, oriC, were transcribed more often when sdiA dosage was high, as were 12 genes within 1 min of a terminus of replication, terB. Transcription of the acrABDEF genes, mapping in three widely spaced loci, was elevated significantly, while several genes involved in DNA repair and replication (e.g., nei, recN, mioC, and mcrC) were moderately elevated in expression. Such global analysis provides a link between septation and the response to DNA-damaging agents.
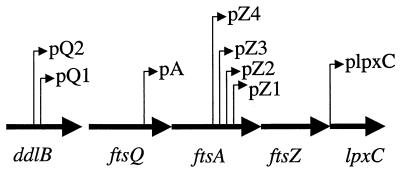
A key early element in cell division is the formation of a septation ring composed of FtsZ polypeptides (19). FtsZ levels must be buffered; overproduction in Escherichia coli leads to a hyperdivision phenotype, while an FtsZ deficiency leads to filamentation (19). Transcription of ftsZ is regulated in a complex manner (see the schematic in Fig. 1 [34]). The pQ1 promoter is rpoS regulated, while the pQ2 promoter is activated by the sdiA-encoded protein. The pA promoter is responsive to the RcsB-RcsC two-component system (13). SdiA is homologous to LuxR (34, 38), the quorum-sensing positive activator of Vibrio fischeri luminescence (21).
FIG. 1.
Transcriptional organization of the ftsZ region.
Besides the ftsQAZ genes, the sdiA gene product of Salmonella enterica serovar Typhimurium positively regulates expression of several genes of unknown function resident in an operon on its virulence plasmid (1), suggesting that its action is pleiotropic. Such pleiotropy has also been suggested by the findings that multicopy plasmids harboring sdiA overcome the inhibitory action of mitomycin C (40), a DNA-damaging agent that intercalates and forms adducts with the genetic material (36). Together, these results suggest definition of the sdiA controlled modulon (23) as a worthwhile exercise since interconnections between cell division, virulence, and DNA metabolism may be unearthed. To this end, we have utilized a recently described E. coli whole-genome, high-density microarray method (39) for obtaining comprehensive expression profiles of strains with either a normal or an elevated dose of sdiA.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this work were E. coli K-12 derivatives (Table 1). They were grown at 37°C in Luria-Bertani (LB) broth (8). When necessary, 150 μg of ampicillin per ml was included in the medium. Soft-agar plates contained 0.3% agar. Liquid cultures were aerated by rotary shaking at 250 rpm. Strains DPD2668 and DPD2669 were grown overnight in LB broth with ampicillin before subculturing by a 250-fold dilution in the same medium. The cells were rapidly collected for total RNA extraction (39) when the culture reached an optical density at 600 nm of 0.45. Electrotransformation was used to introduce plasmids into host strains (32).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| E. coli | ||
| RFM443 | rpsL200 galK2 lacΔ74 | 22 |
| DPD2668 | RFM443/pUC19 | 40 |
| DPD2669 | RFM443/pDEW140 | 40 |
| Plasmid | ||
| pDEW140 | pUC19 plus sdiA (EcoRI) | 40 |
RNA purification, cDNA labeling, and hybridization to DNA microarrays.
These methods have been described previously (39). Total RNA was purified from cell pellets using Qiagen RNeasy Mini Columns (Qiagen, Inc., Valencia, Calif.), with slightly revised protocols. Next, 25 μg of purified total RNA was used as a template for cyanine (Cy3)-labeled cDNA synthesis using random hexamers as primers. cDNA synthesis incorporating Cy5-labeled nucleotide and hybridization with an E. coli whole-genome, high-density microarray were performed as described elsewhere (39).
Determination of specific transcripts levels by quantitative, real-time PCR following reverse transcription.
Bulk cDNA samples were synthesized from total RNA derived from strains DPD2668 and DPD2669 using TaqMan Reverse Transcription Reagents (PE Applied Biosystems, Foster City, Calif.) and random hexamers as primers. Specific primer pairs were designed with the ABI PRISM Primer Express software (PE Applied Biosystems) for several genes as listed in Table 2. The genetic nomenclature of Blattner et al. (6) was followed. A real-time PCR reaction was performed with each specific primer pair using SYBR Green PCR Master Mix (PE Applied Biosystems). Equal amounts of cDNA (0.55 μg), derived from each bulk RNA sample, were used as the initial template in amplification reactions. The reactions were run on an ABI PRISM 7700 Sequence Detection System (PE Applied Biosystems), during which the fluorescence signal from SYBR Green intercalation was monitored to quantify the double-stranded DNA product formed after each PCR cycle. The threshold cycle (Ct) is the first cycle for which a statistically significant increase in the amount of the PCR product is detected. Ct values are thus inversely proportional to the amount of the RNA species in the original bulk RNA sample. The Ct was determined for each amplification reaction. ΔCt between samples derived from DPD2668 and DPD2669 was calculated for each tested gene. Since PCR products double with each amplification cycle, the fold difference in the initial concentration of each transcript equals 2ΔCt.
TABLE 2.
Primer pairs used to compare transcript levels
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| acrA | CAGGCGCAGGAAGTCGTT | CAACCGTGCAGCAACTTGAT |
| acrB | CAGGGTGCTTCGCTGCTT | GCCTTGGTCACGAAAAACTGA |
| acrR | GCGTCGCGCAGCAATTAT | GGCAAAGAGCCAGTTTTCCA |
| b0517 | GCCTGCTGCACAAAATAAGAGA | TTGCGGTGGTCGGTATCAG |
| b1606 | CGCGGTAGCGACAAACATATT | ACGAGCGGGTCATATTATCCA |
| b1707 | AGCATGACGCGGGTGAA | AGGTAATTTGGGAGCAGGCAAT |
| dniR | GGCTGTTAAGCGGCGTATTG | GGCTTCAGGCGAAATTGCT |
| feoA | TCGGCGACCCCATTCAT | GGCCAGATCTTTTTTGCGTAAT |
| gusR | TGACGATCACGGAATTTGTTG | CTGGGCTTATCTCGCCTTTTT |
| icdA | CCGGCACAAGGCAAGAAG | AGGGATAATCGGATTTTCAGGAA |
| mdh | GCCCATAGACAGGGTTGCA | AACGCGGGTACTGAAGTGGTT |
| mioC | AGGAGAAAGGTTGTCCGGAATAT | CAGGGATCTGGCTGGTTATCA |
| nei | GATGTGCTGGATCCGAATCTG | AAAGCGCGGCGACAATAA |
| sdiA | TGCAACGGGAAAAGGACAA | GCGGTGTCACTCAGTATTTAATGC |
| uvrC | GAACGCCACACCAATAACGTC | ACGCGTCACCGAAAAACAAT |
| uvrY | CTGTGGAACGCGCGATTT | ACATGAGTATGCCGGGCATT |
| ybgI | GAACAGCGCGGCGATTAG | CACCGTCTCTTTGCCTTCCA |
| ybgJ | CAGGCGGACCGGATTTG | CAACAACCTGTTTTTCGCTCAA |
| ybgK | GGTTTTCGCCAGTCGGGTAT | ATGCGTAACGCGGGCATA |
| ybgL | GATTCGTGCAGGCAAGCAA | CCGCGATCGGCAAACA |
Motility tests.
Fresh single colonies of strain DPD2668 or strain DPD2669 were picked and stabbed to the center of LB soft-agar plates supplemented with ampicillin. The plates were incubated at 37°C. The sizes of the swarm zones were compared both after 8 h and after overnight incubation.
Determination of MICs in liquid medium.
Cultures were grown in LB medium supplemented with 100 μg ampicillin per ml to ca. 3 × 108 cells/ml. After a 1:1,000 dilution in the same medium, 50-μl aliquots (ca. 15,000 CFU) were inoculated into the wells of a microtiter plate seeded with an equal volume of the same medium containing a second drug whose MIC was to be determined. Tetracycline, nalidixic acid, rifampin, kanamycin sulfate, chloramphenicol, and spectinomycin were tested in duplicate, twofold dilution series yielding the final concentration ranges of 10 to 0.078, 20 to 0.16, 50 to 0.39, 25 to 0.2, 20 to 0.16, and 100 to 0.78 μg/ml, respectively. The negative controls contained medium that was not supplemented with a second drug. After a static, overnight incubation in a humidified chamber, the absorbance of each well at 590 nm was read using a PE HTS 7000 Plus BioAssay Reader (Perkin-Elmer, Boston, Mass.) running the program HTSoft1.0. In this assay, MICs were defined as the lowest concentration of a drug reducing the final culture absorbance by a factor of 2.
RESULTS AND DISCUSSION
Amplification of sdiA has global impact on gene expression.
DPD2668 has a single, chromosomal copy of sdiA and harbors pUC19, while DPD2669 bears sdiA on both the chromosome and pDEW140 (40), a pUC19-based, high-copy plasmid. The growth rates of the two strains were indistinguishable (data not shown). The comprehensive transcript profiles of the two strains from exponential-phase, broth-grown cultures were compared. The presence of sdiA in high copy elevated the expression of ca. 9% of the E. coli protein encoding genes (open reading frames [ORFs]) by a factor of at least 2 while more than 2% of the ORFs appeared to be repressed by a factor of at least 2. Table 3 lists genes whose expression was elevated at least threefold due to the sdiA plasmid, while Table 4 indicates genes that were repressed threefold or more by the presence of a high sdiA copy. These tables follow the functional groupings proposed by Riley and Labedan (28) and used previously in analyses of microarray experiments (39).
TABLE 3.
Transcript levels elevated at least threefold by sdiA amplification as determined by probing of microarrays
| Functional group and gene | Fold increase |
|---|---|
| Cell division | |
| ftsA | 10.0 |
| ftsQ | 8.8 |
| ftsZ | 11.0 |
| sdiA | 30.0 |
| Transport of amino acids, carbohydrates, and cations | |
| glnH | 4.0 |
| xylF | 3.8 |
| trkG | 4.1 |
| DNA degradation | |
| mcrC | 3.5 |
| uvrC | 9.3 |
| DNA replication, repair, restriction, and modification | |
| gidA | 4.1 |
| mioC | 7.0 |
| nei | 8.6 |
| recN | 3.6 |
| Metabolism of small molecules | |
| argA | 3.4 |
| lysR | 3.6 |
| gst | 3.1 |
| Central intermediary metabolism | |
| agaI | 3.3 |
| agaD | 4.5 |
| speC | 9.5 |
| gcvA | 4.6 |
| pntA | 10.4 |
| pntB | 8.2 |
| Degradation of carbon compounds | |
| galE | 3.8 |
| galK | 4.1 |
| galT | 4.9 |
| gusR | 8.0 |
| Energy metabolism (carbon) | |
| dniR | 5.8 |
| pdhR | 4.4 |
| Drug or analog sensitivity | |
| acrA | 6.8 |
| acrD | 3.0 |
| acrE | 14.0 |
| acrF | 6.3 |
| acrR | 4.5 |
| Structural elements | |
| ddlB | 4.6 |
| lpxC | 3.4 |
| Not classified, uvrY | 11.9 |
| ORFs of unknown function | |
| sprT | 3.8 |
| b0135 | 6.4 |
| b0141 | 4.6 |
| b0517 | 11.2 |
| b0530 | 6.5 |
| ybgI | 5.2 |
| ybgJ | 4.7 |
| ybgK | 6.4 |
| ybgL | 3.8 |
| b0767 | 3.3 |
| b1069 | 3.8 |
| b1113 | 3.8 |
| b1498 | 3.7 |
| b1542 | 3.2 |
| b1545 | 4.4 |
| b1601 | 4.0 |
| b1606 | 8.0 |
| b1657 | 4.7 |
| b1707 | 30.0 |
| b1721 | 6.5 |
| b1756 | 3.2 |
| b1875 | 3.4 |
| b1955 | 5.1 |
| b1956 | 14.0 |
| b1967 | 6.6 |
| b2015 | 3.6 |
| b2016 | 3.5 |
| b2017 | 3.8 |
| b2301 | 3.8 |
| b2302 | 7.5 |
| b2420 | 3.2 |
| b2632 | 4.2 |
| b2642 | 15.0 |
| b2649 | 3.0 |
| b2874 | 3.3 |
| b3395 | 4.6 |
| b3397 | 3.5 |
| b3513 | 5.3 |
| b3888 | 3.4 |
| b4191 | 5.1 |
| b4221 | 5.0 |
TABLE 4.
Transcript levels decreased at least threefold by sdiA amplification as determined by probing of microarrays
| Functional group and gene | Fold decrease |
|---|---|
| Taxis and mobility | |
| air | 4.6 |
| cheA | 3.7 |
| cheB | 3.3 |
| cheW | 5.3 |
| cheY | 4.6 |
| cheZ | 4.0 |
| tar | 5.3 |
| tsr | 5.9 |
| Transport of amino acids, carbohydrates, or cations | |
| sdaC | 3.9 |
| glpF | 3.6 |
| malE | 3.7 |
| treB | 5.3 |
| fecA | 3.5 |
| fecE | 3.2 |
| kdpA | 3.5 |
| Metabolism of small molecules, glpK | 3.5 |
| Degradation of amino acids | |
| sdaB | 3.2 |
| tnaA | 4.0 |
| Energy metabolism (carbon) | |
| ackA | 3.1 |
| aldA | 3.3 |
| Cell exterior constituents, nanA | 3.7 |
| Surface structures | |
| flgB | 14.3 |
| flgC | 16.7 |
| flgD | 16.7 |
| flgE | 16.7 |
| flgF | 7.1 |
| flgG | 12.5 |
| flgH | 5.9 |
| flgI | 5.6 |
| flgJ | 6.3 |
| flgK | 6.3 |
| flgL | 11.1 |
| flgM | 7.1 |
| flgN | 5.6 |
| flhA | 3.1 |
| fliA | 10.0 |
| fliC | 14.3 |
| fliD | 4.6 |
| fliE | 5.9 |
| fliF | 11.1 |
| fliG | 9.1 |
| fliH | 5.0 |
| fliI | 3.9 |
| fliJ | 7.1 |
| fliK | 4.8 |
| fliL | 7.7 |
| fliM | 12.5 |
| fliN | 5.0 |
| fliO | 4.4 |
| fliP | 6.3 |
| fliR | 3.1 |
| fliS | 5.9 |
| fliT | 5.6 |
| fliZ | 8.3 |
| ORFs of unknown function | |
| b0290 | 3.5 |
| b1194 | 5.0 |
| b1880 | 3.6 |
| yjiZ | 3.3 |
| yjbP | 4.0 |
| yhjF | 4.8 |
Elevated level of sdiA transcripts due to gene amplification.
The presence of pDEW140, the pUC19 derivative harboring the sdiA promoter and ORF, resulted in a 30-fold elevation in the detection of sdiA transcripts. Thus, an increased gene dosage elevated the amount of the cognate mRNA.
Enhanced expression of genes near sdiA.
Amplification of sdiA elevated the expression of two genes downstream of sdiA; the uvrY transcript was increased 12-fold, while expression of uvrC increased by a factor of 9. These two genes are transcribed in the same direction as sdiA (6). uvrC specifies a subunit of an excision nuclease that removes bulky lesions (30), while uvrY encodes a cognate response regulator to the BarA sensor kinase (D. Georgillis, A. K. Pernestig, S. J. Normark, and O. Melefors, Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000, abstr. H89, p. 368). Both BarA and UvrY are needed for the production of extracellular siderophores (Georgillis et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000). Within the large region (751 bp) between sdiA and uvrYC is predicted to be an intervening gene, yecF, of opposite transcriptional polarity (6). The expression of yecF was unaffected by sdiA amplification. The lack of a heightened yecF hybridization signal in response to sdiA amplification suggests that sdiA and uvrYC are not cotranscribed. In analogy to the adjacent genomic locations of regulatory gene, target operon pairs such as lacI with lacZYA (4), and araBAD with araC (33), SdiA may directly activate uvrYC transcription.
Elevated expression of the ftsZ-containing operon.
sdiA encodes a positive activator that drives transcription of the ftsQAZ genes from promoter Q2 located within the upstream gene ddlB (34). It is not known where the transcript from the SdiA-dependent promoter ends. Increased quantities of RNA corresponding to this operon were thus expected. In strain MG1655 grown to exponential phase in LB broth, these transcripts range in quantity (the fraction of a particular transcript/summed transcripts hybridizing to all ORFs on the microarrays) from 240 to 540 ppm relative to the total ORF-specifying mRNA population (39). Amplification of sdiA, due to its presence on a multicopy plasmid, elevated the expression of transcripts hybridizing to the ddlB, ftsQ, ftsA, and ftsZ genes 5-, 9-, 10-, and 11-fold, respectively, relative to the strain that harbored pUC19 (Table 3). lpxC, the gene 101 bp downstream of ftsZ (6), also appeared to be induced fourfold (Table 3). Since the lpxC transcript is elevated in the pDEW140-containing strain and since a transcriptional terminator is not annotated between lpxC and ftsZ (6), it is reasonable to presume that lpxC is cotranscribed with ftsQAZ from the SdiA-dependent promoter. As expected, expression of the operon whose transcription is known to be activated by SdiA was highly elevated in a strain carrying an sdiA-containing, high-copy plasmid.
The complex structure of this operon (Fig. 1) suggests that the signal hybridizing to ddlB arises from a transcript that lacks the 5′ end of this coding sequence and is thus nonfunctional. This measurement indicates one limitation of microarray analyses using entire ORFs as the immobilized, capture reagents. This signal is misleading since it most likely does not lead to productive expression of DdlB at the protein level. Such inaccuracies may be avoided by subdivision of the ORFs serving as capture agents, a practice that is routinely used in an alternative methodology of microarray analysis (18). Moreover, this result indicates that transcript profiles will be most informative when interpreted within the context of previous studies.
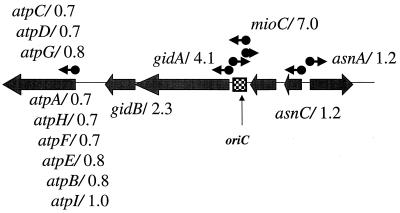
Actions near the origin and a terminus of replication.
Figure 2 depicts the genetic organization of the oriC region, the origin of DNA replication. The three genes immediately flanking oriC were overtranscribed relative to the vector-containing control strain. The cellular content of the mioC transcript was elevated sevenfold, whereas the expression of gidA and gidB were elevated four- and twofold, respectively. This effect was localized since the expression of flanking genes was not increased (Fig. 2).
FIG. 2.
oriC region. Genes are indicated by broad arrows placed on the central line representing a small portion of the circular chromosome. The box indicates the origin of replication. Arrows above the line indicate annotated promoters (6). The atp operon is indicated by the leftmost, broad arrow. The numbers following each gene indicate the fold elevation of gene expression caused by sdiA amplification.
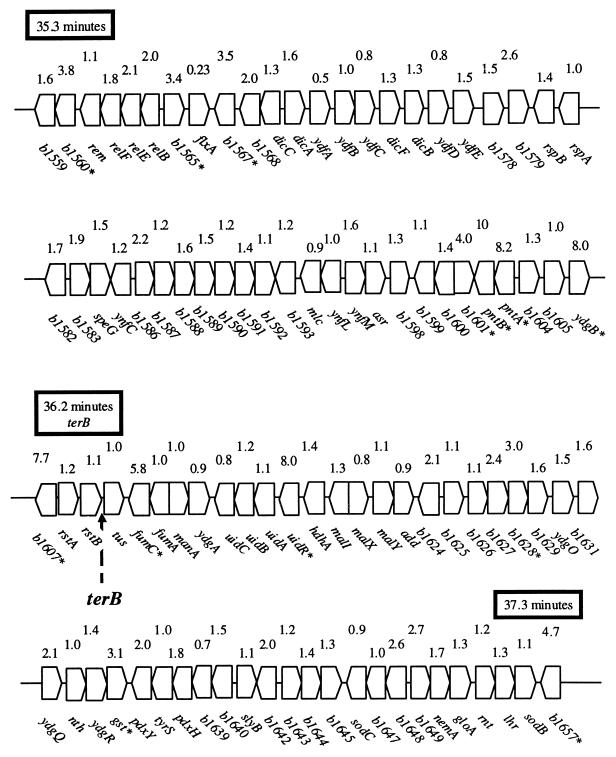
Figure 3 depicts the region (min 35.3 to 37.3) surrounding terB, a terminus of DNA replication. The transcription of 38 genes within this 88-gene region was elevated more than twofold by the presence of pDEW140; The expression of 12 genes was enhanced more than threefold. Unlike the action observed around the origin, oriC, the stimulation seen in the vicinity of terB resulted in a mosaic pattern relative to the gene order. Interestingly, the expression of tus, encoding the terminus-binding factor, was not affected.
FIG. 3.
terB region. The 2-min region of the chromosome surrounding terB (36.2 min) is depicted. The fold elevation, due to sdiA amplification as determined by comprehensive transcript profiling, is indicated above each gene. Genes whose transcripts were elevated >3-fold are marked with an asterisk.
Since SdiA enhances the expression of cell division genes (38), it may also directly or indirectly modulate DNA structure around the origin and terminus so that these regions are more exposed for the mechanical operations resulting in proper chromosomal segregation. The oriC site is attached to the cell membrane (19). mioC, gidA, and gidB, stimulated by sdiA amplification, are involved in DNA replication (26) and clustered with the chromosomal replication origin, oriC (5). If the role of SdiA is to stimulate the expression of genes required for septation during the cell division cycle, coordinate expression of the ftsZ-containing operon with those of mioC and gidAB might be built into the cellular regulatory logic. Alternatively, local structural alterations of the chromosome could be the cause of the observed changes in gene expression. It is possible that the elevated expression level of these three genes was the result of an SdiA-enhanced replication of the origin relative to the remainder of the chromosome, a copy number effect. Another possibility is that the elevated SdiA titer “relaxed” the structure of the oriC region, providing RNA polymerase with greater access to the gidAB and mioC promoters, an unmasking scenario. Thus, the precise mechanism of enhanced mioC and gidAB expression remains to be determined.
The transcript content of most genes surrounding the terB replication terminus also appeared to be induced moderately when sdiA was amplified. This region is subjected to aberrant recombination and replication arrest (11). Such DNA “gymnastics” could also make the region accessible to RNA polymerase when replication is enhanced by sdiA amplification. Garrido et al. observed that transcription of ftsZ peaked at the time of both the initiation and the termination of replication (9). This observation, which is consistent with the transcript profiling described here, is suggestive of coordination between SdiA action and the timing of cell division.
Heightened expression of acr genes.
Loss-of-function acr mutants (5) display enhanced sensitivity to a broad range of inhibitory agents. The acr genes of E. coli are organized into five operons, acrC (4.6 min), acrAB (10.4 min), acrR (10.4 min), acrD (55.6 min), and acrEF (73.5 min [5]). acrC encodes a transmembrane protein (5), while acrD specifies an aminoglycoside efflux pump (29). Interestingly, acrE mutants form filamentous chains of cells indicative of a septation defect, while acrF encodes a lipoprotein (5). The acrAB operon (25)-encoded proteins are components of a major drug efflux pump (2) that, together with TolC, a protein that forms an outer membrane porin, constitute an efflux channel from the cytoplasm out to the culture medium (2, 24, 25). Perhaps, an increased SdiA concentration within the cell, like increased levels of the transcriptional activator Rob (35), leads to elevated AcrA, AcrB, and TolC titers and hence efflux pores for mitomycin C expulsion. This hypothesis is consistent with the heightened sensitivity of a tolC mutant to mitomycin C (7).
In addition to the dramatic, 14-fold change in expression of acrE in response to sdiA amplification, other acr structural genes also displayed an increase in transcript quantity. The acrA, acrD, and acrF transcripts were elevated seven-, three-, and sixfold by multiple copies of sdiA. acrB, however, is 1 of 61 genes not present in the E. coli microarray (39). The inability to detect increased expression of this gene is thus expected. Another method, however, indicated that acrB expression was also elevated (see below). Evidence for elevated expression of acr genes in each locus was found as indicated by the fold expression reported in Table 3. tolC expression was also heightened by a factor of 2.6 due to the presence of pDEW140 (data not shown).
Enhanced expression of other operons that may contribute to the mitomycin C resistance phenotype.
Elevated transcription of the gal operon genes (galEKTM) at min 17 (with basal expression levels of 221, 285, 140, and 198 ppm, respectively) was observed in the strain bearing the sdiA amplification. These genes, which were moderately expressed when strain MG1655 was grown in LB broth (average ppm = 210; ranked 841st, 599th, 1,512th, and 963rd, respectively [39]) were elevated 3.8-, 4.9-, 4.1-, and 2.3-fold, respectively. The expression of other sugar catabolic genes was not induced. Since galactose is incorporated into the K-12 lipopolysaccharide core (27), gal operon elevation could reflect an altered envelope structure.
At min 16 is the ybgIJKL-nei region. sdiA amplification elevated expression of these genes 5.2-, 4.7-, 6.4-, 3.8-, and 8.6-fold. These genes, transcribed in the same orientation, are organized into an operon (10) since the ORFs are densely packed, at times overlapping (6) without internal promoters (10). nei encodes endonuclease VIII, an enzyme that removes oxidized pyrimidine bases from DNA (16, 31), but the functions of the other genes in this operon are unknown. Since reactive oxygen species are implicated in mitomycin C-mediated cell killing (14) and mutagenesis (17), elevated expression of this operon could contribute to the mitomycin C resistance of cells harboring the sdiA amplification (40).
Elevated expression of other genes.
ORF b1707 expression was elevated most drastically, displaying a 30-fold induction. In addition, speC, b0517, b1606 and b2642 were all induced at least eightfold (see Table 3), comparable to the fold induction of the ftsQAZ mRNA. The expression of b2015 and b2016, two adjacent genes at min 45, was elevated 3.6- and 3.5-fold, respectively. b2015 is predicted to be a LysR-type transcriptional regulator (6), and the putative b2016 gene product has low overall homology (E value = 10−5 by the BLASTn program [3]) to hexose 4,6-dehydratase from several gram-positive bacteria (6). Similarly, the expression of two other linked genes, b4221 and b4222, of unknown function was elevated 5.0- and 4.8-fold while transcripts of b2301 and b2302 were elevated 3.8- and 7.5-fold, respectively. The latter pair of ORFs encodes putative glutathione S-transferases (6). Six putative fimbria-like protein genes (6) (b0135, b0136, b0137, b0138, b0141, and b0530) were induced ca. fivefold, among which the first four appeared to be in an operon. Thus, the first experimental evidence for the cotranscription of at least five potential operons was provided by the microarray analysis; elevated transcripts of uvrYC, b2015-b2016, b2301-b2302, b4221-b4222, and b0135-b0136-b0137-b0138 were each observed upon sdiA amplification.
Downregulation of motility-related genes.
Amplification of sdiA caused the expression of 62 genes to decrease by a factor of 3 or more; of these, 41 were involved in motility and chemotaxis. If a more-stringent cutoff of fivefold was imposed, then expression of 34 the genes were downregulated, 30 of which function in chemotaxis or motility (cheW, flgBCDEFGHIJKLMN, fliACEFGHJLMNPSTZ, tar, and tsr). Interestingly, the expression of master regulator genes flhC and flhD (basal levels of 230 and 70 ppm, respectively [39]), controlling flagellum operon expression, was lowered by only about one-third.
Motility defect associated with multiple copies of sdiA.
Since many genes involved in flagellum biosynthesis, chemotaxis, and motility were dramatically repressed in the sdiA overproducing strain, a motility defect was predicted. The swarming of strains having single or multiple copies of sdiA was examined by spotting four single-colony isolates of each strain onto semisolid medium. After 8 h of incubation at 37°C, DPD2668 harboring pUC19 had swarmed (diameter, 32 ± 2.5 mm), while DPD2669 containing pDEW140 had not (diameter, 3.2 ± 0.4 mm). After 23 h of incubation, DPD2668 had filled the petri plate (diameter, 100 mm), while DPD2669 had expanded to a lesser extent, covering about one-half of each plate. There was no obvious difference in the growth rate in LB broth supplemented with ampicillin between the two strains when monitored by absorbance. Thus, the leaky motility phenotype described above could be explained by either (i) plasmid loss allowing swarming of a revertant (sdiA+ haploid) population as ampicillin was exhausted from the medium or (ii) sdiA amplification only partially compromising motility. To distinguish between these possibilities, the site of inoculation and the edge of the swarm after 23 h were streaked for single colonies to an ampicillin-containing LB agar plate. Massive sdiA+ plasmid loss from cells at the edge of the swarm was not observed, suggesting that the motility defective phenotype was not an absolute one.
Comparison of the transcript profiles of the sdiA overexpression strain and its control indicated that sdiA overexpression may result in a deficiency in movement, as confirmed by motility tests. It is not clear how genes involved in chemotaxis and movement are regulated by SdiA. It is possible that control cascades through the regulatory genes flhCD. Since the quantity of these transcripts in cells grown in LB broth is relatively low (39), the small reduction in flhCD expression might be enough to limit the cells' capacity to swarm.
Independent method confirms elevation of gene expression by sdiA amplification.
Comprehensive transcript profiling is well suited to surveying the global changes associated with an altered genotype. An alternative method, quantitative real-time RT-PCR, was used to verify the expression level changes of representative genes caused by increased sdiA dosage. For these confirmatory experiments, total RNA samples, extracted from fresh cultures, were used.
The expression of acrB, 17 genes scored as inducible in the comprehensive transcript profiling analysis (Table 3) and 2 control genes (icdA and mdh), was quantified. The results are summarized in Table 5. A twofold increase in acrB expression was observed upon sdiA amplification, a result similar to the 2.3-fold elevation of acrA expression determined by the same method. Discrepancies in the measurements of the b0157 (<1.5-fold increase in reverse transcriptase PCR (RT-PCR) versus an ∼11-fold increase in the comprehensive microarray experiment) and acrR (decreased 1.2-fold as measured by RT-PCR versus a 4.5-fold increase as monitored by the microarray-based approach) transcripts were observed between the two methods. Since AcrR is a negative regulator of acrAB operon expression (20), we postulate that the microarray measurement of the acrR transcript was erroneous. Reassuringly, the expression changes of the other 15 genes, scored as elevated in the comprehensive transcript profile, were confirmed by the RT-PCR method; they showed the same trend and a similar magnitude of change with both methodologies. Overall, smaller changes were routinely observed with RT-PCR, perhaps attributable to high background fluorescence associated with SYBR Green.
TABLE 5.
Fold induction of specific transcripts attributed to sdiA amplification as determined by probing of microarrays and amplification of cDNA samples
| Gene | Fold changea
|
|
|---|---|---|
| Amplification | Probing | |
| sdiA | 12.7 | 30.0 |
| b1707 | 8.9 | 30.0 |
| b1606 | 4.8 | 8.0 |
| uvrY | 4.9 | 11.9 |
| uvrC | 4.1 | 9.3 |
| gusR | 3.2 | 8.0 |
| nei | 3.2 | 8.6 |
| ybgL | 2.7 | 5.2 |
| ybgK | 2.4 | 4.7 |
| ybgJ | 2.4 | 6.4 |
| ybgI | 2.1 | 3.8 |
| acrA | 2.3 | 6.8 |
| acrB | 2.0 | ND |
| dniR | 2.0 | 5.8 |
| feoA | 2.0 | 5.8 |
| mioC | 1.9 | 7.0 |
| b0517 | 1.4 | 11.2 |
| acrR | 0.8 | 4.5 |
| icdA | 1.1 | <1.5 |
| mdh | 1.1 | <1.5 |
Amplification results were obtained by reverse transcription of bulk RNA followed by PCR amplification. Probing results were obtained with fluorescently labeled cDNA hybridized to DNA microarrays. ND, not determined.
sdiA in high copy conferred resistance to antibiotics with different modes of action.
The responses of a control strain, DPD2668, and DPD2669 harboring pDEW140, the sdiA plasmid, to the antibiotics rifampin, chloramphenicol, nalidixic acid, tetracycline, spectinomycin, and kanamycin were examined in liquid medium. The MICs were identical between the two strains when challenged with rifampin, chloramphenicol, spectinomycin, and kanamycin, having values of 6.3, 2.5, 12.5, and 3.1 μg/ml, respectively. The presence of the sdiA plasmid in DPD2669, however, raised the MIC for tetracycline from 0.6 to 5 μg/ml and that for nalidixic acid from 1.3 to 10 μg/ml. These results are consistent with the role of AcrA and AcrB in efflux (2, 24). They differ, however, in some details compared to studies of acr loss-of-function mutants (25). This may reflect differences in information garnered from genetic alterations causing either loss or gain of function (15).
Summation.
E. coli high-density microarrays have been successfully used to quantify the entire complement of individual mRNA transcripts (39). It has also been used to profile the gene expression level changes upon chemical treatment (T. K. Van Dyk et al., unpublished results; Y. Wei and R. A. LaRossa, unpublished results; M. Zheng et al., unpublished results; Y. Wei, D. G. Söll, and R. A. LaRossa, unpublished). This work showed its utility in determining the global effects of gene dosage amplification. Expression of sdiA is very low (15 ppm) in broth-grown, exponential-phase E. coli (39). The 30-fold elevation of the sdiA transcript in DPD2669 containing sdiA in high copy, compared with that in DPD2668, reflected the amplification of the sdiA gene due to its location on both the chromosome and a multicopy plasmid. As a consequence of sdiA hyperexpression, transcription of the ′ddlB-ftsQAZ-lpxC operon, the only E. coli genes previously known to be activated by SdiA in E. coli (38), was greatly increased. In addition, the expression of genes falling into a few other functional categories (e.g., cell division, DNA replication and repair, drug sensitivity, and macromolecular metabolism) was raised significantly by sdiA amplification. Perhaps SdiA serves as a positive activator of these genes, as well as a few other genes whose functions are obscure.
We previously found that sdiA in high copy conferred resistance to mitomycin C upon E. coli (40). This phenotype (40) is not dependent upon the SOS response (37). When examining the expression profiles from microarray experiments, we found that many genes involved in DNA replication, degradation, repair, transposition, and the stability of chromosomal structure in addition to nei and uvrC were induced moderately. This suggests increased capacity for chromosomal replication and repair upon sdiA overexpression. This may explain why the amplification of sdiA confers mitomycin C resistance.
Alternatively, acr mutations cause sensitivity to acriflavines, molecules that intercalate into double-stranded DNA containing monotonic runs of base pairs (12). Most acr mutants display a defect in acridine efflux; moreover, they are often pleiotropic, being hypersensitive to a wide variety of chemicals (2, 24). Thus, hyperexpression of these genes in a strain harboring an sdiA-bearing multicopy plasmid could lead to mitomycin C expulsion and therefore result in the observed resistance to this DNA-damaging agent. It may also explain why sdiA in multicopy could confer resistance to a broad spectrum of antibiotics. Thus, microarray methods, like other technologies, become part of the scientific method, generating hypotheses requiring further study.
ACKNOWLEDGMENTS
Discussions with J. A. Rafalski, T. Van Dyk, and A. Vollmer were most helpful during the progress of this work.
REFERENCES
- 1.Ahmer B M M, van Reeuwijk J, Timmers C D, Valentine P J, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun M N, Levy S B. Bacterial drug resistance: response to survival threats. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 323–366. [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckwith J. The operon: an historical account. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1227–1231. [Google Scholar]
- 5.Berlyn M K, Low K B, Rudd K E. Linkage map of Escherichia coli K-12. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ed. 9. Washington, D.C.: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 6.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Davidov Y, Rozen R, Smulski D R, Van Dyk T K, Vollmer A C, Elsemore D A, LaRossa R A, Belkin S. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat Res. 2000;46:97–107. doi: 10.1016/s1383-5718(99)00233-8. [DOI] [PubMed] [Google Scholar]
- 8.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 9.Garrido T, Sanchez M, Palacios P, Aldea M, Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gifford C M, Wallace S S. The genes encoding endonuclease VIII and endonuclease III in Escherichia coli are transcribed as the terminal genes in operons. Nucleic Acids Res. 2000;28:762–769. doi: 10.1093/nar/28.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill T M. Features of the chromosomal terminus region. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1602–1614. [Google Scholar]
- 12.Hutchinson F. Mutagenesis. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2218–2235. [Google Scholar]
- 13.Joseleau-Petit D, Vinella D, D'Ari R. Metabolic alarms and cell division in E. coli. J Bacteriol. 1999;181:9–14. doi: 10.1128/jb.181.1.9-14.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishna M C, DeGraff W, TAmura S, Gonzalez F J, Samuni A, Russo A, Mitchell J B. Mechanisms of hypoxic and aerobic cytotoxicity of mitomycin C in Chinese hamster V79 cells. Cancer Res. 1991;51:6622–6628. [PubMed] [Google Scholar]
- 15.LaRossa R A. Mutant selections linking physiology, inhibitors, and genotypes. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2527–2587. [Google Scholar]
- 16.Laval J. Role of DNA repair enzymes in the cellular resistance to oxidative stress. Pathol Biol. 1996;44:14–24. [PubMed] [Google Scholar]
- 17.Levin D E, Holllstein M, Christman M F, Schwiers E A, Ames B N. A new Salmonella tester strain (TA102) with AT base pairs at the site of mutation detects oxidative mutagens. Proc Natl Acad Sci USA. 1982;79:7445–7449. doi: 10.1073/pnas.79.23.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittman M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 19.Lutkenhaus J, Mukherjee A. Cell division. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1615–1626. [Google Scholar]
- 20.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 21.Meighen E A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for β-galactosidases activity. Anal Biochem. 1989;181:40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- 23.Neidhardt F C, Savageau M F. Regulation beyond the operon. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1310–1324. [Google Scholar]
- 24.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikaido H. Outer membrane. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 29–47. [Google Scholar]
- 26.Ogawa T, Okazaki T. Cell cycle-dependent transcription from the gid and mioC promoters of Escherichia coli. J Bacteriol. 1994;176:1609–1615. doi: 10.1128/jb.176.6.1609-1615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rick P D, Silver R P. Enterobacterial common antigen and capsular polysaccharides. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 104–122. [Google Scholar]
- 28.Riley M, Labedan B. Escherichia coli gene products: physiological functions and common ancestries. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2118–2202. [Google Scholar]
- 29.Rosenberg E Y, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182:1754–1756. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rupp W D. DNA repair mechanisms. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2277–2294. [Google Scholar]
- 31.Saito Y, Uraki F, Nakajima S, Asaeda A, Ono K, Kubo K, Yamamot K. Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J Bacteriol. 1997;179:3783–3785. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schleif R. Two positively regulated systems, ara and mal. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1300–1309. [Google Scholar]
- 34.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T, Horii T, Shibayama K, Sato K, Ohsuka S, Arakawa Y, Yamaki K, Takagi K, Ohta M. robA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol Immunol. 1997;41:697–702. doi: 10.1111/j.1348-0421.1997.tb01913.x. [DOI] [PubMed] [Google Scholar]
- 36.Tomasz M, Palom Y. The mitomycin bioreductive antitumor agents: cross-linking and alkylation as the molecular basis of their activity. Pharmacol Ther. 1997;76:73–87. doi: 10.1016/s0163-7258(97)00088-0. [DOI] [PubMed] [Google Scholar]
- 37.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 38.Wang X D, de Boer P A, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei Y, Lee J-M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. High-density miroarrray mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Y, Vollmer A C, LaRossa R A. In vivo titration of mitomycin C action by four Escherichia coli genomic regions on multicopy plasmids. J Bacteriol. 2001;183:2259–2264. doi: 10.1128/JB.183.7.2259-2264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]