Abstract
Cell membrane affinity chromatography has been widely applied in membrane protein (MP)-targeted drug screening and interaction analysis. However, in current methods, the MP sources are derived from cell lines or recombinant protein expression, which are time-consuming for cell culture or purification, and also difficult to ensure the purity and consistent orientation of MPs in the chromatographic stationary phase. In this study, a novel in situ synthesis membrane protein affinity chromatography (iSMAC) method was developed utilizing cell-free protein expression (CFE) and covalent immobilized affinity chromatography, which achieved efficient in situ synthesis and unidirectional insertion of MPs into liposomes in the stationary phase. The advantages of iSMAC are: 1) There is no need to culture cells or prepare recombinant proteins; 2) Specific and purified MPs with stable and controllable content can be obtained within 2 h; 3) MPs maintain the transmembrane structure and a consistent orientation in the chromatographic stationary phase; 4) The flexible and personalized construction of cDNAs makes it possible to analyze drug binding sites. iSMAC was successfully applied to screen PDGFRβ inhibitors from Salvia miltiorrhiza and Schisandra chinensis. Micro columns prepared by in-situ synthesis maintain satisfactory analysis activity within 72 h. Two new PDGFRβ inhibitors, salvianolic acid B and gomisin D, were screened out with KD values of 13.44 and 7.39 μmol/L, respectively. In vitro experiments confirmed that the two compounds decreased α-SMA and collagen Ӏ mRNA levels raised by TGF-β in HSC-T6 cells through regulating the phosphorylation of p38, AKT and ERK. In vivo, Sal B could also attenuate CCl4-induced liver fibrosis by downregulating PDGFRβ downstream related protein levels. The iSMAC method can be applied to other general MPs, and provides a practical approach for the rapid preparation of MP-immobilized or other biological solid-phase materials.
Key words: Affinity chromatography, In situ synthesis of membrane proteins, Unidirectional insertion, PDGFRβ, Drug screening, Ligand–protein interaction, Antihepatic fibrosis
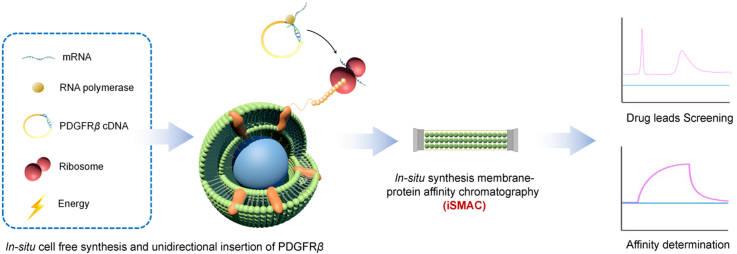
Graphical abstract
In-situ synthesis and unidirectional insertion of membrane proteins in liposome-immobilized silica stationary phase for drug screening and affinity determination.
1. Introduction
Membrane proteins (MPs) are responsible for the exchange of external materials and information with the cell, and various functions, such as intercellular contact, surface recognition, signal transduction, enzyme activity, and transport1. Therefore, they are the most promising potential drug targets. It is estimated that 60% of drug targets are MPs2. However, the focus of drug discovery on MPs is rather difficult due to their low abundance and complex structure. Three-dimensional structures of only 1% of these proteins have been successfully resolved3. The difficulty of purifying and simulating the natural conformation in vitro are bottlenecks in drug screening and interaction analysis with ligands of MPs.
The lipid bilayer environment is necessary for MPs to maintain their natural conformation and physiological activity4. Biochromatography, e.g., cell membrane chromatography (CMC) and cellular membrane affinity chromatography (CMAC), have been extensively applied in research on MPs5, 6, 7. CMC is an efficient method for screening the active components of membrane receptors by directly extracting the cell membrane and fixing it on silica gel in the stationary phase for chromatography8,9. Our research group has developed and applied a series of CMC screening systems to screen for active components in more than 30 herbal medicines10, 11, 12, 13. Further, stationary phase modification technology and protein overexpression strategies have been successfully combined with CMC to stabilize the lipids on columns to achieve specific screening purposes14,15. In one study, target MPs obtained directly from the cell membrane or followed by reconstitution into immobilized artificial membranes (IAMs) were immobilized in a stationary phase and used for drug discovery16. Recently, CMAC columns containing target proteins have been extended to characterization of the relative extent and type of binding of drug candidates as well as target binding17, 18, 19, 20.
However, CMC and CMAC have a number of limitations: directly obtaining the cell membrane as the source of MPs leads to low specificity and abundance, which is unsuitable for screening and interaction analysis of single target proteins; during the process of reconstitution into the membrane and immobilization in a carrier in the stationary phase, MPs have a random orientation, so exposure of the active binding sites cannot be controlled; and preparation of sufficient recombinant MPs is expensive, labor-intensive, and time-consuming.
An in vitro expression system, cell-free expression (CFE), may provide an ideal route for the expression of MPs. The principle involves taking the DNA or mRNA of the target protein as the template and expressing it in vitro in the presence of amino acids, RNA polymerase, and an energy source in the cell extract21. CFE circumvents the physiological limitations of living cells, greatly improves the efficiency and yield of protein synthesis, and decreases protein misfolding and aggregation22. It is particularly suitable for application to MPs23, as the provision of an artificial biomimetic membrane in the expression system allows MPs to be directly expressed, co-translated, and inserted into the phospholipid membrane during synthesis24,25. In addition, the protein shows vectorial insertion into solid-supported biomimetic membranes in the process of synthesis. Sinner group25,26 confirmed that OR5 synthesized by CFE can be inserted into a tethered membrane surface in a functional and oriented form. CFE has been used in high-throughput protein expression, screening, and synthesis of protein drugs. Protein in situ arrays can be quickly and effectively generated by CFE and have been used in the study of protein–protein interaction networks on microchips27,28. CFE is a practical method for obtaining sufficient functional MPs with consistent orientation. However, it has not been applied to porous silica for use in MP-immobilized bioaffinity chromatography.
Therefore, for the unidirectional immobilization of specific MPs on a silica gel in the stationary phase to allow the rapid preparation of bioaffinity chromatography, we developed a novel method, in situ synthesis membrane-protein affinity chromatography (iSMAC), integrating the advantages of CFE and 3-mercaptopropyltrimethoxysilane (MPTS)-modified bioaffinity chromatography29. This method allows the in situ cell-free synthesis and unidirectional insertion of MPs into a liposome-immobilized silica carrier. We successfully used it for high-efficiency and accurate screening of PDGFRβ inhibitors, as well as characterization of the interaction between PDGFRβ and its ligands.
Hepatic fibrosis underlies many complex diseases in the end-stage of liver disease, and is a reversible pathological process30,31. Therefore, reversing hepatic fibrosis is an important goal to block the development of liver disease. It is generally recognized that the activation of hepatic stellate cells (HSCs) is the central link in the occurrence and development of hepatic fibrosis32, so clinical treatment mainly involves inhibition of HSCs activation by interferon and nucleoside analogs33. PDGF is the most powerful mitogen known to promote HSCs division34. When hepatic fibrosis occurs, PDGFRβ is the main subtype on the surface of HSCs mediating multiple signaling pathways, such as MAPK (p38), AKT and Ras/ERK35, 36, 37. Therefore, PDGFRβ has become one of the most important targets in drug development for liver fibrosis. Most PDGFRβ inhibitors developed to date have been kinase inhibitors with unsatisfactory specificity38. The development of drugs capable of acting on the extracellular ligand-binding domain of PDGFRβ is expected to yield higher specificity and lower toxicity. Salvia miltiorrhiza and Schisandra chinensis are herbal medicines with clinical antihepatic fibrosis effects39, 40, 41. In this study, we applied iSMAC to screen active compounds that target PDGFRβ to reverse liver fibrosis, that is, to identify compounds with high specificity and good curative effects for the treatment of liver fibrosis. For general MPs, iSMAC is a promising rapid analysis system for the screening of important compounds and the study of drug–protein interactions.
2. Materials and methods
2.1. Reagents and instruments
Silica gel (5 μm, 200 Å) was obtained from Qingdao Meigao Chemical Co., Ltd. (Qingdao, China). MPTS, N-(4-maleimide butyryl oxide) succinimide (GMBS), dimethyl sulfoxide (DMSO) were purchased from Sigma Co. (St. Louis, MO, USA). S. chinensis and S. miltiorrhiza were purchased from Leiyunshang Pharmacy Co., Ltd. (Shanghai, China). Escherichia coli S30 T7 High Yield Protein Expression System was purchased from Promega Corporation (Biotech Co., Ltd., Beijing, China). Dioleoylphosphatidylcholine (DOPC), dioleoyl phosphoethanolamine (DOPE), NBD-dioleoyl phosphatidylglycerole (NBD-DOPG) were purchased from Avanti Polar Lipids (Alabasta, AL, USA). Salvianolic acid B (Sal B), gomisin D (Gom D) and dexamethasone (DXMS) were purchased from Yuanye Biotech Co. Ltd., (Shanghai, China) (purities≥98%). PDGF-BB and antibodies were purchased from Abcam (Cambridge, Britain); HPLC-grade acetonitrile and MS-grade ammonia acetate were purchased from Sigma Co. (St. Louis, MO, USA).
2.2. Preparation of liposomes and plasmid
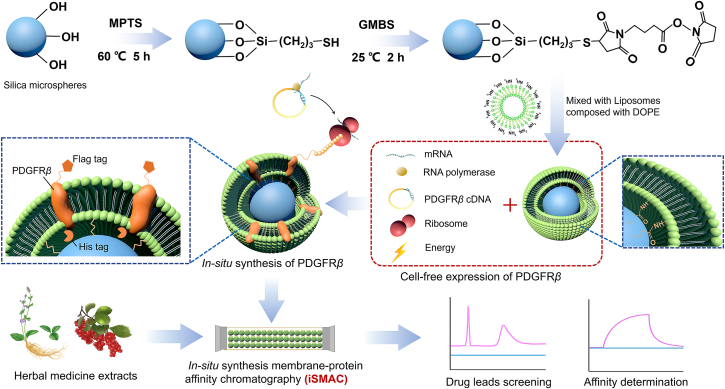
DOPC, DOPE, NBD-DOPG were dissolved in chloroform/MeOH (1:1, v/v) at 80:20:1 mol/L ratio to concentration of 20 mg/mL in a 25 mL flask. The solvent was evaporated by vacuum distillation. Then the thin lipid film was suspended with ultrapure water to obtain a 20 mg/mL lipid mix. The liposome vesicles were formed under ultrasound for 10 min at 400 W and then the liposome extruder (ATS Engineering Ltd., Suzhou, China) was used to extrude 3–4 times to form vesicles with a diameter of less than 100 nm.
The plasmid of PDGFRβ was constructed in Shanghai Jikai gene Chemical Technology Co., Ltd. (Shanghai, China). PDGFRβ was cloned to GV147 and named GV147-PDGFRβ, with a Flag-tag placed at N-terminal, a fusion fluorescent tag “cherry” and a 6His-tag at C-terminal (detailed plasmid vector was shown in Supporting Information Fig. S1).
2.3. In situ synthesis of PDGFRβ stationary phase
Pattern diagram of cell-free in situ synthesis of PDGFRβ was shown in Fig. 1. The principle of cell-free synthesis of membrane proteins is to take cDNA sequence of specific protein as the template, mixing it with cell extracts to provide synthesis raw materials including mRNA, RNA polymerase, ribosome and amino acids, and add phospholipid membrane to provide membrane environment for proteins. Then, membrane proteins can be synthesized rapidly on phospholipid membrane by imitating the synthesis form in cells.
Figure 1.
Pattern diagram of cell-free in situ synthesis of PDGFRβ and applications.
Firstly empty-protein stationary phase was prepared by mixing MPTS-modified silica gel15 (120 °C activation before use) and liposome by vortex then incubating overnight at 4 °C. PDGFRβ was expressed with bacteria lysates (E. coli S30 T7 High Yield Protein Expression System, L1110, Promega, USA) following the manufacturer's protocols. In detail, cDNA of PDGFRβ and membrane coated silica gel was simultaneously added to expression system to synthetize PDGFRβ at condition of 37 °C and 300 rpm shaking in a floor incubator shaker. PDGFRβ inserted stationary phase was then obtained by 3000×g centrifugation and washed for 3 times.
2.4. Characterization of PDGFRβ on stationary phase
2.4.1. Western blotting analysis
PDGFRβ on stationary phase was examined by Western blot. In detail, the stationary phase in 50 μL reaction system was lysed by RIPA (Beyotime Biotechnology, China) for 30 min and supernatant was collected by centrifugation at 3000×g. All was separated by SDS-PAGE and analyzed against primary antibody of PDGFRβ (Affinity Biosciences, Cat# AF6133, RRID: AB_2835016). The bands were scanned by Odyssey infrared Imaging System (Licor, USA) and quantified though Quantity One (Bio-Rad).
2.4.2. Scanning electron microscope (SEM) and confocal microscope analysis
The SEM data were obtained with an Phenom ProX (Phenom Scientific Instruments, Shanghai, China). The prepared liposome-silica samples were washed 3 times and smeared onto the conductive carbon tape mounted on the aluminum stub. The specimens were sputter coated with gold after they were completely dried. Magnification of the electron microscope is 20,000 times.
Fluorescent images were obtained by an Olympus FluoView FV1000 confocal laser scanning microscope working under a Multi Ar laser (458, 488, 514 nm) and a red HeNe laser (633 nm). The sample suspension was dropped onto the slide and scanned under 60× objective. Image was introduced to LAS AF LITE 2.6.1 for analysis.
2.5. Packing of microaffinity columns
Micro columns (400 μm, i.d.) were packed by Sp-403k column loader (nanobaume, USA). The detailed operation is as follows: 5 mg prepared stationary phase was suspended in 500 μL PBS and then placed in the sample bottle, with magnetic stirring at a constant speed of 15 rpm. A fiter union with aperture of 0.25 μm (Valco Instruments Co. Inc., Houston, USA) is installed at one end of the column. High-pressure nitrogen was used as the pressure source, fill the stationary phase suspension with an inner diameter of 400 at a pressure of 5 MPa.
2.6. “Heart-cutting”-offline-UPLC‒QTOF/MS microaffinity chromatography analysis
The PDGFRβ micro column was loaded on Agilent 1200 series equipped with capillary pump (Agilent Technologies, Palo Alto, CA, USA) and ultraviolet detector (Agilent Technologies). The mobile phase was 1 mmol/L PBS (pH 7.4) with a flow rate of 10 μL/min with a column oven of 37 °C. The detection wavelength ranged from 210 to 280 nm. The injection volume was 0.1 μL. The chromatography data were collected by ChemStation B.04.03 and introduced to Origin 8.0 for mapping.
Fractions of S. miltiorrhiza and S. chinensis were collected into 96-well plates every 0.5 min. Non-retention fractions and retention fractions from the chromatogram were separately combined and evaporated with a pressured gas blowing concentrator. After that, samples were dissolved with 20 μL methanol and introduced to Agilent 1290 UPLC‒QTOF/MS for analysis. A XBridge™ C18 column (100 mm × 2.1 mm i.d., 2.5 μm, Waters, Ireland) was used, and the mobile phase was composed of solvent A (0.1% formic acid) and solvent B (acetonitrile) at 0.8 mL/min by a linear gradient elution program. Components were identified by following methods our group used before42,43.
2.7. Frontal affinity chromatography (FAC) assay
Gom D and Sal B were dissolved in DMSO to 50 mmol/L as stock solutions. In FAC experiment, both Gom D and Sal B were further diluted in PBS to a gradient concentration of 100, 50, 25, 10, 5, 1 μmol/L to obtain the chromatography mobile phase. The flow rate was 10 μL/min under micro mode. The chromatography columns were first equilibrated with 1 mmol/L PBS, and then mobile phase with a certain concentration of compound was passed through the column as feed solution until a stable breakthrough curve was formed. Between two different concentrations, the column was washed with PBS until the ultraviolet absorption was returned to baseline. In addition, a control column filled with PDGFRβ-absent stationary phase was used to eliminate the non-specific interactions and obtain dead time t0.
When non-specific interactions are negligible in comparison with specific ones, the total amount of compounds captured (defined as q) and ligand concentrations (defined as [L]) can be written as one Equation44. KD was calculated by plotting reciprocal graph 1/q versus 1/[L].
2.8. Affinity analysis by SPR
PDGFRβ protein was fixed on a Biacore CM5 sensor chip (GE Healthcare, USA). Sal B and Gom D were dissolved in 5% DMSO PBS at a series of concentrations. Then analytes were injected with a flow rate of 30 μL/min. The dissociation time was 120 s. Affinity fitting was performed using Biacore T200 evaluation software (GE Healthcare, USA) and global fitting using by steady-state affinity model (1:1) to get the affinity constant.
2.9. Cell culture
HSC-T6 cell line was obtained from FuHeng Cell Center (fh0402, Shanghai, China). Cells were grown in high glucose-Dulbecco's medified Eagle's medium (DMEM, Gibco) with 10% fetal bovine serum (FBS; Gibco) and maintained at 37 °C incubator with 5% CO2. TGF-β was used to activate HSC-T6 cells. And cells were cultured into 6-well plates in the absence or presence of Sal B and Gom D.
2.10. PDGFRβ small interfering RNA transfection
RNA interfering experiment was performed to downregulate the PDGFRβ expression in HSC-T6 cells using small interfering RNAs (siRNA) (RiboBio, Guangzhou, China). After cells were seeded into 6-well plates at 37 °C for 24 h, cells were transfected with 5 μL 20 μmol/L PDGFRβ siRNAs or siControl using by riboFECTTM CP Reagent (RiboBio, Guangzhou, China). The sequences of PDGFRβ siRNAs are presented in Supporting Information Table S1. Transfection efficiency was assessed after 48 h.
2.11. Real-time quantitative PCR analysis
Cellular total RNA was isolated by Trizol reagent (Invitrogen) as the standard protocol. The RNA expressions of PDGFRβ, α-SMA and collagen I were measured by real-time PCR. GAPDH served as housekeeping gene for normalization. Data were analyzed using the 2−ΔΔCt method. The primer sequences are showed in Supporting Information Table S2.
2.12. Western blotting analysis
Cellular total protein was isolated using RIPA buffer and quantified by the BCA protein assay kit (Beyotime Biotechnology, Shanghai, China). Proteins were separated by an SDS-PAGE gel, transferred to polyvinylidene difluoride (PVDF) membranes, and blocked in 5% skimmed milk. Then the membranes were incubated with different primary antibodies against phospho-p38 MAPK (Cell Signaling Technology, Cat# 4511, RRID: AB_2139682), phospho-AKT (Cell Signaling Technology, Cat# 4060, RRID: AB_2315049), phospho-ERK (Cell Signaling Technology, Cat# 4370, RRID: AB_2315112) or GAPDH (Affinity Biosciences, Cat# AF7021, RRID: AB_2839421) overnight at 4 °C, and the secondary antibody (Affinity Biosciences, Cat# S0001, RRID: AB_2839429) for 1 h at room temperature. Finally, the membranes were determined by Odyssey infrared Imaging System and the bands were quantified though Quantity One.
2.13. Animals and in vivo experiment
Male BALB/c mice (6‒8 weeks-old, 18–22 g) were obtained from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The mice were maintained at constant temperature (21 ± 2 °C) and humidity (50 ± 10%) with 12 h light–dark cycles. All the experimental procedures were concordant with the Animal Ethics Committee of Shanghai 9th People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China (Approval ID: 2018172).
Mice were randomly divided into three groups: control, liver fibrosis model and 50 mg/kg Sal B group, each of which were 6 mice. 1 mL/kg 10% CCl4 peanut oil was given intraperitoneally into mice 3 times/week for 4 weeks to provide eminent liver fibrosis model. For Sal B group, mice were orally received 50 mg/kg Sal B every day for 4 weeks. The mice in control group were simultaneously acquired equivalent amount of peanut oil and 0.9% saline. Thereafter, serum samples and liver tissues were collected and stored at −80 °C for further analysis.
2.14. Serum biochemical evaluation
The activities of serum aspartate transaminase (AST) and alanine transaminase (ALT) were detected by automatic biochemical analyzer (Chemray 240, Kayto, Shenzhen, China) according to the manufacturer's instructions.
2.15. Histological analysis
A portion of fresh liver tissues were fixed in 10% neutral formalin solution, embedded in paraffin, sectioned, and then stained with hematoxylin and eosin (H&E) or Masson's trichrome staining according to a standard protocol. And the H&E or Masson-stained liver sections were examined using a fluorescence microscope (Nikon Eclipse Ni, Japan).
2.16. Immunofluorescence staining
The fixed liver tissue sections were deparaffinized and blocked with bovine serum albumin (BSA), then treated with antibodies targeted to phospho-p38 MAPK, phospho-AKT and phospho-ERK overnight at 4 °C. Next day, the liver tissue sections were incubated with fluorescence-conjugated secondary antibodies and counter-stained with 40, 6-diamidino-2-phenylindole (DAPI) for nuclei. Then the liver specimens were digitized with the Pannoramic MIDI Scanner (3D histech, Hungary) and analysed by Case Viewer 2.4 (3D histech, Hungary).
2.17. Statistical analysis
The results are shown as mean ± SD. One-way ANOVA and Student-Newman-Keuls post hoc test were used to perform multiple comparisons. GraphPad Prism 8.0 software (GraphPad Software Incorporated, USA) was used to prepare related graphs.
3. Results and discussion
3.1. Optimization of the PDGFRβ CFE system
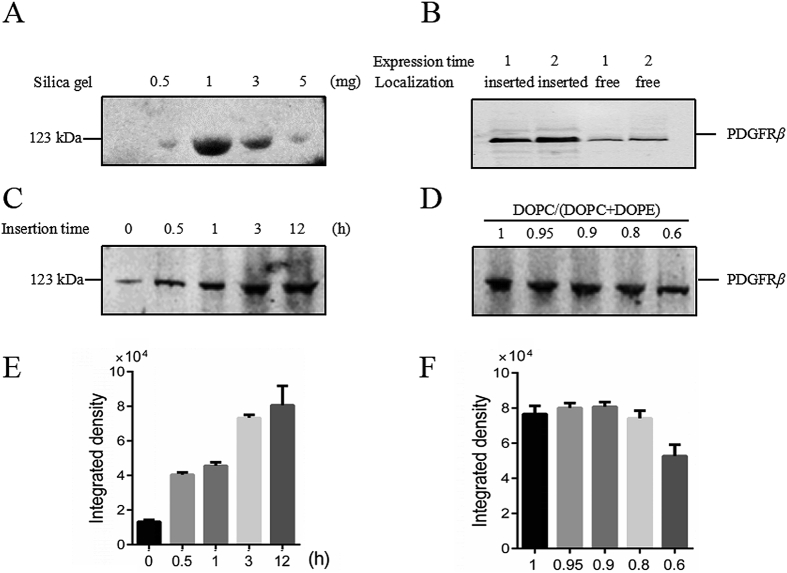
First, the amount of silica gel added to the CFE system was investigated to achieve the maximum expression efficiency. As shown in Fig. 2A, 1 mg silica gel per 50 μL expression system achieved the maximum PDGFRβ expression. The addition of excess silica caused a marked decrease in expression, and little protein expression was observed in the presence of 5 mg silica.
Figure 2.
Optimization of expression conditions: (A) investigation of added silica gel in per 50 μL system, (B) investigation of expression time: observe the amount of inserted and not-inserted PDGFRβ at 1 or 2 h expression time, (C) investigation of insertion time given to PDGFRβ, (D) investigation of maximum proportion of DOPE, (E) quantification analysis of bands in (C), (F) quantification analysis of bands in (D).
Based on the expression time recommended by the expression kit, the levels of PDGFRβ synthesis at 1 and 2 h were investigated. The levels of both total PDGFRβ expression and insertion in the membrane were higher with an expression time of 2 h compared to 1 h (Fig. 2B). Therefore, an expression time of 2 h was adopted in further experiments.
As shown in Fig. 2B, a portion of the PDGFRβ had failed to insert into the membrane at an expression time of 2 h. We assume that it takes some time for insertion of the protein. Therefore, next, we investigated insertion times of 0.5, 1, 3, and 12 h. The results are shown in Fig. 2C and E. An insertion time of 12 h was selected.
DOPC was chosen as the basic lipid for the preparation of liposomes. Because DOPE was added during the preparation of liposomes, its effects on PDGFRβ expression were investigated. As shown in Fig. 2D and F, DOPE had no effect on PDGFRβ expression at concentrations below 20%, but decreased PDGFRβ expression at 40%. Therefore, a DOPC:DOPE ratio of 4:1 was used in this work.
PDGFRβ expression was quantified using an enzyme-linked immunosorbent assay (ELISA); it was about 3.52 μg PDGFRβ per column. Next, a Kinase-Lumi™ Luminescent Kinase Assay was used to examine the kinase activity of PDGFRβ expressed by CFE. In a reaction tube with 0.1 μmol/L ATP, 1 mg inserted PDGFRβ in the stationary phase consumed 28.3% ATP in 15 min, thus confirming the kinase activity of the PDGFRβ prepared by CFE.
3.2. Characterization of PDGFRβ in the stationary phase
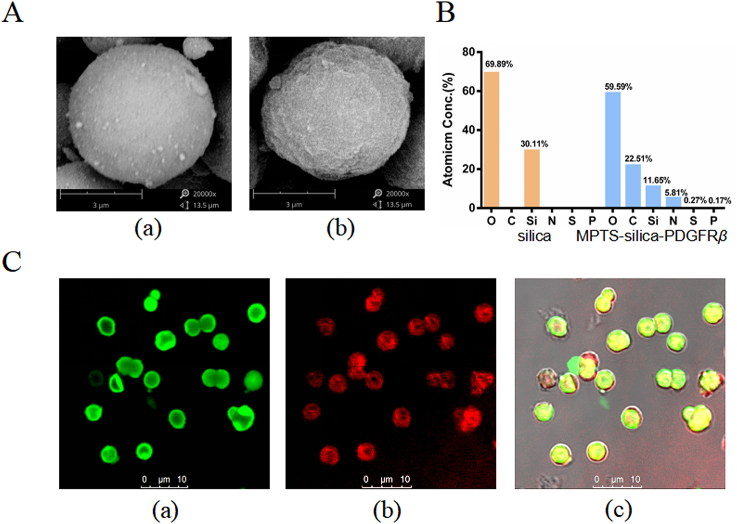
Observation by SEM showed successful surface modification of the silica gel and phospholipid coverage (Fig. 3A). The detection of nitrogen, phosphorus, and sulfur on the surface indicated attachment of the lipid membrane and proteins in the stationary phase (Fig. 3B).
Figure 3.
(A) SEM images of (a) silica; (b) PDGFRβ inserted MPTS-silica, magnification of the electron microscope is 20,000 times, (B) quantification of Atomic Conc. on the surface of stationary phase in (A) by atomic energy spectrum, (C) fluorescence observed by laser confocal microscope of (a) fluorescence from NBD-DOPG; (b) fusion fluorescence on PDGFRβ; (c) colocalization.
The PDGFRβ expression and lipid membrane binding were further examined by laser scanning confocal microscopy (Fig. 3C). Obvious fluorescence was observed, which confirmed that liposomes were wrapped in the gel and that PDGFRβ was expressed. Moreover, the results confirmed the insertion of PDGFRβ into the lipid bilayer membrane.
3.3. Evaluation of PDGFRβ micro columns
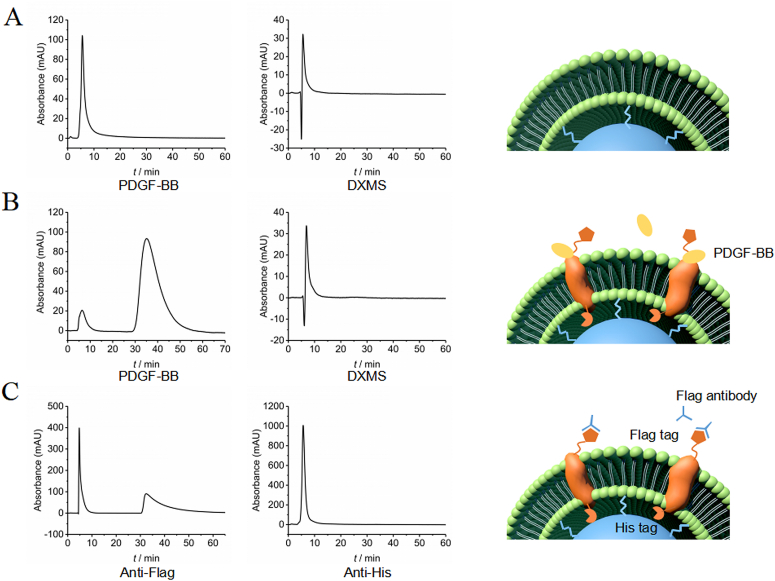
Next, the effectiveness and selectivity of the prepared PDGFRβ columns were evaluated. PDGF-BB, the ligand that binds to an extracellular site of PDGFRβ, was selected as a positive control and DXMS was used as a negative control. As shown in Fig. 4A and B, PDGF-BB showed strong retention behavior, reaching a peak at 35.4 min in the PDGFRβ column while with no retention in the negative control column, and DXMS was barely retained in either column. These observations indicate that the expressed PDGFRβ recognized and bound to its ligand. The prepared columns had satisfactory selectivity, and remained stable for 3 days (Supporting Information Fig. S2).
Figure 4.
Investigation of effectiveness, specificity and protein orientation of PDGFRβ micro columns: (A) retention behaviour of PDGF-BB and DXMS on PDGFRβ-empty column, (B) retention behaviour of PDGF-BB and DXMS on PDGFRβ-inserted column, (C) retention behaviour of antibody of Flag-tag and His-tag on PDGFRβ inserted column.
Previous studies have shown that, in CFE, membrane receptors adopt a consistent orientation in liposomes, with the N-terminus 100% exposed to the outside26. The mechanism of unidirectional insertion involves that the cell-free protein synthesis simulates the synthesis in vivo, following the translation sequence from N-terminal to C-terminal. The membrane receptors started to be synthesized from the N-terminal outside the silica-liposome carrier. When the hydrophobic sequence of the trans-membrane domain is synthesized, the hydrophobic structure is inserted into the liposome phospholipid bilayer spontaneously through hydrophobic interaction. For the electrostatic interaction induced by hydrophobic sequence and the charge of the hydrophobic region of liposomes has a single direction preference, the receptors present a consistent orientation of N-terminal outward on the stationary phase carrier45,46. To examine whether the expressed PDGFRβ was unidirectionally inserted and presented in the stationary phase, a Flag-tag was introduced at the N-terminus and a 6His-tag was introduced at the C-terminus. Their relative accessibilities to antibodies were examined according to their retention times in the column. Following unidirectional insertion, the N-terminus of PDGFRβ exposed on the surface in the stationary phase, with the C-terminus inserted into the membrane. As shown in Fig. 4C, the antibody to the Flag-tag was retained in the column, while the antibody to the 6His-tag showed no retention, indicating that the Flag-tag bonded to the antibody but the 6His-tag did not. Therefore, PDGFRβ was inserted into the lipid bilayer membrane in a unidirectional manner. These observations confirm that our screening involves the extracellular domain of PDGFRβ affinity components.
3.4. Screening of PDGFRβ affinity components
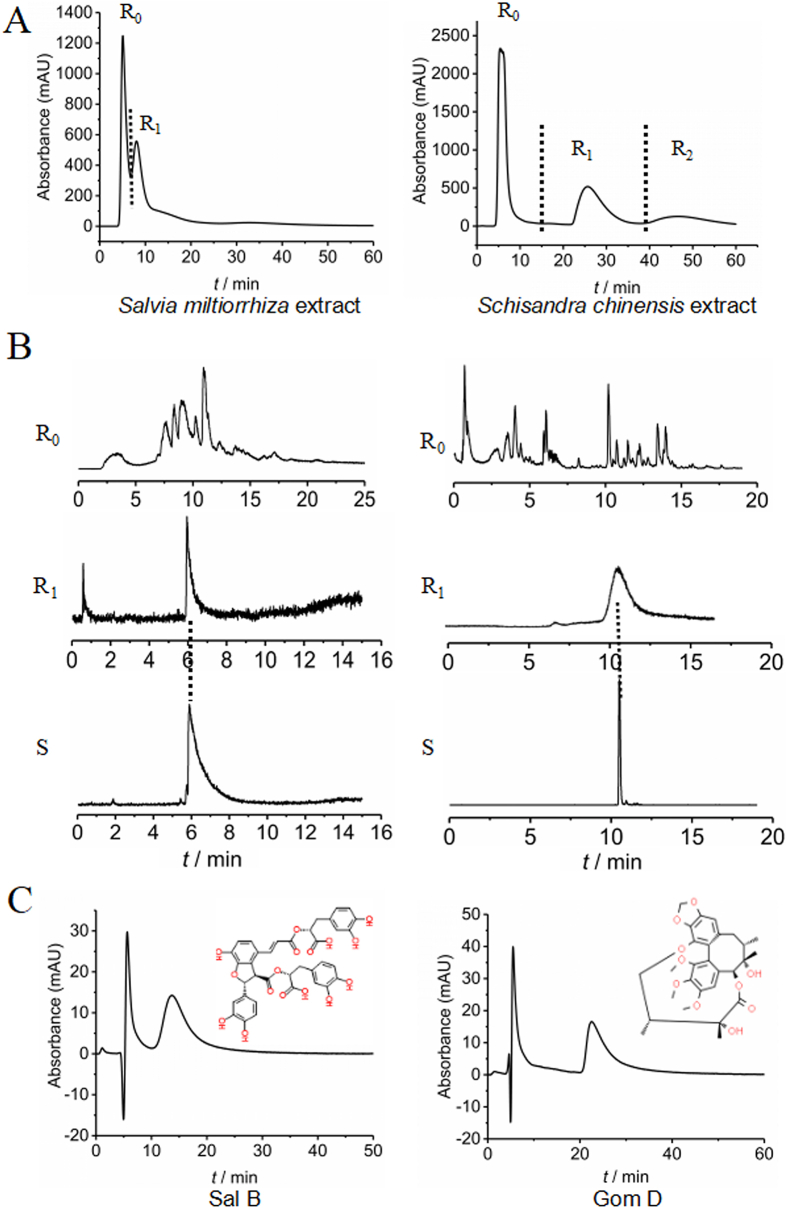
Next, the affinity components of PDGFRβ were screened from S. chinensis and S. miltiorrhiza. The chromatogram of S. miltiorrhiza is shown in Fig. 5A. Fraction R1 showed obvious retention in the column, while R0 did not. R0 and R1 were collected and analyzed by UPLC‒QTOF/MS. By matching with the established chemical composition library of S. miltiorrhiza and using standard compounds, the main retention component was identified as Sal B (Fig. 5B). Similarly, R1 in the chromatogram of S. chinensis was identified as Gom D. For further verification, standards of Sal B and Gom D were analyzed using PDGFRβ columns, and both showed retention, with peak times of 13.48 min and 22.52 min, respectively (Fig. 5C). Here, the R2 component of S. chinensis showed strong affinity with PDGFRβ. However, there are no relevant reports regarding the liver-protective effects of the potential active compound in R2. Further pharmacological research is being carried out by our group to determine its activity. This study mainly focused on the pharmacological effects and possible mechanisms of action of Sal B and Gom D.
Figure 5.
Spectrogram of affinity components of PDGFRβ from S. miltiorrhiza and S. chinensis by micro PDGFRβ-offline-UPLC/MS systerm: (A) UV chromatogram of S. miltiorrhiza and S. chinensis extracts on PDGFRβ column, (B) Mass spectrum of retention and non-retention components on PDGFRβ column, (C) UV chromatogram of standards of Sal B and Gom D on PDGFRβ column.
3.5. Determination of KD by FAC and SPR
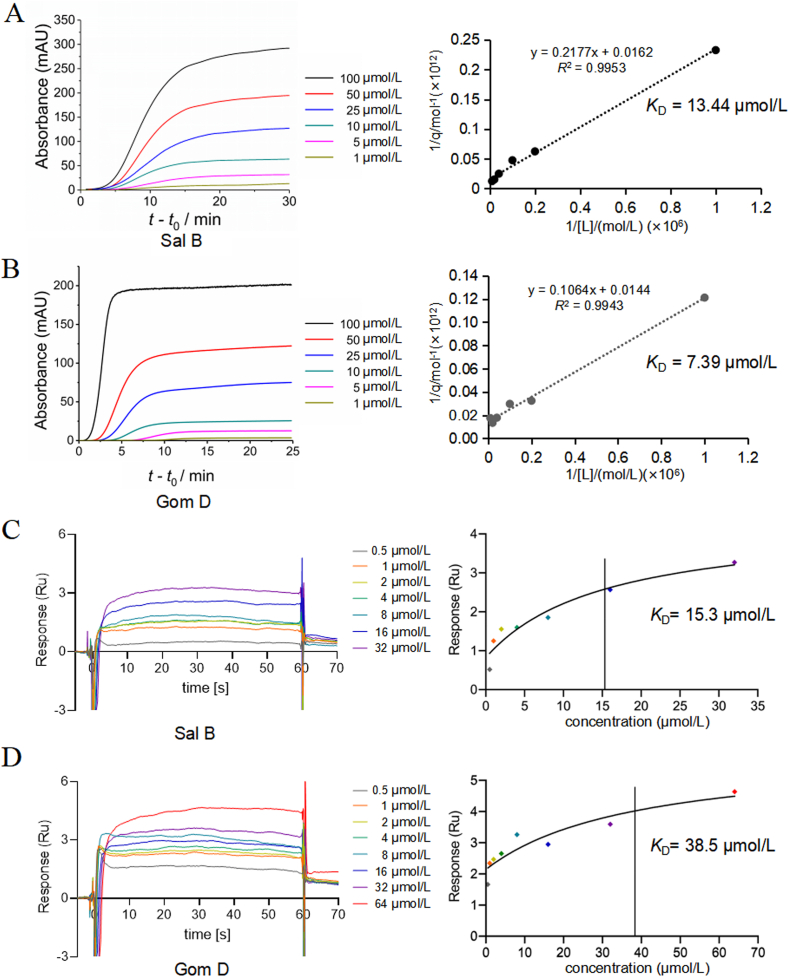
FAC was used to study the interactions between PDGFRβ and affinity compounds. In this approach, increasing concentrations of solutions of compounds were used as the mobile phase and infused into the affinity column. The binding interaction between the compounds and PDGFRβ was characterized by an increase in breakthrough time with decreasing concentration. If there were no specific interactions, the breakthrough time would remain constant regardless of the ligand concentration. Therefore, experiments in PDGFRβ-absent columns were carried out to detect potential nonspecific binding. The results are shown in Supporting Information Fig. S3. The breakthrough time was constant for both Sal B and Gom D regardless of the concentration, indicating negligible nonspecific interactions. The breakthrough curves of Sal B and Gom D on PDGFRβ columns were shown in Fig. 6A and B. The KD values of Sal B and Gom D were calculated to be 13.44 and 7.39 μmol/L, respectively, confirming that they have strong affinity to the extracellular domain of PDGFRβ. In addition, SPR affinity analysis was performed to determine the direct binding of Sal B and Gom D to PDGFRβ. Their respective KD values were 15.3 and 38.5 μmol/L (Fig. 6C and D). The KD values obtained by the two affinity determination methods were within an order of magnitude, which confirms the accuracy of FAC by iSMAC. Thus, CFE can provide sufficient PDGFRβ with a consistent orientation to determine the affinity between proteins and test compounds, which has promise for application to any MPs.
Figure 6.
Affinity analysis of the interaction of Sal B, Gom D with PDGFRβ. (A) FAC of Sal B on PDGFRβ columns: frontal affinity breakthrough curves and calculation of KD, (B) FAC of Gom D on PDGFRβ columns: frontal affinity breakthrough curves and calculation of KD, (C) SPR analysis of Sal B, (D) SPR analysis of Gom D.
3.6. Sal B and Gom D attenuate HSCs activation via the PDGFRβ pathway
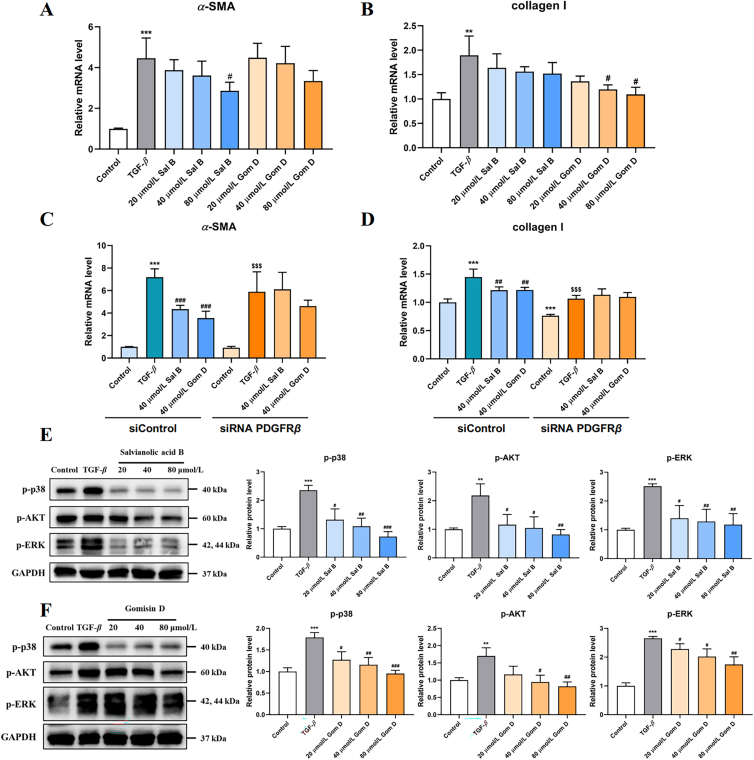
Next, the pharmacological activities of Sal B and Gom D in HSCs activation were examined. As shown in Fig. 7A and B, Sal B and Gom D concentration-dependently (20, 40, and 80 μmol/L) decreased the levels of α-SMA and collagen I mRNA expression in HSC-T6 cells. To validate whether Sal B and Gom D attenuate HSCs activation via PDGFRβ, siRNA was used to downregulate PDGFRβ expression. Real-time quantitative PCR showed a marked decrease in PDGFRβ mRNA expression in HSC-T6 cells transfected with PDGFRβ siRNAs (Supporting Information Fig. S4). The siRNA PDGFRβ-2 was chosen for subsequent experiments. After transfection with PDGFRβ siRNA, the inhibitory effects of Sal B and Gom D on α-SMA and collagen I expression disappeared (Fig. 7C and D). PDGFRβ downstream related proteins were also examined to determine whether Sal B and Gom D influenced PDGFRβ-associated signaling pathways. Both of them decreased activated p-p38, p-AKT, and p-ERK protein levels. Therefore, Sal B and Gom D attenuate HSCs activation by inhibiting the PDGFRβ pathway.
Figure 7.
Sal B and Gom D attenuate HSCs activation via PDGFRβ pathway. The mRNA levels of α-SMA (A) and collagen Ӏ (B) in HSC-T6 cells treated with Sal B and Gom D, n = 3–4. (C, D) The effect of siRNA PDGFRβ on Sal B and Gom D′ inhibition in HSC-T6 cellular activation, n = 4–5. The protein levels associated PDGFRβ pathway in HSC-T6 cells treated with Sal B (E) and Gom D (F), n = 3. Data are shown as mean ± SD. ∗∗P < 0.01, ∗∗∗P < 0.001 versus control group; #P < 0.05, ##P < 0.01, ###P < 0.001 versus TGF-β group. $$$P < 0.001 versus siRNA PDGFRβ-Control group.
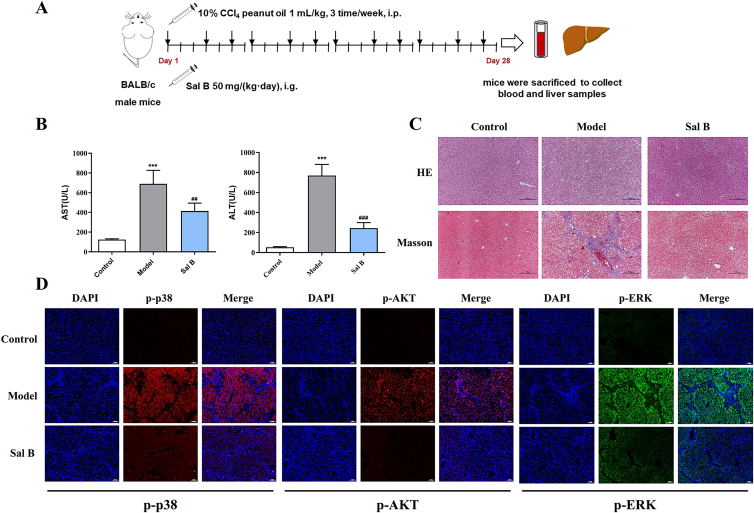
3.7. Sal B improves CCl4-induced liver fibrosis via the PDGFRβ pathway in BALB/c mice
As shown in Fig. 7, Sal B had a more significant effect in downregulating the downstream proteins of PDGFRβ than Gom D. An important phenolic acid in S. miltiorrhiza, Sal B is well absorbed in vivo and has been examined in several clinical trials47; so it has good prospects for use as a drug. Therefore, in this study, Sal B was selected and investigated as a potential antihepatic fibrosis compound.
BALB/c mice were used to determine whether Sal B could attenuate CCl4-induced liver fibrosis via the PDGFRβ pathway (Fig. 8A). As shown in Fig. 8B, Sal B markedly decreased the serum AST and ALT levels in CCl4-treated model mice. H&E and Masson staining results showed that Sal B significantly reduced inflammatory cell infiltration and collagen deposition, and attenuated CCl4-induced injury and fibrosis (Fig. 8C). Furthermore, the results of immunofluorescence analysis showed that the protein levels of p-p38, p-AKT, and p-ERK were increased in the CCl4-induced liver fibrosis model, and decreased after Sal B treatment (Fig. 8D). Taken together, these results suggest that Sal B improves CCl4-induced liver fibrosis via the PDGFRβ pathway in BALB/c mice.
Figure 8.
Sal B improves CCl4-induced liver fibrosis via PDGFRβ pathway in BALB/c mice. (A) Schematic presentation of experimental regimen. (B) The activities of serum AST and ALT after Sal B treatment, n = 6. (C) Representative pictures of H&E and Masson-stained liver section. (D) Immunofluorescence images for p-p38 (red), p-AKT (red), p-ERK (green) and DAPI (blue) in liver sections. Data are shown as mean ± SD. ∗∗∗P < 0.001 versus control group; ##P < 0.01, ###P < 0.001 versus model group.
4. Conclusions
A novel in situ synthesis MP affinity chromatography method designated iSMAC was proposed and successfully applied to construct a PDGFRβ affinity chromatography model. iSMAC performs significant advantages comparing to previous membrane receptor affinity chromatography: membrane proteins can be obtained rapidly and efficiently by cell-free synthesis without complex purification. Moreover, cell-free synthesis well simulates the intracellular protein synthesis, so the activity and transmembrane structure of membrane proteins are as close as possible to the natural conformation. The labeled proteins can be synthesized by flexibly constructing cDNA to facilitate the detection of the yield and function of target protein. What's more important, iSMAC can fix the single, stable and N-terminal outward unidirectional orientation of target membrane protein on the stationary phase, which can realize accurate drug screening and drug‒protein interaction analysis for the extracellular region of specific membrane proteins.
In this study, by constructing specific tags on cDNA, it was confirmed that PDGFRβ was unidirectionally inserted into the liposome membrane, which facilitated accurate screening and interaction analysis of binding sites. iSMAC was combined with micro-bioaffinity chromatography analysis to examine the potential for screening of PDGFRβ inhibitors and determining drug–ligand affinity interactions with PDGFRβ extracellular sites. The results identified two components, Sal B and Gom D from S. miltiorrhiza and S. chinensis, respectively. Their respective KD values with PDGFRβ were 13.44 and 7.39 μmol/L in FAC and 15.3 and 38.5 μmol/L in SPR, which confirmed the accuracy of iSMAC for affinity determination. Further in vitro experiments confirmed that the two compounds decreased α-SMA and collagen I mRNA levels that were increased by TGF-β in HSC-T6 cells through the PDGFRβ pathway. In vivo, Sal B also attenuated CCl4-induced liver fibrosis by downregulating the levels of PDGFRβ downstream related proteins. These results suggest that Sal B and Gom D could be useful leading compounds for the development of drugs against hepatic fibrosis. More importantly, iSMAC can be theoretically extended to interaction analysis of other MPs, thus providing a new concept and technical platform for the accurate screening and activity evaluation of MP-targeting drugs. With a slight regret, due to the N-terminal outward orientation of proteins on chromatography stationary phase, this system focuses on the study of active domains in extracellular region of membrane proteins. The research of intracellular active domains cannot be carried out. The development of ideal chromatography column carriers such as monolithic columns is expected to achieve flexible control of protein orientation so as to further achieve screening and affinity analysis targeting to specific active extracelluar or intracelluar domains of MPs.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 82073814, 81973275, 82003909, 81973291, 82122066, 81803815) and Rising-Star Program of Shanghai Science and Technology Committee (19QA1411500).
Author contributions
Yanqiu Gu: investigation, validation, data curation, writing-original draft. Rong Wang: investigation, project administration. Panpan Chen: investigation, data curation. Shengnan Li: validation, formal analysis. Xinyi Chai and Chun Chen: resources. Yue Liu, Yan Cao and Diya Lv: visualization. Zhanying Hong and Zhenyu Zhu: supervision, writing-review & editing. Yifeng Chai, Yongfang Yuan and Xiaofei Chen: conceptualization, methodology, software, writing-review & editing, funding acquisition.
Conflicts of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2022.04.010.
Contributor Information
Yifeng Chai, Email: yfchai@smmu.edu.cn.
Yongfang Yuan, Email: nmxyyf@126.com.
Xiaofei Chen, Email: xfchen2010@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chadli M., Rebaud S., Maniti O., Tillier B., Cortes S., Girard-Egrot A. New tethered phospholipid bilayers integrating functional G-protein-coupled receptor membrane proteins. Langmuir. 2017;33:10385–10401. doi: 10.1021/acs.langmuir.7b01636. [DOI] [PubMed] [Google Scholar]
- 2.Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y., Wang K., Yan N. The recombinant expression systems for structure determination of eukaryotic membrane proteins. Protein Cell. 2014;5:658–672. doi: 10.1007/s13238-014-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee A.G., Michelangeli F., East J.M. Tests for the importance of fluidity for the function of membrane proteins. Biochem Soc Trans. 1989;17:962–964. doi: 10.1042/bst0170962. [DOI] [PubMed] [Google Scholar]
- 5.Hou X., Sun M., Bao T., Xie X., Wei F., Wang S. Recent advances in screening active components from natural products based on bioaffinity techniques. Acta Pharm Sin B. 2020;10:1800–1813. doi: 10.1016/j.apsb.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Jin H., Xia D., Shao H., Peng K., Liu X., et al. Biomimetic polymer-based method for selective capture of C-reactive protein in biological fluids. ACS Appl Mater Interfaces. 2018;10:41999–42008. doi: 10.1021/acsami.8b15581. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q., Zhang Q., Huang H., Zhao P., Sun L., Peng K., et al. Fabrication and application of zwitterionic phosphorylcholine functionalized monoliths with different hydrophilic crosslinkers in hydrophilic interaction chromatography. Anal Chim Acta. 2020;1101:222–229. doi: 10.1016/j.aca.2019.12.016. [DOI] [PubMed] [Google Scholar]
- 8.He L., Wang S., Yang G., Zhang Y., Wang C., Yuan B., et al. Progress in cell membrane chromatography. Drug Discov Ther. 2007;1:104–107. [PubMed] [Google Scholar]
- 9.Hou X., Wang S., Zhang T., Ma J., Zhang J., Zhang Y., et al. Recent advances in cell membrane chromatography for traditional Chinese medicines analysis. J Pharm Biomed Anal. 2014;101:141–150. doi: 10.1016/j.jpba.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Ding X., Chen X., Cao Y., Jia D., Wang D., Zhu Z., et al. Quality improvements of cell membrane chromatographic column. J Chromatogr A. 2014;1359:330–335. doi: 10.1016/j.chroma.2014.07.071. [DOI] [PubMed] [Google Scholar]
- 11.Chen X., Cao Y., Zhang H., Zhu Z., Liu M., Liu H., et al. Comparative normal/failing rat myocardium cell membrane chromatographic analysis system for screening specific components that counteract doxorubicin-induced heart failure from Acontium carmichaeli. Anal Chem. 2014;86:4748–4757. doi: 10.1021/ac500287e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia D., Chen X., Cao Y., Wu X., Ding X., Zhang H., et al. On-line comprehensive two-dimensional HepG2 cell membrane chromatographic analysis system for charactering anti-hepatoma components from rat serum after oral administration of Radix scutellariae: a strategy for rapid screening active compounds in vivo. J Pharm Biomed Anal. 2016;118:27–33. doi: 10.1016/j.jpba.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Chen C., Gu Y., Wang R., Chai X., Jiang S., Wang S., et al. Comparative two-dimensional GPC3 overexpressing SK-Hep1 cell membrane chromatography/C18/time-of-flight mass spectrometry for screening selective GPC3 inhibitor components from Scutellariae Radix. J Chromatogr B Analyt Technol Biomed Life Sci. 2021;1163:122492. doi: 10.1016/j.jchromb.2020.122492. [DOI] [PubMed] [Google Scholar]
- 14.Ding X., Cao Y., Yuan Y., Gong Z., Liu Y., Zhao L., et al. Development of APTES-decorated HepG2 cancer stem cell membrane chromatography for screening active components from Salvia miltiorrhiza. Anal Chem. 2016;88:12081–12089. doi: 10.1021/acs.analchem.6b02709. [DOI] [PubMed] [Google Scholar]
- 15.Gu Y., Chen X., Wang Y., Liu Y., Zheng L., Li X., et al. Development of 3-mercaptopropyltrimethoxysilane (MPTS)-modified bone marrow mononuclear cell membrane chromatography for screening anti-osteoporosis components from Scutellariae Radix. Acta Pharm Sin B. 2020;10:1856–1865. doi: 10.1016/j.apsb.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moaddel R., Wainer I.W. The preparation and development of cellular membrane affinity chromatography columns. Nat Protoc. 2009;4:197–205. doi: 10.1038/nprot.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moaddel R., Jozwiak K., Yamaguchi R., Wainer I.W. Direct chromatographic determination of dissociation rate constants of ligand-receptor complexes: assessment of the interaction of noncompetitive inhibitors with an immobilized nicotinic acetylcholine receptor-based liquid chromatography stationary phase. Anal Chem. 2005;77:5421–5426. doi: 10.1021/ac0504464. [DOI] [PubMed] [Google Scholar]
- 18.Moaddel R., Calleri E., Massolini G., Frazier C.R., Wainer I.W. The synthesis and initial characterization of an immobilized purinergic receptor (P2Y1) liquid chromatography stationary phase for online screening. Anal Biochem. 2007;364:216–218. doi: 10.1016/j.ab.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Q., Jia L., Zhang X., Zhu A., Wang S., Xie X. Accurate construction of cell membrane biomimetic graphene nanodecoys via purposeful surface engineering to improve screening efficiency of active components of traditional Chinese medicine. Acta Pharm Sin B. 2022;12:394–405. doi: 10.1016/j.apsb.2021.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bu Y., Zhang X., Zhu A., Li L., Xie X., Wang S. Inside-out-oriented cell membrane biomimetic magnetic nanoparticles for high-performance drug lead discovery. Anal Chem. 2021;93:7898–7907. doi: 10.1021/acs.analchem.1c00567. [DOI] [PubMed] [Google Scholar]
- 21.Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. Cell-free protein synthesis: applications come of age. Biotechnol Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makino S., Beebe E.T., Markley J.L., Fox B.G. Cell-free protein synthesis for functional and structural studies. Methods Mol Biol. 2014;1091:161–178. doi: 10.1007/978-1-62703-691-7_11. [DOI] [PubMed] [Google Scholar]
- 23.Junge F., Haberstock S., Roos C., Stefer S., Proverbio D., Dotsch V., et al. Advances in cell-free protein synthesis for the functional and structural analysis of membrane proteins. Nat Biotechnol. 2011;28:262–271. doi: 10.1016/j.nbt.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Arslan Yildiz A., Yildiz U.H., Liedberg B., Sinner E.K. Biomimetic membrane platform: fabrication, characterization and applications. Colloids Surf B Biointerfaces. 2013;103:510–516. doi: 10.1016/j.colsurfb.2012.10.066. [DOI] [PubMed] [Google Scholar]
- 25.Robelek R., Lemker E.S., Wiltschi B., Kirste V., Naumann R., Oesterhelt D., et al. Incorporation of in vitro synthesized GPCR into a tethered artificial lipid membrane system. Angew Chem Int Ed Engl. 2007;46:605–608. doi: 10.1002/anie.200602231. [DOI] [PubMed] [Google Scholar]
- 26.Ritz S., Hulko M., Zerfass C., May S., Hospach I., Krasteva N., et al. Cell-free expression of a mammalian olfactory receptor and unidirectional insertion into small unilamellar vesicles (SUVs) Biochimie. 2013;95:1909–1916. doi: 10.1016/j.biochi.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Lee K.H., Kim D.M. Recent advances in development of cell-free protein synthesis systems for fast and efficient production of recombinant proteins. FEMS Microbiol Lett. 2018;365:fny174. doi: 10.1093/femsle/fny174. [DOI] [PubMed] [Google Scholar]
- 28.May S., Andreasson-Ochsner M., Fu Z., Low Y.X., Tan D., de Hoog H.P., et al. In vitro expressed GPCR inserted in polymersome membranes for ligand-binding studies. Angew Chem Int Ed Engl. 2013;52:749–753. doi: 10.1002/anie.201204645. [DOI] [PubMed] [Google Scholar]
- 29.Chen X., Wu Y., Chen C., Gu Y., Zhu C., Wang S., et al. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm Sin B. 2021;11:222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez-Gea V., Friedman S.L. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 31.Kisseleva T., Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151–166. doi: 10.1038/s41575-020-00372-7. [DOI] [PubMed] [Google Scholar]
- 32.Parola M., Pinzani M. Liver fibrosis: pathophysiology, pathogenetic targets and clinical issues. Mol Aspect Med. 2019;65:37–55. doi: 10.1016/j.mam.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Mahdinloo S., Kiaie S.H., Amiri A., Hemmati S., Valizadeh H., Zakeri-Milani P. Efficient drug and gene delivery to liver fibrosis: rationale, recent advances, and perspectives. Acta Pharm Sin B. 2020;10:1279–1293. doi: 10.1016/j.apsb.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borkham-Kamphorst E., Weiskirchen R. The PDGF system and its antagonists in liver fibrosis. Cytokine Growth Factor Rev. 2016;28:53–61. doi: 10.1016/j.cytogfr.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Andrae J., Gallini R., Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah R., Reyes-Gordillo K., Arellanes-Robledo J., Lechuga C.G., Hernandez-Nazara Z., Cotty A., et al. TGF-beta1 up-regulates the expression of PDGF-beta receptor mRNA and induces a delayed PI3K-, AKT-, and p70(S6K)-dependent proliferative response in activated hepatic stellate cells. Alcohol Clin Exp Res. 2013;37:1838–1848. doi: 10.1111/acer.12167. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F., Ni C., Kong D., Zhang X., Zhu X., Chen L., et al. Ligustrazine attenuates oxidative stress-induced activation of hepatic stellate cells by interrupting platelet-derived growth factor-beta receptor-mediated ERK and p38 pathways. Toxicol Appl Pharmacol. 2012;265:51–60. doi: 10.1016/j.taap.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Shiha G.E., Abu-Elsaad N.M., Zalata K.R., Ibrahim T.M. Tracking anti-fibrotic pathways of nilotinib and imatinib in experimentally induced liver fibrosis: an insight. Clin Exp Pharmacol Physiol. 2014;41:788–797. doi: 10.1111/1440-1681.12286. [DOI] [PubMed] [Google Scholar]
- 39.Nagappan A., Kim J.H., Jung D.Y., Jung M.H. Cryptotanshinone from the Salvia miltiorrhiza bunge attenuates ethanol-induced liver injury by activation of AMPK/SIRT1 and Nrf2 signaling pathways. Int J Mol Sci. 2019;21:265. doi: 10.3390/ijms21010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang B.E. Treatment of chronic liver diseases with traditional Chinese medicine. J Gastroenterol Hepatol. 2000;15(Suppl):E67–E70. doi: 10.1046/j.1440-1746.2000.02100.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang W., Zhu Y., Zhang Q., Ma L., Yang L., Guo W., et al. Research progress in application and mechanism of Schisandrae chinensis fructus for prevention and treatment of liver diseases. Zhongguo Zhongyao Zazhi. 2020;45:3759–3769. doi: 10.19540/j.cnki.cjcmm.20200513.601. [DOI] [PubMed] [Google Scholar]
- 42.Chen X., Cao Y., Lv D., Zhu Z., Zhang J., Chai Y. Comprehensive two-dimensional HepG2/cell membrane chromatography/monolithic column/time-of-flight mass spectrometry system for screening anti-tumor components from herbal medicines. J Chromatogr A. 2012;1242:67–74. doi: 10.1016/j.chroma.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 43.Zheng L., Chen S., Cao Y., Zhao L., Gao Y., Ding X., et al. Combination of comprehensive two-dimensional prostate cancer cell membrane chromatographic system and network pharmacology for characterizing membrane binding active components from Radix et Rhizoma Rhei and their targets. J Chromatogr A. 2018;1564:145–154. doi: 10.1016/j.chroma.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 44.Lecas L., Hartmann L., Caro L., Mohamed-Bouteben S., Raingeval C., Krimm I., et al. Miniaturized weak affinity chromatography for ligand identification of nanodiscs-embedded G-protein coupled receptors. Anal Chim Acta. 2020;1113:26–35. doi: 10.1016/j.aca.2020.03.062. [DOI] [PubMed] [Google Scholar]
- 45.Zapf T., Tan C.W., Reinelt T., Huber C., Shaohua D., Geifman-Shochat S., et al. Synthesis and functional reconstitution of light-harvesting complex II into polymeric membrane architectures. Angew Chem Int Ed Engl. 2015;54:14664–14668. doi: 10.1002/anie.201506304. [DOI] [PubMed] [Google Scholar]
- 46.Kalmbach R., Chizhov I., Schumacher M.C., Friedrich T., Bamberg E., Engelhard M. Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. J Mol Biol. 2007;371:639–648. doi: 10.1016/j.jmb.2007.05.087. [DOI] [PubMed] [Google Scholar]
- 47.Liu L., Li J., Zhang Y., Zhang S., Ye J., Wen Z., et al. Salvianolic acid B inhibits platelets as a P2Y12 antagonist and PDE inhibitor: evidence from clinic to laboratory. Thromb Res. 2014;134:866–876. doi: 10.1016/j.thromres.2014.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.