Abstract
Molecular glues can specifically induce aggregation between two or more proteins to modulate biological functions. In recent years, molecular glues have been widely used as protein degraders. In addition, however, molecular glues play a variety of vital roles, such as complex stabilization, interactome modulation and transporter inhibition, enabling challenging therapeutic targets to be druggable and offering an exciting novel approach for drug discovery. Since most molecular glues are identified serendipitously, exploration of their systematic discovery and rational design are important. In this review, representative examples of molecular glues with various physiological functions are divided into those mediating homo-dimerization, homo-polymerization and hetero-dimerization according to their aggregation modes, and we attempt to elucidate their mechanisms of action. In particular, we aim to highlight some biochemical techniques typically exploited within these representative studies and classify them in terms of three stages of molecular glue development: starting point, optimization and identification.
KEY WORDS: Small molecule, Molecular glue, Dimerization, Polymerization, Protein–protein interaction
Graphical abstract
Through rational design, optimization and identification, molecular glues can specifically induce proteins’ homo-dimerization, homo-polymerization or hetero-dimerization to regulate their functions, which is a potential therapeutic strategy.
1. Introduction
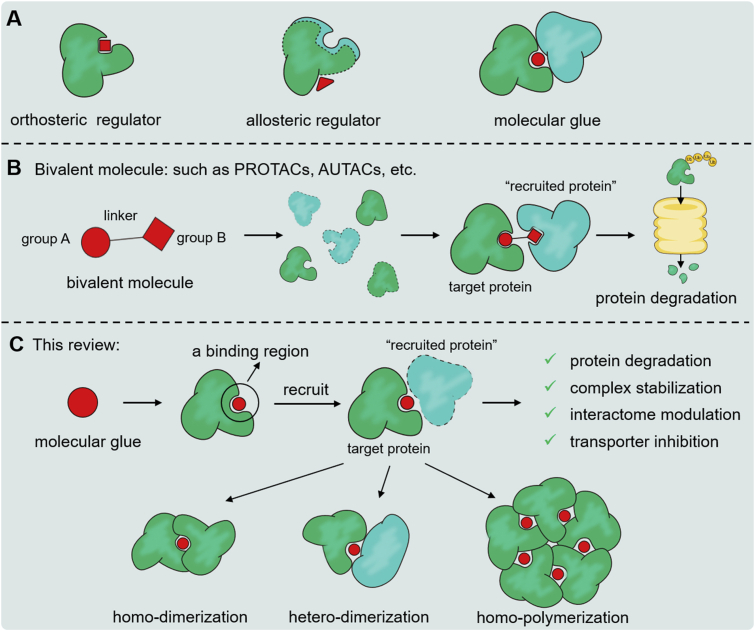
Since the publication of the first successful cases in the early 1990s, the last three decades have witnessed rapid advances of protein structure-based drug design1, 2, 3. With the current advances in proteomics, molecular biology and computational engineering, numerous excellent protein targets have been identified, leading to a boom in structure-based drug design and the success of many novel drug discoveries4. In the human body, target proteins commonly exert biological activities by binding to endogenous ligands, such as small molecules, proteins, or nucleic acids. An orthosteric site is defined as the target protein's surface that directly interacts with endogenous ligands, and an allosteric site is defined as the target protein's region that can affect the conformation of the target protein to modulate functions (Fig. 1A)5. The strategy to design small molecules that occupy orthosteric or allosteric sites of the target protein has dominated the discovery of small-molecular drugs over the past thirty years. However, on account of the lack of suitable binding pockets and propriate chemical entities, more than 80 percent of the human proteome is undruggable6. To break through this bottleneck, in recent years, researchers have attempted to regulate the functions of targets by developing small molecules that can recruit other proteins (Fig. 1B and C)7, 8, 9. Classified by their characteristics, there are two types of small molecules that induce the target proteins to bind to “recruited proteins”: bivalent molecules and molecular glues. A bivalent molecule possesses three components, including two functional groups and a linker (Fig. 1B). Functional group A is used to bind the target protein, and functional group B is used to bind the “recruited protein”. The strategy to develop bivalent molecules has recently focused on discovering small molecules of target protein degraders, such as proteolysis targeting chimeras (PROTACs)10, lysosome targeting chimeras (LYTACs)11,12, chaperone-mediated autophagy (CMA)13, and autophagy targeting chimeras (AUTACs)14,15. There are a vast number of systematic reviews on those studies. Even though bivalent molecules have achieved superior bioactivities, a major drawback of bivalent molecules is their sheer size16. Not only may those bivalent molecules be orally unavailable due to solubility problems, but they also tend to have poor membrane permeability, resulting in adverse pharmacokinetics. Moreover, most important therapeutic proteins such as transcription factors lack ligand binding sites, and thus using PROTAC approach is challenging17, 18, 19.
Figure 1.
(A) Difference of orthosteric regulator, allosteric regulator and molecular glue. (B) Schematic diagram of divalent molecular interaction mode. (C) Schematic diagram of molecular interaction mode and classification of protein polymerization patterns induced by molecular glues.
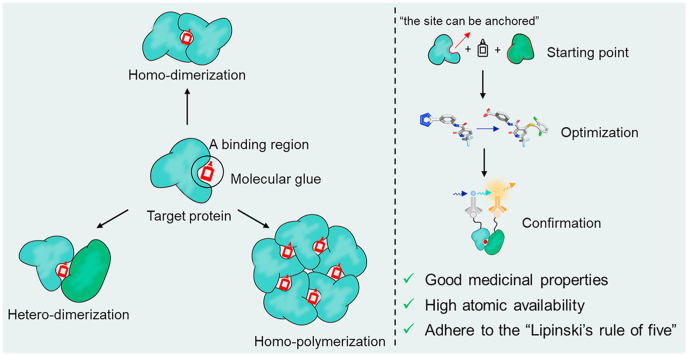
In contrast, molecular glues refer to monovalent small molecules that rely on the secondary structural characteristics of target protein to mediate protein–protein interactions (PPIs, Fig. 1C). When a molecular glue binds the target protein, the molecule itself and some residues of the target protein can form a binding region together, which mediates the bond between the target protein and the “recruited protein(s)”. Subsequently, such polynary complexes cause changes in a series of biological functions, such as protein degradation, complex stabilization, interactome modulation or transporter inhibition. In contrast with bivalent molecules, a molecular glue behaves as an indivisible structure, rather than two protein-binding fragments linked by a connectome with an appropriate length. Therefore, molecular glues are much smaller, possess good medicinal properties, have high atomic availability, and more readily adhere to “Lipinski's rule of five”20. In recent years, due to their fascinating pharmaceutical properties, the discovery of molecular glues has received increasing research interest. However, a number of challenges for drug discovery remain, including the fact that the design of molecular glues still depends on retrospective elucidation of modes of action and serendipitous discovery, and to date, only a few have been successfully discovered21,22.
This review discusses representative cases of molecular glues and analyzes the binding mode of the molecular glue–protein complex. Based on the aggregation mode of molecular glues mediating protein–protein interactions, three mediating modes including homo-dimerization, homo-polymerization and hetero-dimerization are reviewed in detail (Fig. 1C). Overall, we hope to explain the general law of molecular glue mediating mechanisms, exploit the strategies and approaches of their design, optimization and identification, and finally summarize the potentials and prospects from the perspective of pharmaceutical chemistry.
2. Mediating homo-dimerization
2.1. PD-L1 dimerization
During the immune response, immune checkpoints can modulate antigen recognizing of the T-cell receptor (TCR) and repress the immune system from targeting self-antigens under normal conditions23. Unfortunately, cancer cells suppress T-cell activation by hijacking the checkpoint pathway, and thus avoiding immune attack24. The crucial checkpoint molecule, programmed cell death-1 (PD-1, express on the surface of activated T lymphocytes) binds to programmed cell death-ligand 1 (PD-L1, overexpression on the surface of cancer cells), causing tumor immune escape and promoting tumor progression25, 26, 27. Therefore, regulation of the PD-1/PD-L1 interaction represents a considerable strategy for tumor immunotherapy.
To date, U.S. Food and Drug Administration (FDA) have approved seven PD-1/PD-L1 monoclonal antibodies for clinical use28,29. However, due to the drawbacks of antibodies, such as high manufacturing cost, lack of oral activity and immune-related side effects, it is necessary to design PD-1/PD-L1 small molecule inhibitors as alternatives for monoclonal antibodies. Nevertheless, the flat and large PPI interface between PD-1 and PD-L1 makes the design of small molecular inhibitors difficult.
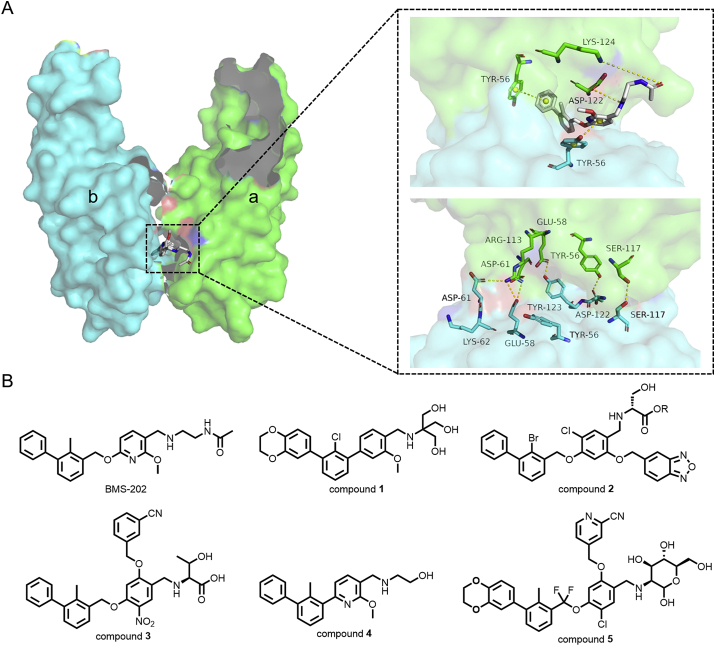
In 2015, several compounds possessing hydrophobic biphenyl scaffolds were disclosed by Bristol-Myers-Squibb (BMS) as the first PD-1/PD-L1 nonpeptidic PPI inhibitors in their patents30,31. Through a homogeneous time-resolved fluorescence (HTRF) assay, these compounds were identified to inhibit the formation of PD-1/PD-L1 complex, while the detailed mechanism remained ambiguous. In 2017, Zak et al.32 performed NMR experiments and found that BMS-202 directly binds to its target protein PD-L1, but not to PD-1. Moreover, the results also indicated that BMS-202 induces formation of an approximately 30 kDa complex, which is consistent with the mass of the 2:1 complex of PD-L1 (2) and BMS-202 (1). Further crystal structural analysis confirmed that molecular glue BMS-202 mediated the dimerization of PD-L132. As shown in Fig. 2A, aAsp122 and aLys124 form hydrogen bonds with BMS-202, the bGlu58, bASP61, bAsp122 and bTyr123 residues form hydrogen bonds with aTyr56, aGlu58, aAsp61 and aArg113. There is a π‒π stacking interaction between the biphenyl ring of BMS-202 and aTyr56 residue, which locks the BMS-202 molecule and PD-L1 dimer in place to generate a stable ternary complex. These findings contributed to the recent discoveries of PD-L1 dimer molecular glues reported by Muszak33, Liu34, Ouyang35, Wang36 and Song37 (compounds 1–5, Fig. 2B).
Figure 2.
(A) The molecular glue BMS-202 mediates the homo-dimerization of PD-L1 (monomer a: green, monomer b: cyan, PDB:5J89). (B) Compounds 1–5 were the best compounds reported by Muszak33, Liu34, Ouyang35, Wang36 and Song37, respectively.
2.2. REV1 dimerization
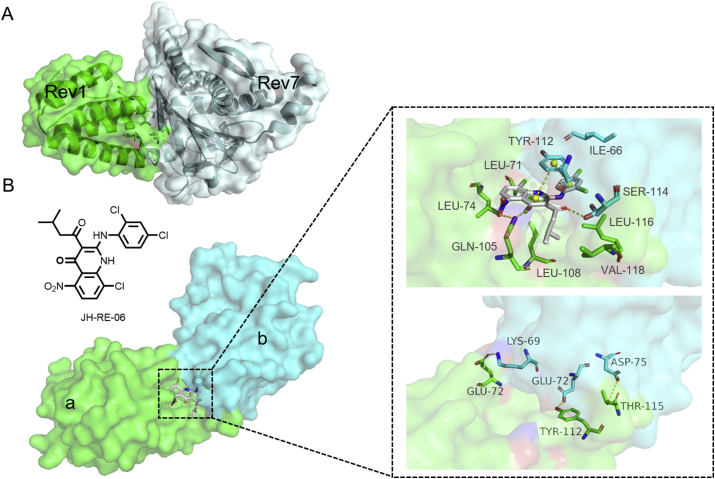
Mutagenic translesion synthesis (TLS) can lead to chemotherapy resistance; therefore targeting TLS is a powerful way to sensitize tumors to chemotherapy drugs38. TLS occurs in two steps in mammalian cells, first by inserting a TLS DNA polymerase (like REV1) to introduce the nucleotide opposite the lesion, and then extending TLS DNA polymerase (such as REV7) to extend the 3′ terminus39. The REV1 C-terminal domain (CTD) recruits POL-ζ by interacting with REV7 constituents, and the complex plays an essential role in harmonizing TLS (Fig. 3A)38. Inhibition of REV1‒REV7 interactions has received much attention, while the shallow binding interface limits the development of small molecules.
Figure 3.
(A) REV1 CTD–REV7 complex (REV7: gray, REV1: green, PDB:4FJO). (B) The molecular glue JH-RE-06 mediates homo-dimerization of cREV1 CTD (monomer a: green, monomer b: cyan, PDB:6C8C).
In 2019, Wojtaszek et al.40,41 screened approximately 10,000 structurally diverse compounds using a special enzyme linked immunosorbent assay (ELISA), and found that compound JH-RE-06 acts as a molecular glue to potently hinder the REV1‒REV7 PPI interaction. Further cocrystal structure studies revealed that JH-RE-06 induces REV1 dimerization. In the chimeric REV1 CTD (cREV1 CTD) dimer, two REV7-binding pockets face each other to form a deep cavity for JH-RE-06 binding (Fig. 3B). The CTD of monomer A is led out by the acyl chain of JH-RE-06 and a large 1,4-dihydroquinolinone group, and there are a lot of hydrophobic interactions between cREV1 CTD and JH-RE-06 (e.g., bIle66, aLeu71, aLeu74, aLeu108, bTyr112, aLeu116 and aVal118; Fig. 3B). The sidechains of aGln105 and bSer114 form direct polar interactions with the hydrogen bond receptor groups of the 1,4-dihydroquinolinone moiety of JH-RE-06. The C-terminal tails of the two monomers pair with each other to form an antiparallel β sheet for the binding of JH-RE-06. Specifically, bLys69, bGlu72 and bAsp75 residues form hydrogen bonds with aGlu75, aTyr112 and aThr115 to stabilize the cREV1 CTD dimer.
The formation of JH-RE-06 induced dimer shields the REV7-binding surface, thus blocking the interaction between REV7 and REV1 CTD. Further in vitro and in vivo pharmacodynamics studies showed that JH-RE-06 combined with cisplatin enhanced the toxicity of cisplatin to various cell lines, and reduced the volume of the xenograft human melanoma in mice42.
2.3. MDMX dimerization
Murine double minute 2 (MDM2) and MDMX, the negative regulator of the P53 tumor suppressor, are promising targets for cancer therapy43. MDM2 antagonists alone may cause tumor cells to overexpress MDMX to inhibit the P53 pathway, therefore, the identification of MDM2/MDMX dual antagonists has been intensively studied recently44,45.
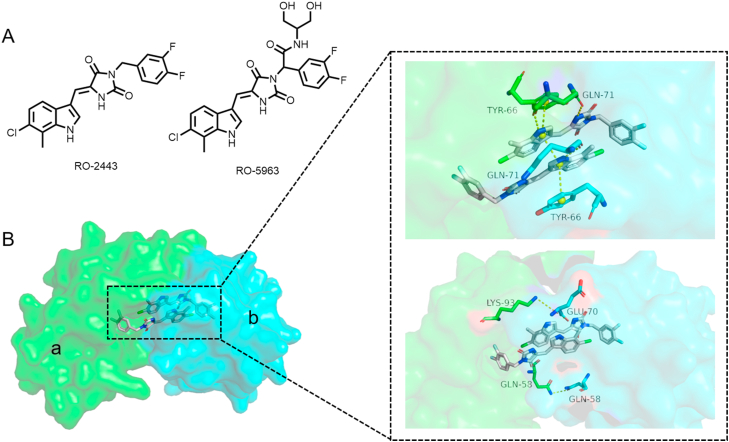
Through small molecules library screening, RO-2443 was found to exhibit similar inhibitory activity against both P53‒MDMX and P53‒MDM2 interactions (Fig. 4A)46. In the mechanistic study examining the interaction between RO-2443 and MDMX, the results from NMR spectroscopy, static light scattering (SEC-SLS) and kinetic analysis revealed that RO-2443 may act as a molecular glue to induce MDMX dimerization. Co-crystals of MDMX/RO-2443 with high resolution at 1.8 Å confirmed this prediction (Fig. 4B), RO-2443 occupies the P53 binding pocket of MDMX to form a dimer and be incorporated into a central cavity, in which the two RO-2443 molecules are surrounded by the interaction of the two proteins, and the four monomers are arranged in a pair of dimers. The four-level π-sandwich interaction between the two chloroindole groups of RO-2443 and Tyr66 of the corresponding protein monomer forms an extensive aromatic stacking interaction, which is the key interaction of polymer formation. In addition, RO-2443 forms two hydrogen bond interactions with the Gln71 residue. Because the molecular glue pulls the two MDMX dimers closer, the aLys93 residue interacts with the bGlu70 residue, and aGlu58 and bGlu58 form hydrogen bond interactions that stabilize the dimer.
Figure 4.
(A) Structures of the reported molecular glues mediating MDMX dimerization. (B) The molecular glue RO-2443 mediates the homo-dimerization of MDMX (monomer a: green, monomer b: cyan, PDB:3U15).
The molecular glue RO-2443 and its derivative RO-5963 may provide an effective treatment for MDMX-overexpressing cancers by mediating the activation of P53 apoptotic activity, while their poor water solubility and pharmacological properties require further optimization.
3. Mediating homo-polymerization
3.1. BCL6 polymerization
B-cell lymphoma 6 (BCL6), a transcriptional repressor protein, is a potential drug target for non-Hodgkin's lymphoma47,48. Several small molecule inhibitors and peptides of BCL6 with in vivo efficacy can only work at high concentrations, limiting their clinical application49, 50, 51.
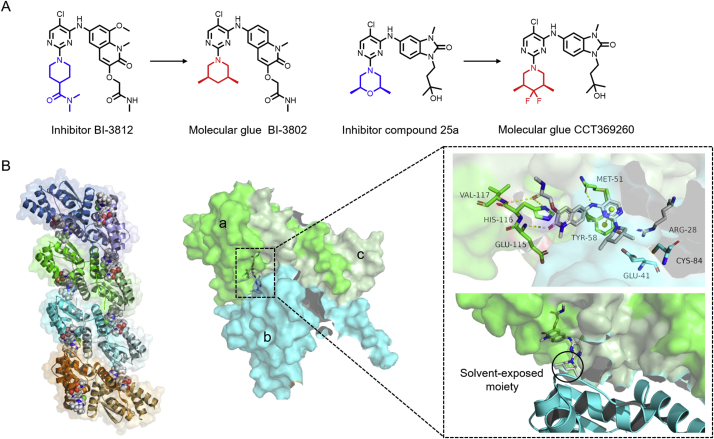
Kerres et al.52 showed that the broad-complex, tramtrack and bric-à-brac (BTB) domain of BCL6 is highly druggable, the compounds targeting this region may lead to development of potential BCL6 inhibitors. They screened a library of 1.7 million compounds using a fluorescence polarization (FP) assay, and identified a series of 4-amino-5-chloropyrimidine compounds. Among them, BI-3802, a close analogue of the BCL6 inhibitor BI-3812 (Fig. 5A), was unexpectedly found to mediate protein aggregation. Cryo-EM structural analysis showed that the key residues aTyr58, cArg28, bGlu41 and bCys84 between BCL6 dimers form a groove for BI-3802 to bind53. The aMet51, aVal117, aHis116 and aGlu115 residues also form hydrogen bonds with BI-3802 (Fig. 5B). This complex structure is modeled as a polymer by ordering dimers in the same binding pattern, which can be identified by the non-cullin E3 ubiquitin ligase seven in absentia homolog 1 (SIAH1) for ubiquitination and degradation (Fig. 5B).
Figure 5.
(A) Structures of molecular glues BI-3802, CCT369260 and their corresponding inhibitors. (B) BI-3802 mediates the homo-polymerization of BCL6 (monomer a: green, monomer b: cyan, monomer c: palegreen, PDB: 6XMX).
A large part of the degrading compound BI-3802 locates at the BTB domain, and the dimethylpiperidine ring remains solvent exposed that can be used to bind with another monomer, which provides an idea for the design of small molecule protein aggregation inducers by modifying the different substituted groups of small molecules when they are exposed to solvent. In 2020, a series of BCL6 inhibitors structurally similar to BI-3802 were reported, the researchers speculated that a similar molecular glue might be obtained by modifying the structure of the solvent exposed region54. Ultimately, a subset of these inhibitors, such as CCT369260 (Fig. 5A), was found to form BCL6 polymers, which can be recognized and degraded by SIAH1.
3.2. MERS-CoV polymerization
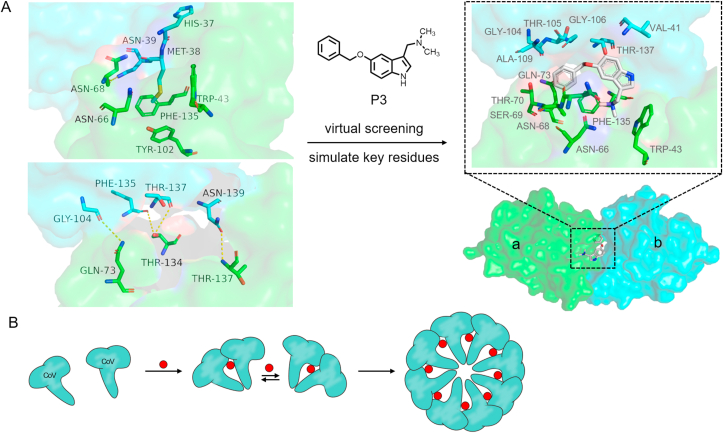
Middle East respiratory syndrome coronavirus (MERS-CoV) can cause severe acute respiratory syndrome (SARS) with a high fatality rate55,56. The strongly related novel coronavirus corona virus disease 2019 (COVID-19) is ravaging the world, further highlighting the global public health risks posed by coronaviruses57,58. Development of small molecules inducing the polymerization of CoV nucleocapsid (N) protein is a viable strategy for the exploitation of antiviral drugs59. The CoV N protein consists of N-terminal domains (NTDs) and CTDs, and CoV N-CTD is responsible for the oligomerization of the N protein through protein–protein interactions60. Structural studies have shown that His37, Met38 and Trp43 residues play important roles in the formation of hydrophobic vesicles containing carrier fusion residues and mediate the formation of CoV N-NTD dimers (Fig. 6A)61. As shown in Fig. 6, the side chain of BMet38 interacts with a conserved hydrophobic pocket composed of aTrp43, aAsn66, aAsn68, aTyr102, and aPhe135. bHis37 and bAsn39 are packed against aTrp43 and aPhe135, and contribute to this hydrophobic interaction. The primary chain oxygens of bGly104, bPhe135, bThr137 and aThr137 and the side chains of aGln73, aThr134 and bAsn139 form hydrogen bonds, respectively.
Figure 6.
(A) The conservative hydrophobic interactions of CoV N-NTD dimers are shown on the upper left and hydrogen bonds on the lower left (monomer a: green, monomer b: cyan, PDB:4UD1). P3 mediates the homo-polymerization of MERS-CoV N-NTD (right, PDB:6KL6). (B) Diagram of molecular glue P3-mediated polymerization of viral proteins.
It is possible to substitute small molecules for carrier fusion residues to stabilize the PPIs of CoV N-NTD dimers. Using structure-based screening to target the key Trp43 residue in the template where His37 and Met38 were removed, Lin et al.61 attempted to search compounds that could replace these residues and thus act as molecular glues. They identified 5-benzoxyglycine P3 as a novel positive N protein PPI stabilizer with N-NTD protein stabilizing and antiviral activity, which can be used in drug discovery for anti-CoV diseases. X-ray crystallography revealed that P3 along with residues of two monomers generates a large hydrophobic driving force simultaneously, which stabilizes the dimeric conformation of N-NTD, and the residues composition is aTrp43, aAsn66, aAsn68, aSer69, aThr70, aGln73, aPhe135, bVal41, bGly104, bThr105, bGly106, bAla109 and bThr137. Small-angle X-ray scattering (SAXS) indicated that P3 mediates the polymerization of N proteins to form an eight-membered circular structure (Fig. 6B), which blocks the CTD associated with viral transcriptional assembly, producing an antiviral effect.
4. Mediating hetero-dimerization
Molecular glues can not only regulate polymerization of proteins themselves, but also mediate target proteins by hijacking the functions of foreign proteins. For example, by inducing interactions with E3 ubiquitin ligases or enhancing pre-existing PPIs, molecular glues can trigger the ubiquitination and degradation of target proteins, and thus those molecules are commonly referenced as “molecular glue degraders”. Although the design of molecular glue degraders still depends on accidental discovery, some systems have been well studied and created tremendous sensation in the field of chemical biology. In the next sections, we focus on the molecular glues that mediate the dimerization of heterologous proteins, most of which are molecular glue degraders.
4.1. DDB1CRBN-substrate
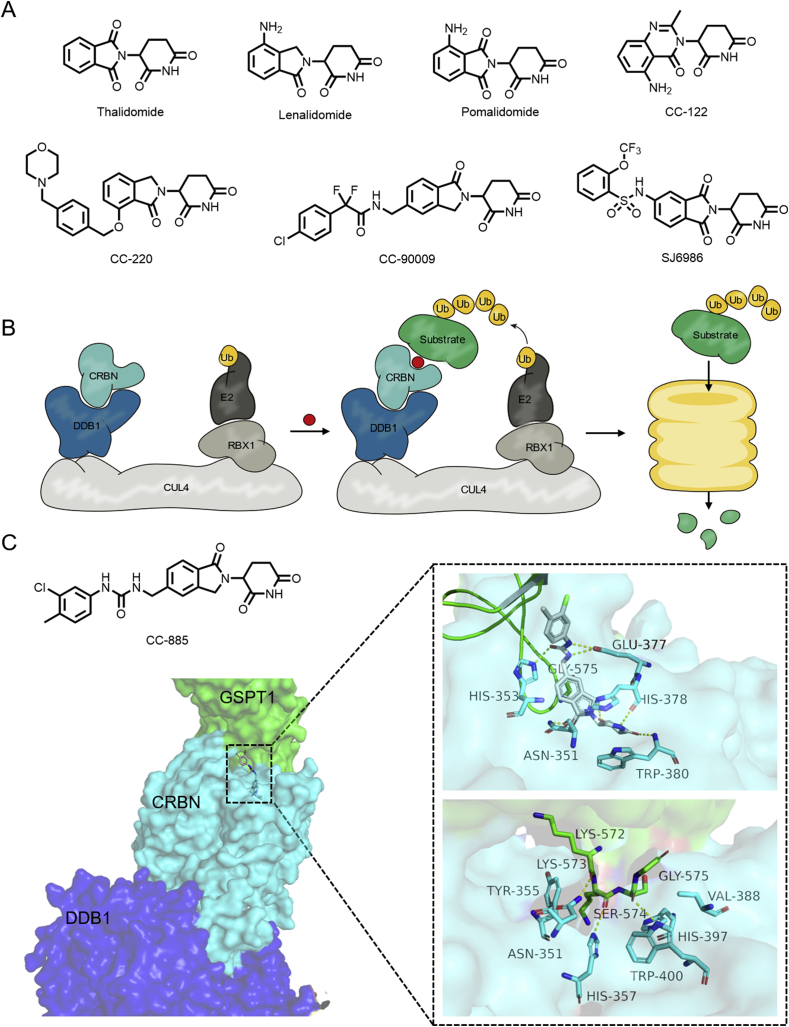
The immunomodulatory drugs (IMiDs) thalidomide and its analogs bind to the substrate receptor cereblon (CRBN) of E3 ligase complex and modulate the binding region of CRBN to recruit the “neo-substrate” protein, causing degradation of the “neo-substrate” protein and exerting immunosuppressant activity (Fig. 7A,B)62,63.
Figure 7.
(A) Structures of CRBN-binding drugs. (B) Diagram of molecular glues mediated hetero-dimerization of DDB1CRBN and substrate. (C) CC-885 mediates the hetero-dimerization of CRBN‒GSPT1 (DDB1 colored in blue, CRBN colored in cyan, GSPT1 colored in green; PDB: 5HXB). Hydrogen bonds are shown as yellow dotted lines.
The transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) are the primary neo-substrate of lenalidomide-bound CRL4CRBN64,65. Compared with lenalidomide or pomalidomide, CC-122 and CC-220 have a higher affinity for CRBN and stronger ability to induce IKZF1/3 degradation, and the therapeutic effect of CC-220 on systemic lupus erythematosus has been studied in 2018 as well. Matyskiela et al.66 synthetized a series of compounds that are bound to CRBN, among which CC-885 exhibits particularly significant anti-proliferative activity against acute myeloid leukemia (AML) (Fig. 7C). Subsequent immunoprecipitation experiments revealed that CC-885 recruits GSPT1 (G1 to S phase transition 1/eukaryotic peptide chain release factor GTP-binding subunit ERF3A) to form dimers67. To date, molecular glues CC-90009 and SJ6986 with a similar mechanism have also been reported, and CC-90009 has entered clinical development68,69. Other neo-substrates reported include the protein kinase CK1α, the transcription factor ZBTB1670, the putative ubiquitin ligase ZFP9171, the kinase CSNK1A172, and the P63 isoforms ΔNp63α and TAp63α (related to teratogenicity)73. Related studies are ongoing.
Although each IMiD displays distinct patterns of substrate specificity, a comparison of IMiDs and closely related analogs revealed that most of these compounds possess a common C2H2 zinc finger recognition degron motif. These reports also indicated that a β-hairpin loop with a critical glycine is found within this motif. In addition, CRBN-ligand-GSPT1 binding is mainly mediated by hydrogen bonds formed by the neo-substrate backbone70, which is crucial to understand the principle and basis of how ligand modifies the interaction interface of the neo-substrate and CRBN. Taking CC-885 as an example, we will introduce the mechanism of IMiDs-mediated hetero-dimerization of CRBN and their substrate. CC-885 forms hydrogen bonds with Trp380, His378, Asn351, Glu377 and His353 residues, while the binding pattern of isoindolinone ring to CRBN is similar to lenalidomide (Fig. 7C). The terminal substituted benzene ring of CC-885 is located near the β-hairpin loop of GSPT1, and CC-885 works as an interaction hotspot in CRBN through planer hydrophobic group. GSPT1 forms protein–protein interactions with CRBN Asn351, His357 and Trp400 residues, all directly contributing to forming the hydrogen bonds with oxygen atoms from GSPT1 Lys572, Lys573, and Ser574. In addition, GSPT1 has a special folding structure around Gly575 that binds CC-885 and CRBN. The side chains of CRBN residues Tyr355, His397 and Val388 provide further van der Waals interactions.
4.2. DDB1DCAF15-RBM39
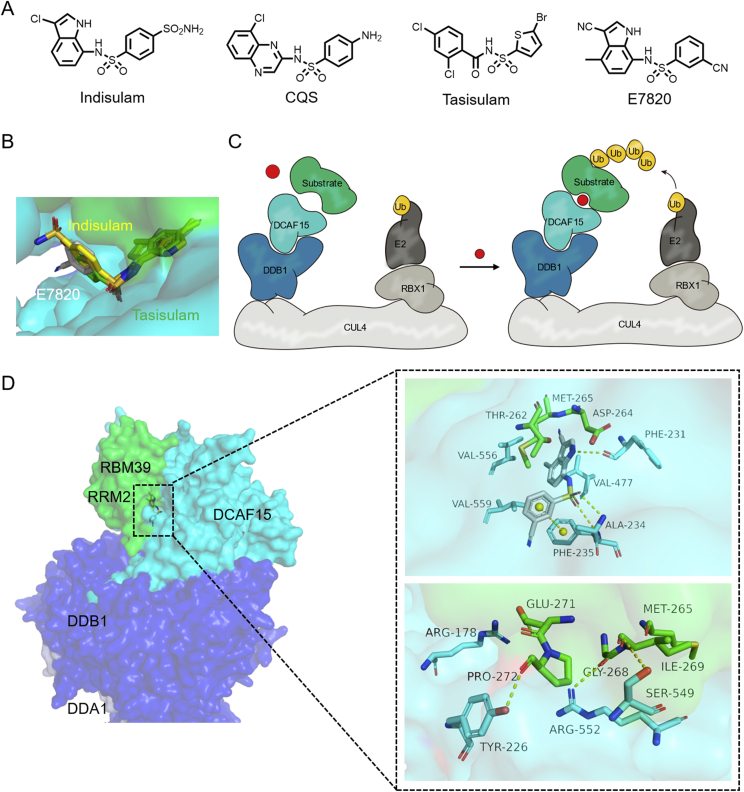
Indisulam74 and E782075 have significant anti-proliferative effects on many hematopoietic and lymphoid cancer cell lines (Fig. 8A). In 2017, Han et al.76 found that RNA binding motif protein 39 (RBM39) is the target of these anticancer molecules. The indisulam-induced inactivation of RBM39 leads to abnormal pre-mRNA splicing, contributing to its lethal effect on cancer cells. Using CRISPR/Cas9 engineering, they subsequently demonstrated that indisulam may function as a molecular glue that induces the recruitment of RBM39 to the CUL4-DCAF15 E3 ubiquitin ligase, ultimately resulting in RBM39 ubiquitination and degradation (Fig. 8B and C)76.
Figure 8.
(A) Chemical structures of aryl-sulfonamides molecular glues. (B) A combination of E7820 and indisulam on the surface of DCAF15 and RBM39 (indisulam PDB:6Q0W). (C) Schematic diagram of the action mechanism of aryl sulfonamides molecular glues. (D) DDB1-DCAF15-E7820-RBM39 co-crystal structure (DDB1 colored in blue, DCAF15 colored in cyan, RBM39 colored in green, DDA1 colored in lightblue; PDB:6Q0R). Hydrogen bonds are shown as yellow dotted lines.
Du et al.77 solved the crystal structure of the DCAF15–E7820–RBM39 complex in 2019, Faust et al.78 presented a more detailed cryo-EM structure at a resolution of 4.4 Å in 2020. They both showed that E7820 binds to the shallow pocket on DCAF15 surface, and forms hydrogen bonds with the Ala234, Phe235 and Phe231 residues with good geometry (Fig. 8D). A key π‒π stacking interaction is formed between the Phe235 residue of DCAF15 and the benzonitrile ring of E7820. The E7820-modified DCAF15 interface interacts with RBM39 through the α-helix of the second RNA recognition motif (RRM2) domain. The side chains of Arg552, Ser549, and Tyr226 of DCAF15 form hydrogen bonds with Gly268, Ile269, and Pro272 of the RRM2 domain, respectively. There are also some salt bridges and van der Waals interactions.
Kinetic studies revealed that DCAF15 has a certain affinity with RBM39 itself, and the combination of aryl sulfonamides results in a formation of the ternary complex with higher affinity77. While IMiDs have nanomolar affinity for CRBN, there have been no reports suggesting neo-substrate will interact with CRBN in the absence of IMiD79. Moreover, for the IMiDs/CRBN system, β hairpin requires less sequence specificity, and most IMiDs are structurally similar and have many neo-substrates. However, the interaction between α-helix of RBM39 and DCAF15 is more specific, making it difficult to find neo-substrate. Nevertheless, they both support the concept that molecular glues depend on the secondary structural characteristics of target proteins, rather than the classically defined ligand binding pocket, representing a potential method for expanding the druggable space in the proteome.
4.3. DDB1-CDK12/cyclin K
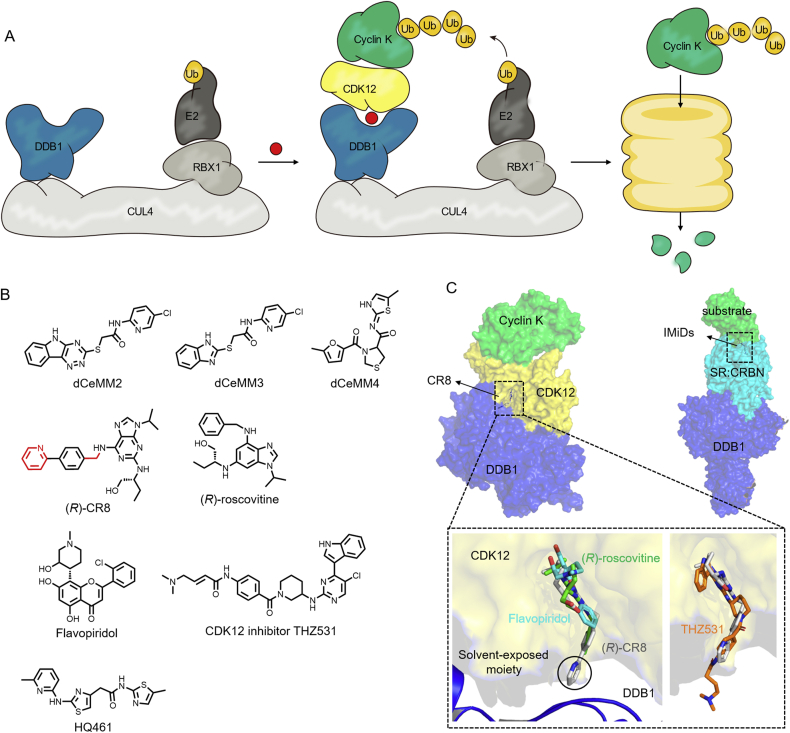
The molecular glue degraders mentioned above target the substrate receptors (such as CRBN and DCAF15) of E3 ubiquitin ligases to recruit substrates80, 81, 82. Three types of small molecules that induce the hetero-dimeric target complex CDK12‒cyclin K and DDB1 binding will be briefly described in the following paragraphs. It is noteworthy that, unlike the cases described above, the interaction of CDK12‒cyclin K induced by these molecular glues is functionally independent of the substrate receptors (Fig. 9A). Molecular glues may not necessarily bind directly to the target (cyclin K in this case) but rather may use an intermediate protein (CDK12 in this case), and the transfer of ubiquitin is largely dependent on the spatial location of the E2 and substrate protein. The molecular glue toolkit is expanding to bypass the requirement in consideration of whether the target tissues/cells express specific DCAF and thus has a wider range of applications.
Figure 9.
(A) Schematic diagram of the action mechanism of molecular glue CR8. (B) Structures of three types of DDB1 and CDK12/cyclinK molecular glues. (C) Comparison of protein complex structures of CR8 (PDB:6TD3) and IMiDs molecular glues. A partial enlargement of the CR8 solvent exposure area compared with (R)-roscovitine (PDB:2A4L), flavopiridol (PDB:3BLR) and THZ531 (PDB:5ACB).
Mayor-Ruiz et al.83 revealed that dCeMM2/3/4 (Fig. 9B) prompt the interaction between CDK12-cyclin K and a CRL4 ligase complex. Similarly, the new type compound HQ461 was discovered and designed by phenotype-based high-throughput screening and biochemical reconstitution, which can directly occupy and modify the ATP binding pocket of CDK12 for DDB1 binding84.
Likewise, CR8, discovered through systematically mining databases, is a new type molecular glue that induces proximity of CDK12‒cyclin K and DDB1‒CUL4‒RBX1 E3 ligase core (Fig. 9C)85. To achieve this, CR8 locates at the ATP-binding pocket of CDK12 and bridges the CDK12‒DDB1 interface. More importantly, the localization of 2-pyridyl in the surface-exposed moieties of CR8 confers its gain-of-function glue activity by forming discrete contacts with Ile909, Arg928 and Arg947 residues in DDB1 β-propeller domain C (BPC, also involved in DCAF binding). Słabicki et al.85 demonstrated that modification of surface-exposed regions of the inhibitor can develop the target-binding molecules into molecular glues, the same as the modification of BI-3802. The parent compound (R)-roscovitine86 and natural product flavopiridol87 both stimulate CDK12‒cyclin K interacting with DDB1, albeit with lower binding affinity than CR8 (Fig. 9C). In contrast, the exposure volume of the covalent CDK12 inhibitor THZ531 is bulky, which is predicted to have steric hindrance with DDB1, losing the ability to recruit CDK12–DDB1 instead88.
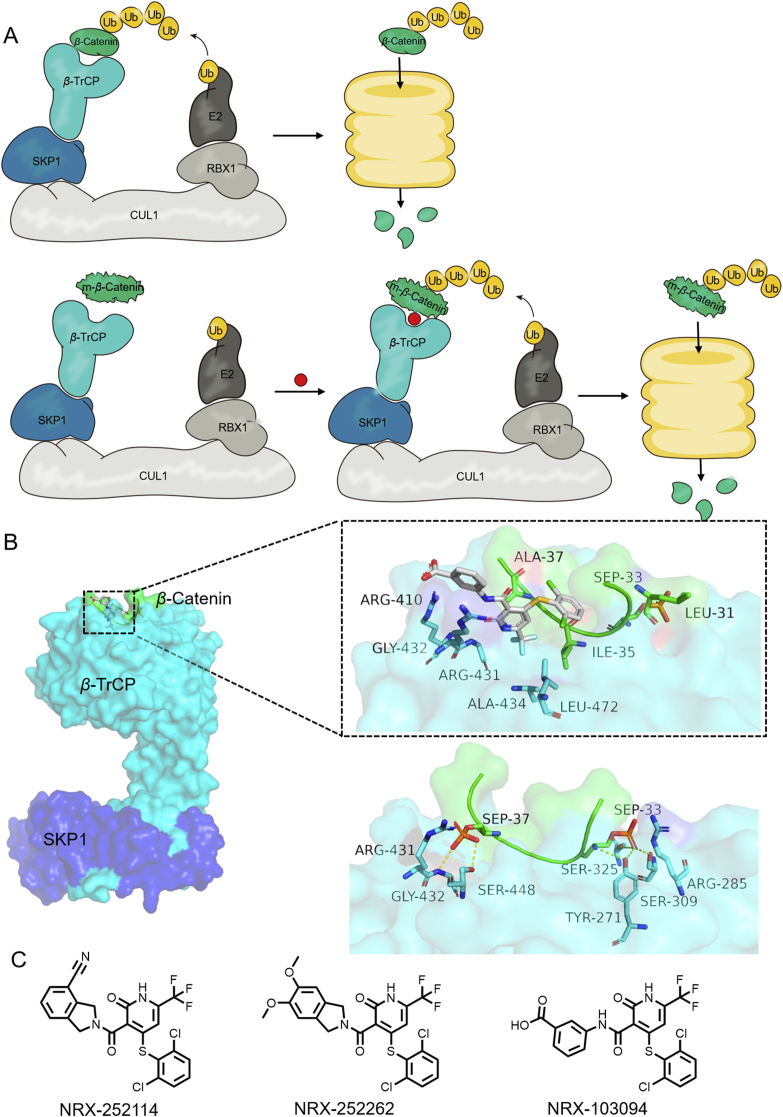
4.4. SCFβ-TrCP‒β-catenin
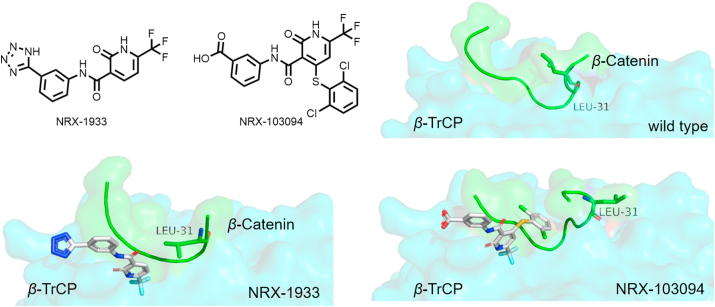
Simonetta et al.89 reported the rational design and identification of potent molecular glues for the oncogenic transcription factor, β-catenin, and its homologous RING-E3 ligase, SCFβ-TrCP. Under normal conditions, the phosphorylated β-catenin binds to β-TrCP at Ser33 and Ser37, leading to ubiquitylation and degradation (Fig. 10A)90. However, mutations in these β-catenin phosphodegron attenuate β-catenin‒β-TrCP interactions, leading to an enhanced oncogenic transcriptional program. Phosphorylated residue Ser37 mediates key electrostatic and hydrogen bonding in PPI interactions, and the removal of the phosphate of Ser37 leads to a reduction in binding affinity, while simultaneously forming a small hydrophobic pocket between the PPI interface (Fig. 10B). Using the concept of molecular glues, this research group discovered that NRX-103094 can occupy the mutated interface (Fig. 10C). As shown in the complex of the S37A β-catenin mutation and β-TrCP, the Ala434 and Leu472 residues of β-TrCP and Leu31 and Ile35 residues of the β-catenin form hydrophobic pockets, which are filled by the trifluoromethyl substituent of NRX-103094.
Figure 10.
(A) Schematic diagram of the action mode of molecular glue NRX-103094. (B) NRX-103094 mediate the hetero-dimerization of β-catenin‒β-TrCP (up, PDB:6M91), and comparison with wild-type (down, PDB:1P22). (C) Structures of NRX series as molecular glues.
4.5. LC3-mHTT
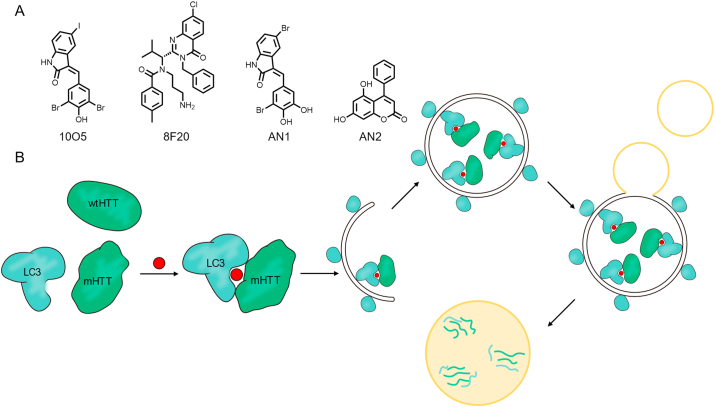
Intracellular protein degradation mainly occurs through the ubiquitin‒proteasome system (UPS) or autophagy–lysosome pathway91,92. Although most current studies focus on E3 ligases of UPS, Li et al.93 first applied the concept of autophagosome-tethering compounds (ATTEC), which eliminates pathogenic proteins by mediating target proteins to the autophagosome protein microtubule-associated protein 1A/1B light chain 3 (LC3) for autophagic degradation. Mutant huntingtin (mHTT) protein causes a neurodegenerative disorder disease, Huntington's disease (HD)94,95. mHTT contains an expansion of the polyglutamine (polyQ) tract (more than 35 glutamine residues) and forms aggregates in neurons96. Such aggregates or large proteins can resist being degraded by proteasomes, so PROTACs is an inefficient approach in this condition.
Fei's group93 developed a high-throughput drug screening platform based on small molecule microarray (SMM) and oblix-incidence reflectivity difference (OI-RD) technology, that can quickly, sensitively and without labeling find small molecules binding to target proteins from thousands of small molecule compounds. They selected four compounds that tether mHTT and LC3 to incorporate mHTT into double-membrane autophagosomes for further degradation through autolysosomes (Fig. 11A). Such compounds specifically interact with mHTT but not with wide-type HTT (wtHTT) by distinguishing the emergent conformation of the expanded polyQ tract, which is distinct from the short polyQ stretch in wtHTT, achieving allele-selective. LC3 is polymerized and amplified to form a membrane structure, which wraps the mHTT to form a complete autophagosome during the process of autophagy. After fusion with the lysosome, the encapsulated substances can be degraded (Fig. 11B). However, to parse the interactive interface of the compound and LC3, comprehensive pharmacochemical and structural studies still need to be performed.
Figure 11.
(A) Structures of ATTEC. (B) Schematic diagram of the action mode of ATTEC.
4.6. GLP-1–GLP-1R
Although molecular glue degraders are the most common molecular glues mediating hetero-dimerization of target proteins identified in recent studies, it should be noted that in addition to acting as a degrader, molecular glues can also be used to increase ligase-substrate binding affinity by stabilizing hetero-dimerization, acting as PPI enhancers or stabilizers to augment substrate–ligase interactions. Such molecular glues can be therapeutically useful against targets previously considered undruggable, such as the PPI between the target protein and its natural substrate, which may be weakened by mutations.
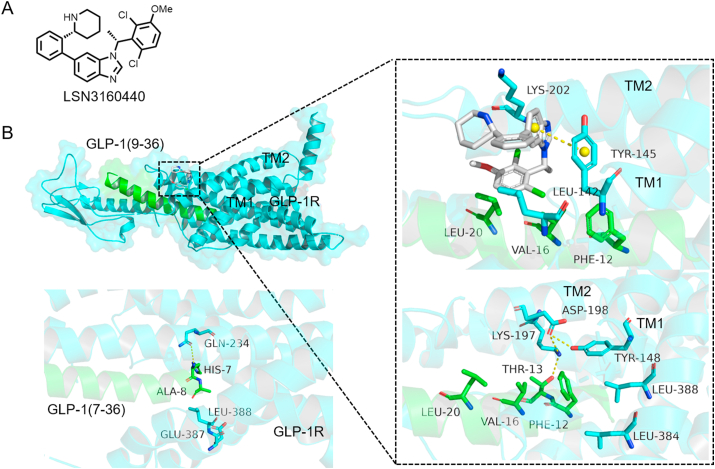
The glucagon-like peptide 1 receptor (GLP-1R) acts as an important target of type 2 diabetes97, 98, 99, but the half-life of its endogenous ligand GLP-1(7–36) is only 2 min, followed by hydrolysis to remove the N-terminal dipeptide His7-Ala8 to produce metabolite GLP-1(9–36)100. His7 and Ala8 are key residues of GLP-1(7–36) in the interaction between GLP-1(7–36) and GLP-1R, as His7 forms a hydrogen bond with Gln234, and Ala8 generates van der Waals interactions with Glu387 and Leu388. And therefore, compared with GLP-1(7–36), GLP-1(9–36) has a weaker affinity for GLP-1R. Bueno et al.101 reported the discovery of a molecular glue called LSN3160440 (Fig. 12A), which participates in and enhances the interactions between GLP-1R and GLP-1(9–36). LSN3160440 binds between the TM1 and TM2 interfaces of GLP-1R, and the benzimidazole group of LSN3160440 interacts with GLP-1R in a variety of ways, such as van der Waals contacts with Leu142, π‒π stacking with Tyr145, and frequent water bridging with Lys202 (Fig. 12B). In the absence of His7 and Ala8 residues, LSN3160440 can act as a molecular glue to establish new bridging contacts between Val16 and Leu20 residues of the GLP-1(9–36) and the GLP-1R, enhancing the affinity of PPI.
Figure 12.
(A) Structure of LSN3160440. (B) GLP-1(9–36)–LSN3140660–GLP-1R complex (right, GLP-1R: cyan, GLP-1: green; PDB:6VCB), and comparison with GLP-1(7–36)–GLP-1R complex (left, PDB:5VAI).
5. Biochemical techniques for the development of molecular glues
The development of biochemical techniques has made a significant contribution to the discovery of molecular glues, and this review lists most of the discovery strategies discussed above (Table 1)102,103. The following paragraphs primarily introduce the relatively latest technologies or methods and divide them into three stages of molecular glue development: starting point, optimization and confirmation.
Table 1.
The discovery strategies of molecular glues and the specific techniques exploited within these studies.
| Target protein | Molecular glues | Discovery strategy | Techniques | Refs. |
|---|---|---|---|---|
| PD-L1 | BMS-202 | Serendipity | NMR, DSF, crystal | 32 |
| REV1 | JH-RE-06 | Serendipity | Crystal, SDS-PAGE | 40 |
| MDMX | RO-2443, RO-5986 | Serendipity | NMR, SEC-SLS, crystal, TR-FRET | 46 |
| BCL6 | BI-3802 | Serendipity | IF, Cryo-EM, WB, TR-FRET | 53 |
| CCT369260 | Serendipity | Nano-BRET, IF, MSD | 54 | |
| MERS-CoV | P3 | Virtual screening | SAXS, IF, crystal, thermostability | 61 |
| DDB1CRBN–substrate | Thalidomide, lenalidomide, pomalidomide | Serendipity | Proteomic, LC‒MS/MS, crystal, TR-FRET | 62,63 |
| CC-122, CC-220 | Rational design | Crystal, WB, TR-FRET | 64,65 | |
| CC-885 | Rational design | IP, crystal, pull-down, IF, LC‒MS/MS, SPPIER | 67 | |
| CC-90009 | Rational design | WB, proteomic, crystal | 69 | |
| SJ6986 | Rational design | WB, MD simulation | 68 | |
| DDB1DCAF15–RBM39 | Indisulam, E7820 | Serendipity | IP, LC‒MS/MS, CRISPR/Cas9, pull-down, Cryo-EM, TR-FRET | 77,78 |
| DDB1–CDK12/cyclinK | dCeMM2, dCeMM3, dCeMM4 | Chemical screening | WB, CRISPR/Cas9, pull-down | 83 |
| (R)-CR8 | Data mining | Co-IP, ITC, crystal, TR-FRET, nano-BRET | 85 | |
| HQ461 | HTS | CRISPR/Cas9, Co-IP, WB, pull-down, CXMS, LC‒MS/MS, DSF, thermostability | 84 | |
| SCFβ-TrCP–β-catenin | NRX-252114, NRX-252262, NRX-103094 | HTS, rational design | FP, crystal, TR-FRET, pull-down | 89 |
| LC3-mHTT | 10O5, 8F20, AN1, AN2 | HTS | SMM, OI-RD, HTRF, MST, pull-down, IF | 93 |
| GLP-1–GLP-1R | LSN3160440 | HTS, rational design | Crystal, MD simulation, IF | 101 |
5.1. Starting point: Rational design
Although most molecular glues have been discovered by chance, or as secondary development of existing compounds, we can still obtain some insights into the direction of screening and rational design from some of the reports.
Some key residues on the PPI interfaces contribute a lot to the total binding energy of the interaction, termed “hot spots”104, 105, 106. For potential PPI or PPI with weak binding affinity, molecular glues can function as “hot spots” to mediate protein–protein binding by occupying the protein sites that can be anchored. Therefore, screening small molecules that can interact well with these sites can be regarded as the starting point for the identification of molecular glues. The screening strategy can be divided into structural screening based on shape complementarity and hydrophobic interactions, and pharmacophore screening based on identifying the key characteristics of interacting proteins. For example, a large number of novel thalidomide analogs were discovered and developed through phenotypic screening67, 68, 69, and antiviral molecule P3 was obtained by structure-based virtual screening61.
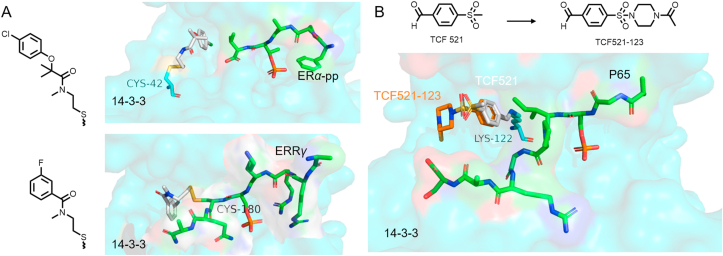
For existing protein–protein interactions, the tethering strategy107 may capture the fragments with an inherent binding affinity for pockets, which can be used as the starting point for the development of molecular glues107,108. The host protein 14-3-3 binds with the phosphorylated motifs of client proteins to play a critical role in disease-related signaling networks, for example, the stabilization of 14-3-3 and estrogen receptor α derived phosphopeptide (ERα-pp) is an effective strategy for the treatment of ERα-positive breast cancer109. Based on the tethering strategy, Ottmann's group109 introduced a cysteine at position 42 of 14-3-3 to act as a handle and identified several fragments that binding to the complex of 14-3-3 and ERα-pp with increased binding affinity (Fig. 13A). While inverting the strategy with the reactive handle on the estrogen related receptor gamma (ERRγ), a client-derived peptide of 14-3-3, they identified several fragments that stabilize the complex of 14-3-3 and ERRγ by targeting the Cys180 residue of ERRγ (Fig. 13A), which provide the starting point for the discovery of molecule glues110.
Figure 13.
(A) Crystallographic overlay of 14-3-3(C42)-ERα (PDB:6HMT); 14-3-3-ERRγ(C180) (PDB:6Y3W) and the structures of fragments they screened out. (B) Crystallographic overlay of 14-3-3/P65 with TCF521 (PDB:6YOW) and 14-3-3/P65 with TCF521-123 (PDB:6YPY).
Inspired by cysteine, lysine has been used in covalent drug targeting methods as well111,112. Wolter et al.113 reported an unprecedented concept for identifying initial fragments of molecular glues based on lysine covalent imine formation. Lys122 is located at the PPI interface between 14-3-3 and the NF-κB subunit derived peptide P65, which is an ideal target residue for molecular glues screening (Fig. 13B). Based on this, they screened several fragments containing aldehyde groups, and grew these fragments to obtain molecular glue TCF521-123. The methods of disulfide capture or covalent anchoring reported are applicable to the early discovery of molecular glues, and further structural optimization and sufficient confirmation are required to fully validate the utility108.
5.2. Optimization: Structural modification
When the starting point for a structure-enabled design campaign is successfully finished, such as an active fragment or small molecule lead compound was achieved, to improve both the potency and cooperativity of the molecular glues, structural modification is highly desirable. According to different systems, corresponding optimization strategies need to be adapted to local conditions, but there are also some unique methods for reference.
BI-3802 targeting BCL6 protein was designed by exposing small groups in the solvent region, suggesting that modification of molecular structures in the exposed regions of the PPI interface or the target binding pocket surface could lead to the discovery of new molecular glues53. Likewise, molecules such as CR8 were optimized using this strategy. However, it's noteworthy that large groups should be avoided in the solvent-exposed region, which may result in steric hindrance and adversely affect the action of molecular glues, such as the CDK12 inhibitor THZ53185.
Analysis of the existing binding pockets of small molecules and targets to find space for further action also provides a strategy for molecular glues optimization. NRX-1933 was identified from a high-throughput screen (HTS), and the crystal structure of NRX-1933‒β-catenin showed that the N-terminus of β-catenin has the potential for movement. Therefore, Simonetta et al.89 proposed such a modification strategy: designing compounds to rearrange of β-catenin Leu31 and, as a result, inducing a larger, more druggable binding pocket (Fig. 14). Further optimization of NRX-1933 gave NRX-103094, a molecular glue that mediates SCFβ-TrCP‒β-catenin dimerization.
Figure 14.
Structural optimization strategy of NRX-103094. β-TrCP‒NRX-1933‒β-catenin complex (PDB:6M93), other proteins PDB have been mentioned above.
5.3. Confirmation: Identification method
Biophysical techniques can be used to verify the mechanism of molecular glues, and some specifical experimental tools allow for monitoring protein polymerization, such as X-ray crystallography, Förster resonance energy transfer, polyacrylamide gel electrophoresis, and differential scanning fluorimetry (DSF). Here, we focus on some burgeoning and distinctive biophysical and biochemical techniques that have been adapted for the identification and confirmation of molecular glues.
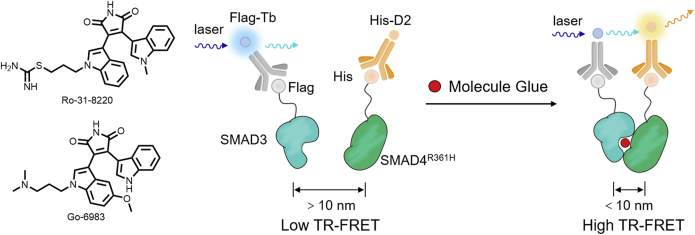
Time-resolved fluorescence resonance energy transfer (TR-FRET) utilizes the energy transfer of two fluorescence groups for detection purposes. With the help of the TR-FRET assay, Tang et al.114 reported that the bisindolylmaleimide derivatives, Ro-31-8220 and Go-6983 acted as molecular glues to restore SMAD4R361H PPI with SMAD3. When the molecular glues induce proximity of SMAD4R361H and SMAD3, Flag-Tb and His-D2 fluorophores are brought closer together, thus achieving efficient energy transfer and generating TR-FRET signals (Fig. 15).
Figure 15.
Schematic illustration of TR-FRET assay and the structures of the resulting molecular glues Ro-31-8220 and Go-6983.
The in situ proximity ligation assay (PLA) allows the observation of intracellular PPI at the endogenous level. The antibodies bind to the target proteins, and are recognized by secondary antibodies that are coupled with short single-stranded oligonucleotide pairs. When the target proteins are adjacent (<40 nm), oligonucleotides hybridize and form a continuous circular DNA structure, and amplification of the signal can be detected by a fluorescence microscope115. Separation of phases-based protein interaction reporter (SPPIER) is an assay based on fluorophore phase transition, and can dynamically display protein–protein binding induced by small molecules in living cells. The protein encoded by green fluorescent protein (GFP) forms high-fluorescent GFP droplets when protein polymerization is induced, and thus emitting a detectable signal. Chung et al.116 successfully utilized this technique to detect the interaction between CRBN and GSPT1 induced by IMiDs such as CC-885.
6. Conclusions
Molecular glues play physiological roles by stabilizing existing PPIs, establishing new protein interactions, and occupying the binding sites of original pathogenic proteins and inhibit their subsequent pathways. Therefore, molecular glues are presented as potential strategies of drug design that will be utilized in the treatment of diseases involved in immunology, neurology, cancer and others. Importantly, molecular glues depend on the secondary structural characteristics of targets and not necessarily on the classically defined ligand binding pockets, hence this feature broadens the range of drug therapeutic targets and makes drugs available for previously undruggable targets, bringing hope and paving a new path for drug development.
However, many challenges remain in subsequent drug development and modification. Molecular glues have mostly been discovered by accident or by secondary sifting through previous databases. Therefore, most current researches pay close attention to the screening strategies and the establishment of mechanism verification methods. It is essential to understand the action mechanism of molecular glues to screen and identify molecular glues according to different modes of action. Although this review classifies the different types of molecular glues mediating protein polymerization into homo-dimerization, hetero-dimerization and homo-polymerization, molecular glues can be essentially classified into two types according to their functional processes. First, a molecular glue binds to a target protein, and forms interaction surface (mostly known as “hot spots”) that can mediate the target protein binding with the “recruited protein”, resulting the aggregation of the target protein with “recruited protein”. Without such molecular glue, the target protein will not polymerize with the “recruited protein”. The cases such as IMiDs, CR8, or BI-3802, function in this mode. On the other hand, a molecular glue can bind to the establishing PPI interface, enhancing or stabilizing their interaction affinity. However, it is worth noting that the existing protein–protein interaction may only have a weak binding force. For example, E7820 and LSN3160440 act in this mode.
For pre-existing PPI complementary surface, there have been some successes with structure-based screening or structural optimization targeting this region. Additionally, more compatible interfaces than expected may emerge between unrelated proteins. For unformulated protein targets, we may be able to use docking software to find complementary interfaces between two proteins and design the novel molecular glues.
For molecular glues, whether accidently discovered, or obtained through fragment-based drug discovery, subsequent structural modifications are needed. Since molecular glues often regulate protein interactions through the solvent exposure region, new molecular glues may be obtained by structural modification from it. Meanwhile, as the PPI interface is usually narrow, the design of molecular glues that can induce migration or rearrangement of residues on protein–protein binding surfaces and thus inducing larger pockets suitable for molecular glues, is also an excellent strategy for consideration.
Although this review focuses on small molecules, it should be noted that other macromolecule, such as macrolides and peptides may also have potential application prospects as molecular glues. Macrolides were the first molecular glues to be discovered21,117, which paved the way for the discovery of subsequent molecular glues. Cyclosporine exerts immunosuppressant activity by inducing the binding of cyclophilin and calcineurin simultaneously118. The similar FK506/rapamycin systems have been well studied and verified, and other macrolide molecular glues have also been developed119, 120, 121. As early as 1992, Schreiber et al.122 discovered that antigenic peptides act as glues between major histocompatibility complex (MHC) presenter proteins and T-cell receptors to establish an immune response. In recent years, cyclic peptides and helical peptides have been widely used in the development and design of PPI inhibitors123, 124, 125, 126, 127, 128, 129, demonstrating their ability to target the surfaces of undruggable proteins. These peptides possess a large binding interface and satisfactory binding affinity, making them promising candidates for recruiting other proteins, and thus, it is reasonable to speculate that cyclic peptides and helical peptides may also be developed as molecular glues in the future.
Taken together, with advances in crystallography and biochemical technology, the class of molecular glues is constantly expanding, and researchers are gaining a deeper understanding of their mechanisms of action. Rational design, optimization strategies and detection technologies are continuing to develop, which bring hope for breakthroughs in undruggable targets.
Acknowledgments
This study was supported from the National Natural Science Foundation of China (Nos. 82173672, 82173679, 81903446 and 81973167, China), Natural Science Foundation of Jiangsu Province (BK20190564, China), “Double First-Class” University project of China Pharmaceutical University (CPU2018GY04, China), and Free exploration basic research project of Shenzhen Virtual University Park (2021Szvup162, China) for financial support.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Hong Yao, Email: hyao1989@sina.cn.
Shengtao Xu, Email: cpuxst@126.com.
Jinyi Xu, Email: jinyixu@china.com.
Author contributions
Hong Yao conceived the formulation of overarching research goals and superintended the whole study. Hongyu Wu collected the data and drafted the manuscript. Chen He, Yilin Jia, Shengtao Xu, Zheying Zhu, Dahong Li and Jinyi Xu revised the manuscript. All the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- 1.Roberts N.A., Martin J.A., Kinchington D., Broadhurst A.V., Craig J.C., Duncan I.B., et al. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 2.Erickson J., Neidhart D.J., VanDrie J., Kempf D.J., Wang X.C., Norbeck D.W., et al. Design, activity, and 2.8 Å crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease. Science. 1990;249:527–533. doi: 10.1126/science.2200122. [DOI] [PubMed] [Google Scholar]
- 3.Dorsey B.D., Levin R.B., McDaniel S.L., Vacca J.P., Guare J.P., Darke P.L., et al. L-735,524: the design of a potent and orally bioavailable HIV protease inhibitor. J Med Chem. 1994;37:3443–3451. doi: 10.1021/jm00047a001. [DOI] [PubMed] [Google Scholar]
- 4.Anderson A.C. The process of structure-based drug design. Chem Biol. 2003;10:787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Wenthur C.J., Gentry P.R., Mathews T.P., Lindsley C.W. Drugs for allosteric sites on receptors. Annu Rev Pharmacol Toxicol. 2014;54:165–184. doi: 10.1146/annurev-pharmtox-010611-134525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neklesa T.K., Winkler J.D., Crews C.M. Targeted protein degradation by PROTACs. Pharmacol Ther. 2017;174:138–144. doi: 10.1016/j.pharmthera.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 7.Testa A., Hughes S.J., Lucas X., Wright J.E., Ciulli A. Structure-based design of a macrocyclic PROTAC. Angew Chem Int Ed. 2020;59:1727–1734. doi: 10.1002/anie.201914396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie S., Sun Y., Liu Y., Li X., Li X., Zhong W., et al. Development of alectinib-based PROTACs as novel potent degraders of anaplastic lymphoma kinase (ALK) J Med Chem. 2021;64:9120–9140. doi: 10.1021/acs.jmedchem.1c00270. [DOI] [PubMed] [Google Scholar]
- 9.Imaide S., Riching K.M., Makukhin N., Vetma V., Whitworth C., Hughes S.J., et al. Trivalent PROTACs enhance protein degradation through cooperativity and avidity. Nat Chem Biol. 2021;17:1157–1167. doi: 10.1038/s41589-021-00878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai A.C., Crews C.M. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banik S.M., Pedram K., Wisnovsky S., Ahn G., Riley N.M., Bertozzi C.R. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature. 2020;584:291–297. doi: 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitworth C., Ciulli A. New class of molecule targets proteins outside cells for degradation. Nature. 2020;584:193–194. doi: 10.1038/d41586-020-02211-w. [DOI] [PubMed] [Google Scholar]
- 13.Kaushik S., Cuervo A.M. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi D., Arimoto H. Targeting selective autophagy by AUTAC degraders. Autophagy. 2020;16:765–766. doi: 10.1080/15548627.2020.1718362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi D., Moriyama J., Nakamura T., Miki E., Takahashi E., Sato A., et al. AUTACs: cargo-specific degraders using selective autophagy. Mol Cell. 2019;76:797–810. doi: 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Churcher I. Protac-induced protein degradation in drug discovery: breaking the rules-or just making new ones?. J Med Chem. 2018;61:444–452. doi: 10.1021/acs.jmedchem.7b01272. [DOI] [PubMed] [Google Scholar]
- 17.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., et al. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 18.Liu J., Chen H., Kaniskan HUm, Xie L., Chen X., Jin J., et al. TF-PROTACs enable targeted degradation of transcription factors. J Am Chem Soc. 2021;143:8902–8910. doi: 10.1021/jacs.1c03852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samarasinghe K.T., Jaime-Figueroa S., Burgess M., Nalawansha D.A., Dai K., Hu Z., et al. Targeted degradation of transcription factors by TRAFTACs: transcription factor targeting chimeras. Cell Chem Biol. 2021;28:648–661. doi: 10.1016/j.chembiol.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tinworth C.P., Young R.J. Facts, patterns, and principles in drug discovery: appraising the rule of 5 with measured physicochemical data. J Med Chem. 2020;63:10091–10108. doi: 10.1021/acs.jmedchem.9b01596. [DOI] [PubMed] [Google Scholar]
- 21.Schreiber S.L. The rise of molecular glues. Cell. 2021;184:3–9. doi: 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Kozicka Z., Thomä N.H. Haven't got a glue: protein surface variation for the design of molecular glue degraders. Cell Chem Biol. 2021;28:1032–1047. doi: 10.1016/j.chembiol.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlino M.S., Larkin J., Long G.V. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398:1002–1014. doi: 10.1016/S0140-6736(21)01206-X. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y., Song G., Xie S., Jiang W., Chen X., Chu M., et al. The roles of PD-1/PD-L1 in the prognosis and immunotherapy of prostate cancer. Mol Ther. 2021;29:1958–1969. doi: 10.1016/j.ymthe.2021.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sehgal A., Whiteside T.L., Boyiadzis M. Programmed death-1 checkpoint blockade in acute myeloid leukemia. Expet Opin Biol Ther. 2015;15:1191–1203. doi: 10.1517/14712598.2015.1051028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahoney K.M., Rennert P.D., Freeman G.J. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 28.Lin X., Lu X., Luo G., Xiang H. Progress in PD-1/PD-L1 pathway inhibitors: from biomacromolecules to small molecules. Eur J Med Chem. 2020;186:111876. doi: 10.1016/j.ejmech.2019.111876. [DOI] [PubMed] [Google Scholar]
- 29.Mullard A. FDA approves 100th monoclonal antibody product. Nat Rev Drug Discov. 2021;20:491–495. doi: 10.1038/d41573-021-00079-7. [DOI] [PubMed] [Google Scholar]
- 30.Miller M.M., Mapelli C., Allen M.P., Bowsher M.S., Boy K.M., Gillis E.P., et al. inventors; Bristol-Myers Squibb Company, assignee . 2017 Dec 26. Macrocyclic inhibitors of the PD-1/PD-L1 and CD80(B7-1)/PD-L1 protein/protein interactions. US patent: 9,850,283. [Google Scholar]
- 31.Chupak L.S., Zheng X., inventors . 2015 Mar 12. Bristol-myers Squibb Company, assignee. Compounds useful as immunomodulators. Patent application: WO2015034820 A1. [Google Scholar]
- 32.Zak K.M., Grudnik P., Guzik K., Zieba B.J., Musielak B., Dömling A., et al. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1) Oncotarget. 2016;7:30323–30335. doi: 10.18632/oncotarget.8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muszak D., Surmiak E., Plewka J., Magiera-Mularz K., Kocik-Krol J., Musielak B., et al. Terphenyl-based small-molecule inhibitors of programmed cell death-1/programmed death-ligand 1 protein‒protein interaction. J Med Chem. 2021;64:11614–11636. doi: 10.1021/acs.jmedchem.1c00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L., Yao Z., Wang S., Xie T., Wu G., Zhang H., et al. Syntheses, biological evaluations, and mechanistic studies of benzo [c][1,2,5] oxadiazole derivatives as potent PD-L1 inhibitors with in vivo antitumor activity. J Med Chem. 2021;64:8391–8409. doi: 10.1021/acs.jmedchem.1c00392. [DOI] [PubMed] [Google Scholar]
- 35.OuYang Y., Gao J., Zhao L., Lu J., Zhong H., Tang H., et al. Design, synthesis, and evaluation of O-(biphenyl-3-ylmethoxy) nitrophenyl derivatives as PD-1/PD-L1 inhibitors with potent anticancer efficacy in vivo. J Med Chem. 2021;64:7646–7666. doi: 10.1021/acs.jmedchem.1c00370. [DOI] [PubMed] [Google Scholar]
- 36.Wang T., Cai S., Wang M., Zhang W., Zhang K., Chen D., et al. Novel biphenyl pyridines as potent small-molecule inhibitors targeting the programmed cell death-1/programmed cell death‒ligand 1 interaction. J Med Chem. 2021;64:7390–7403. doi: 10.1021/acs.jmedchem.1c00010. [DOI] [PubMed] [Google Scholar]
- 37.Song Z., Liu B., Peng X., Gu W., Sun Y., Xing L., et al. Design, synthesis, and pharmacological evaluation of biaryl-containing PD-1/PD-L1 interaction inhibitors bearing a unique difluoromethyleneoxy linkage. J Med Chem. 2021;64:16687–16702. doi: 10.1021/acs.jmedchem.1c01422. [DOI] [PubMed] [Google Scholar]
- 38.Vaisman A., Woodgate R. Translesion DNA polymerases in eukaryotes: what makes them tick?. Crit Rev Biochem Mol Biol. 2017;52:274–303. doi: 10.1080/10409238.2017.1291576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto K., Cho Y., Yang I.Y., Akagi J.I., Ohashi E., Tateishi S., et al. The vital role of polymerase ζ and REV1 in mutagenic, but not correct, DNA synthesis across benzo [a] pyrene-dG and recruitment of polymerase ζ by REV1 to replication-stalled site. J Biol Chem. 2012;287:9613–9622. doi: 10.1074/jbc.M111.331728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wojtaszek J.L., Chatterjee N., Najeeb J., Ramos A., Lee M., Bian K., et al. A small molecule targeting mutagenic translesion synthesis improves chemotherapy. Cell. 2019;178:152–159. doi: 10.1016/j.cell.2019.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taniguchi T. REV1-POL ζ inhibition and cancer therapy. Mol Cell. 2019;75:419–420. doi: 10.1016/j.molcel.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee N., Whitman M.A., Harris C.A., Min S.M., Jonas O., Lien E.C., et al. REV1 inhibitor JH-RE-06 enhances tumor cell response to chemotherapy by triggering senescence hallmarks. Proc Natl Acad Sci U S A. 2020;117:28918–28921. doi: 10.1073/pnas.2016064117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade M., Wang Y.V., Wahl G.M. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S., Lou J., Li Y., Zhou F., Yan Z., Lyu X., et al. Recent progress and clinical development of inhibitors that block MDM4/p53 protein–protein interactions. J Med Chem. 2021;64:10621–10640. doi: 10.1021/acs.jmedchem.1c00940. [DOI] [PubMed] [Google Scholar]
- 45.Carvajal L.A., Neriah D.B., Senecal A., Benard L., Thiruthuvanathan V., Yatsenko T., et al. Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aao3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graves B., Thompson T., Xia M., Janson C., Lukacs C., Deo D., et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci U S A. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mlynarczyk C., Fontán L., Melnick A. Germinal center-derived lymphomas: the darkest side of humoral immunity. Immunol Rev. 2019;288:214–239. doi: 10.1111/imr.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leeman-Neill R.J., Bhagat G. BCL6 as a therapeutic target for lymphoma. Expert Opin Ther Targets. 2018;22:143–152. doi: 10.1080/14728222.2018.1420782. [DOI] [PubMed] [Google Scholar]
- 49.Ai Y., Hwang L., MacKerell A.D., Jr., Melnick A., Xue F. Progress toward B-cell lymphoma 6 BTB domain inhibitors for the treatment of diffuse large B-cell lymphoma and beyond. J Med Chem. 2021;64:4333–4358. doi: 10.1021/acs.jmedchem.0c01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoull W., Abrams R.D., Anderson E., Blades K., Barton P., Box M., et al. Discovery of pyrazolo [1,5-a] pyrimidine B-cell lymphoma 6 (BCL6) binders and optimization to high affinity macrocyclic inhibitors. J Med Chem. 2017;60:4386–4402. doi: 10.1021/acs.jmedchem.7b00359. [DOI] [PubMed] [Google Scholar]
- 51.Pearce A.C., Bamford M.J., Barber R., Bridges A., Convery M.A., Demetriou C., et al. GSK137, a potent small-molecule BCL6 inhibitor with in vivo activity, suppresses antibody responses in mice. J Biol Chem. 2021;297:100928. doi: 10.1016/j.jbc.2021.100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kerres N., Steurer S., Schlager S., Bader G., Berger H., Caligiuri M., et al. Chemically induced degradation of the oncogenic transcription factor BCL6. Cell Rep. 2017;20:2860–2875. doi: 10.1016/j.celrep.2017.08.081. [DOI] [PubMed] [Google Scholar]
- 53.Słabicki M., Yoon H., Koeppel J., Nitsch L., Burman S.S.R., Genua C.D., et al. Small-molecule-induced polymerization triggers degradation of BCL6. Nature. 2020;588:164–168. doi: 10.1038/s41586-020-2925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellenie B.R., Cheung K.-M.J., Varela A., Pierrat O.A., Collie G.W., Box G.M., et al. Achieving in vivo target depletion through the discovery and optimization of benzimidazolone BCL6 degraders. J Med Chem. 2020;63:4047–4068. doi: 10.1021/acs.jmedchem.9b02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mubarak A., Alturaiki W., Hemida M.G. Middle East respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J Immunol Res. 2019;2019:6491738. doi: 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park Y.J., Walls A.C., Wang Z., Sauer M.M., Li W., Tortorici M.A., et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat Struct Mol Biol. 2019;26:1151–1157. doi: 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin S.Y., Liu C.L., Chang Y.M., Zhao J., Perlman S., Hou M.H. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin S.M., Lin S.C., Hsu J.N., Chang C.K., Chien C.M., Wang Y.S., et al. Structure-based stabilization of non-native protein–protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J Med Chem. 2020;63:3131–3141. doi: 10.1021/acs.jmedchem.9b01913. [DOI] [PubMed] [Google Scholar]
- 62.Chamberlain P.P., Cathers B.E. Cereblon modulators: low molecular weight inducers of protein degradation. Drug Discov Today Technol. 2019;31:29–34. doi: 10.1016/j.ddtec.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Kim S.A., Go A., Jo S.H., Park S.J., Jeon Y.U., Kim J.E., et al. A novel cereblon modulator for targeted protein degradation. Eur J Med Chem. 2019;166:65–74. doi: 10.1016/j.ejmech.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 64.Krönke J., Udeshi N.D., Narla A., Grauman P., Hurst S.N., McConky M., et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fischer E.S., Böhm K., Lydeard J.R., Yang H., Stadler M.B., Cavadini S., et al. Structure of the DDB1–CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matyskiela M.E., Lu G., Ito T., Pagarigan B., Lu C.C., Miller K., et al. A novel cereblon modulator recruits GSPT1 to the CRL4 CRBN ubiquitin ligase. Nature. 2016;535:252–257. doi: 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- 67.Sperling A.S., Burgess M., Keshishian H., Gasser J.A., Bhatt S., Jan M., et al. Patterns of substrate affinity, competition, and degradation kinetics underlie biological activity of thalidomide analogs. Blood. 2019;134:160–170. doi: 10.1182/blood.2019000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishiguchi G., Keramatnia F., Min J., Chang Y., Jonchere B., Das S., et al. Identification of potent, selective, and orally bioavailable small-molecule GSPT1/2 degraders from a focused library of cereblon modulators. J Med Chem. 2021;64:7296–7311. doi: 10.1021/acs.jmedchem.0c01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen J.D., Correa M., Alexander M., Nagy M., Huang D., Sapienza J., et al. CC-90009: a cereblon E3 ligase modulating drug that promotes selective degradation of GSPT1 for the treatment of acute myeloid leukemia. J Med Chem. 2021;64:1835–1843. doi: 10.1021/acs.jmedchem.0c01489. [DOI] [PubMed] [Google Scholar]
- 70.Matyskiela M.E., Zhu J., Baughman J.M., Clayton T., Slade M., Wong H.K., et al. Cereblon modulators target ZBTB16 and its oncogenic fusion partners for degradation via distinct structural degrons. ACS Chem Biol. 2020;15:3149–3158. doi: 10.1021/acschembio.0c00674. [DOI] [PubMed] [Google Scholar]
- 71.An J., Ponthier C.M., Sack R., Seebacher J., Stadler M.B., Donovan K.A., et al. pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4CRBN ubiquitin ligase. Nat Commun. 2017;8:1–11. doi: 10.1038/ncomms15398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krönke J., Fink E.C., Hollenbach P.W., MacBeth K.J., Hurst S.N., Udeshi N.D., et al. Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Asatsuma-Okumura T., Ando H., De Simone M., Yamamoto J., Sato T., Shimizu N., et al. p63 is a cereblon substrate involved in thalidomide teratogenicity. Nat Chem Biol. 2019;15:1077–1084. doi: 10.1038/s41589-019-0366-7. [DOI] [PubMed] [Google Scholar]
- 74.Owa T., Yoshino H., Okauchi T., Yoshimatsu K., Ozawa Y., Sugi N.H., et al. Discovery of novel antitumor sulfonamides targeting G1 phase of the cell cycle. J Med Chem. 1999;42:3789–3799. doi: 10.1021/jm9902638. [DOI] [PubMed] [Google Scholar]
- 75.Semba T., Funahashi Y., Ono N., Yamamoto Y., Sugi N.H., Asada M., et al. An angiogenesis inhibitor E7820 shows broad-spectrum tumor growth inhibition in a xenograft model: possible value of integrin α2 on platelets as a biological marker. Clin Cancer Res. 2004;10:1430–1438. doi: 10.1158/1078-0432.ccr-0109-03. [DOI] [PubMed] [Google Scholar]
- 76.Han T., Goralski M., Gaskill N., Capota E., Kim J., Ting T.C., et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356 doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- 77.Du X., Volkov O.A., Czerwinski R.M., Tan H., Huerta C., Morton E.R., et al. Structural basis and kinetic pathway of RBM39 recruitment to DCAF15 by a sulfonamide molecular glue E7820. Structure. 2019;27:1625–1633. doi: 10.1016/j.str.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 78.Faust T.B., Yoon H., Nowak R.P., Donovan K.A., Li Z., Cai Q., et al. Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nat Chem Biol. 2020;16:7–14. doi: 10.1038/s41589-019-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 80.Baek K., Scott D.C., Schulman B.A. NEDD8 and ubiquitin ligation by cullin‒RING E3 ligases. Curr Opin Struct Biol. 2021;67:101–109. doi: 10.1016/j.sbi.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song Q., Feng S., Peng W., Li A., Ma T., Yu B., et al. Cullin‒ring ligases as promising targets for gastric carcinoma treatment. Pharmacol Res. 2021;170:105493. doi: 10.1016/j.phrs.2021.105493. [DOI] [PubMed] [Google Scholar]
- 82.Petroski M.D., Deshaies R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 83.Mayor-Ruiz C., Bauer S., Brand M., Kozicka Z., Siklos M., Imrichova H., et al. Rational discovery of molecular glue degraders via scalable chemical profiling. Nat Chem Biol. 2020;16:1199–1207. doi: 10.1038/s41589-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lv L., Chen P., Cao L., Li Y., Han T. Discovery of a molecular glue promoting CDK12‒DDB1 interaction to trigger cyclin K degradation. Elife. 2020;9 doi: 10.7554/eLife.59994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sabicki M., Kozicka Z., Petzold G., Li Y.D., Manojkumar M., Bunker R.D., et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature. 2020;585:293–297. doi: 10.1038/s41586-020-2374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meijer L., Borgne A., Mulner O., Chong J.P., Blow J.J., Inagaki N., et al. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 87.Sedlacek H., Czech J., Naik R., Kaur G., Worland P., Losiewicz M., et al. Flavopiridol (L86 8275; NSC 649890), a new kinase inhibitor for tumor therapy. Int J Oncol. 1996;9:1143–1168. doi: 10.3892/ijo.9.6.1143. [DOI] [PubMed] [Google Scholar]
- 88.Zhang T., Kwiatkowski N., Olson C.M., Dixon-Clarke S.E., Abraham B.J., Greifenberg A.K., et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol. 2016;12:876–884. doi: 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simonetta K.R., Taygerly J., Boyle K., Basham S.E., Padovani C., Lou Y., et al. Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction. Nat Commun. 2019;10:1402. doi: 10.1038/s41467-019-09358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu G., Xu G., Schulman B.A., Jeffrey P.D., Harper J.W., Pavletich N.P. Structure of a beta-TrCP1‒Skp1‒beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin Ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 91.Bonam S.R., Wang F., Muller S. Lysosomes as a therapeutic target. Nat Rev Drug Discov. 2019;18:923–948. doi: 10.1038/s41573-019-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alabi S., Crews C. Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs. J Biol Chem. 2021;296:100647. doi: 10.1016/j.jbc.2021.100647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Z., Wang C., Wang Z., Zhu C., Li J., Sha T., et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019;575:203–209. doi: 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- 94.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang N., Gray M., Lu X.H., Cantle J.P., Holley S.M., Greiner E., et al. Neuronal targets for reducing mutant huntingtin expression to ameliorate disease in a mouse model of Huntington's disease. Nat Med. 2014;20:536–541. doi: 10.1038/nm.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ast A., Buntru A., Schindler F., Hasenkopf R., Schulz A., Brusendorf L., et al. mHTT seeding activity: a marker of disease progression and neurotoxicity in models of Huntington's disease. Mol Cell. 2018;71:675–688. doi: 10.1016/j.molcel.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 97.Simó R., Hernández C. GLP-1R as a target for the treatment of diabetic retinopathy: friend or foe? Diabetes. 2017;66:1453–1460. doi: 10.2337/db16-1364. [DOI] [PubMed] [Google Scholar]
- 98.Baggio L.L., Huang Q., Brown T.J., Drucker D.J. A recombinant human glucagon-like peptide (GLP)-1-albumin protein (albugon) mimics peptidergic activation of GLP-1 receptor-dependent pathways coupled with satiety, gastrointestinal motility, and glucose homeostasis. Diabetes. 2004;53:2492–2500. doi: 10.2337/diabetes.53.9.2492. [DOI] [PubMed] [Google Scholar]
- 99.Drucker D.J., Nauck M.A. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368:1696–1705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- 100.Mentlein R., Gallwitz B., Schmidt W.E. Dipeptidyl-peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon-like peptide-1(7-36)amide, peptide histidine methionine and is responsible for their degradation in human serum. Eur J Biochem. 2010;214:829–835. doi: 10.1111/j.1432-1033.1993.tb17986.x. [DOI] [PubMed] [Google Scholar]
- 101.Bueno A.B., Sun B., Willard F.S., Feng D., Ho J.D., Wainscott D.B., et al. Structural insights into probe-dependent positive allosterism of the GLP-1 receptor. Nat Chem Biol. 2020;16:1105–1110. doi: 10.1038/s41589-020-0589-7. [DOI] [PubMed] [Google Scholar]
- 102.Thabault L., Liberelle M., Frédérick R. Targeting protein self-association in drug design. Drug Discov Today. 2021;26:1148–1163. doi: 10.1016/j.drudis.2021.01.028. [DOI] [PubMed] [Google Scholar]
- 103.Dong G., Ding Y., He S., Sheng C. Molecular glues for targeted protein degradation: from serendipity to rational discovery. J Med Chem. 2021;64:10606–10620. doi: 10.1021/acs.jmedchem.1c00895. [DOI] [PubMed] [Google Scholar]
- 104.Clackson Tim, Wells James, A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 105.Sitani D., Giorgetti A., Alfonso-Prieto M., Carloni P. Robust principal component analysis-based prediction of protein‒protein interaction hot spots. Proteins. 2021;89:639–647. doi: 10.1002/prot.26047. [DOI] [PubMed] [Google Scholar]
- 106.Martino E., Chiarugi S., Margheriti F., Garau G. Mapping, structure and modulation of PPI. Front Chem. 2021;9:718405. doi: 10.3389/fchem.2021.718405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Erlanson D.A., Wells J.A., Braisted A.C. Tethering: fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 108.Wolter M., Valenti D., Cossar P.J., Hristeva S., Levy L.M., Genski T., et al. An exploration of chemical properties required for cooperative stabilization of the 14-3-3 interaction with NF-κB—utilizing a reversible covalent tethering approach. J Med Chem. 2021;64:8423–8436. doi: 10.1021/acs.jmedchem.1c00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sijbesma E., Hallenbeck K.K., Leysen S., De Vink P.J., Skóra L., Jahnke W., et al. Site-directed fragment-based screening for the discovery of protein‒protein interaction stabilizers. J Am Chem Soc. 2019;141:3524–3531. doi: 10.1021/jacs.8b11658. [DOI] [PubMed] [Google Scholar]
- 110.Sijbesma E., Somsen B.A., Miley G.P., De Gevel I.A.-L., Brunsveld L., Arkin M.R., et al. Fluorescence anisotropy-based tethering for discovery of protein‒protein interaction stabilizers. ACS Chem Biol. 2020;15:3143–3148. doi: 10.1021/acschembio.0c00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hacker S.M., Backus K.M., Lazear M.R., Forli S., Correia B.E., Cravatt B.F. Global profiling of lysine reactivity and ligandability in the human proteome. Nat Chem. 2017;9:1181–1190. doi: 10.1038/nchem.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cossar P.J., Wolter M., van Dijck L., Valenti D., Levy L.M., Ottmann C., et al. Reversible covalent imine-tethering for selective stabilization of 14-3-3 hub protein interactions. J Am Chem Soc. 2021;143:8454–8464. doi: 10.1021/jacs.1c03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolter M., Valenti D., Cossar P.J., Levy L.M., Hristeva S., Genski T., et al. Fragment-based stabilizers of protein–protein interactions through imine-based tethering. Angew Chem Int Ed. 2020;132:21704–21708. doi: 10.1002/anie.202008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tang C., Mo X., Niu Q., Wahafu A., Yang X., Qui M., et al. Hypomorph mutation-directed small-molecule protein‒protein interaction inducers to restore mutant SMAD4-suppressed TGF-β signaling. Cell Chem Biol. 2021;28:636–647. doi: 10.1016/j.chembiol.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vallet C., Aschmann D., Beuck C., Killa M., Meiners A., Mertel M., et al. Functional disruption of the cancer-relevant interaction between survivin and histone H3 with a guanidiniocarbonyl pyrrole ligand. Angew Chem Int Ed. 2020;59:5567–5571. doi: 10.1002/anie.201915400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chung C.I., Zhang Q., Shu X. Dynamic imaging of small molecule induced protein‒protein interactions in living cells with a fluorophore phase transition based approach. Anal Chem. 2018;90:14287–14293. doi: 10.1021/acs.analchem.8b03476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gerry C.J., Schreiber S.L. Unifying principles of bifunctional, proximity-inducing small molecules. Nat Chem Biol. 2020;16:369–378. doi: 10.1038/s41589-020-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu J., Farmer J.D., Lane W.S., Friedman J., Schreiber S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 119.Sabatini D.M., Erdjument-Bromage H., Lui M., Tempst P., Snyder S.H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 120.Sabers C.J., Martin M.M., Brunn G.J., Williams J.M., Dumont F.J., Wiederrecht G., et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 121.Schreiber S.L., Crabtree G.R. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1993;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 122.Schreiber S.L. Immunophilin-sensitive protein phosphatase action in cell signaling pathways. Cell. 1992;70:365–368. doi: 10.1016/0092-8674(92)90158-9. [DOI] [PubMed] [Google Scholar]
- 123.Hart P., Hommen P., Noisier A., Krzyzanowski A., Schüler D., Porfetye A.T., et al. Structure based design of bicyclic peptide inhibitors of RbAp48. Angew Chem Int Ed. 2021;60:1813–1820. doi: 10.1002/anie.202009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.de Araujo A.D., Lim J., Wu K.C., Xiang Y., Good A.C., Skerlj R., et al. Bicyclic helical peptides as dual inhibitors selective for Bcl2A1 and Mcl-1 proteins. J Med Chem. 2018;61:2962–2972. doi: 10.1021/acs.jmedchem.8b00010. [DOI] [PubMed] [Google Scholar]
- 125.Mabonga L., Kappo A.P. Protein‒protein interaction modulators: advances, successes and remaining challenges. Biophys Rev. 2019;11:559–581. doi: 10.1007/s12551-019-00570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lian W., Upadhyaya P., Rhodes C.A., Liu Y., Pei D. Screening bicyclic peptide libraries for protein‒protein interaction inhibitors: discovery of a tumor necrosis factor-alpha antagonist. J Am Chem Soc. 2013;135:11990–11995. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yokoo H., Ohoka N., Naito M., Demizu Y. Design and synthesis of peptide-based chimeric molecules to induce degradation of the estrogen and androgen receptors. Bioorg Med Chem. 2020;28:115595–115600. doi: 10.1016/j.bmc.2020.115595. [DOI] [PubMed] [Google Scholar]
- 128.Tsomaia N. Peptide therapeutics: targeting the undruggable space. Eur J Med Chem. 2015;94:459–470. doi: 10.1016/j.ejmech.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 129.Klein M. Stabilized helical peptides: overview of the technologies and its impact on drug discovery. Expet Opin Drug Discov. 2017;12:1117–1125. doi: 10.1080/17460441.2017.1372745. [DOI] [PubMed] [Google Scholar]