Abstract
Objective
Transfer RNA-derived small RNAs (tsRNAs) are a novel type of non-coding RNA with various regulatory functions. They are associated with oxidative stress in various diseases, but their potential functions in radiation-induced lung injury (RILI) remain uncertain.
Methods
To explore the role of tsRNAs in RILI, we used X-rays to irradiate human bronchial epithelial cells and examined the expression profile of altered tsRNAs by RNA sequencing and bioinformatics analysis. Sequencing results were verified by qRT-PCR. tsRNA functions were explored using several methods, including CCK-8, reactive oxygen species (ROS) assays, cell transfection, and western blotting.
Results
Eighty-six differentially expressed tRNA-derived fragments (tRFs) were identified: 64 were upregulated, and 22 were downregulated. Among them, the regulation of tRF-Gly-GCC, associated with oxidative stress, may be mediated by the inhibition of cell proliferation, promotion of ROS production, and apoptosis in the occurrence and development of RILI. A Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis suggested that the underlying molecular mechanism may involve the PI3K/AKT and the FOXO1 signaling pathways.
Conclusion
Our findings provide new insights into the molecular mechanisms underpinning RILI, advancing the clinical prevention and treatment of this disease.
Keywords: tRNA-derived small RNAs, radiation-induced lung injury, tRNA-derived fragment, tRF-Gly-GCC, oxidative stress
Introduction
Radiation-induced lung injury (RILI) is one of the most commonly observed complications of chest radiotherapy and a major obstacle to improving the overall outcome of patients with thoracic malignancies. 1 Newly developed radiotherapeutic equipment and techniques, such as proton heavy-ion linear accelerators (linac) and stereotactic body radiation therapy, increase the delivery precision of the irradiation dose to the tumor and surrounding normal tissues, reducing the occurrence of RILI. 2 Nevertheless, RILI occurs at a high rate in approximately half of the cases,3,4 and oxidative stress caused by ionizing radiation exposure plays a key role in the occurrence and development of this condition. Currently, preventing RILI is difficult, as the underlying molecular mechanism is not fully understood. Thus, exploring the mechanisms of RILI and identifying novel therapeutic targets are essential.
Numerous transcriptomic studies have identified an increasing number of RNA subtypes, such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), that have been extensively analyzed to determine their unique, diverse biological functions. Due to their relevance in gene regulation, non-coding RNAs have been widely studied in various fields, and several studies reported roles for lncRNAs and miRNAs in RILI. Transfer RNA-derived small RNAs (tsRNAs) are a newly discovered subtype of non-coding RNA that has attracted increasing attention. Fragments of tRNA derived from mature tRNAs or their precursors function as tRNA degradation products and play regulatory roles in many pathophysiological processes.5-7 They can be grouped into tRNA-derived fragments (tRFs) and tRNA halves (tiRNAs) based on cleavage sites. Each group has particular molecular dimensions, nucleotide composition, and biological functions. 8 Some studies have reported that tRFs are associated with oxidative stress in diabetes, cancer, cardiovascular disease, and other diseases. 9 However, a correlation with oxidative stress in the occurrence and development of RILI has not yet been reported.
In this study, we focused on a new small non-coding RNA subtype, tRFs, using X-rays to irradiate human bronchial epithelial cells and investigating the expression profile of altered tsRNAs by RNA sequencing and bioinformatics analyses. Our main objective was to evaluate the biological function of tRFs associated with oxidative stress in the process of RILI, providing a theoretical basis for the clinical prevention and treatment of this disease.
Methods
Cell Culture and Irradiation
Normal human lung bronchial epithelial cells (BEAS-2B) were obtained from the cell bank of Central South University. Cells were cultured in Dulbecco’s modified Eagle medium, supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin, at 37°C, 5% CO2, and 95% humidity. Cells were divided into irradiation (IR) and control groups. The IR group was irradiated using a Varian linac (Varian Medical System, Palo Alto, CA, USA) with a 6 MV X-ray photon beam source skin distance at a dose of 400 cGy min−1. The control group was unirradiated. Three replicates were performed in each group.
Library Construction and RNA Sequencing
Total RNA was extracted from each group at 72 h post-irradiation using the RNAiso Plus kit (Takara Bio, Kyoto, Japan). Total RNA purity and concentration were tested using a NanoDrop ND-1000. Next, we preprocessed the tsRNA and selected the sequencing library size for the RNA biotype to be sequenced using an automated gel cutter. Libraries were identified and quantified in absolute terms using an Agilent2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Finally, standard small RNA sequencing was performed on an Illumina NextSeq instrument.
Sequencing Data and Pathway Analyses
After generating the original sequencing data, intron sequences were removed, and “CCA” was added at the 3′ ends to generate a mature tRNA library. Sequencing quality was refined using FastQC, and the trimmed reads (pass Illumina quality filter, trimmed 5′, 3′-adaptor bases by cutadapt) were aligned. The expression profiles of tRFs and tiRNAs were computed according to the number of reads mapped. Differentially expressed tRFs and tiRNAs were screened based on the count value using the R package “edgeR.” The tsRNA target genes were predicted from the Miranda database (miranda_score ≥140, miranda_energy ≤ −10) and targetScan database (context_plus_score ≤ −.1). KEGG analysis of tRFs was used to predict target genes using the online website DAVID (https://david.ncifcrf.gov/). Based on Fisher’s test of hypergeometric distribution, a P-value < .05 was set as the criterion for significant enrichment of a pathway. Fold change (cutoff 1.5) and P-value (cutoff value .05, performed only for multiple replicates) were used to screen for pathways significantly enriched in differential genes.
Cell Transfection
The tRF-Gly-GCC mimic and mimic negative control (NC), inhibitor, and inhibitor NC were designed and synthesized by Ruibo, Guangzhou, China. The sequences are listed in Table 1. Lipofectamine 3000 (Invitrogen, USA) was used to transfect BEAS-2B cells according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were harvested for subsequent experimental analysis.
Table 1.
Specific Sequences of tRF-Gly-GCC Mimics, Inhibitors, and Negative Controls (NC).
| Name of the Sequence | Specific Sequence |
|---|---|
| tRF-Gly-GCC mimic | GCAUGGCUGGUUCACUGGUAGAAUUGUC |
| tRF-Gly-GCC mimic NC | CGAGUGAUGGUAUUCGAUGCGAUCGUGU |
| tRF-Gly-GCC inhibitor | GAGAAUUCUAGCAGACAACCAGCCAUGC |
| tRF-Gly-GCC inhibitor NC | ACACGAUCGCAUCGAAUACCAUCACUCG |
Proliferation Assay
A cell counting kit (CCK-8; Tongren Institute of Chemistry, Japan) was used to determine cell proliferation. Transfected cells (1 × 103 cells/well) were seeded into a 96-well plate for 24 h, and 10% CCK-8 working solution was added to each well 1–5 d after irradiation. The 96-well plate was incubated at 37°C for 2 h, and the absorbance at 450 nm was measured using a Bio-Rad microplate reader at each time point (Synergy H1M; BioTek, USA). Cell viability was calculated based on the measured optical density at 450 nm using the following formula: cell vitality (%) = [As(radiation)–Ab(blank)]/[As(control)–Ab(blank)] × 100%, where As represents the absorbance of wells with cells and CCK8 solution and Ab represents the absorbance of wells with medium and CCk8 solution without cells. These experiments were repeated in triplicate.
AV/PI Apoptosis Assay
An Annexin V-FITC/PI apoptosis kit (BestBio, Shanghai, China) was used to detect cell apoptosis. Transfected cells (1 × 105 cells/well) were seeded into a 6-well plate for 24 h, collected, and analyzed by flow cytometry 72 h after irradiation. The experiment was repeated thrice.
ROS Assay
Reactive oxygen species was detected using a ROS assay kit (Beyotime, Shanghai, China). Transfected cells (1 × 105 cells/well) were seeded into a 6-well plate for 24 h, treated with 10 µmol/L DCFH-DA 72 h after irradiation, and incubated at 37°C for 30 min. After washing three times with phosphate-buffered saline, cells were visualized under a fluorescence microscope (IX73, Olympus, Japan).
qRT-PCR
The transfection efficiency of tRF-Gly-GCC was verified by qRT-PCR. After transfection, RNA was extracted from cells using TRIzol reagent and reverse transcribed to cDNA. SYBR Premix Ex Taq (Takara Bio, Kusatsu, Shiga, Japan) was used to prepare reactions for qRT-PCR analysis. The expression of oxidative stress-related genes (NOX2, NOX4, and PGC-1α) was also examined by qRT-PCR. The primer sequences are as follows: NOX2-F: 5′-TGCGATTCACACCATTGCAC-3’; NOX2-R: 5′-ACAGCGTGATGACAACTCCA-3’; NOX4-F: 5′-CTGCATGGTGGTGGTGCTAT-3’; NOX4-R: 5′-GCCCTCCTGAAACATGCAAC-3’; PGC-1α-F: 5′-TCGGAAGACACCCTCTTCTCTT-3’; PGC-1α-R: 5′-TCCATGGGGCTCCAATTTTACC-3’.
Western Blot Analysis
Cell lysates were prepared using RIPA lysis buffer. A bicinchoninic acid protein detection kit (Beyotime, Shanghai, China) was used to determine protein concentration. Cell lysates (30 μg per sample) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After transfer, the membrane was blocked with 5% skim milk, incubated with a specified primary antibody at 4°C overnight, and followed by incubation with HRP-conjugated anti-rabbit or mouse IgG secondary antibody. Antibody binding was detected by chemiluminescence using BeyoECL Plus (Beyotime) and visualized using a gel imager. The following antibodies were used in this study: AKT, p-AKT, PI3K, p-FOXO1, Bcl-2, Bax, and GAPDH (Affinity, Suzhou, China). All primary antibodies were used at a 1:1000 dilution.
Statistical Analyses
SPSS software (version 22.0, SPSS Inc. Chicago, IL, USA) was used to analyze the data. Values are represented as mean ± standard deviation. A paired t-test was used to analyze differences in tsRNA expression between two groups, and a chi-square test or unpaired Student’s t-test was used to assess statistical significance, which was set at P<.05.
Results
Irradiation-Induced Oxidative Stress in Human Bronchial Epithelial Cells
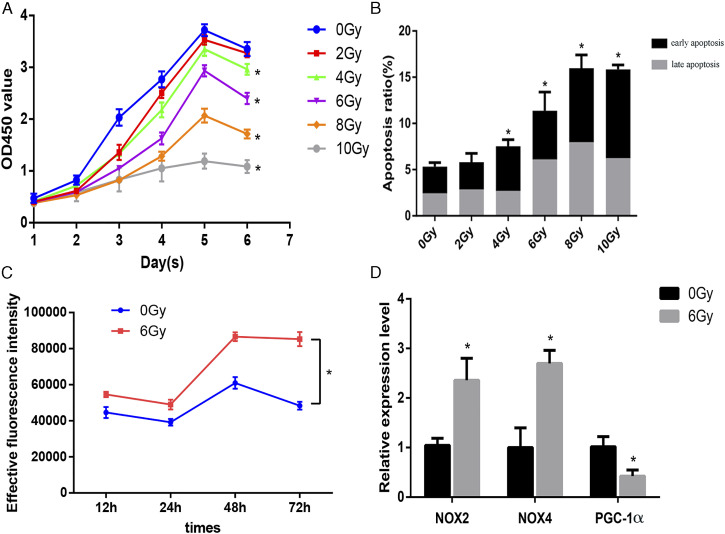
The appropriate radiation dose for this study was selected based on cell proliferation and apoptosis data under different irradiation doses. The data showed statistical differences between each dose group and the non-irradiation group (P < .05). Proliferative ability began to decline on the fifth day after irradiation and gradually decreased with increasing irradiation doses. The cell viability in the 6 Gy group was 51.64% at 72 h post-irradiation (Figure 1A), and the apoptosis rate in the 6 Gy group was remarkably different from that in the control group, including early and late apoptosis (Figure 1B). Therefore, 6 Gy was used as an appropriate irradiation dose for the in vitro model of radiation-induced damage in subsequent experiments. Next, we examined ROS levels at different time intervals following irradiation; the results revealed that ROS levels in the 6 Gy irradiation group were higher than in the 0 Gy control group, indicating that irradiation caused oxidative stress. Cellular ROS levels showed a cumulative increase over time. Increased levels were observed at 48 h and 72 h after exposure (Figure 1C). Furthermore, we observed alterations in the expression of oxidative stress-related genes, such as NOX2, NOX4 and PGC-1α, at 72 h after 6 Gy irradiation (Figure 1D).
Figure 1.
(A) Growth curves of dose groups. (B) Apoptosis rates in dose groups. (C) Reactive oxygen species (ROS) quantification in irradiated and control groups at different time intervals. (D) mRNA expression levels of oxidative stress-related genes 72 h post-irradiation in the irradiation and control groups. *P < 0.05.
tsRNAs Expression Profile After Irradiation
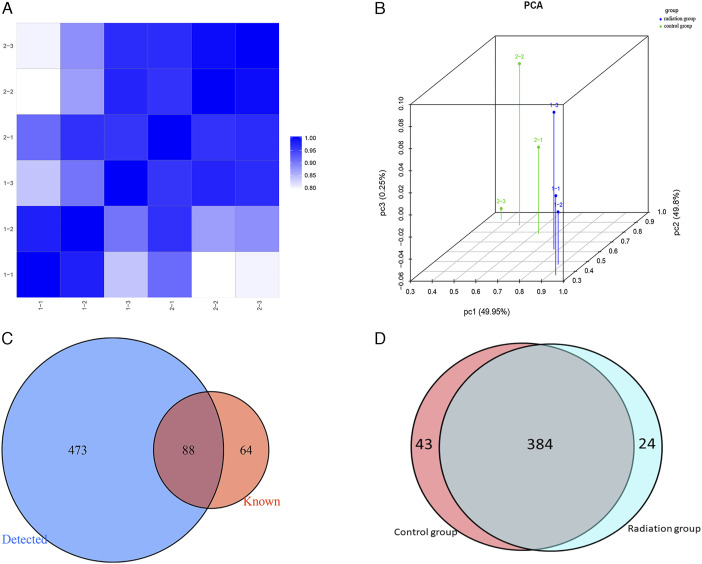
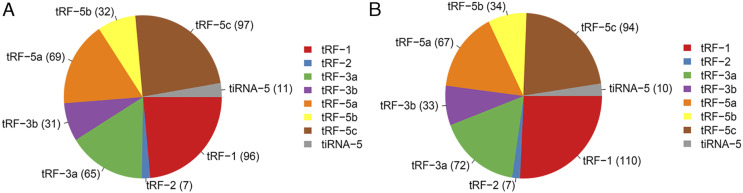
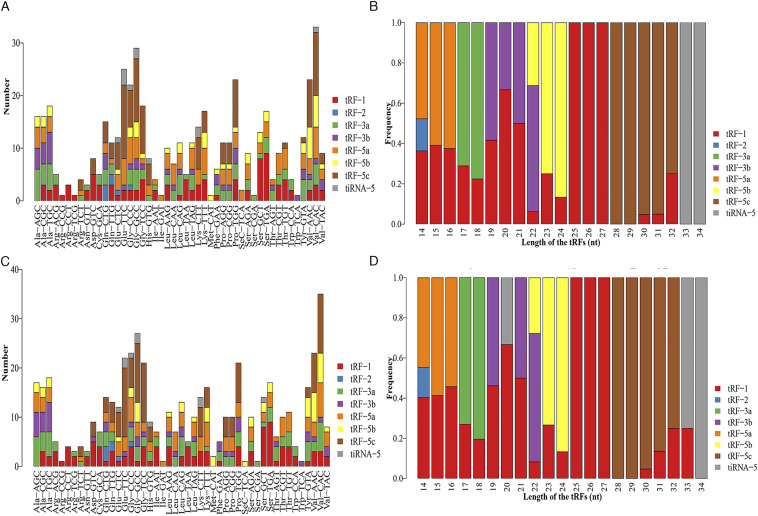
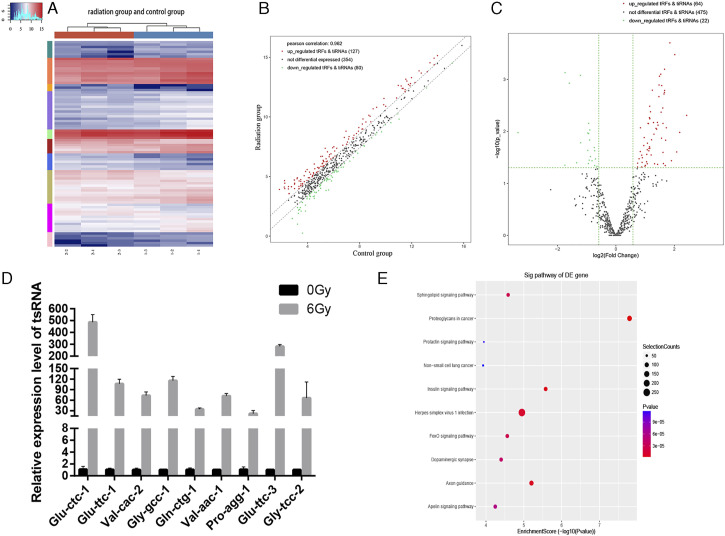
To determine the expression profiles of tsRNAs in our in vitro model of radiation-induced damage, we harvested RNA from irradiated and control cells to perform RNA sequencing. After the initial processing of raw data, we calculated the correlation coefficient between any two RNA transcripts in all samples according to the expression level of each transcript (Figure 2A). Principal component analysis was used for tRF and tiRNA expression profiling (Figure 2B). We identified 561 types of tsRNAs in our RNA sequencing data, of which 88 overlapped with the GtRNAdb and tRNAscan-SE database, and 473 were novel (Figure 2C). Thus, our sequencing results represent a substantial enrichment of the tsRNA databases. Moreover, 24 and 43 differentially expressed tsRNAs were identified in the irradiation and control groups, respectively (Figure 2D). The distribution of tRF and tiRNA subtypes in the irradiation and control groups are shown in Figures 3A and 3B. Based on their mapped positions, the tsRNAs can be grouped into five subtypes: tRF-5, tRF-3, tRF-1, tRF-2, and tiRNA. Among these tRF and tiRNA subtypes, the expression level of each subtype varied greatly. The stacked graph shows that several tRF and tiRNA subtypes originated from the same anticodon tRNA (Figure 4A and 4C). Figure 4B and 4D show the relationship between the subtype frequency and the length of tRFs and tiRNAs. The results yielded only a few types of tiRNA; thus, we did not focus on the tiRNAs.
Figure 2.
(A) Heatmap of correlation coefficient from all samples. (B) Principal coordinate analysis of tRNA-derived fragment (tRFs) and tRNA halves (tiRNAs) expression profiles. (C) Venn diagram based on the number of known and detected tRFs and tiRNAs. (D) Venn diagram based on the number of commonly and specifically expressed tRFs and tiRNAs.
Figure 3.
Pie graph for each tRNA-derived fragment (tRF) and tRNA halves (tiRNA) subtype. Pie graph of tRF and tiRNA subtype distribution in the irradiation (A) and control group (B).
Figure 4.
Stacked bar graph of the number distribution of subtypes in the irradiation (A) and control group (C). The horizontal coordinate represents the tRNA isodecoders, and the vertical axis represents the number of all tsRNA subtypes. The tsRNA subtypes are represented by different colors. Stacked bar graph of the length distribution of subtypes in the irradiation (B) and control group (D). The horizontal coordinate represents the length of tsRNAs, the vertical axis represents the frequency of the subtype, and different colors represent different tsRNA subtypes.
Identification of Irradiation-Related Differentially Expressed tRFs
As shown in the cluster heatmap, differentially expressed tRFs between the two groups were similar, indicating a minor difference between the two groups; the sequencing results were relatively accurate (Figure 5A). A total of 561 tsRNAs were detected in the two groups, and 86 tRFs were differentially expressed (| log2FC| ≥ 1.5 and P ≤ .05). The volcano plot shows that 64 of the 86 tRFs were upregulated, and 22 were downregulated in the irradiation group (Figure 5B). Associations between the identified tRFs and tiRNAs in the radiation and control groups are shown in a scatter plot (Figure 5C). In the irradiation and control groups, the top ten most upregulated or downregulated tRFs after irradiation are listed in Table 2. To further verify the accuracy of the sequencing results, we randomly selected nine upregulated tRFs with significant differential expression and high expression abundance in each sample as candidate tRFs for qRT-PCR (Figure 5D). We found that the nine differentially expressed tRFs were remarkably upregulated after irradiation, consistent with the sequencing data, indicating the reliability of the sequencing results. The list of predicted target genes is shown in the Supplementary Table 1. KEGG pathway analysis indicated that the target genes of differentially expressed tsRNA were mainly enriched in proteoglycans in cancer, non-small cell lung cancer, sphingolipid signaling pathway, insulin signaling pathway, FOXO signaling pathway, dopaminergic synapses, and axonal mediations in cancer (Figure 5E).
Figure 5.
(A) Unsupervised hierarchical clustering heatmap for tRNA-derived small RNA (tsRNA). Scatter plot (B) and volcano plot (C) of differentially expressed tsRNAs. (D) Expression levels of nine tRFs in the irradiation and control groups assessed using qRT-PCR. (E) KEGG analysis of differentially expressed tRFs.
Table 2.
Top Ten Upregulated and Downregulated tRNA-Derived Small RNAs, According to the Fold Change Values After Irradiation.
| tsRNA | Type | Length | Fold Change | P-Value | Regulation |
|---|---|---|---|---|---|
| tRF-1:29-Gln-CTG-3 | tRF-5c | 29 | 5.37096702 | .004938995 | Up |
| tRF-56:75-Gln-CTG-1-M2 | tRF-3b | 20 | 4.543641115 | .010538749 | Up |
| tRF-1:14-Gln-TTG-1-M3 | tRF-5a | 14 | 4.236362485 | .025019764 | Up |
| tRF-1:29-Pro-TGG-1 | tRF-5c | 29 | 4.043167132 | .000336076 | Up |
| tRF-1:29-Gln-CTG-4-M2 | tRF-5c | 29 | 3.735291472 | .036817666 | Up |
| tRF-1:28-Lys-CTT-1-M4 | tRF-5c | 28 | 3.601944031 | .000198995 | Up |
| tRF-1:24-Phe-GAA-1-M3 | tRF-5b | 24 | 3.308082602 | .008576172 | Up |
| tRF-1:16-SeC-TCA-1 | tRF-5a | 16 | 3.261123182 | .014809955 | Up |
| tRF-1:29-Thr-TGT-4-M2 | tRF-5c | 29 | 3.223592177 | .045735715 | Up |
| tRF-1:29-Pro-AGG-1-M6 | tRF-5c | 29 | 3.183104021 | .001634106 | Up |
| tRF-1:22-chrM.Gln-TTG | tRF-5b | 22 | .098002312 | .010584441 | Down |
| tRF-+1:T18-Ile-AAT-5-2 | tRF-1 | 18 | .297904629 | .044944438 | Down |
| tRF-+1:T14-Arg-CCT-4 | tRF-1 | 14 | .299330866 | .000746315 | Down |
| tRF-+1:T25-Leu-CAG-1-6 | tRF-1 | 25 | .331200255 | .001190312 | Down |
| tRF-+1:T31-Gly-CCC-1-2 | tRF-1 | 31 | .394082304 | .040217829 | Down |
| tRF-60:76-Tyr-GTA-1-M5 | tRF-3a | 17 | .430803677 | .000840835 | Down |
| tRF-+1:T14-Lys-TTT-3-2 | tRF-1 | 14 | .432898914 | .016572525 | Down |
| tRF-28:41-Gln-CTG-1-M7 | tRF-2 | 14 | .512817363 | .023194956 | Down |
| tRF-69:86-Leu-TAA-1 | tRF-3a | 18 | .512921961 | .010797955 | Down |
| tRF-+1:T15-Leu-AAG-2-4 | tRF-1 | 15 | .519592713 | .041541137 | Down |
Verification of tRF-Gly-GCC Transfection Efficiency
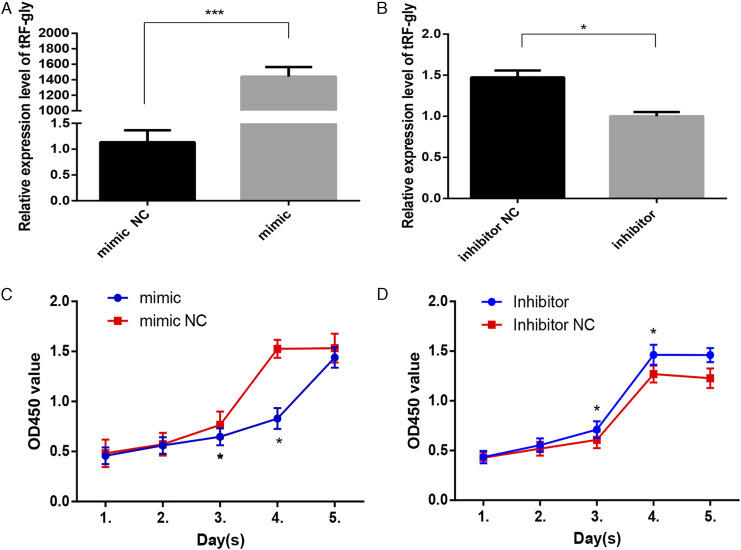
The significance of 5′-tRF-Gly-GCC has been extensively reported in many biological processes. Therefore, we selected tRF-Gly-GCC for further analysis. To explore the function of tRF-Gly-GCC, we first evaluated the transfection efficiency. In BEAS-2B cells transfected with a tRF-Gly-GCC mimic or mimic NC, tRF-Gly-GCC expression was significantly higher 24 h after transfection in the mimic compared with the mimic NC group, displaying an increase in expression of more than 1000 times (Figure 6A). tRF-Gly-GCC expression in cells transfected with a tRF-Gly-GCC inhibitor was significantly lower than that of cells transfected with a tRF-Gly-GCC inhibitor NC, as shown in Figure 6B (P < .05).
Figure 6.
Transfection efficiency of tRF-Gly-GCC mimic (A) and inhibitor (B) in BEAS-2B cells. Effects of transfection of tRF-Gly-GCC mimic (C) and inhibitor (D) on cell proliferation. *P < 0.05,*** P < 0.001.
Effect of tRF-Gly-GCC on Cell Proliferation
Cell proliferation in the tRF-Gly-GCC mimic group was significantly decreased compared with the tRF-Gly-GCC mimic NC group (Figure 6C), whereas the proliferative ability of cells transfected with the tRF-Gly-GCC inhibitor was significantly higher than that of the tRF-Gly-GCC inhibitor NC group (Figure 6D) (both P < .05). These results suggest that tRF-Gly-GCC can inhibit cell proliferation.
Effect of tRF-Gly-GCC on Apoptosis
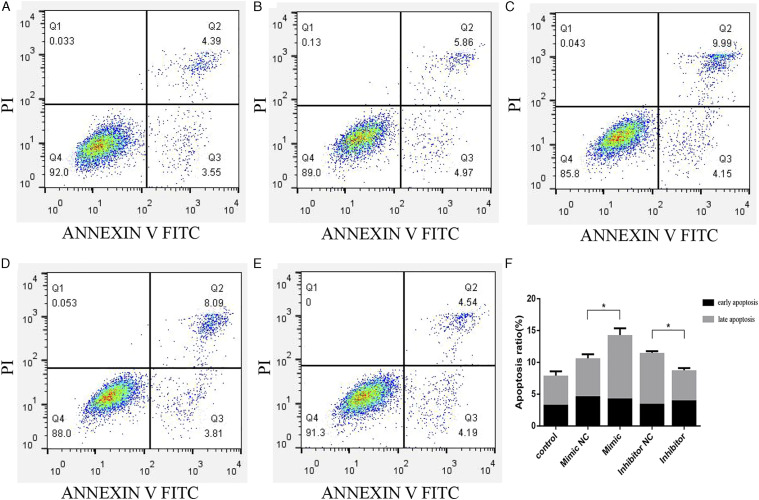
Flow cytometry results indicated that the apoptosis rate in BEAS-2B cells transfected with the tRF-Gly-GCC mimic was significantly higher than that of cells transfected with the mimic NC, while the results after transfection with the tRF-Gly-GCC inhibitor and inhibitor NC were correspondingly inverted. The proportion of apoptotic cells, including both early and late apoptosis, was lower in the tRF-Gly-GCC inhibitor compared with the inhibitor NC group (Figure 7A-7F). These data suggest that tRF-Gly-GCC may increase apoptosis during RILI.
Figure 7.
Effects of transfection of tRF-Gly-GCC mimic and inhibitor on cell apoptosis in control (A), tRF-Gly-GCC mimic negative control (NC) (B), tRF-Gly-GCC mimic (C), tRF-Gly-GCC inhibitor NC (D), and tRF-Gly-GCC inhibitor groups (E). Histogram of apoptosis in each group (F). *P < 0.05.
ROS Detection
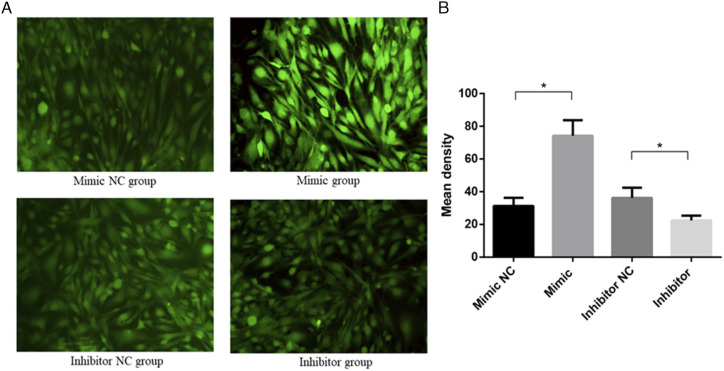
The ROS assay results showed that ROS levels in cells transfected with a tRF-Gly-GCC mimic were significantly higher than in cells transfected with the mimic NC, while ROS levels were significantly lower in the presence of the tRF-Gly-GCC inhibitor compared with the inhibitor NC (P < .05), as shown in Figure 8A-8B. These results indicate that tRF-Gly-GCC may promote oxidative stress, leading to RILI.
Figure 8.
Representative images of reactive oxygen species detected by fluorescence microscopy in each group (A) and histogram of mean density (B). *P < 0.05.
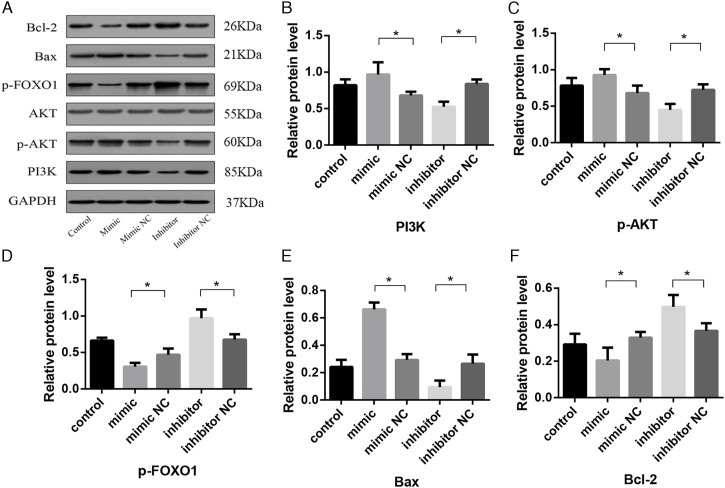
Effect of tRF-Gly-GCC on the Expression of Proteins in the PI3K/AKT and FOXO1 Pathways
We next analyzed the effect of tRF-Gly-GCC on the expression of proteins involved in PI3K/AKT and FOXO1 signaling using western blot analysis, with signal intensity displayed in the form of a bar chart (Figure 9A). PI3K (Figure 9B) and p-AKT (Figure 9C) expression increased in cells expressing the tRF-Gly-GCC mimic but decreased in the presence of the tRF-Gly-GCC inhibitor. When we examined the expression of downstream proteins in the pathway, the results showed that p-FOXO1 expression decreased in the tRF-Gly-GCC mimic group and increased in the inhibitor group (Figure 9D). When we assessed the levels of apoptosis-related proteins, we observed that Bcl-2 expression decreased and Bax expression increased in the tRF-Gly-GCC mimic group. In contrast, Bcl-2 expression increased, and Bax expression decreased in the inhibitor group (Figure 9E and 9F). Thus, the underlying molecular mechanisms of tRF-Gly-GCC function may be related to the PI3K/AKT and the FOXO1 signaling pathways.
Figure 9.
Western blot analysis showing protein levels of AKT, p-AKT, PI3K, p-FOXO1, Bcl-2, and Bax in BEAS-2B cells (A). Protein levels quantified by densitometry normalized to GAPDH are shown in bar graphs (B–F). Data are represented as the mean ± SD. *P < 0.05.
Discussion
As high flux, second-generation sequencing technology gradually advances, this technique is becoming widely available to many researchers. Unlike preceding techniques, such as microarray analysis, RNA sequencing allows for in-depth analysis, revealing many unique features of small non-coding RNAs, and their roles in the occurrence and development of many diseases. Studies have shown that tsRNAs (tRFs and tiRNAs) derived from small fragments of tRNA might be new potential molecular targets, as they participate in various cellular physiological processes and play a key role in the pathogenesis and development of certain diseases.10-12 tRF-03357 was reported to promote cell proliferation, migration, and invasion by regulating HMBOX1 in high-grade serous ovarian cancer. 13 TRF-Leu-CAG was found to stimulate the cell cycle and proliferation in non-small cell lung carcinoma. 14 Studies have reported that inhibiting Leu-CAG3′ tsRNA can trigger apoptosis in tumor cells but not in normal liver cells. 15 Another study demonstrated that tRF-315 protects prostate cancer cells from cisplatin-induced mitochondria-dependent apoptosis. 16 To explore the role of tsRNAs in RILI, we used X-rays to irradiate human bronchial epithelial cells and determined the expression profile of altered tsRNAs by RNA sequencing and bioinformatics analyses. Our results showed that ionizing radiation could alter the expression profile of tsRNAs, with 86 (64 upregulated and 22 downregulated) differentially expressed tsRNAs in the irradiation group compared with the control.
In this study, we did not observe differences in the proportions of tRFs in the irradiation and control groups, in agreement with similar results in the literature. Huang et al. reported no significant differences in the proportions of various types of tRFs in three breast cancer cell lines, as evidenced by high-throughput sequencing. 17 A similar finding was reported in murine models of choroidal neovascularization disease. 18 We speculate that tRFs may be produced in a specific manner that defines the ratios of tRFs produced. In addition, we found that tRF-5 accounted for the highest proportion of the tRF types. tRF-5 originates from the 5′ end of mature tRNA, and its occurrence is mainly dependent on the activity of the Dicer protein.19,20 Among the tRF-5 fragments, tRF-5c comprises the highest proportion of these RNAs. The tRF that we selected for further analysis, tRF-Gly-GCC-1, is derived from bases 1–28 of tRNA-Gly and belongs to the tRF-5c family. Among the few tRFs that have been functionally identified, 5′-tRF-Gly-GCC is the most studied. Hua et al. reported that 5′-tRF-Gly-GCC downregulation might lead to poor sperm development and early embryo abnormalities. 21 In addition, 5′-tRF-Gly-GCC reportedly inhibits endogenous reverse transcription factor MERVL-related genes in the zygote and during late development. 22 5′-tRF-Gly-GCC has also been associated with the metastatic progression of breast and lung cancers.23,24 A recent study reported that ALKBH3 upregulation could lead to increased expression of 5′-tRF-Gly-GCC, subsequently promoting tRNA cleavage to produce tRFs, potentially representing a novel biomarker for colorectal cancer diagnosis. 25 Furthermore, Zhong et al. reported that tRF-Gly-GCC contributes to oxidative stress-induced lipid metabolism in the alcoholic fatty liver. 26 Based on these findings, in the current study, we chose to explore the involvement of tRF-Gly-GCC in the biological functions of oxidative stress, including cell proliferation, apoptosis, and intracellular ROS production, which are implicated in the development of RILI.
Studies on tsRNA and oxidative stress have reported that Gly-tRF is associated with the oxidative pathway of hepatic lipid metabolism, promoting adipogenesis and inhibiting fatty acid β-oxidation by regulating the SIRT1 signal transduction pathway. 26 Reports have implicated tsRNAs in cardiovascular diseases caused by cardiac pathologic conditions, such as aging, oxidative stress, and metabolic disorders. 9 Our findings indicate that tRF-Gly-GCC may promote ROS production, suggesting that tsRNA has a potential regulatory function in the oxidative stress-associated development of RILI.
In the current study, pathway enrichment analysis revealed that the predicted target genes of differentially expressed tRFs were enriched in the PI3K/AKT and FOXO signaling pathways. These pathways are key signaling mediators of cellular responses against oxidative stress and inflammation. Research has shown that in explosion-induced lung injury, CD28 deficiency can reduce PI3K/AKT phosphorylation and increase that of FOXO1 through the PI3K/AKT/FOXO1 signaling pathway to improve lung inflammation and oxidative stress, ultimately reversing the effects of explosion-induced lung injury. 27 In addition, Venkatesan et al. reported that the effects of H2O2 exposure in mesangial cells of patients with diabetic nephropathy were mediated by the PI3K/AKT pathway and resulted in the negative regulation of FOXO1, with FOXO1 upregulation significantly alleviating the effects of oxidative stress. 28 This study revealed that a tRF mimic was able to promote the PI3K and p-AKT expression, inhibiting the levels of the downstream protein p-FOXO1, which is consistent with the results of studies reporting that increasing p-FOXO1 can improve the response to oxidative stress. In addition, tRF-Gly-GCC can inhibit the expression of the anti-apoptotic gene Bcl-2 and promote the expression of pro-apoptotic Bax. Oxidative stress-induced chondrocyte apoptosis can be triggered by the activation of the PI3K/AKT and caspase pathways in the early stages of osteoarthritis. 29 Our findings suggest that tRF-Gly-GCC may downregulate FOXO1 expression through the PI3K/AKT pathway and mediate oxidative stress-induced apoptosis. Therefore, we posit that tRF-Gly-GCC may affect p-FOXO1 expression through the PI3K/AKT pathway and participate in the regulation of oxidative stress during RILI.
Our study does have a few limitations. Although we successfully showed that our selected tRF, tRF-Gly-GCC, may be involved in the oxidative stress underlying RILI, it is also possible that some of the other 85 differentially expressed tRFs that we identified might be involved in the pathophysiology of RILI. Further research into these fragments is warranted to gain a better understanding of the role played by these RNAs in oxidative stress. In addition, we did not perform in vivo experiments to validate our results. Therefore, we plan to carry out in vivo experiments in future research.
In summary, we analyzed the expression profile of altered tsRNA caused by ionizing radiation, showing that the function of tRF-Gly-GCC associated with oxidative stress may inhibit cell proliferation and promote ROS production and apoptosis during the development of RILI. Our finding provides new insights into the molecular mechanisms underpinning RILI, advancing the clinical prevention and treatment of this disease.
Supplemental Material
Supplemental Material for Potential Functions of the tRNA-Derived Fragment tRF-Gly-GCC Associated With Oxidative Stress in Radiation-Induced Lung Injury by Lin Deng, Housheng Wang, Ting Fan, Liuyin Chen, Zhiling Shi, JingLin Mi, WeiMei Huang, Rensheng Wang, Kai Hu in Dose-Response
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (82060019, Natural Science Foundation of Guangxi (2018JJA140869) and Guangxi Medical and Health Appropriate Technology Development and Application Project (S2018097)
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
References
- 1.Hanania AN, Mainwaring W, Ghebre YT, Hanania NA, Ludwig M. Radiation-induced lung injury: Assessment and management. Chest[J]. 2019;156(1):150-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giuranno L, Ient J, De Ruysscher D, Vooijs MA. Radiation-Induced Lung Injury (RILI). Front Oncol[J]. 2019;9:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park YH, Kim JS. Predictors of radiation pneumonitis and pulmonary function changes after concurrent chemoradiotherapy of non-small cell lung cancer. Radiat Oncol J[J]. 2013;31(1):34-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arroyo-Hernández M, Maldonado F, Lozano-Ruiz F, Munoz-Montano W, Nunez-Baez M, Arrieta O. Radiation-induced lung injury: current evidence. BMC Pulm Med[J]. 2021;21(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balatti V, Nigita G, Veneziano D, et al. tsRNA signatures in cancer. Proc Natl Acad Sci U S A[J].2017, 114(30):8071-8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol[J]. 2018;19(1):45-58. [DOI] [PubMed] [Google Scholar]
- 7.Farina NH, Scalia S, Adams CE, et al. Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer. J Cell Physiol[J].2020, 235(6):5318-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pliatsika V, Loher P, Telonis AG, Rigoutsos I. MINTbase: a framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics[J]. 2016;32(16):2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J, Cowan DB, Wang DZ. tRNA-Derived Small RNAs and Their Potential Roles in Cardiac Hypertrophy. Front Pharmacol[J]. 2020;11:572941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng LL, Xu WL, Liu S, et al. tRF2Cancer: A web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res[J]. 2016;44(W1):W185-W193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang P, Zhang X, Chen S, et al. A novel serum tsRNA for diagnosis and prediction of Nephritis in SLE. Front Immunol[J]. 2021;12:735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin C, Xu PP, Zhang X, et al. Pathological significance of tRNA-derived small RNAs in neurological disorders. Neural Regen Res[J]. 2020;15(2):212-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Li F, Wang J, et al. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. Onco Targets Ther[J]. 2019;12:6371-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao Y, Sun Q, Liu X, Wang P, Wu R, Ma Z. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des[J].2017, 90(5):730-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HK, Fuchs G, Wang S, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature[J]. 2017;552(7683):57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Lee M, Song G, Lim W. tRNA(Lys)-Derived Fragment Alleviates Cisplatin-Induced Apoptosis in Prostate Cancer Cells. Pharmaceutics[J]. 2021;13(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Ge H, Zheng M, et al. Serum tRNA-derived fragments (tRFs) as potential candidates for diagnosis of nontriple negative breast cancer. J Cell Physiol[J]. 2020;235(3):2809-2824. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Liu S, Wang JH, et al. Differential expressions of microRNAs and Transfer RNA-derived Small RNAs: Potential targets of choroidal neovascularization. Curr Eye Res[J]. 2019;44(11):1226-1235. [DOI] [PubMed] [Google Scholar]
- 19.Telonis AG, Loher P, Honda S, et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget[J]. 2015;6(28):24797-24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev[J]. 2008;22(20):2773-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua M, Liu W, Chen Y, et al. Identification of small non-coding RNAs as sperm quality biomarkers for in vitro fertilization. Cell Discov[J]. 2019;5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma U, Conine CC, Shea JM, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science[J]. 2016;351(6271):391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Yang Y, Tan X, et al. Identification of tRNA-Derived Fragments Expression Profile in Breast Cancer Tissues. Curr Genomics[J]. 2019;20(3):199-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Qi M, Shen B, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res[J]. 2019;47(5):2533-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Yang X, Jiang G, et al. 5'-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis. Genome Med[J]. 2021;13(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong F, Hu Z, Jiang K, et al. Complement C3 activation regulates the production of tRNA-derived fragments Gly-tRFs and promotes alcohol-induced liver injury and steatosis. Cell Res[J]. 2019;29(7):548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Tong C, Xu Y, et al. CD28 deficiency ameliorates blast exposure-induced lung inflammation, oxidative stress, apoptosis, and T cell accumulation in the lungs via the PI3K/AKT/FOXO1 signaling pathway. Oxid Med Cell Longev[J]. 2019;2019:4848560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesan B, Mahimainathan L, Das F, Ghosh-Choudhury N, Ghosh Choudhury G. Downregulation of catalase by reactive oxygen species via PI 3 kinase/AKT signaling in mesangial cells. J Cell Physiol[J]. 2007;211(2):457-467. [DOI] [PubMed] [Google Scholar]
- 29.Li D, Ni S, Miao KS, Zhuang C. PI3K/AKT and caspase pathways mediate oxidative stress-induced chondrocyte apoptosis. Cell Stress Chaperones[J]. 2019;24(1):195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Potential Functions of the tRNA-Derived Fragment tRF-Gly-GCC Associated With Oxidative Stress in Radiation-Induced Lung Injury by Lin Deng, Housheng Wang, Ting Fan, Liuyin Chen, Zhiling Shi, JingLin Mi, WeiMei Huang, Rensheng Wang, Kai Hu in Dose-Response