Abstract
Bacteriophage T4 middle-mode transcription requires two phage-encoded proteins, the MotA transcription factor and AsiA coactivator, along with Escherichia coli RNA polymerase holoenzyme containing the ς70 subunit. A motA positive control (pc) mutant, motA-pc1, was used to select for suppressor mutations that alter other proteins in the transcription complex. Separate genetic selections isolated two AsiA mutants (S22F and Q51E) and five ς70 mutants (Y571C, Y571H, D570N, L595P, and S604P). All seven suppressor mutants gave partial suppressor phenotypes in vivo as judged by plaque morphology and burst size measurements. The S22F mutant AsiA protein and glutathione S-transferase fusions of the five mutant ς70 proteins were purified. All of these mutant proteins allowed normal levels of in vitro transcription when tested with wild-type MotA protein, but they failed to suppress the mutant MotA-pc1 protein in the same assay. The ς70 substitutions affected the 4.2 region, which binds the −35 sequence of E. coli promoters. In the presence of E. coli RNA polymerase without T4 proteins, the L595P and S604P substitutions greatly decreased transcription from standard E. coli promoters. This defect could not be explained solely by a disruption in −35 recognition since similar results were obtained with extended −10 promoters. The generalized transcriptional defect of these two mutants correlated with a defect in binding to core RNA polymerase, as judged by immunoprecipitation analysis. The L595P mutant, which was the most defective for in vitro transcription, failed to support E. coli growth.
Bacteriophage T4 uses three stages of transcription: early, middle, and late. This temporal progression is achieved by using three classes of promoters and by modifying the host RNA polymerase. Middle transcription requires two phage-encoded proteins, the MotA transcription factor and a coactivator, AsiA (for a review, see reference 37). AsiA binds to the ς70 subunit of host RNA polymerase and inhibits transcription from most Escherichia coli promoters. MotA binds to T4 middle promoters at the mot box, a consensus sequence centered at −30 (13, 30). The C- and N-terminal domains of MotA were solved by nuclear magnetic resonance and X-ray crystallography, respectively (9, 10). The C-terminal domain binds DNA, while the N-terminal domain is involved in transcriptional activation (10, 11). Based on the structural data, two residues (D30A and F31A) on an acidic, hydrophobic surface patch of the N-terminal domain were altered, resulting in a motA positive control mutant (motA-pc1). The mutant protein (full-length) binds normally to its DNA site but does not efficiently activate transcription (10).
MotA probably interacts directly with the ς70 subunit of host RNA polymerase during activation of middle-mode transcription. MotA and ς70 form a complex with unique mobility in native polyacrylamide gels, although this was tested in the absence of DNA and the other RNA polymerase subunits (11). Furthermore, MotA binds specifically to the −30 region of middle promoters, thus acting like a class II activator (see reference 16 for a review). Other class II activators generally interact with the 4.2 region of ς70, which has a predicted helix-turn-helix motif and recognizes the −35 sequence of E. coli promoters (see reference 12 for a review). Various amino acid substitutions within the 4.2 region affect activation by transcription factors, including FNR, AraC, CRP, and PhoB (20, 21, 23, 24). The 4.2 region has also been shown to interact with phage λ cI repressor protein, which also binds at a site near the −30 region to activate transcription (22).
Additionally or alternatively, MotA may interact with the AsiA coactivator. AsiA has been shown to bind to ς70 in the 4.2 region (6, 32, 34, 35), implying that AsiA is in close proximity to DNA-bound MotA. Upon binding to ς70, AsiA blocks transcription from host promoters requiring the −35 region (6, 33). Thus, AsiA blocks the 4.2 domain from recognizing the −35 region and possibly interacts directly with MotA to activate T4 middle promoters.
Using native polyacrylamide gel electrophoresis, Gerber and Hinton (11) detected a ternary MotA-AsiA-ς70 complex. This complex was detected in the absence of promoter DNA and other RNA polymerase subunits but it may well reflect interactions that occur during initiation of middle-mode transcription. Since MotA and AsiA can each bind separately to ς70, the existence of this ternary complex does not clarify whether MotA and AsiA directly interact with each other.
To investigate these interactions, we conducted genetic selections to try to identify substitutions in AsiA and ς70 that suppress the activation defect caused by motA-pc1. In this report, we describe two asiA mutants and five rpoD (ς70) mutants that gave moderate suppression in vivo. In vitro analysis of the mutant proteins failed to recapitulate the suppression but did reveal a novel class of ς70 mutants.
MATERIALS AND METHODS
Media and chemicals.
L broth consisted of NaCl (10 g/liter), Bacto Tryptone (Difco; 10 g/liter), and yeast extract (Difco; 5 g/liter). Hershey plates contained NaCl (8 g/liter), Bacto agar (10 g/liter), sodium citrate (2 g/liter), glucose (1.3 g/liter) and Bacto Tryptone (13 g/liter). Hershey top agar had the same composition except that glucose was at 3 g/liter and Bacto agar at 6.5 g/liter. Oligonucleotides were prepared by the Duke University DNA Core Facility.
Bacterial and phage strains.
E. coli and phage strains are listed in Table 1. T4 denAB motA-pc1 was created by homologous recombination (marker rescue) with plasmid pMotA-pc1. Progeny phage with motA-pc1 were identified by their inability to grow on E. coli TabG, and the presence of the two mutations was confirmed by sequencing. K10 motA-pc1 was created by a genetic cross between T4 denAB motA-pc1 and K10 motAΔ.
TABLE 1.
Genotypes of E. coli and phage T4 strains useda
| Strain | Genotype | Reference |
|---|---|---|
| E. coli | ||
| MCS1 | supD araD139 Δ(ara-leu7697) ΔlacX74 galU galK hsdR rpsL pro (uncharacterized proline auxotrophy) | 19 |
| TabG | E. coli B, nonsuppressing, hsdR gal met tabG | 29 |
| BE-BS | E. coli B, nonsuppressing | 31 |
| Phage T4 | ||
| T4 denAB | denA (nd28) [denB-rII]Δ (rIIPT8) | 2 |
| T4 denAB motAΔ | T4 denAB motAΔ | 2 |
| T4 denAB motA-pc1 | T4 denAB motA-D30A/F31A | This study |
| K10 | T4 denAB 38 (amB262) 51 (amS29) | 31 |
| K10 motAΔ | K10 motAΔ | 2 |
| K10 motA-pc1 | K10 motA-D30A/F31A | This study |
The denA and denB mutations inhibit host DNA degradation, and the 38am and 51am mutations block phage growth unless an amber suppressor is present.
Construction of plasmids.
Plasmids are listed in Table 2. Plasmid pMPC43, which expresses MotA-pc1, was used in the AsiA suppressor selection. This plasmid was created by inserting the 1.1-kb SalI/BamHI fragment from pMotA-pc1 into SalI/BamHI-cleaved pSU18 (25). Plasmid pMPC34 was created as follows. The small ori(34)-containing HindIII/SalI fragment from pKK061-1 (26) was inserted into HindIII/SalI-cleaved pSU18, and then the resulting plasmid was cleaved with HindIII and ligated to the 4.9-kb rpoD-containing HindIII fragment of pJH62 (kindly provided by D. Siegele, Texas A & M University). After isolation of a clone with the insert in the proper orientation, most of the dnaG gene (originally from pJH62) was removed by cleaving with HpaI and religating, which removes a 1.7-kb fragment. The resulting plasmid, pMPC34, contains T4 ori(34) and the plasmid vector origin of replication, along with rpoD, which is apparently transcribed from the upstream cam gene promoter.
TABLE 2.
Plasmids used
| Plasmid | Vector backbone | Cloned gene | Reference(s) |
|---|---|---|---|
| pAsiA | pET21a | asiA | 14 |
| pAsiA-S22F | pET21a | asiA-S22F | This study |
| pBSPLO+ | pBR322 | supF | 31 |
| pBSPLO+-AsiA | pBR322 | supF asiA | This study |
| pMotA-pc1 | pET3b | motA-pc1 | 10 |
| pMPC34 | pSU18 | rpoD ori(34) | This study |
| pMPC43 | pSU18 | motA-pc1 | This study |
| pJH62 | ColE1 | rpoD operon | 15 |
| pGEX-ς70 | pGEX-2T | rpoD | 7, 23 |
The wild-type glutathione S-transferase (GST)-ς70 fusion plasmid, pGEX-ς70, contains residues 8 to 613 of rpoD, in frame and downstream of the GST reading frame (7, 23). Each GST-ς70 mutant was cloned by purifying a 980-bp NcoI fragment from the appropriate pMPC34-derived plasmid and ligating it into NcoI-cleaved pGEX-ς70. The proper insert orientation was determined by restriction mapping, and the 4.2 region was sequenced to confirm the appropriate mutation.
The pBSPLO+-AsiA plasmid used in the asiA suppressor selection was created by ligating the 300-bp NdeI/BamHI fragment from pAsiA (14) (kindly provided by D. Hinton, National Institutes of Health) to NdeI/BamHI-cleaved pBSPLO+ (31). The AsiA overexpression plasmid, pAsiA, contains the asiA gene cloned into pET-21A vector between the NdeI and BamHI sites (14). A comparable AsiA-S22F overexpression plasmid was created by amplifying the asiA-S22F gene from the T4 genome using PCR primers containing flanking NdeI and BamHI sites. After digestion with NdeI and BamHI, the fragment was ligated into NdeI/BamHI-cut pET-21A vector. The insert was verified by sequencing.
Proteins.
MotA and MotA-pc1 were overexpressed and purified as described earlier (10). RNA polymerase holoenzyme was purchased from Boehringer Mannheim. RNA polymerase core was purchased from Epicentre Technologies, except for the experiment in Fig. 7, in which core was purified as previously described (36). The GST-ς70 proteins were purified as described before (23).
FIG. 7.
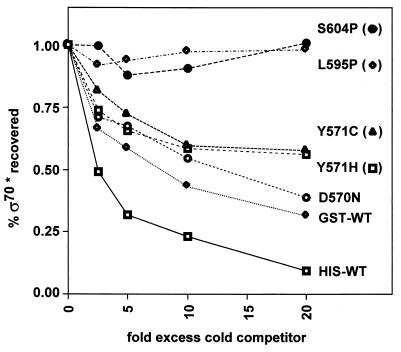
Competition assay for binding of ς70 mutants to core RNA polymerase. Wild-type His-ς70 labeled with [γ-32P]ATP and kinase was added to nonradioactive core RNA polymerase and Sepharose beads coated with polyclonal antibodies against the core. The amount of radioactivity bound to the beads was measured as a function of increasing concentrations of cold competitor proteins (0, 0.25, 0.5, 1, and 2 μM). Each point in this figure shows the average of three determinations.
Overexpression and purification of AsiA proteins followed the procedures previously described (14). Briefly, cells were harvested, frozen at −80°C, resuspended in AsiA sonication buffer (20 mM Tris-Cl [pH 8], 1 mM EDTA, 10% glycerol, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaCl), and sonicated until lysed. After centrifugation at 100,000 × g for 90 min, ammonium sulfate (60% saturation) was added to the supernatant and the protein was collected by centrifugation. The protein was resuspended in and dialyzed against sonication buffer and then purified sequentially by column chromatography on Q-Sepharose, Affi-Gel Blue, and Sephadex G-50.
Isolation of AsiA suppressor mutants.
The first method involved mutagenesis of T4 denAB motAΔ with 2-hydroxylamine following the protocol of Drake and Ripley (8). Mutagenized phage (roughly 107 PFU) were plated on TabG cells harboring pMotA-pc1, and plaques were found at a frequency of about 10−5. Phage from plaques were recovered and streaked on lawns of TabG and TabG pMotA-pc1 to verify MotA-dependent growth (no growth on TabG) and to isolate individual plaques (on TabG pMotA-pc1). The asiA gene from candidate mutants was amplified by PCR and sequenced.
In the second method, the asiA gene was mutagenized by PCR, cloned into a plasmid, and then rescued into the phage genome by homologous recombination. PCR mixtures contained 20 mM Tris-Cl (pH 8.4), 50 mM KCl, 5 mM MgCl2, 150 μM (each) dATP, dGTP, TTP, and dCTP, 50 pmol each of primer A (5′-CCCGACGCATATGAATAAAAACATTGATACAGTTCG-3′) and primer B (5′-GCCGGATCCAGAATATTAGGAAGGGCTA-3′), 10 U of Taq polymerase (Gibco BRL), and 50 ng of pBSPLO+-AsiA DNA. Roughly 15 to 20% of the PCR products should contain at least one point mutation after 30 amplification cycles (39). The PCR primers introduced NdeI and BamHI restriction sites, which were cleaved prior to ligation to NdeI/BamHI-cleaved pBSPLO+. The DNA was electroporated into MCS1 cells harboring plasmid pMPC43, resulting in >50,000 viable transformants (determined by plating a small aliquot on selective plates). The pool of transformants was grown in L broth containing chloramphenicol (10 μg/ml) and ampicillin (40 μg/ml) and then infected with K10 motAΔ at a multiplicity of infection (MOI) of 3. The pBSPLO+-AsiA plasmid inserted into the phage genome by homologous recombination at a frequency of about 3.5 × 10−4 during this infection. About 5,000 to 10,000 integrant plaques, selected by growth on a nonsuppressing host strain (BE-BS), were pooled. The integrated plasmid in the phage genome is lost at a high frequency by homologous recombination, frequently leaving behind any mutations in the cloned segment (asiA in this case). Phage with suppressors of the motA-pc1 defect were recovered by plating on TabG pBSPLO+ cells at a frequency of 5.5 × 10−4, about twice that measured in a parallel procedure using unmutagenized pBSPLO+-AsiA. After verifying that each candidate mutant could grow on TabG pMPC43 pBSPLO+ (to test motA-pc1 suppression) but not on BE-BS pMPC43 (to test for loss of the integrated plasmid), the asiA gene from the mutant was PCR amplified and sequenced.
Isolation of ς70 suppressor mutants.
Plasmid pMPC34, which contains rpoD, was mutagenized by growing MCS1 cells harboring the plasmid in L broth containing chloramphenicol (10 μg/ml) and N4-aminocytidine (50 μg/ml; causes both transitions and transversions [38]). Plasmid DNA was isolated from the mutagenized cells and transformed into TabG pMotA-pc1. A pool consisting of about 106 transformants (determined by plating a small aliquot) was incubated for a 2-h outgrowth without selection and then grown in L broth containing chloramphenicol (10 μg/ml) and ampicillin (40 μg/ml) to an optical density at 560 mm of 0.4. The cells were infected with T4 denAB motAΔ (MOI = 3), and the resulting lysate was used to transduce MCS1 cells to chloramphenicol resistance. Plasmid DNA from the transductant pool was isolated and used to repeat the selection scheme (i.e., transformation into TabG pMotA-pc1, infection with T4 denAB motAΔ, and transduction into MCS1 cells). After the second round of selection, the pool of plasmid DNA was used to transform TabG pMotA-pc1. Individual transformants were tested for the ability to support plaque formation by T4 denAB motAΔ. Transformants harboring a suppressor mutation in rpoD allowed small plaques to form, while comparable cells harboring the wild-type plasmid did not allow plaque formation. The rpoD gene from each suppressor plasmid was subcloned to map the causative mutation(s), which in each case was within the BglI-XhoI fragment (contains the C-terminal 85 codons of rpoD).
Plaque spot tests.
To test suppression by the asiA mutants, TabG cells harboring pBSPLO+ were mixed with Hershey top agar and overlaid on a square Hershey plate. Appropriate dilutions of the indicated phage were then spotted on the plate in 3-μl aliquots, and the plate was incubated at 37°C overnight. To test suppression by the rpoD mutants, TabG cells harboring pMotA-pc1 and the indicated ς70-expression plasmid were mixed with Hershey top agar, overlaid on a square Hershey plate, and spotted with the indicated phage as described above.
Burst size experiments.
Burst experiments were conducted as described previously (10) with the following exceptions. For the suppressor mutants in rpoD, TabG cells harboring pMPC34 expressing either wild-type or mutant ς70 were infected with T4 denAB motA-pc1 at an MOI of 0.1, and samples were taken at 90 min postinfection. To measure burst size of the suppressor mutations in asiA, TabG cells harboring pBSPLO+ were infected at an MOI of 0.1 with K10 motA-pc1 strains containing either wild-type or mutant asiA, and samples were taken at 90 min postinfection. Progeny phage titers were determined on MCS1 cells, and the burst size was corrected for free (unattached) phage as previously described (10).
In vitro transcription.
The transcription buffer contained 175 mM KCl, 10 mM Tris-Cl [pH 7.9], 10 mM MgCl2, 1 mM dithiothreitol, and bovine serum albumin at 50 μg/ml. Transcription assays (10 μl) also contained 400 μM (each) rATP, rGTP, and rCTP, 40 μM UTP, 4 U of RNasin (Promega), 25 to 50 fmol of DNA template, and 0.7 μM [α-32P]UTP (3,000 Ci/mmol), along with the indicated proteins. For the T4 middle-mode transcription assays, MotA+ or MotA-pc1 (2 pmol) was preincubated with the DNA and ribonucleoside triphosphates in 5 μl of transcription buffer for 5 min at 37°C. Similarly, RNA polymerase (core or holoenzyme; 0.05 pmol), AsiA (2 pmol), and GST-ς70 (0.5 pmol) were preincubated in another 5-μl aliquot of transcription buffer for 5 min at 37°C. The two preincubations were then mixed to initiate the reaction. For E. coli promoter transcription assays, RNA polymerase core enzyme (0.05 pmol) was preincubated with the indicated GST-ς70 protein (0.5 pmol) in 5 μl of transcription buffer for 5 min at 37°C before addition of the DNA substrate (also in 5 μl of transcription buffer). After the preincubations, the mixed reactions (10 μl) were incubated at 37°C for 30 min and then placed on ice. The reactions were terminated by adding 10 μl of stop solution (95% [vol/vol] formamide, 2 mM Na2-EDTA, 0.05% bromphenol blue). Samples were heated at 95°C for 4 min and subjected to electrophoresis through a denaturing 8% polyacrylamide gel. Electrophoretic bands were quantitated with an AMBIS direct radioisotope counting system.
T4-modified DNA templates containing middle promoters PuvsY and Pori(34) were prepared from plasmids pGJB1 and pGJB4, respectively (26), as previously described (30). The PuvsY template was cleaved with SspI and EcoRV to yield a 221-base runoff transcript, while the Pori(34) template was cleaved with AseI and EcoRV to yield a 320-base runoff. For the E. coli transcription assays, the tac promoter template was prepared by purifying the 370-bp SspI/BamHI fragment of pPH310 (4), resulting in a 320-base runoff transcript. The unmodified PuvsY DNA template was a gel-purified 467-bp SspI/EcoRV fragment of pGJB1, which yields a 221-base runoff. The KAB promoter templates were obtained from plasmid pRW50 derivatives (kindly provided by S. Minchin, University of Birmingham, United Kingdom) for promoters KAB-TG and KAB-TT (1). The gel-purified 1.5-kb HpaI/SacII fragment yields a runoff transcript of about 900 bases.
ς-core binding assay.
Binding of the GST-ς70 mutants to core RNA polymerase was analyzed essentially as described by Sharp et al. (36). Wild-type or mutant GST-ς70 was added in increasing concentrations in a competition against wild-type (His-tagged) ς70, labeled by a kinase reaction as previously described (36). The buffer for this experiment contained 10 mM Tris-Cl [pH 8], 5% glycerol, 1 mM β-mercaptoethanol, 0.3 M NaCl, 10 mM MgCl2, and bovine serum albumin at 200 μg/ml. Siliconized Eppendorf tubes were used to decrease nonspecific binding. Each reaction contained 100 nM labeled His-ς70 and 30 nM core, along with 0, 0.25, 0.5, 1, or 2 μM cold ς70 competitor. After a 1-h incubation at 37°C, the reaction mix was added to 100 μl of preequilibrated protein A-Sepharose beads (10% volume) coupled to polyclonal anti-core antibody (Animal Pharm Services Inc., Healdsburg, Calif.). The beads were rocked at 4°C for 1 h, washed with 1 ml of buffer, collected by centrifugation, aspirated to remove residual liquid, resuspended in water, and finally added to scintillation fluid for counting. Coimmunoprecipitation of the labeled ς70 is dependent on the presence of core RNA polymerase (data not shown).
Test of GST-ς70 mutants for in vivo function in E. coli.
The ptrp-rpoD gene (23) was introduced into the chromosome of E. coli strain MCS1 by P1 transduction, using the linked chloramphenicol-resistance marker for selection. Wild-type and mutant GST-ς70 fusion plasmids (pGEX-ς70 and derivatives) were transformed into this strain and incubated for 1 h at 37°C in L broth for phenotypic expression. The cells were then serially diluted and spotted onto chloramphenicol-ampicillin L broth plates either with or without 0.2 mM indole-3-acrylic acid (Aldrich). The plates did not contain isopropyl-β-d-thiogalactopyranoside (IPTG), because the tac promoter on the GST-ς70 plasmids is leaky enough to allow sufficient ς70 expression for cell growth without induction (23). Plates were incubated at 37°C for 18 h.
RESULTS AND DISCUSSION
Selection of asiA and rpoD mutants that suppress motA-pc1.
The pc1 mutation in motA prevents T4 growth in E. coli TabG but not in wild-type E. coli (10). All motA point mutants, including amber and temperature-sensitive mutants, have this same characteristic, but a motA deletion mutant is lethal even in wild-type E. coli strains (2). Apparently, a very small amount of MotA activity is sufficient for growth in wild-type E. coli, but the mutation in TabG somehow increases this requirement (2, 10, 29, 37). Although the motA-pc1 mutant grows in wild-type E. coli, the MotA-pc1 protein is defective for in vitro transcription with RNA polymerase purified from wild-type (non-TabG) E. coli (10). Furthermore, the pc1 mutation decreases T4 origin-dependent plasmid replication in wild-type E. coli (5). Thus, while motA-pc1 defects can be observed without the mutation in the TabG strain, we used this host strain for genetic selections of suppressor mutants.
Two methods were used to isolate suppressor mutations in the T4 asiA gene (also see Materials and Methods). The first was to mutagenize a T4 denAB motAΔ strain with hydroxylamine and select for growth in the presence of plasmid-produced MotA-pc1 protein. The second method involved PCR mutagenesis of the asiA gene on a plasmid, substituting the mutagenized gene into a T4 denAB motAΔ phage genome by homologous recombination and selecting for phage that grow in the presence of plasmid-produced MotA-pc1. Many of the isolated suppressor mutants from both procedures did not contain a mutation in asiA; these were not pursued. We did identify two asiA substitution mutations (S22F and Q51E) that each allowed growth with MotA-pc1. Each mutation was substituted into the asiA gene of an unmutagenized K10 motA-pc1 genome using the T4 insertion/substitution system (31), and this recapitulated the suppressor phenotype.
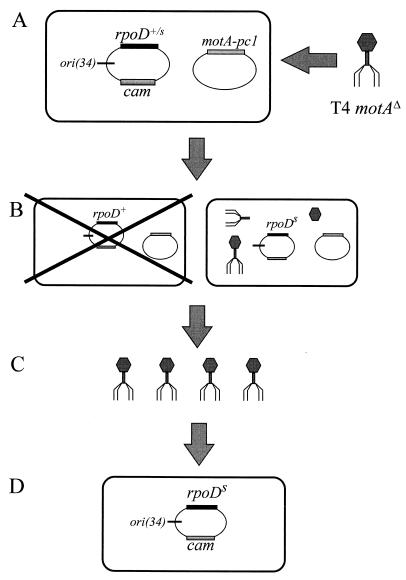
The selection to isolate suppressors in rpoD modified a scheme originally used by Kreuzer and Alberts (18, 19) to isolate T4 replication origins. Plasmid pMPC34 was constructed with rpoD and the MotA-dependent T4 origin, ori(34) (Fig. 1). This plasmid was randomly mutagenized and then introduced into TabG cells that also contained a plasmid expressing MotA-pc1. The TabG cells harboring both plasmids were infected with T4 denAB motAΔ, and the lysate was collected. Since only MotA-pc1 is present, almost all infected cells undergo an abortive infection. However, a productive infection could occur in any cell that contains a plasmid with an rpoD mutation that suppresses the MotA defect. The mutant pMPC34 plasmid would replicate due to the T4 origin of replication, and the resulting concatemers of plasmid DNA would get packaged into phage heads and thereby produce plasmid-transducing particles in the lysate. The lysate was used to transduce cells that were restrictive for growth of T4 denAB motAΔ (so that the transductants would not be killed by phage growth), and transductants were selected by growth in the presence of chloramphenicol. This procedure enriches for plasmids that contain a mutant rpoD gene that suppresses the MotA-pc1 activation defect, and individual candidate plasmids were analyzed after two rounds of enrichment (see Materials and Methods). Selected plasmids were subcloned to localize the rpoD mutation and were sequenced to identify the mutation. Five point mutations were identified, and each affects the 4.2 region of ς70: Tyr-571 to Cys (Y571C), Tyr-571 to His (Y571H), Leu-595 to Pro (L595P), Ser-604 to Pro (S604P), and Asp-570 to Asn (D570N).
FIG. 1.
Schematic for ς70 suppressor selection. (A) TabG cells contain a MotA-pc1 expression plasmid and mutagenized plasmid pMPC34, which contains the ς70 gene (rpoD) and the T4 origin, ori(34). The cells are infected with T4 denAB motAΔ. (B) Most infected cells contain a wild-type rpoD gene (or rpoD mutations that do not suppress) and therefore lead to an abortive infection (left cell). A viable infection should occur only if a mutant ς70 is expressed from the plasmid and suppresses the MotA-pc1 defect (right cell). The suppressing mutation in rpoD is designated rpoDS in this diagram. The viable infection allows T4 growth and replication of the pMPC34, which then forms long concatemers of plasmid DNA. (C) The concatemeric plasmid DNA is packaged into some of the phage heads to produce transducing phage in the lysate. The lysate is then used to transduce E. coli cells, and transductants are selected by the cam marker on pMPC34 (D).
The 4.2 region has also been shown to be involved in activation by a number of other transcription factors, including λ cI protein, FNR, PhoB, CRP, and AraC (16). Each of these activators has a binding site very near −35, similar to the location of the T4 mot box. Interestingly, different substitutions in residues D570 and Y571 affect activation by PhoB (17, 24) and by MotA (this work). The 4.2 region of ς70 is also important in the interaction with AsiA. Fragments of ς70 containing either residues 568 to 600 or residues 547 to 603 bind AsiA, and furthermore, AsiA protein creates a hydroxyl radical footprint at residues 572 to 588 of ς70 (6, 32, 34, 35). With one exception (S604P), the suppressor mutations that we isolated (D570N, Y571C, Y571H, and L595P) are within the ς70 fragments that bind AsiA and are from one to seven residues from the footprinted region.
Particularly given their locations, there are several possible explanations for suppression by the ς70 suppressor mutations. For example, MotA and ς70 may interact directly, with the MotA-pc1 substitutions weakening this interaction and the suppressor substitutions restoring it. Alternatively, MotA may interact predominantly with AsiA, with the ς70 suppressor mutations acting indirectly (e.g., inducing a conformational change in AsiA that allows better interaction with MotA-pc1).
In vivo tests for suppression by asiA and rpoD mutants.
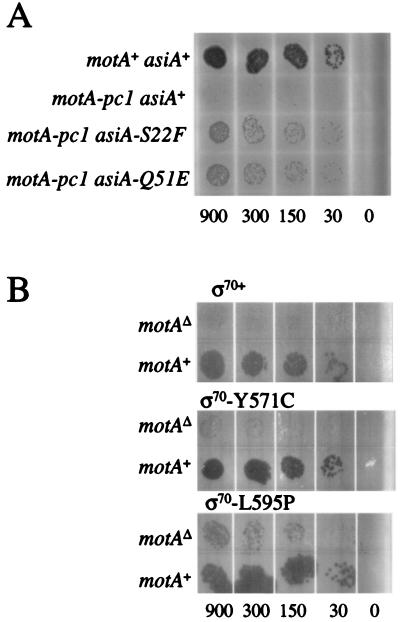
We tested the ability of each isolated mutant to restore growth in the presence of MotA-pc1 by observing plaque formation on E. coli strain TabG. To test suppression by the asiA mutants, spot tests were performed with T4 strains containing motA-pc1 and either the wild-type or mutant asiA gene. The presence of each asiA mutation led to plaque formation, but the plaque size was still smaller than that of the motA+ control phage (Fig. 2A). Therefore, the asiA mutations partially suppress the growth defect caused by motA-pc1.
FIG. 2.
Plaque spot tests to visualize suppression. (A) The plate contained a lawn of TabG pBSPLO+ cells, and the indicated T4 K10 strain was spotted on the lawn in 3-μl aliquots. The aliquots contained approximately the number of phage indicated below the plate. The positive control was T4 strain K10 (motA+ asiA+), which resulted in full growth (top row), while the negative control K10 motA-pc1 (asiA+) showed very little growth (second row). (B) The plates contained lawns of TabG cells with the indicated ς70 expression plasmid and a MotA-pc1 expression plasmid. Phage T4 denAB motAΔ (top row) or T4 denAB (motA+; bottom row) were spotted on the lawn in 3-μl aliquots that contained approximately the number of phage indicated below the plate. All plates were incubated at 37°C overnight.
Spot tests were also performed with cells containing a MotA-pc1 expression plasmid along with each mutant ς70 expression plasmid (Fig. 2B). Each mutant ς70 allowed small to moderate-sized plaques, with the L595P and S604P mutants giving the strongest suppression, Y571C and Y571H intermediate suppression, and D570N weak suppression (Fig. 2B and data not shown). However, none of the mutant ς70 proteins returned growth to the level achieved by the control motA+ strain, again indicating partial suppression.
We next quantitated in vivo suppression by each asiA mutation by measuring burst size (average number of phage progeny produced from each infected cell). Each of the asiA mutations caused an increase in burst size compared to the wild-type asiA strain, but the burst sizes were still quite low compared to the motA+ infection (Table 3). To measure changes in burst size with the rpoD mutants, cells harboring either wild-type or mutant ς70 expression plasmid were infected with T4 motA-pc1. As with the asiA suppressors, each mutant ς70 caused a several-fold increase in burst size, but it was still lower than that of a motA+ infection (Table 3). The ς70 in vivo assays may have been affected by the presence of wild-type ς70 protein expressed from the genomic copy of rpoD. (For unknown reasons, we were unable to construct a TabG strain in which the trp promoter controlled expression of the genomic copy of rpoD.) In summary, plaque and burst size measurements demonstrate that the two asiA mutations and the five rpoD mutations allow partial suppression of the motA-pc1 defect in vivo.
TABLE 3.
Burst size analysis of suppressor mutantsa
| T4 strain | Plasmid gene | Burst size |
|---|---|---|
| K10 (motA+ asiA+) | supF | 45 |
| K10 motA-pc1 (asiA+) | supF | 0.4 |
| K10 motA-pc1 asiA-S22F | supF | 2.0 |
| K10 motA-pc1 asiA-Q51E | supF | 1.3 |
| T4 denAB (motA+ asiA+) | rpoD+ | 73 |
| T4 denAB motA-pc1 | rpoD+ | 0.5 |
| T4 denAB motA-pc1 | rpoD-D570N | 2.6 |
| T4 denAB motA-pc1 | rpoD-Y571C | 4.6 |
| T4 denAB motA-pc1 | rpoD-Y571H | 5.5 |
| T4 denAB motA-pc1 | rpoD-L595P | 2.8 |
| T4 denAB motA-pc1 | rpoD-S604P | 2.3 |
The host cells for the infections with K10 and its derivatives contained plasmid pBSPLO+, which carries supF and thereby allows suppression of the amber mutations in this phage background. The host cells for the infections with T4 denAB and its derivatives contained a plasmid encoding either wild-type or mutant ς70 (rpoD gene), along with the normal (wild-type) chromosomal rpoD gene.
Transcription from T4 middle-mode promoters.
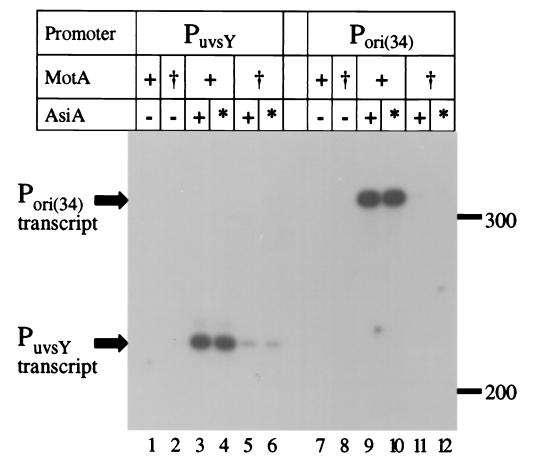
We next attempted to measure suppression of the MotA-pc1 defect with in vitro transcription assays using purified proteins (see Materials and Methods). Activation was tested using T4-modified (glucosylated hydroxymethyl-cytosine-containing) templates with either of two T4 middle-mode promoters (PuvsY and Pori(34)) in the presence of RNA polymerase holoenzyme, wild-type or mutant MotA, and wild-type or S22F mutant AsiA. As expected from previous work (28), transcription did not occur in the absence of AsiA protein (Fig. 3, lanes 1, 2, 7, and 8). Wild-type MotA induced a large amount of transcription with either AsiA protein, indicating that the S22F substitution does not affect activation with wild-type MotA (lanes 3, 4, 9, and 10). Transcription was dramatically decreased in the presence of MotA-pc1 and wild-type AsiA (lanes 5 and 11), as previously reported (10). However, we did not detect an increase in transcription when the AsiA-S22F was present with MotA-pc1 (lanes 6 and 12).
FIG. 3.
In vitro transcription from T4 middle promoters with AsiA or AsiA-S22F. Transcription assays contained RNA polymerase holoenzyme, wild-type MotA (+) or MotA-pc1 (†) (2 pmol each), and wild-type AsiA (+) or AsiA-S22F (∗) (2 pmol each) as indicated. Each T4-modified DNA template contained the middle promoter indicated at the top. Size standards (in bases) are indicated to the right of the gel.
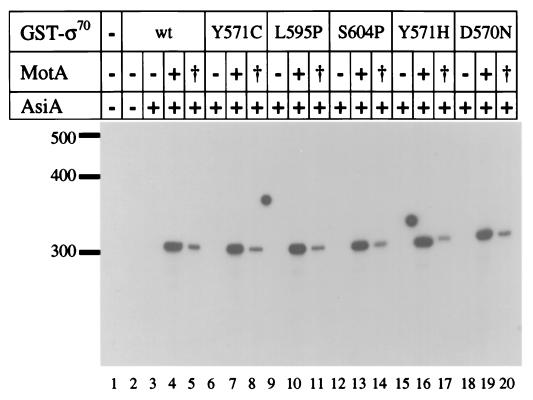
We also analyzed transcription with the mutant ς70 proteins. Wild-type and each mutant ς70 were overexpressed and purified as GST fusions (see Materials and Methods). GST-ς70 fusions have been analyzed in previous studies and found to behave much like native ς70 (7). Transcription was tested using the Pori(34) substrate with wild-type or mutant GST-ς70, core RNA polymerase, MotA (wild type or pc1) and wild-type AsiA (Fig. 4). Comparable amounts of transcription were induced with each ς70 protein and wild-type MotA protein, indicating that the mutations in ς70 do not affect activation with wild-type MotA (compare lane 4 with lanes 7, 10, 13, 16, and 19). Also, transcription with each GST-ς70 still required MotA (lanes 3, 6, 9, 12, 15, and 18). As above, transcription was reduced with MotA-pc1 and wild-type ς70 (lane 5). Once again we did not observe a noticeable suppression of the transcription defect with any mutant ς70 compared to wild type (compare lane 5 with lanes 8, 11, 14, 17, and 20). We also failed to observe suppression with a template containing the PuvsY middle promoter and in assays that contained AsiA-S22F and either ς70-L595P or ς70-D570N (data not shown).
FIG. 4.
In vitro transcription with ς70 suppressor mutants from the T4 ori(34) promoter. Reactions contained RNA polymerase core enzyme, AsiA (2 pmol, where indicated), wild-type MotA (+) or MotA-pc1 (†) (2 pmol each), and the indicated GST-ς70 fusion protein (0.5 pmol). Runoff transcripts (320 bases) were synthesized from a linear T4-modified DNA template containing the ori(34) middle promoter. Size standards (in bases) are indicated to the left of the gel.
The transcription assay may not be able to measure suppression for any number of reasons. First, the composition of the transcription buffer may prevent the mutations from suppressing the activation defect, especially if suppression occurs by a weak protein-protein interaction (although we failed to detect suppression with a variety of salt concentrations; data not shown). Second, the unmodified RNA polymerase may be lacking a T4-directed modification required to obtain suppression, even though unmodified polymerase is adequate for observing MotA-dependent activation from middle promoters. RNA polymerase undergoes several modifications during a T4 infection, including ADP ribosylation of the α subunits and binding of T4-encoded RpbA and Alc proteins (for review, see reference 37). Third, suppression may require RNA polymerase from strain TabG, since that strain was used for in vivo selections and suppression tests (TabG apparently contains a mutation within or closely linked to the gene for the β subunit of RNA polymerase [29]). Fourth, suppression may occur only at a subset of middle promoters, and we tested only two. Middle promoters are used to express a number of essential replication proteins, and a large suppression effect at one or two promoters may be adequate to cause the in vivo phenotype. Fifth, suppression may not affect transcription per se, but rather activation of DNA replication at T4 origins (which contain middle-mode promoters). Indeed, restoration of origin function was critical for the ς70 suppressor selection.
Transcription of E. coli promoters with mutant ς70 proteins.
To better understand the nature of the ς70 mutants, transcription assays were performed using E. coli promoters in the absence of T4 proteins. We first analyzed transcription from the tac promoter, which has strong −10 and −35 consensus elements (but no extended −10 element). Transcription by core RNA polymerase was strictly dependent on ς70 (Fig. 5A, lanes 1 and 2), and nearly normal transcription occurred with the Y571C, Y571H, and D570N mutants (lanes 3, 6, and 7; see figure legend). However, the L595P and S604P substitutions caused dramatic defects in transcription from Ptac (<1 and 15%, respectively, compared to wild-type ς70; lanes 4 and 5). The defects caused by the proline substitutions were surprising since these substitutions did not reduce transcription from T4 middle promoters.
FIG. 5.
In vitro transcription with ς70 mutants from the tac and T4 uvsY promoters. Reactions contained RNA polymerase core enzyme and the indicated GST-ς70 protein (0.5 pmol). Runoff transcripts were synthesized from a linear template containing either the tac promoter (A) (320-base transcript) or the T4 uvsY promoter (B) (221-base transcript, indicated by the arrow; the template was not T4 modified). Size standards (in bases) are indicated to the left of the gel. The heavy spot at the bottom left corner (just above the numeral 2) is a film artifact.
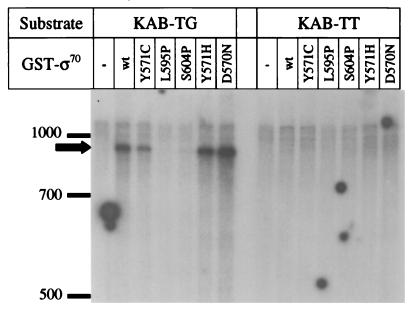
The two proline substitutions are at the C-terminal end of the 4.2 region, which recognizes the −35 sequence of promoters and is predicted to form a helix-turn-helix structure. The proline substitutions may simply change the structural conformation of the domain and block the ability of ς70 to recognize the −35 sequence, thus preventing transcription from Ptac. We analyzed this possibility by measuring transcription from extended −10 promoters. These promoters contain a 5′-TG-3′ positioned at −15 and −14 and do not require a −35 sequence for activity (3). If the proline substitutions block recognition solely of the −35 sequence, normal transcription should occur with an extended −10 promoter. The T4 uvsY promoter contains an extended −10 sequence and no consensus −35 sequence (37). As predicted from previous experiments (30), transcription from PuvsY occurred without MotA when AsiA was not included in the reaction and the template was not T4 modified (Fig. 5B, lane 2). Results with the ς70 substitution mutants were nearly identical to those with the tac promoter (compare Fig. 5A and B). Most importantly, the L595P and S604P substitutions greatly decreased transcription (<1 and 10%, respectively, compared to wild type [lanes 2, 4, and 5]).
This result was confirmed using a second substrate, KAB-TG, which contains a semisynthetic promoter that is dependent on its extended −10 sequence (1). The proline substitutions greatly decreased transcription from this extended −10 promoter (Fig. 6, left lanes). As a control, we also tested a second substrate, KAB-TT, which has a T nucleotide substituted at position −14, thus eliminating the extended −10 element. As expected (1), transcription did not occur with the KAB-TT substrate with any ς70 protein, demonstrating that efficient transcription of KAB-TG depends on the extended −10 sequence (Fig. 6, right lanes). The main conclusion is that the two proline substitutions, at least in the context of these GST fusions, cause a major defect in transcription from both standard E. coli promoters and those with an extended −10 and yet do not block MotA-activated T4 middle-mode transcription.
FIG. 6.
In vitro transcription with ς70 mutants from KAB substrates. Reactions contained RNA polymerase core enzyme and the indicated GST-ς70 fusion protein (0.5 pmol). The substrates contained either the extended −10 promoter (KAB-TG, left side) or a variant without the extended −10 element (KAB-TT, right side). The arrow indicates the runoff transcripts, roughly 900 bases in length. With the extended −10 substrate, GST-ς70 Y571C produced about half as many transcripts as wild-type ς70 and D570N produced about twice as many as wild type. Transcripts were not detected with any of the ς70 proteins from the KAB-TT substrate. Size standards (in bases) are indicated to the left of the gel, and the round spots are film artifacts.
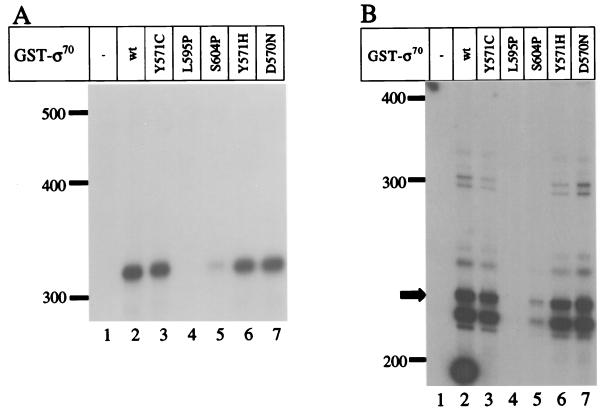
The proline substitutions affect the interaction of sigma with core.
Previous studies have implicated 4.2 as one of several ς70 regions that are involved in the interaction of the protein with core RNA polymerase (27, 36). At present, it is not clear which subunit(s) of core polymerase interacts with the 4.2 region. One possible model to explain the transcriptional defect of the proline substitution mutants is that the altered ς70 is unable to interact with core RNA polymerase. We used a competitive binding assay to determine the relative binding affinity of each mutant ς70 compared to the wild-type protein. His-tagged ς70* labeled with 32P (by a kinase reaction) was competed against increasing concentrations (250 nM to 2 μM) of nonradioactive wild-type or mutant ς70 for binding to a limiting amount of core. The complexes were separated by immunoprecipitation with an antibody directed against core and then analyzed for the amount of radioactive ς70 bound. As expected (36), nonradioactive His-WT competed strongly, with nearly complete competition at a 20-fold molar excess (Fig. 7). The GST-tagged wild-type ς70 also competed well, though not as strongly as the His-tagged protein, consistent with results from previous competition experiments (M. M. Sharp and C. A. Gross, unpublished data). The D570N, Y571C, and Y571H mutant proteins all competed nearly as well as the wild-type GST fusion protein, consistent with their ability to transcribe E. coli promoters. Interestingly, however, the L595P and S604P proteins both showed strong core binding defects (Fig. 7). These two proteins were essentially unable to compete under the conditions of this assay. The lack of competition of these two mutant proteins cannot be attributed to inactive protein because each induces MotA-dependent transcription from T4 middle-mode promoters normally (Fig. 4).
The binding assay can be quantified by comparing the concentrations of wild-type and mutant ς70 required for equivalent competition. We can reliably quantify our results in the range of 25 to 90% of ς70* competed, thus setting upper and lower limits for the assay (36). Since 2 μM concentrations of the GST-S604P and GST-L595P proteins were unable to compete but 250 nM GST-WT showed significant competition, the mutants exhibit a specific core-binding defect of greater than 10-fold.
The transcriptional defects of the S604P and L595P proteins may therefore be caused by an inability to bind or alteration in binding to core polymerase. Unlike the proline substitution, the alanine substitution of S604 did not suppress the motA-pc1 mutation, nor did it exhibit a core-binding defect (data not shown). This suggests that the proline substitutions may be disrupting the region 4.2 helix, causing localized misfolding of a region required to interact with the core. Nonetheless, the mutant 4.2 region must still be able to interact with MotA and/or AsiA, because the proline substitutions do not block MotA-dependent transcription from T4 middle-mode promoters (Fig. 4). The proficiency in T4 middle-mode transcription also demonstrates that the S604P and L595P proteins can interact productively with the rest of the transcription complex under some conditions and that the proline substitutions do not result in gross misfolding of ς70. The presence of the T4 activator proteins apparently overcomes the generalized defect in the proline-substitution mutants. Perhaps AsiA and or MotA also contacts the core, replacing the defective interaction, or alters the structure of ς70 so that it can interact productively with the core.
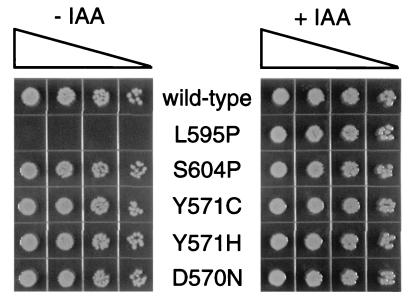
ς70 L595P does not support E. coli growth.
Finally, we also tested whether the mutant ς70 proteins are sufficient for E. coli viability. An E. coli strain with the chromosomal rpoD under the control of a trp promoter (and no other source of ς70) is inviable on rich media unless the trp repressor is inactivated by the antagonist indole-3-acrylic acid (23). As expected from the past study, a plasmid expressing the wild-type GST-ς70 rescued survival in the absence of indole-acrylic acid (Fig. 8, top row). We found that the L595P fusion protein was not able to rescue survival, correlating to its gross defect in transcription. However, expression of any of the other four mutants, including S604P, allowed apparently normal growth (Fig. 8). The small amount of residual transcription by the S604P mutant (Fig. 5) is apparently sufficient for cell viability.
FIG. 8.
E. coli viability assay for GST-ς70 mutants. Wild-type and mutant GST-ς70 expression plasmids were transformed into MCS1 ptrp-rpoD cells (see Materials and Methods). After outgrowth, 4-μl aliquots of fourfold serial dilutions of each transformation were spotted onto L broth plates that contained ampicillin to select for the plasmid, either with or without 0.2 mM indole-3-acrylic acid (IAA) to induce ς70 expression from the chromosomal ptrp-rpoD (provides wild-type ς70).
Summary.
We isolated two asiA and five rpoD mutations that suppress the motA-pc1 activation defect. Each of the seven mutations caused partial suppression in vivo, but suppression was not detected with in vitro transcription assays. Nonetheless, two of the ς70 mutants, L595P and S604P, had novel biochemical properties. Both proteins showed greatly reduced transcription from E. coli promoters (both normal and extended −10) and defective binding to core polymerase, yet were fully functional for MotA-dependent transcription from T4 middle-mode promoters.
ACKNOWLEDGMENTS
We thank Wilma Ross for important discussions and for sharing unpublished results. K.N.K. and M.P.C. were supported by grant GM34622 from the NIH, and M.P.C. was supported in part by Training Grant T32 CA09111. C.A.G. and M.M.S. were supported by grant GM30477 from the NIH.
REFERENCES
- 1.Barne K A, Bown J A, Busby S J W, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase ς70 subunit is responsible for the recognition of the ‘extended −10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson K H, Kreuzer K N. Role of MotA transcription factor in bacteriophage T4 DNA replication. J Mol Biol. 1992;228:88–100. doi: 10.1016/0022-2836(92)90493-4. [DOI] [PubMed] [Google Scholar]
- 3.Bown J, Barne K, Minchin S, Busby S. Extended −10 promoters. Nucleic Acids Mol Biol. 1997;11:41–52. [Google Scholar]
- 4.Carles-Kinch K, George J W, Kreuzer K N. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 1997;16:4142–4151. doi: 10.1093/emboj/16.13.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cicero M P. Macromolecular interactions required for activation of bacteriophage T4 middle promoters. Ph.D. thesis. Durham, N.C: Duke University; 1999. [Google Scholar]
- 6.Colland F, Orsini G, Brody E N, Buc H, Kolb A. The bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol Microbiol. 1998;27:819–829. doi: 10.1046/j.1365-2958.1998.00729.x. [DOI] [PubMed] [Google Scholar]
- 7.Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription initiation factor ς70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 8.Drake J W, Ripley L S. Induced mutagenesis and isolation of T4 mutants. In: Karam J D, editor. Molecular biology of bacteriophage T4. Washington, D.C.: ASM Press; 1994. pp. 447–451. [Google Scholar]
- 9.Finnin M S, Hoffman D W, White S W. The DNA binding domain of the MotA transcription factor from bacteriophage T4 shows structural similarity to the TATA-binding protein. Proc Natl Acad Sci USA. 1994;91:10972–10976. doi: 10.1073/pnas.91.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finnin M S, Cicero M P, Davies C, Porter S J, White S W, Kreuzer K N. MotA transcription factor from bacteriophage T4 has an acidic activation domain. EMBO J. 1997;16:1992–2003. doi: 10.1093/emboj/16.8.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerber J S, Hinton D H. An N-terminal mutation in the bacteriophage T4 motA gene yields a protein that binds DNA but is defective for activation of transcription. J Bacteriol. 1996;178:6133–6139. doi: 10.1128/jb.178.21.6133-6139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross C A, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 13.Hinton D M. Transcription from bacteriophage T4 middle promoter using T4 MotA protein and phage-modified RNA polymerase. J Biol Chem. 1991;266:18034–18044. [PubMed] [Google Scholar]
- 14.Hinton D M, March-Amegadzie R, Gerber J S, Sharma M. The bacteriophage T4 middle transcription system: T4-modified RNA polymerase, AsiA (sigma-70 binding protein), and the transcriptional activator MotA. Methods Enzymol. 1996;274:43–57. doi: 10.1016/s0076-6879(96)74007-7. [DOI] [PubMed] [Google Scholar]
- 15.Hu J C, Gross C A. Mutations in rpoD that increase expression of genes in the mal regulon of Escherichia coli K-12. J Mol Biol. 1988;203:15–27. doi: 10.1016/0022-2836(88)90087-3. [DOI] [PubMed] [Google Scholar]
- 16.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S-K, Makino K, Amemura M, Nakata A, Shinagawa H. Mutational analysis of the role of the first helix of region 4.2 of the ς70 subunit of Escherichia coli RNA polymerase in transcriptional activation by activator protein PhoB. Mol Gen Genet. 1995;248:1–8. doi: 10.1007/BF02456607. [DOI] [PubMed] [Google Scholar]
- 18.Kreuzer K N, Alberts B M. A defective phage system reveals bacteriophage T4 replication origins that coincide with recombination hot spots. Proc Natl Acad Sci USA. 1985;82:3345–3349. doi: 10.1073/pnas.82.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreuzer K N, Alberts B M. Characterization of a defective phage system for the analysis of bacteriophage T4 replication origins. J Mol Biol. 1986;188:185–198. doi: 10.1016/0022-2836(86)90303-7. [DOI] [PubMed] [Google Scholar]
- 20.Kuldell N, Hochschild A. Amino acid substitutions in the −35 recognition motif of ς70 that result in defects in phage λ repressor-stimulated transcription. J Bacteriol. 1994;176:2991–2998. doi: 10.1128/jb.176.10.2991-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Grimes B, Fujita N, Makino K, Malloch R A, Hayward R S, Ishihama A. Role of the sigma70 subunit of Escherichia coli RNA polymerase in transcription activation. J Mol Biol. 1994;235:405–413. doi: 10.1006/jmbi.1994.1001. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Moyle H, Susskind M M. Target of transcriptional activation function of phage λ cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 23.Lonetto M A, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase ς70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 24.Makino K, Amemura M, Kim S-K, Nakata A, Shinagawa H. Role of the ς70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 1993;7:149–160. doi: 10.1101/gad.7.1.149. [DOI] [PubMed] [Google Scholar]
- 25.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 26.Menkens A E, Kreuzer K N. Deletion analysis of bacteriophage T4 tertiary origins. J Biol Chem. 1988;263:11358–11365. [PubMed] [Google Scholar]
- 27.Nagai H, Shimamoto N. Regions of the Escherichia coli primary sigma factor ς70 that are involved in interaction with RNA polymerase core enzyme. Genes Cells. 1997;2:725–734. doi: 10.1046/j.1365-2443.1997.1600357.x. [DOI] [PubMed] [Google Scholar]
- 28.Ouhammouch M, Adelman K, Harvey S R, Orsini G, Brody E D. Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc Natl Acad Sci USA. 1995;92:1451–1455. doi: 10.1073/pnas.92.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pulitzer J F, Coppo A, Caruso M. Host-virus interactions in the control of T4 prereplicative transcription. II. Interaction between tabC(rho) mutants and T4 mot mutants. J Mol Biol. 1979;135:979–997. doi: 10.1016/0022-2836(79)90523-0. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt R P, Kreuzer K N. Purified MotA protein binds the −30 region of a bacteriophage T4 middle promoter and activates transcription in vitro. J Biol Chem. 1992;267:11399–11407. [PubMed] [Google Scholar]
- 31.Selick H E, Kreuzer K N, Alberts B M. The bacteriophage T4 insertion/substitution vector system. J Biol Chem. 1988;263:11336–11347. [PubMed] [Google Scholar]
- 32.Severinov K, Muir T W. Expressed protein ligation, a novel method for studying protein-protein interactions in transcription. J Biol Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 33.Severinova E, Severinov K, Darst S A. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J Mol Biol. 1998;279:9–18. doi: 10.1006/jmbi.1998.1742. [DOI] [PubMed] [Google Scholar]
- 34.Severinova E, Severinov K, Fenyo D, Marr M, Brody E N, Roberts J W, Chait B T, Darst S A. Domain organization of the Escherichia coli RNA polymerase ς70 subunit. J Mol Biol. 1996;263:637–647. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- 35.Sharma U K, Ravishankar S, Shandil R K, Praveen P V, Balganesh T S. Study of the interaction between bacteriophage T4 asiA and Escherichia coli ς70, using the yeast two-hybrid system: neutralization of asiA toxicity to E. coli cells by coexpression of a truncated ς70 fragment. J Bacteriol. 1999;181:5855–5859. doi: 10.1128/jb.181.18.5855-5859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp M M, Chan C L, Lu C Z, Marr M T, Nechaev S, Merritt E W, Severinov K, Roberts J W, Gross C A. The interface of ς with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stitt B, Hinton D. Regulation of middle-mode transcription. In: Karam J D, editor. Molecular biology of bacteriophage T4. Washington, D.C.: ASM Press; 1994. pp. 142–160. [Google Scholar]
- 38.Watanabe M, Nohmi T, Ohta T. Effects of umuDC, mucAB, and samAB operons on the mutational specificity of chemical mutagenesis in Escherichia coli: II. Base substitution mutagenesis. Mutat Res DNA Repair. 1994;314:39–49. doi: 10.1016/0921-8777(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, Zhang X, Ebright R H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]