Abstract
The coccoid cyanobacterium Chroococcidiopsis dominates microbial communities in the most extreme arid hot and cold deserts. These communities withstand constraints that result from multiple cycles of drying and wetting and/or prolonged desiccation, through mechanisms which remain poorly understood. Here we describe the first system for genetic manipulation of Chroococcidiopsis. Plasmids pDUCA7 and pRL489, based on the pDU1 replicon of Nostoc sp. strain PCC 7524, were transferred to different isolates of Chroococcidiopsis via conjugation and electroporation. This report provides the first evidence that pDU1 replicons can be maintained in cyanobacteria other than Nostoc and Anabaena. Following conjugation, both plasmids replicated in Chroococcidiopsis sp. strains 029, 057, and 123 but not in strains 171 and 584. Both plasmids were electroporated into strains 029 and 123 but not into strains 057, 171, and 584. Expression of PpsbA-luxAB on pRL489 was visualized through in vivo luminescence. Efficiencies of conjugative transfer for pDUCA7 and pRL489 into Chroococcidiopsis sp. strain 029 were approximately 10−2 and 10−4 transconjugants per recipient cell, respectively. Conjugative transfer occurred with a lower efficiency into strains 057 and 123. Electrotransformation efficiencies of about 10−4 electrotransformants per recipient cell were achieved with strains 029 and 123, using either pDUCA7 or pRL489. Extracellular deoxyribonucleases were associated with each of the five strains. Phylogenetic analysis, based upon the V6 to V8 variable regions of 16S rRNA, suggests that desert strains 057, 123, 171, and 029 are distinct from the type species strain Chroococcidiopsis thermalis PCC 7203. The high efficiency of conjugative transfer of Chroococcidiopsis sp. strain 029, from the Negev Desert, Israel, makes this a suitable experimental strain for genetic studies on desiccation tolerance.
Desiccation damages membranes, proteins, and nucleic acids and is lethal to the majority of organisms. Some organisms, the anhydrobiotes, withstand the physiological constraints which result from multiple cycles of drying and wetting and/or prolonged desiccation, and they resume metabolism when water becomes available. How they do so poses provocative questions (7). Desiccation tolerance of cyanobacteria is of particular interest because these phototrophs produce intracellular oxygen that can generate reactive oxygen species (30). Two cyanobacteria, Nostoc commune and Chroococcidiopsis, are the subjects of studies aimed at an understanding of desiccation tolerance (29). Chroococcidiopsis is characteristically the dominant photosynthetic form in microbial habitats of extreme arid cold and hot deserts, and in the most extreme of these environments, it is the sole photosynthetic microorganism. These microbial communities live in airspaces of porous rocks or in microscopic fissures of weathering rocks or form biofilms at the stone-soil interface under pebbles in desert pavements (16). Most of the time, the cells are desiccated or frozen. Long-term measurements show that in the ice-free Ross desert of Antarctica, cells are wetted and metabolically active for a total of 500 to 800 h per year (17). In the most arid areas of hot deserts such as the Negev Desert, Israel, the number of metabolically active hours per year is probably even less than this (E. I. Friedmann and C. P. McKay, personal communication).
Despite an interest in the strategies employed by anhydrobiotes, desiccation tolerance is still poorly understood. What is clear is that desiccation tolerance reflects numerous different structural, physiological, and molecular mechanisms (3, 7, 29, 30). One mechanism shared by anhydrobiotes is the accumulation of trehalose and sucrose, two nonreducing sugars which replace the structural water of cellular components, thus circumventing lethal damage during drying (7). The production of both these compounds has been reported for several cyanobacteria, including Chroococcidiopsis spp. (22) and Nostoc commune (29). Chroococcidiopsis spp. and N. commune share several features that may account for their extreme tolerance of desiccation. Both produce abundant exocellular polysaccharides which may play a central role in desiccation tolerance of cells by regulating the loss and uptake of water (8, 23, 30). Ultrastructural and cytological studies of laboratory- and field-dried cells of Chroococcidiopsis spp. suggested that the amounts of acid-, sulfate-, and beta-linked polysaccharides in the cell envelope increase in response to water deficit (18, 20). In dried cells of N. commune the extracellular polysaccharide (a complex glycan) provides an immobilization matrix in which secreted enzymes remain active (36) and where UV-absorbing pigments accumulate for photoprotection (29, 30). d-Ribose and 3-O-lactyl glucuronic acid in the glycan of N. commune DHR1 are thought to influence the rheological properties of the extracellular matrix upon desiccation and rehydration (21). The presence of abundant Fe-superoxide dismutase in dried cells of N. commune CHEN1986 (36) and in a desert strain of a Chroococcidiopsis sp. (19) suggests that an important mechanism in desiccation tolerance is the minimization of the risk of hydroxyl radical formation (30). A capacity to withstand γ-radiation is thought to be an incidental consequence of the ability to repair DNA damage that results from desiccation (30). The ability of desert strains of Chroococcidiopsis to withstand exposure to 5 kGy of X-rays (1 kGy = 0.1 megarad), with survival reduced by 1 or 2 orders of magnitude, emphasizes their capacity for DNA repair (1).
It is likely that desiccation tolerance involves the action of a large number of genes possibly acting in parallel pathways. Understanding the molecular basis for desiccation tolerance is therefore a significant challenge. Although sophisticated genetic systems are available for the analysis of cyanobacteria, only a few strains are currently the subject of genetic manipulations (37); none of these strains shows vigor in response to extremes of water deficit. It is not yet clear whether this situation is attributable to the successes achieved with these strains or the fact that many strains are simply unsuitable for genetic manipulation (37). We investigated the possibility of gene transfer via conjugation and electroporation in different strains of Chroococcidiopsis spp. isolated from geographically remote deserts. Chroococcidiopsis sp. strain 029 from the Negev Desert, Israel, was identified as a conjugally efficient strain, and the possibility of using this strain to represent desert populations was assessed through phylogenetic analysis (based upon variable regions V6 to V8 of 16S rRNA genes [32]).
MATERIALS AND METHODS
Microorganisms and growth conditions.
Five Chroococcidiopsis sp. strains from hot and cold deserts were obtained from the Culture Collection of Microorganisms from Extreme Environments (CCMEE) at Florida State University (now located at the University of Oregon, Eugene) (Table 1). Chroococcidiopsis sp. strain PCC 7203 was obtained from the American Type Culture Collection (ATCC 27900). Anabaena sp. strain PCC 7120 (from Jeff Elhai) was used for control purposes.
TABLE 1.
Chroococcidiopsis sp. strains
| Strain no.a | Strain history |
|---|---|
| 029 (CCMEE N6904) | Chroococcidiopsis sp. strain; cryptoendolithic, found in airspaces of porous rocks, forming in a zone a few millimeters wide under the rock crust in Nubian sandstone, Negev Desert, Makhtesh Ramon, Israel |
| 057 (CCMEE S 6e) | Chroococcidiopsis sp. strain; chasmoendolithic, found in microscopic fissures of weathering rocks, near the surface in granite, Sinai Desert, Egypt |
| 123 (CCMEE CH-7) | Chroococcidiopsis sp. strain; chasmoendolithic, found in granite boulder, coastal desert, Guanaqueros Peninsula, Coquimbo, Chile |
| 171 (CCMEE A789-2) | Chroococcidiopsis sp. strain; cryptoendolithic; found in Beacon sandstone, Ross Desert (McMurdo Dry Valleys) University Valley, Antarctica |
| 584 (CCMEE Gobi 91-19) | Chroococcidiopsis sp. strain; hypolithic; found under pebbles of a desert pavement, forming biofilms at the stone-soil interface under stones of desert pavement, Gobi Desert near the Tian Shan mountain range of Mongolia |
| ATCC 27900 | C. thermalis (see reference 31), from soil in East Germany (PCC 7203) |
For the five strains obtained from the CCMEE, the CCMEE reference number is given in parentheses.
All cyanobacterial strains were grown at room temperature, in liquid BG-11 medium (31), with a photon flux density of 90 μmol m−2 s−1 provided by fluorescent cool-white lamps. Escherichia coli strains were grown in Luria-Bertani (LB) medium (34) supplemented with ampicillin (100 μg ml−1), kanamycin sulfate (20 μg ml−1), or chloramphenicol (25 μg ml−1), as appropriate.
Conjugation.
Cargo plasmids pDUCA7 and pRL489 (Table 2) intended for transfer to the cyanobacterial host were first replicated in an E. coli strain, HB101, bearing the helper plasmid pRL528, which carries genes encoding DNA methylases (Table 2). Triparental mating was performed as follows: E. coli strain HB101 bearing a conjugative plasmid and E. coli strain HB101 bearing both a cargo plasmid and helper plasmid were grown overnight. Cells from 1-ml aliquots (1 ml for each mating) were washed twice with one volume of LB medium (without antibiotics) and resuspended in 200 μl of LB medium. One-milliliter aliquots of each Chroococcidiopsis sp. culture (ranging in age from 4 weeks to 2 months; 106 to 107 cells ml−1) were centrifuged, and the cells were resuspended in 100 μl of BG-11 medium. A 100-μl volume of the E. coli mixture (prepared as described above) was added to 100 μl of a cyanobacterial cell suspension, and 5-μl aliquots were spotted on Nuclepore REC-85 filters resting on BG-11 agar (1.5%, wt/vol). The donor 1-donor 2-recipient cell ratio was approximately 10:10:1. Alternatively, recipient cyanobacterial cells were diluted serially (1:104) before the E. coli mixture was added. Matings were carried out for 48 h under conditions used for growth of Chroococcidiopsis spp. Triparental spot matings were also performed as described above, using E. coli cells bearing pDUCA7 but lacking the helper plasmid pRL528. In other controls, spot matings were performed using 106 cells of each Chroococcidiopsis sp. strain mixed with 105 cells of E. coli bearing either the conjugative plasmid or the cargo plasmid.
TABLE 2.
Plasmids
| Plasmid(s) | Relevant characteristics | Source (reference) |
|---|---|---|
| pRL528 | Helper plasmid (Cm1), ColK oriV, carries the genes for M.AvaI and M.Eco47II and mob gene from ColK for the mobilization of pMB1-based plasmida | J. Elhai (12) |
| RK2 and RP4 | Conjugative plasmids (Apr Kmr Tcr), RK2 oriV, RK2 oriT, provide tra genes and nicking function from RK2; IncPα | J. Elhai (39) |
| pRL443 | Spontaneous Kms mutant of RP4 (Apr Tcr) | J. Elhai (12) |
| pRK2013 | RK2 derivative (Kmr), colE1 oriV replaces RK2 oriV | J. Elhai (15) |
| pDUCA7 | Cargo plasmid (Kmr), RK2 oriV, RK2 oriT, pDU1 repliconb | J. C. Meeks (4) |
| pRL489c | Cargo plasmid (Kmr), pMB1 oriV, pMB1 oriT, pDU1 replicon,b PpsbA-luxAB | J Elhai (9) |
Derivatives of pMB1, a ColE1-like plasmid, retain bom site (basis of mobility or oriT) but not the mob gene, which encodes a DNA-nicking function. These derivatives can be transferred only when a mob product is provided in trans, by a helper plasmid.
Cyanobacterial origin of replicon from pDU1, a plasmid from Nostoc sp. strain PCC 7524.
PpsbA is the promoter of photosystem II gene psbA, from A. hybridus.
Selection of Chroococcidiopsis transconjugants was performed by transferring the filters to BG-11 agar containing neomycin (400 μg ml−1) under a photon flux density of 90 μmol m−2 s−1. Purification of transconjugants was performed by counterselecting E. coli through the use of BG-11, which does not support its growth (38). One-month-old single green colonies of putative transconjugants were restreaked twice on selective medium before transfer to liquid BG-11 containing neomycin (100 μg ml−1). Aliquots of the liquid cultures were plated on LB medium to detect the presence of E. coli.
Electroporation.
One-milliliter aliquots were harvested from 1-month-old cultures of each Chroococcidiopsis sp. strain. Cells were collected, washed twice with cold 1 mM HEPES buffer, pH 7.4, and resuspended in 100 μl of the same buffer (about 108 total cells). Cargo plasmids (Table 2) were methylated in vivo using E. coli carrying helper plasmid pRL528 (Table 2). Plasmid DNA was extracted using the Wizard Plus Minipreps DNA Purification System (Promega, Madison, Wis.) and added to 100 μl of the cyanobacterial suspension at a final concentration of approximately of 2.5 μg ml−1. The mixture was placed between the electrodes (0.1-mm gap) of a cold electroporation cuvette and pulsed once in a Gene Pulser Controller (Bio-Rad Laboratories, Richmond, Calif.) at 13 kV cm−1 (25 μF and 200 Ω settings; time constant, ca. 3.5). After electroporation, cyanobacterial cells were resuspended in 2 ml of BG-11 medium and allowed to grow for 24 h. After centrifugation and resuspension in 100 μl of BG-11 medium, 5-μl aliquots were spotted on Nuclepore REC-85 filters and incubated until green colonies appeared. Electrotransformants were used to inoculate liquid BG-11 medium containing neomycin (100 μg ml−1). Control electroporations were performed in the absence of plasmid DNA.
Extracellular deoxyribonuclease assay.
One-milliliter aliquots were harvested from 1-month-old cultures of each cyanobacterial strain, and cells were collected and streaked on BG-11 agar (1.5%, wt/vol) containing 0.3 mg of DNA-methyl green (Sigma Chemical Co., St. Louis, Mo.) per ml and 0.05× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as described previously (41). Petri plates were incubated under cyanobacterial growth conditions.
Extraction of genomic DNA.
Total DNAs were extracted from Chroococcidiopsis sp. cells as described previously (2), except that genomic DNA was not purified through cesium chloride buoyant density ultracentrifugation.
Extraction of plasmid DNA.
Cells from 15-ml aliquots of axenic cultures of transconjugants of Chroococcidiopsis sp. strain 029 and electrotransformants obtained from strains 029 and 123 were washed with 1 ml of TE buffer (1 mM EDTA, pH 8.0; 10 mM Tris-HCl, pH 7.4). Cells (107 cells) were subjected to 30 cycles of freezing in liquid nitrogen and thawing at 50°C, and the mixture was then used directly for plasmid DNA extraction using the Wizard Plus Minipreps DNA Purification System (Promega). After the centrifugation of the cell lysate, the supernatant fraction was mixed with 1 volume of binding mix, prepared by resuspending 250 mg of silica (Sigma) in 100 ml of a solution containing 5 M guanidine thiocyanate and 4% (wt/vol) Triton X-100. After two washes with 50% ethanol and drying in a speed vacuum concentrator, plasmid DNA was eluted from silica gel particles with 30 μl of sterile water. Since the yield of plasmid DNAs was undetectable with agarose gel electrophoresis, transformation of E. coli was used routinely to assay plasmids from Chroococcidiopsis sp. transconjugants and electrotransformants. Typically, 10-μl aliquots were used to electroporate 2 × 109 cells of ElectroMAX E. coli DH10B ml−1 (Novagen-LTI, Rockville, Md.). About 10 colonies of E. coli transformants were selected on LB agar plates containing 20 μg of Km ml−1 (cargo plasmids carry the neomycin phosphotransferase gene, which confers resistance to kanamycin and neomycin).
DNA dot blotting and Southern hybridization.
In order to characterize axenic Chroococcidiopsis sp. transconjugants, plasmid pDUCA7 was labeled by nick translation using alkali-labile digoxigenin (DIG)–11-dUTP (Boehringer Mannheim GmbH) and used as probe. Dot blots were performed using genomic DNAs from transconjugants of strains 029, 057, and 123, and parental strains were used as controls. Southern analyses were performed using genomic DNAs extracted from Chroococcidiopsis sp. 029 transconjugants before and after digestion with PstI. Genomic DNA from wild-type Chroococcidiopsis sp. 029 and authentic pDUCA7 were used as controls. Hybridizations were performed according to protocols specified in the Genius System User's Guide for Membrane Hybridization (Boehringer Mannheim GmbH) using the DIG Easy Hyb Granules System (Boehringer Mannheim GmbH). Hybridization blots were visualized with anti-DIG alkaline phosphatase and the chemiluminescent detection system of Boehringer Mannheim GmbH.
Luciferase detection.
Light emission resulting from the oxidation of n-decanal and catalyzed by luciferase was detected in vivo in cells bearing plasmid pRL489 (Table 2). Colonies were exposed for 5 min to the vapor of several μl of n-decanal spread on the inner surface of the top of the petri dish. Bioluminescence was recorded as digital images using the television camera of a 400 Alpha Innotech ChemImager low-light imaging system operated with Alphase 3.3 software, a Dell Pentium computer, and an Optiquest color monitor.
Amplification of 16S rRNA genes and DNA sequencing.
Genomic DNAs extracted from Chroococcidiopsis sp. strains were used with different sets of primers to amplify the almost complete 16S rRNA genes. The positions in E. coli 16S rRNA that are equivalent to these primer sequences are provided for reference. The forward primer was either the universal primer F2C (5′-AGA GTT TGA TCA/C TGG CTC-3′) or the cyanobacterium-specific primer CYA106F (28) corresponding to E. coli nucleotides 8 to 25 and 106 to 127, respectively. The universal primer C (5′-ACG GGC GGT GTG TAC-3′) corresponding to E. coli positions 1406 to 1392 was used as the reverse primer. Partial sequences were obtained by using the forward universal primer AC (5′-CAG CCG CGG TAA TAC-3′) corresponding to E. coli positions 552 to 536 and the reverse universal primer C. Conditions for the PCR assay were 30 cycles of annealing for 1 min at 40°C and elongation for 3 min at 72°C; amplifications were initiated with a 5-min denaturation at 95°C and ended with a 7-min extension at 72°C. PCR products were purified with the QIAquik PCR purification kit (Qiagen Inc., Chatsworth, Calif.) and used as templates in sequencing reactions with the Applied Biosystems PRISM Dye Terminator Cycle Sequencing Ready reaction kit (Perkin-Elmer). Sequencing reactions were obtained by using the forward primers F2C, CYA106F, and AC and the reverse universal primer C and then were analyzed by using an Applied Biosystem 377 DNA sequencer (Perkin-Elmer).
Phylogenetic reconstruction.
The 16S ribosomal DNA (rDNA) sequences of Chroococcidiopsis sp. strains were aligned manually, and sequences from nucleotides 1 to 480 (corresponding to the numbering of the E. coli 16S rRNA) were used for the analysis.
Phylogenetic analysis.
DNA sequences were first aligned using the MEGALIGN feature of version 4.0 of the LaserGene software (DNASTAR Inc., Madison, Wis.). Phylogenetic trees were constructed based upon parsimony analysis (ordinary parsimony) and distance methods using the Phylogenetic Inference Package (PHYLIP, version 3.57c) obtained from J. Felsenstein, Department of Genetics, University of Washington, Seattle. SEQBOOT was used to produce 100 data sets from bootstrap resampling (14). Majority rule strict consensus analysis was performed with CONSENSE, with the Anabaena sp. strain NIVA-CYA 281 sequence arbitrarily designated as the outgroup. Distance matrices were obtained with the Kimura two-parameter model using the default transition/transversion ratio (26) and calculated with the DNADIST and NEIGHBOR (33) programs of PHYLIP (14). Unrooted trees were plotted in DRAWTREE or DRAWGRAM and edited in Adobe Illustrator version 9.0.
Nucleotide sequence accession numbers.
DNA sequences and annotations of Chroococcidiopsis sp. strains 029, 057, 123, and 171 were deposited in GenBank under accession numbers AF279107, AF279108, AF279109, and AF279110, respectively.
RESULTS
Conjugative transfer in Chroococcidiopsis spp.
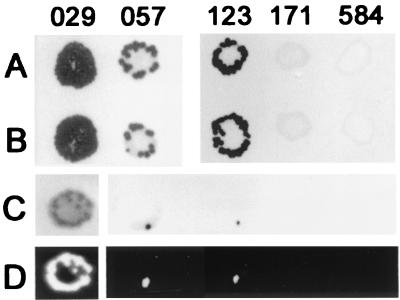
Conjugative transfer of pDUCA7 and pRL489 (Table 2) was achieved in strains 029, 057, and 123 but not in strains 171 and 584 (Table 1). Mobilization of pDUCA7 via pRK2013 yielded green colonies of presumptive transconjugants from Chroococcidiopsis sp. strains 029, 057, and 123, visible on top of a yellow basal deposit of dead cells, about 4 weeks after the start of antibiotic selection (Fig. 1A). The efficiency of conjugation with Chroococcidiopsis sp. strains 057 and 123 was about 10−4 transconjugants per recipient cell, i.e., about 20 to 30 neomycin-resistant colonies out of 105 cells per spot mating (Fig. 1A; Table 3). The efficiency of conjugative transfer of Chroococcidiopsis sp. strain 029 was estimated as about 10−2 transconjugants per recipient cell after serial dilutions of the recipient. No transconjugants were observed in strains 171 and 584 when the number of recipient cells per spot was increased from 105 (Fig. 1A) to 106. The conjugative efficiencies of Chroococcidiopsis sp. strains 029, 057, and 123 were unaffected by the use of E. coli lacking the helper plasmid but bearing plasmid pDUCA7 (Fig. 1B). Plasmid pDUCA7 has an RK2 oriT and requires only the mobilization functions supplied by the conjugative plasmid. Using the same experimental conditions, no neomycin-resistant colonies were obtained from Anabaena sp. strain PCC 7120. Dot blot analysis using pDUCA7 DNA as the probe and total DNA extracted from wild-type Chroococcidiopsis spp. and from axenic liquid cultures obtained from transconjugants of strains 029, 057, and 123 confirmed the presence of pDUCA7 in the transconjugants (not shown). Neomycin-resistant colonies of Chroococcidiopsis sp. strains were never obtained in control experiments in which 105 recipient cells per spot were mixed with 106 E. coli cells bearing either the conjugative plasmid pRK2013 or cargo plasmid pDUCA7. Matings via pRL443 yielded frequencies of transconjugants of Chroococcidiopsis sp. strains 029, 057, and 123 that were 1 order of magnitude lower than those observed using pRK2013. Transconjugants of strain Chroococcidiopsis sp. strain 029 were subjected to further investigation (see below).
FIG. 1.
Typical result of spot matings using Chroococcidiopsis sp. strains 029, 057, 123, 171, and 584. Rows: A, mobilization of plasmid pDUCA7 using conjugative plasmid pK2013 in the presence of helper plasmid pRL528; B, mobilization of plasmid pDUCA7 using conjugative plasmid pK2013 in the absence of helper plasmid pRL528; C, presumptive transconjugants of Chroococcidiopsis cells bearing plasmid pRL489 obtained using conjugative plasmid pRL443 and helper plasmid pRL528; D, luminescence was observed in colonies (see Fig. 1C) obtained from strains 029, 057, and 123 and not in the areas where strains 171 and 584 were used as the recipients. Filters shown were collected after 30 days of incubation.
TABLE 3.
Gene transfer efficienciesa
| Recipient strainb | Conjugative efficiency with:
|
Electroporation efficiency with:
|
||
|---|---|---|---|---|
| pDUCA7c and pRK2013d | pRL489c and pRL443d | pDUCA7c | pRL489c | |
| 029 (CCMEE N6904) | (3 ± 1.2) × 10−2 | (4 ± 0.9) × 10−4 | (3 ± 0.7) × 10−4 | (5 ± 0.4) × 10−4 |
| 057 (CCMEE S6e) | (5 ± 0.6) × 10−4 | (3 ± 0.5) × 10−5 | ND | ND8 |
| 123 (CCME CH-7) | (3 ± 0.4) × 10−4 | (4 ± 0.3) × 10−5 | (5 ± 0.4) × 10−4 | (3 ± 0.7) × 10−4 |
| 171 (CCME A789-2) | ND | ND | ND | ND |
| 584 (CCMEE Gobi 91-19) | ND | ND | ND | ND |
| ATCC 27900 | ND | ND | — | — |
Gene transfer efficiencies are expressed as the numbers of transconjugants or electrotransformants per recipient cell. ND, the result was <10−6 for conjugative efficiencies and <10−8 for electroporation efficiencies; —, not determined. Values are the averages of three or five independent experiments (± the standard error).
For the five strains obtained from the CCMEE, the CCMEE reference number is given in parentheses.
Cargo plasmid.
Conjugative plasmid.
Plasmid pRL489 (Table 2) was mobilized via pRL443 into Chroococcidiopsis sp. strains 029, 057 and 123 but not into strains 171 and 584 (Fig. 1C; Table 3). The efficiency of conjugative transfer of pRL489 into Chroococcidiopsis sp. strain 029 was about 10−4 transconjugants per recipient cell, i.e., 10-fold higher than that into strains 057 and 123 (Fig. 1C; Table 3). When 1-month-old conjugation plates were tested for luciferase activity, luminescence was detected in green neomycin-resistant colonies from strains 029, 057, and 123 (Fig. 1D). Luminescence was not detected in corresponding spots of growth when cells of strain 171 and 584 were used as the recipient cells (Fig. 1D). Neomycin-resistant colonies of Chroococcidiopsis sp. strains were never observed in matings via pRL443 using E. coli cells bearing plasmid pRL489 but lacking plasmid pRL528. Plasmid pRL489 replicates via a pMBI oriT and needs DNA nicking function encoded by the helper plasmid for its mobilization. Aliquots of liquid cultures derived from presumptive transconjugants of Chroococcidiopsis sp. strains 029, 057, and 123 were judged to be axenic after plating on LB medium and incubation at 37°C overnight.
Conjugative transfer of pDUCA7 and pRL489 into Chroococcidiopsis sp. strains 029, 057, and 123 was unaffected by the age (up to 2 months) of the cyanobacterial cells used as the recipients.
Electrotransformation of Chroococcidiopsis spp.
Gene transfer of pDUCA7 and pRL489 into Chroococcidiopsis sp. strains 029 and 123 but not strains 057, 171, and 584 was achieved via electroporation (Table 3). Green colonies of electrotransformants appeared on neomycin-containing BG-11 medium within one month (approximately 10 generations) and were purified further through single-colony isolation. Neomycin-resistant colonies of Chroococcidiopsis strains 029 and 123 were never obtained following electroporation in the absence of plasmid DNA. Electrotransformants of Chroococcidiopsis sp. strains 029 and 123 were subjected to further investigation (see below).
A summary of the efficiencies of transfer of pDUCA7 and pRL489 into Chroococcidiopsis sp. strains via conjugation and electroporation is shown in Table 3. Gene transfer frequencies were evaluated with each cell aggregate or single cell of Chroococcidiopsis sp. considered to be 1 CFU. Though this method may be biased, it is unavoidable due to the complex life cycle of Chroococcidiopsis spp., which is characterized by the occurrence of single cells and multicellular aggregates (see reference 1).
Analysis of Chroococcidiopsis sp. strain 029 transconjugants.
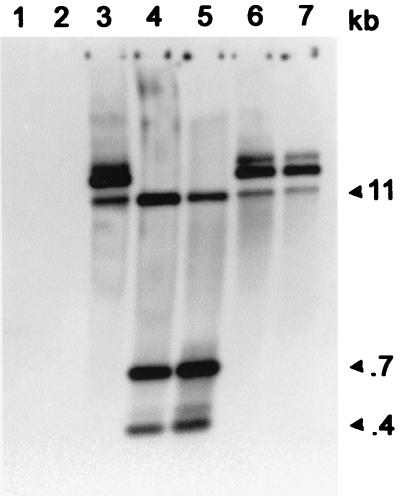
Transconjugants from Chroococcidiopsis sp. strain 029 were investigated further because this strain exhibited the highest efficiency of conjugative transfer (Table 3). Two independent isolates (Chroococcidiopsis sp. strains CH91B1 and CH91B2) were obtained following mobilization of plasmid pDUCA7 via pRK2013 and were analyzed through Southern analysis using pDUCA7 as the probe. Comparable results were obtained in independent trials with each of these isolates. Southern analysis was performed on total DNAs extracted from Chroococcidiopsis sp. strain CH91B1 and Chroococcidiopsis sp. strain 029 (wild type) and plasmid DNA from E. coli strain DH10B transformed with DNA extracted from Chroococcidiopsis sp. strain CH91B1. Hybridization signals were detected from total DNA of Chroococcidiopsis sp. strain CH91B1 (Fig. 2, lane 3), from plasmid DNA of E. coli strain DH10B transformed with DNA extracted from CH91B1 (Fig. 2, lane 6), and from authentic pDUCA7 (Fig. 2, lane 7). DNA extracted from wild-type cells of Chroococcidiopsis sp. strain 029 did not hybridize with the probe (Fig. 2, lane 2). After digestion with PstI the same hybridization pattern was present in total DNA extracted from CH91B1 (Fig. 2, lane 4) and in the authentic pDUCA7 (Fig. 2, lane 5).
FIG. 2.
Southern analysis of Chroococcidiopsis sp. strain 029 transconjugants (isolate CH91B1) obtained after mobilization of pDUCA7. The probe was labelled pDUCA7. Lanes: 1, 1-kb DNA ladder (LTI); 2, total DNA from Chroococcidiopsis sp. strain 029 (wild type); 3, total DNA from Chroococcidiopsis strain 029 isolate CH91B1; 4, total DNA from Chroococcidiopsis strain 029 isolate CH91B1 digested with PstI; 5, authentic pDUCA7 digested with PstI; 6, undigested pDUCA7 from E. coli DH10B transformed with plasmid DNA extracted from Chroococcidiopsis sp. strain 029 isolate CH91B1; 7, authentic pDUCA7.
After digestion with PstI the same pattern of bands was observed in the authentic plasmid pDUCA7 and plasmid extracted from E. coli strain DH10B transformed with extracts obtained from CH91B1 and CH91B2 (not shown). Restriction analysis and Southern analysis confirmed that pDUCA7 replicated autonomously and had not undergone deletion or rearrangement in transconjugants of Chroococcidiopsis sp. strain 029.
Analysis of electrotransformants of Chroococcidiopsis sp. strains 029 and 123.
Identical restriction patterns were found for plasmid pDUCA7 (digested with PstI) and pRL489 (digested with BamHI) replicating in E. coli before and after passage through electrotransformants of Chroococcidiopsis sp. strains 029 and 123 (two independent isolates for each plasmid and strain were analyzed). Restriction analysis confirmed that pDUCA7 and pRL489 replicated in their original form in electrotransformants of Chroococcidiopsis sp. strains 029 and 123.
Extracellular nucleases in Chroococcidiopsis spp.
Nuclease activities were detected in the extracellular fluids of all five Chroococcidiopsis sp. strains (Table 1) as a zone of clearing in agar medium containing DNA-methyl green. After a 10-day incubation, an almost complete clearing was produced by Chroococcidiopsis sp. strain 584 and by Anabaena sp. strain PCC 7120, used as the control, while weak nuclease activities were detected in Chroococcidiopsis sp. strains 029, 057, 123, and 171.
Morphology of Chroococcidiopsis spp.
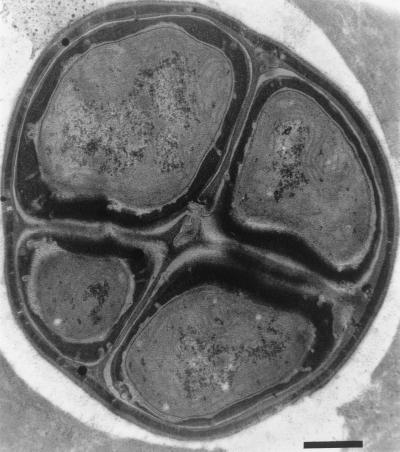
Despite an overall morphological similarity, the five desert strains of Chroococcidiopsis spp. (Table 1) differed in cell size, sheath thickness, and number of cells per aggregate. Chroococcidiopsis sp. strains 029 and 123 showed a similar morphology, while each of the Chroococcidiopsis sp. strains 057, 171, and 584 exhibited different morphologies (not shown). A thick, multilayered envelope surrounding the cells (Fig. 3) characterized 2-month-old cells of Chroococcidiopsis sp. strain 029.
FIG. 3.
Electron micrograph of ultrathin section of 2-month-old Chroococcidiopsis strain 029 showing multicellular aggregate containing cells of different sizes. An electron-translucent layer (white) and a fibrillar electron-dense envelope of capsular polysaccharide (grey) surround the aggregate. Bar = 0.5 μm.
16S rRNA variable regions V6 to V8 and phylogeny of Chroococcidiopsis spp.
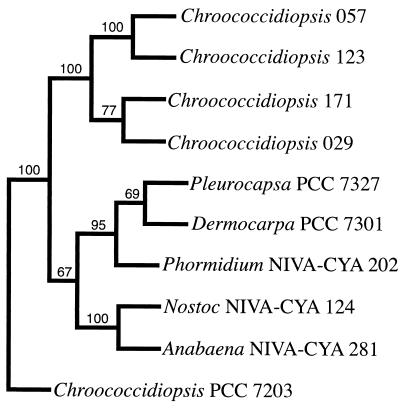
Nearly complete and partial 16S rDNA genes were amplified from Chroococcidiopsis sp. strains 029, 057, 123, and 171 using the primer combinations F2C-C, CYA106-C, and AC-C. High quality sequences were obtained from all the PCR products with sequence similarity to nucleotides 1 to 480 of 16S rRNA of E. coli; these were used for the phylogenetic analysis. The partial 16S rRNA gene sequences of the Chroococcidiopsis sp. strains were compared to those of Chroococcidiopsis thermalis PCC 7203 (EMBL accession no. z82789) and other cyanobacteria. The bottom of the unrooted tree (Fig. 4) is defined by the partial 16S rDNA sequence from Chroococcidiopsis sp. strain PCC 7203. In contrast, the four Chroococcidiopsis sp. strains from the desert form a coherent, distant grouping, which was present in all of the 100 data sets.
FIG. 4.
Phylogenetic analysis of partial 16S rDNA. An unrooted consensus tree is shown. The numbers at the tree forks indicate the number of times out of 100 data sets that the strains to the right of the fork clustered (only numbers greater than 50 are shown).
DISCUSSION
We provide the first report of genetic manipulation of Chroococcidiopsis spp. from hot and cold deserts and the first evidence that pDU1-based replicons can replicate in cyanobacteria of taxonomic section II (31). Until now vectors that include the pDU1 replicon of a plasmid of Nostoc sp. strain PCC 7524 (taxonomic section IV) had not been shown to replicate in strains other than Anabaena and Nostoc strains (35); IncQ plasmids such as pKT210 and pKT230 have been reported to replicate in a variety of unicellular cyanobacteria (37). PpsbA is a chloroplast promoter from Amaranthus hybridus that shares high sequence similarity with the consensus E. coli ς70 promoter and functions as a strong promoter in Anabaena sp. strain PCC 7120 (9). If luxAB gene expression in Chroococcidiopsis spp. is being driven by PpsbA, this promoter may have utility for expressing foreign genes in this cyanobacterium.
It is well known that a major problem in the genetic manipulation of cyanobacteria is the widespread distribution of restriction endonucleases (24). For several cyanobacteria, this problem is overcome by using E. coli strains which carry methylases to first modify plasmid DNA (12). Among the Chroococcidiopsis sp. strains that were amenable to conjugative transfer, strain 029 exhibited the highest frequency of transconjugation. Compared to Chroococcidiopsis sp. strain 029, the conjugative transfer of pDUCA7 via pRK2013 in strains 057 and 123 was 100-fold lower, while that of pRL489 via PRL443, was 10-fold lower. When pDUCA7 was mobilized by pRL443 instead of pRK2013 the efficiency of conjugative transfer was reduced by at least 1 order of magnitude. The higher efficiency of the conjugative transfer of pDUCA7 via pRK2013 might reflect the compatibility between ColE1 oriV present on pRK1023 and RK2 oriV on pDUCA7. This allows both the conjugative and the cargo plasmids to replicate in the same E. coli donor and probably enhances the transfer frequency. The mobilization of pDUCA7 into Nostoc sp. strain PCC 73102 was at least 50-fold improved (relative to that of pRL443) by the use of either pRK2013 or pRK2073 (a Kms Spr derivative of pRK2013 [5, 6]). Despite the compatibility between the origins of replication of conjugative plasmid pRK443 and cargo plasmid pRL489, the efficiencies of its transfer in Chroococcidiopsis sp. strains 029, 057, and 123 were lower than those of pDUCA7 mobilized via pRK2013. Differences in the conjugative transfer of different plasmids in the same Chroococcidiopsis sp. strain and of the same cargo plasmid in different Chroococcidiopsis sp. strains are to be expected since the efficiency of gene transfer depends on both the cargo plasmid and the recipient strain (13, 38).
A correlation between the morphology of a given strain and its efficiency of conjugative transfer was not apparent; Chroococcidiopsis sp. strains 123 and 029 are morphologically identical but differ significantly in their transformability. The reduced conjugative efficiencies of Chroococcidiopsis sp. strains 057 and 123 may reflect the presence of host-specific restriction specificities. The low yield of plasmid DNAs extracted from Chroococcidiopsis transconjugants (see Materials and Methods) prevented us from testing whether it was possible to obtain electrotransformants from strain 057 using plasmid DNA extracted from transconjugants of this strain or to improve the transformation efficiencies of strains 029 and 123 by using plasmid DNAs extracted from the correspondent transconjugants. In Anabaena sp. strain PCC 7120, which contains isoschizomers of AvaI, AvaII, and AvaIII, the efficiency of conjugative transfer decreases as an exponential function of the number of unprotected sites (11). The fact that conjugative transfer of pDUCA7 into Chroococcidiopsis sp. strains 029, 057, and 123 was unaffected by the absence of the helper plasmid pRL528 may suggest that AvaI and Eco47II restriction activities are not present in these three strains. Restriction did not pose a problem in the transformation of Synechocystis sp. strain PCC 6803, Synechococcus sp. strain PCC 7942 (10), or Nostoc sp. strain PCC 73102 (5, 6).
The role of cell envelope composition and structure has never been investigated as a possible barrier for conjugative transfer in cyanobacteria, although selection of conjugation-deficient recipient cells of E. coli yielded mutants which exhibited defects in the outer-membrane OmpA or in lipopolysaccharides (25). Cyanobacterial cell walls, despite their overall gram-negative structure, are characterized by a thicker peptidoglycan layer, by lipopolysaccharides containing a small amount of bound phosphate and often lacking ketodeoxyoctonate, and by the presence of external layers which differ in composition and structure (23). Extracellular polysaccharides and wall layers may prevent efficient conjugation by hindering cell-to-cell contacts. However, aged cells of Chroococcidiopsis spp. are characterized by a thickening of the cell envelope, yet no lowering of conjugative efficiencies was observed when late-stationary-phase 2-month-old (Fig. 3) Chroococcidiopsis sp. strains 029, 057, and 123 were used as recipients. The failure to obtain transconjugants in Chroococcidiopsis sp. strains 171 and 584 might be due to the presence of host-specific restriction endonucleases. Strains 171, 584, and 057 also failed to be transformed via electroporation, which may be due to the production of extracellular nucleases that may represent a barrier (see below).
Unlike conjugation, transformation through electroporation is prone to the problem of extracellular nucleases. The screening of over 150 strains of nostocacean cyanobacteria revealed that the great majority exhibited nuclease activity as demonstrated by the production of halos of clearing in agar medium containing DNA-methyl green (41). Moreover, a sugar-nonspecific nuclease was detected in cultures of Anabaena sp. strain PCC 7120 (27). In the present study, electrotransformation of Chroococcidiopsis sp. strains 029 and 123 with either pDUCA7 or pRL489 occurred at about 10−4 per recipient cell, while no electrotransformants were obtained in strains 057, 171 and 584. The data presented here on the electrotransformation frequencies of Chroococcidiopsis spp. may not be optimal because the same conditions were used for all the strains; settings of field strength and time constant which maximize DNA uptake while minimizing cell killing should be established for each cyanobacterial strain (10). The fact that all five Chroococcidiopsis sp. strains are a source of extracellular deoxyribonuclease(s) suggests that further studies on gene transfer of these forms should rely on conjugation only. The inability to achieve gene transfer in Chroococcidiopsis sp. strains 171 and 584 via electroporation and conjugation emphasizes that successful genetic manipulation of some cyanobacterial strains can be time-consuming; some cyanobacterial strains may indeed be recalcitrant to in vitro gene transfer. The evolutionary consequences of such recalcitrance in field populations of cyanobacteria remain poorly understood.
Through parsimony analysis of 16S rDNA sequences the strains of Chroococcidiopsis (taxonomic section II [31]) from hot and cold deserts represent a coherent group that is divergent from representative strains of sections III (LPP group) and IV (Fig. 4). These Chroococcidiopsis strains also appear to be distinct from Chroococcidiopsis sp. strain PCC 7203, the proposed type strain of species C. thermalis. Chroococcidiopsis sp. strain PCC 7203 was received at the Pasteur Culture Collection as Myxosarcina chroococcoides CCAP (1451/1) but was subsequently assigned to Chroococcidiopsis on the basis of the production of immotile, not motile, baeocytes; in fact it grouped with heterocystous cyanobacteria in one phylogenetic analysis (40). The taxonomic status of Chroococcidiopsis sp. strain PCC 7203 thus remains questionable (Fig. 4B). Chroococcidiopsis sp. strain PCC 7203 is morphologically identical to Chroococcidiopsis sp. strain 584 and conjugative gene transfer failed in both strains (not shown). Since Chroococcidiopsis sp. strain 584 had the greatest sensitivity to ionizing radiation in a previous survey of 10 Chroococcidiopsis strains, including strains 029, 057, and 171 (1), and was resistant to gene transfer, it was not studied further. The taxonomic assignment of Chroococcidiopsis sp. strain 584 is currently under question. Notwithstanding, we observed no correlation between either morphology or position in a phylogenetic tree and capacity for gene transfer. Sequence analysis revealed 95.6% nucleotide conservation in Chroococcidiopsis sp. strains 057 and 123, two strains amenable to conjugative gene transfer, but only the former was transformable via electroporation. Chroococcidiopsis sp. strains 029 and 123 were morphologically similar, showed 86.9% nucleotide conservation and exhibited a comparable suitability to electrotransformation, while efficiency of conjugative transfer of strain 123 was 10- to 100-fold reduced. In contrast, Chroococcidiopsis sp. strains 029 and 171 shared 92.5% nucleotide conservation but only the former was suitable to gene transfer via conjugation and electroporation.
In conclusion, Chroococcidiopsis sp. strain 029 from the Negev Desert, Israel, exhibited the highest efficiency in conjugative gene transfer and electrotransformation. Phylogenetic analysis suggests that this strain is representative of populations of Chroococcidiopsis spp. from hot and cold deserts, and it offers promise as an experimental strain for the elucidation of mechanisms of desiccation tolerance.
ACKNOWLEDGMENTS
This work was supported by NSF grant IBN9513157 (to M.P.), DARPA grant N00173-98-1-G005 (to M.P. and R.F.H.), NASA grant NAGW 40–44, and NSF grant OPP 96-14969 (to E.I.F.).
We thank R. W. Castenholz for providing Chroococcidiopsis strains, J. C. Meeks and J. Elhai for plasmids, and anonymous reviewers for pointing out important omissions and inaccuracies in an early version of the manuscript.
REFERENCES
- 1.Billi D, Friedmann E I, Hofer K G, Grilli Caiola M, Ocampo-Friedmann R. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl Environ Microbiol. 2000;66:1489–1492. doi: 10.1128/aem.66.4.1489-1492.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billi D, Grilli Caiola M, Paolozzi L, Ghelardini P. A method for DNA extraction from the desert cyanobacterium Chroococcidiopsis and its application to identification of ftsZ. Appl Environ Microbiol. 1998;64:4053–4056. doi: 10.1128/aem.64.10.4053-4056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billi D, Potts M. Life without water: responses of prokaryotes to desiccation. In: Storey K B, Storey J M, editors. Environmental stressors and gene responses. Amsterdam, The Netherlands: Elsevier; 2000. pp. 181–192. [Google Scholar]
- 4.Buikema W J, Haselkorn R. Isolation and complementation of nitrogen fixation mutants of the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:1879–1885. doi: 10.1128/jb.173.6.1879-1885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen M F, Meeks J C, Cai Y A, Wolk C P. Transposon mutagenesis of heterocyst-forming filamentous cyanobacteria. Methods Enzymol. 1998;297:3–17. [Google Scholar]
- 6.Cohen M F, Wallis J G, Campbell E L, Meeks J C. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple cellular differentiation alternatives. Microbiology. 1994;140:3233–3240. doi: 10.1099/13500872-140-12-3233. [DOI] [PubMed] [Google Scholar]
- 7.Crowe J H, Crowe L M, Carpenter J E, Petrelski S, Hoekstra F A, de Araujo P, Panek A D. Anhydrobiosis: cellular adaptation to extreme dehydration. In: Dantzler W H, editor. Handbook of physiology. Vol. 2. Oxford, United Kingdom: Oxford University Press; 1997. pp. 1445–1478. [Google Scholar]
- 8.De Philippis R, Vincentini M. Exocellular polysaccharides from cyanobacteria and their possible applications. FEMS Microbiol Rev. 1998;22:151–175. [Google Scholar]
- 9.Elhai J. Strong and regulated promoters in the cyanobacterium Anabaena PCC 7120. FEMS Microbiol Lett. 1993;114:179–184. doi: 10.1111/j.1574-6968.1993.tb06570.x. [DOI] [PubMed] [Google Scholar]
- 10.Elhai J, Thiel T, Pakrasi H B. DNA transfer into cyanobacteria. In: Gelvin S, Schilpoort R, editors. Plant molecular biology manual, A12. Amsterdam, The Netherlands: Martinus Nijhoff; 1990. pp. 1–23. [Google Scholar]
- 11.Elhai J, Vepritskiy A, Muro-Pastor A M, Flores E, Wolk C P. Reduction of conjugative transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elhai J, Wolk C P. Conjugative transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 13.Fatma T, Venkataraman L V. Conjugative gene-transfer in filamentous cyanobacteria. Curr Sci. 1992;63:186–192. [Google Scholar]
- 14.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedmann E I. Extreme environments, limits of adaptation and extinction. In: Guerrero R, Pedrós-Alió C, editors. Trends in microbial ecology. Barcelona, Spain: Spanish Society for Microbiology; 1993. pp. 9–12. [Google Scholar]
- 17.Friedmann E I, Kappen L, Meyer M A, Nienow A J. Long-term productivity in the cryptoendolithic microbial community in the Ross Desert, Antarctica. Microb Ecol. 1993;25:51–69. doi: 10.1007/BF00182129. [DOI] [PubMed] [Google Scholar]
- 18.Grilli Caiola M, Billi D, Friedmann E I. Effect of desiccation on envelopes of the cyanobacterium Chroococcidiopsis sp. (Chroococcales) Eur J Phycol. 1996;31:97–105. [Google Scholar]
- 19.Grilli Caiola M, Canini A, Ocampo-Friedmann R. Iron superoxide dismutase (Fe-SOD) localization in Chroococcidiopsis sp. (Chroococcales, Cyanobacteria) Phycologia. 1996;35:90–94. [Google Scholar]
- 20.Grilli Caiola M, Ocampo-Friedmann R, Friedmann E I. Cytology of long-term desiccation in the desert cyanobacterium Chroococcidiopsis sp. (Chroococcales) Phycologia. 1993;32:315–322. doi: 10.2216/i0031-8884-32-5-315.1. [DOI] [PubMed] [Google Scholar]
- 21.Helm R F, Huang Z, Edwards D, Leeson H, Peery W, Potts M. Structural characterization of the released polysaccharide of desiccation-tolerant Nostoc commune DRH1. J Bacteriol. 2000;182:974–982. doi: 10.1128/jb.182.4.974-982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershkovitz N, Oren A, Cohen Y. Accumulation of trehalose and sucrose in cyanobacteria exposed to matric water stress. Appl Environ Microbiol. 1991;57:645–648. doi: 10.1128/aem.57.3.645-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoiczyk E, Hansel A. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol. 2000;182:1191–1199. doi: 10.1128/jb.182.5.1191-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houmard J, Tandeau de Marsac N. Cyanobacterial genetic tools: current status. Methods Enzymol. 1988;167:808–847. doi: 10.1016/0076-6879(88)67092-3. [DOI] [PubMed] [Google Scholar]
- 25.Ippen-Ihler K A, Minkley E G., Jr The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- 26.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 27.Muro-Pastor A M, Flores E, Herrero A, Wolk C P. Identification, genetic analysis and characterization of a sugar-non-specific nuclease from the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1992;6:3021–3030. doi: 10.1111/j.1365-2958.1992.tb01760.x. [DOI] [PubMed] [Google Scholar]
- 28.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58:755–805. doi: 10.1128/mr.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potts M. Mechanisms of desiccation tolerance in cyanobacteria. Eur J Phycol. 1999;34:319–328. [Google Scholar]
- 31.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 32.Rudi K, Skulberg O M, Larsen F, Jakobsen K S. Strain characterization and classification of oxyphotobacteria in clone cultures on the basis of 16S rRNA sequences from the variable regions V6, V7, and V8. Appl Environ Microbiol. 1997;63:2593–2599. doi: 10.1128/aem.63.7.2593-2599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schmetterer G, Wolk C P. Identification of the region of cyanobacterial plasmid pDU1 necessary for replication in Anabaena sp. strain M-131. Gene. 1988;62:101–109. doi: 10.1016/0378-1119(88)90583-5. [DOI] [PubMed] [Google Scholar]
- 36.Shirkey B, Kovarcik D P, Wright D J, Wilmoth G, Prickett T F, Helm R F, Gregory E M, Potts M. Active Fe-containing superoxide dismutase and abundant sodF mRNA in Nostoc commune (cyanobacteria) after years of desiccation. J Bacteriol. 2000;182:189–197. doi: 10.1128/jb.182.1.189-197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiel T. Genetic analysis of cyanobacteria. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 581–611. [Google Scholar]
- 38.Thiel T, Wolk C P. Conjugative transfer of plasmids to cyanobacteria. Methods Enzymol. 1987;153:232–243. doi: 10.1016/0076-6879(87)53056-7. [DOI] [PubMed] [Google Scholar]
- 39.Thomas C M, Smith C A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- 40.Turner S. Molecular systematics of oxygenic photosynthetic bacteria. In: Bhattacharya D, editor. Origins of algae and their plastids. Vienna, Austria: Springer-Verlag; 1997. pp. 13–52. [Google Scholar]
- 41.Wolk C P, Kraus J. Two approaches to obtaining low, extracellular deoxyribonuclease activity in cultures of heterocyst-forming cyanobacteria. Arch Microbiol. 1982;131:302–307. [Google Scholar]