Abstract
A modified polymerase chain reaction protocol was used to amplify the entire envelope-coding region of HIV-1 directly from brain and lymph node tissue obtained at autopsy from three HIV-l-infected individuals. Molecular analysis of amplified DNA by digestion with 18 restriction endonucleases, singly and in combination, revealed different HIV-1 genotypes in the brain and lymph node compartments in each of the three individuals. This anatomic compartmentalization of HIV-1 populations may reflect different viral genomic sequences that determine tropism or differences in host immune selection pressures in the brain and lymphoid compartments that drive the emergence of distinct viral populations.
Numerous lines of evidence support a causative role for the human immunodeficiency virus (HIV-1) in the neurologic dysfunctions seen in many patients with acquired immunodeficiency syndrome (AIDS; reviewed in references 2 and 31). The presence of HIV-1 can be directly demonstrated in the central nervous system (CNS) by virus isolation and direct hybridization analysis (4,12,13,18,25,39). The major target for HIV-1 infection in the brain appears to be cells of the monocyte/macrophage lineage. Direct viral involvement in the various neurologic dysfunctions including demyelination is difficult to reconcile with the low incidence of neuronal and glial cell infection by HIV-1. Rare low-level infection in some astrocytes and neurons has been observed (39), and the possibility of latent infection of neuronal or glial cells below the limits of detection of in situ molecular hybridization techniques has not been excluded. Watkins et al. (37) have demonstrated HIV-1 replication and resultant destruction of microglial cells in vitro. In neurons, HIV-1 gp120 has also been shown to induce increases in cellular calcium associated with neurotoxicity; this implicates a viral mechanism in neuronal destruction.
As judged by a variety of biologic properties including replication in various cell lines, cytopathogenic capacity, and ability to modulate CD4 antigen, brain isolates of HIV are phenotypically distinct from virus populations of the lymphoid compartments and have been proposed to represent a distinct group of viruses (4). HIV-1 strains isolated from CNS tissue display a preferential cell tropism for macrophages (4,5,19,37), and this altered host-cell tropism is determined by a region of gp120 separate from the CD4-binding domain (27,34). Indeed, although infection of macrophages in vitro appears to proceed via a CD4-dependent route (8,26), it exhibits increased resistance to soluble CD4 compared with HIV-1 infection of T cells (15). Additional support for an alternative mode of entry by HIV-1 is the CD4-independent infection demonstrated in fetal dorsal root ganglion glial cells (20), glioma-derived glial cells (7,11,17,38), and muscle cells (7).
For several reasons we have focused on the envelope open reading frame (ORF) of HIV-1 to study the molecular basis for altered HIV-1 tropism. HIV-1 tropism appears to be determined by events prior to formation of the provirus (3,27,34). We have observed that mutations in the CD4-binding epitope of HIV-1 that restrict the ability of HIV-1 to infect and replicate in T cells (21) do not alter the infectivity of HIV-1 for glial cell lines (S. Haggerty et al., manuscript submitted). Single amino acid changes in HIV-1 envelope glycoprotein alter the tropism of HIV-1 for T cells and monocytes (9), and determinants for cytopathogenicity and tropism in a variety of isolates have been attributed to the envelope glycoprotein (40). Thus, it is possible that if distinct receptor molecules are important for HIV-1 replication in the CNS, one may expect to observe genotypic alterations in the envelope as well as modification of the cell-specific tropism of the virus. Two separate groups of investigators have localized the macrophage tropism-determining region of gp120 spanning 159 amino acids upstream of the CD4-binding epitope (27,34). In view of the fact that these researchers amplified the HIV-1 isolates in tissue culture prior to analysis and that there were not paired lymphoid isolates from the same patients, further delineation of the macrophage-determining regions was not done.
METHODS
We have adapted the polymerase chain reaction (PCR) to allow amplification and characterization of the envelope-coding region of HIV-1 directly from paired tissues (brain and lymph node) of each of three HIV-1-infected individuals. To date, analyses of CNS-derived HIV-1 isolates have been subsequent to propagation of virus in cell culture. Given that in vitro propagation of virus probably favors isolation of adapted variants, PCR amplification of viral genes should allow a more representative analysis of virus populations in vivo. Tissue samples were obtained from three HIV-1 infected individuals post mortem. Relevant clinical observations and HIV-1-related neurologic findings at autopsy are shown in Table 1. Two patients (Ha and Sk) displayed typical cognitive problems and at autopsy were found to have extensive morphologic CNS damage. One individual (Wi) died of causes unrelated to AIDS, and autopsy revealed no evidence of HIV-1-related morphologic changes in the CNS (Table 1).
Table 1.
AIDS-related Clinical Findings and Neurologic Symptoms prior to Death and Morphologic Findings Post Mortem
| Subject | Findings a | |
|---|---|---|
|
| ||
| Ha | Clinical | AIDS, PCP, meningoencephalitis |
| Post mortem | CNS lymphoma, cortical necrosis | |
| Sk | Clinical | AIDS, PCP, Kaposi’s sarcoma, CMV pneumonia, meningoencephalitis |
| Post mortem | Marked PML-induced demyelination | |
| Wi | Clinical | ARC, no neurologic symptoms |
| Post mortem | No morphologic changes | |
PCP, Pneumocytis carinii pneumonia; PML, progressive multifocal leukoencephalopathy; CMV, cytomegalovirus.
Total cellular DNA was extracted from brain and lymph node tissue of each patient as described (6), and the complete envelope-coding region of HIV-1 was amplified by PCR using two rounds of amplification and nested primers (Fig. 1). Both sets of nested primers (set 8/3, followed by set 7/9; Fig. 1A) were directed to highly conserved regions of the HIV-1 genome and specified the retention of two restriction-endonuclease recognition sites to facilitate subsequent subcloning and construction of chimeric virus genomes. We found that titration of MgCl2 (not shown) and of each primer set (7/9, Fig. 1B; data not shown) was critical for efficient amplification of a 3.1-kb region of HIV-1 spanning the entire envelope ORF. This fragment was demonstrated to be envelope specific after transfer of the gel contents to nylon and hybridization to an envelope-specific probe. It had been observed previously that amplification of large regions of DNA by PCR is strongly influenced by primer concentration (S. Dewhurst, personal communication). As shown in Figure 1B, low primer concentrations were required to facilitate 3.1-kb DNA amplification, and optimal amplification was observed at around 5 pM of each primer. The use of nested primers was also essential to amplify the envelope-coding region from a genomic background (Fig. 1C), presumably because of a decrease in competition by spuriously primed DNA for substrates and the resultant increase in specificity and yield.
FIG. 1.
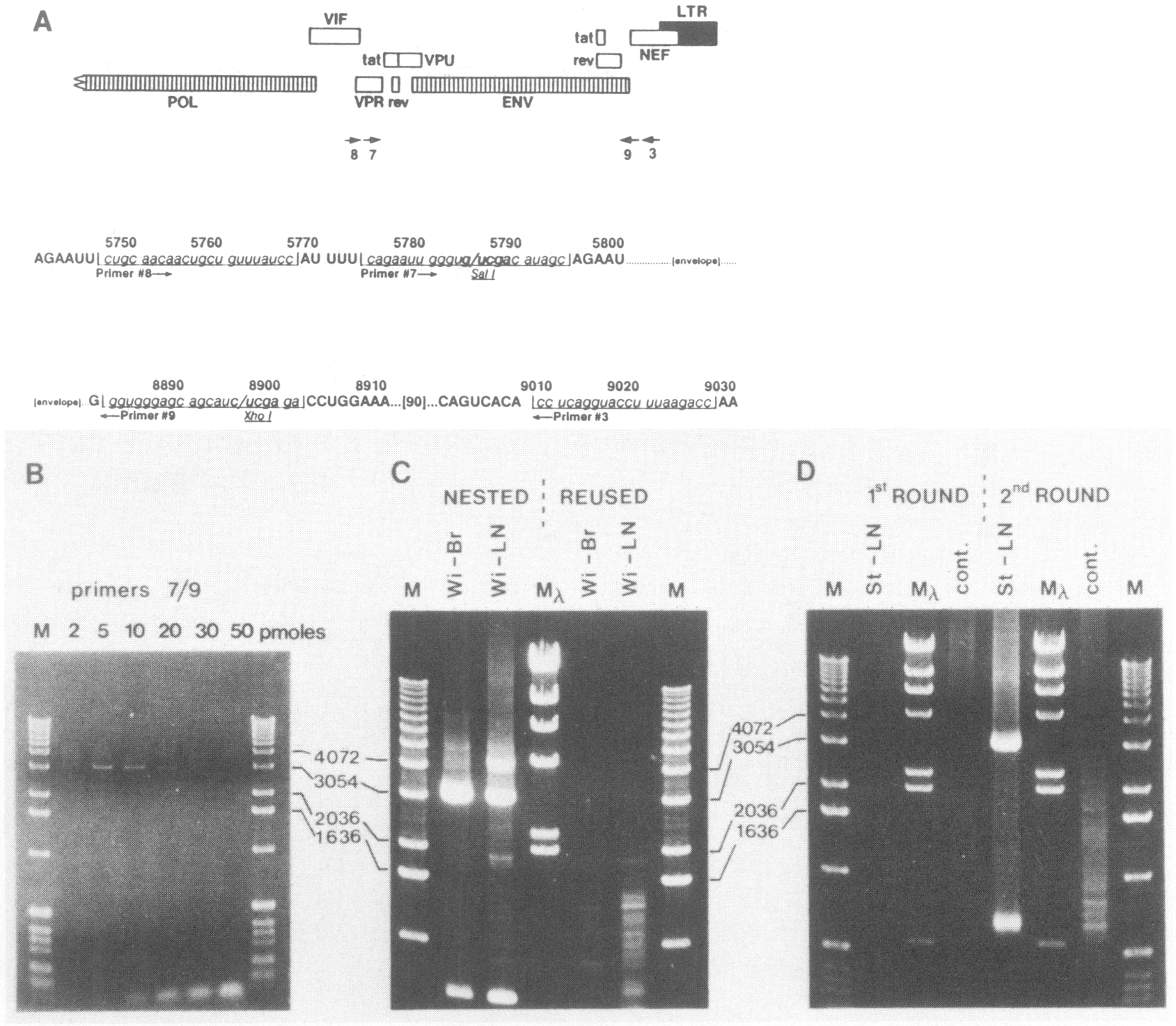
PCR-mediated amplification of complete HIV-1 envelope-coding region. A. Region of the genome targeted by nested PCR primers (numbering according to the HXB2 sequence [30|). DNA sequence of the primer-binding regions is shown below the representation of the ORFs. B. Effect of primer concentration on 3.1-kb env amplification. One picogram of cloned HXB2 DNA (30) was subjected to a preliminary 2-min denaturation step at 95°C and amplified by 30 PCR cycles, each consisting of 95°C for 30 sec (denaturation) followed by 58°C for 30 sec (annealing) and then by 72°C for 2 min (extension). PCR reaction buffer was as described by Perkin-Elmer Cetus except that the MgCl2 concentration was increased to 2.0 mM. Amount of each primer (pM) in the PCR reaction is indicated. C. Comparative efficiency of two rounds of nested PCR amplification (“Nested”; primer 8/3, 1st round followed by 7/9, 2nd round) versus two rounds of amplification with the same primer set (“Reused”; primers 8/3); 0.5 μg cellular DNA from brain (Br) and lymph node (LN) of subject Wi were subjected to PCR as described for Panel B. One fiftieth of the 1st-round PCR products (1 μl) was diluted to 100 μl for 2nd-round amplification. Reaction conditions were identical in both rounds. D. Efficiency of single-v double-round PCR: 0.5 μg of lymph node DNA from subject St or 0.5 μg of cellular DNA from an HI V-l -negative CD4+ T-cell line CEM (“cont.”) were amplified by one round of PCR (primers 8/3) or an additional second round (primers 7/9) as described in Panel C. M, 1-kb DNA ladder (BRL); Mλ, HindIII-digested λ-DNA mol. wt. markers.
Two rounds of amplification with nested primer pairs resulted in the amplification of a ~3-kb envelope-specific DNA band from patient DNA (“St-LN,” Fig. 1D) but not from the HIV-1-negative, CD4-positive cell line, CEM (“cont.,” Fig. 1D). Use of only one round did not result in a significant level of amplification product from these samples (1st ROUND v 2nd ROUND, Fig. 1D). In addition to amplification of a 3.1-kb env-specific fragment, we routinely observed a smaller product of approximately 700 pb after second-round amplification (Fig. 1D), which by molecular hybridization was likewise found to be HIV-1 envelope specific and which may have been the result of spurious priming during PCR amplification.
RESULTS AND DISCUSSION
After two rounds of PCR, the 3.1-kb env-specific DNA fragment was gel purified and mapped using 18 restriction endonucleases in single or double digests, and the restriction pattern was analyzed by Southern blot hybridization. Clear differences were evident in the digestion profile of amplified material from brain and lymph node from individual patients. For example, Figure 2 shows the digestion patterns of PCR-amplified env-specific DNA from brain and lymph node tissue of patient Wi. Several distinct restriction-site differences between the two compartments are indicated. This marked predominance of particular genotypes within tissue compartments was not attributable to selective amplification of these genotypes. The polymorphisms observed were not a result of the PCR procedure nor of partial enzyme digestion, as all polymorphisms were verified using single and double enzyme digests using PCR-amplified DNA prepared in different experiments. The digest patterns for each sample remained consistent through repeated procedures, and mock samples without host DNA were consistently negative. Similar amplification and analysis of the envelope region from cells infected with the HXB2 isolate (30) provided the expected cleavage pattern for that virus (data not shown).
FIG. 2.
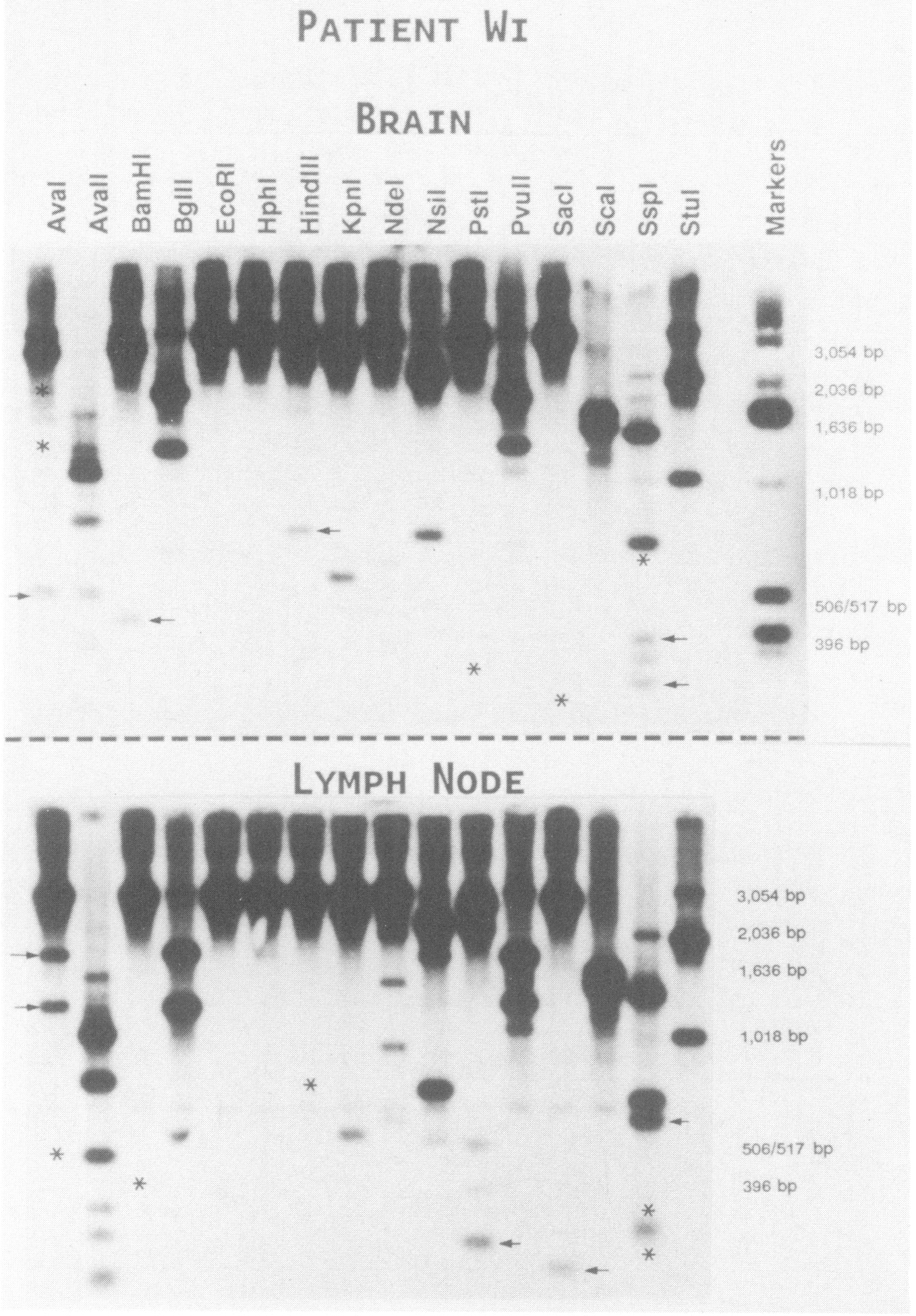
Distinct restriction fragment polymorphisms between PCR-amplified env DNA from brain and lymph node tissue (subject Wi). Total cellular DNA was amplified by two rounds of nested PCR as described in Figure 1C. Then 100 to 200 ng of PCR-amplified, gel-purified, 3.1-kb env DNA was digested overnight with the indicated restriction enzyme, electrophoresed, transferred to nylon membranes, and hybridized to a subgenomic HIV-1 env probe (35). Additional DNA fragments resulting from extra-endonuclease cleavage sites (arrows) and loss of fragments secondary to absence of a cleavage site (asterisk) are shown. Higher mol. wt. bands are overexposed to allow visualization of lower mol. wt. fragments.
The restriction maps of PCR-amplified env DNA from the brain and lymph node of all three patients are shown on Figure 3 together with maps of seven North American and one European isolate. It is important to emphasize that the PCR reaction represents envelope-coding sequences from the population of viruses that existed in that tissue compartment and which would be expected to contain myriad individual (quasispecies) genotypes (23). Thus, the observed polymorphisms predominant in the population of viruses within that compartment are represented by the restriction map; i.e., the major genotype present within that compartment. Because these genotypes were amplified by PCR, the surprising marked predominance of particular genotypes within tissue compartments (Fig. 3) was not attributable to selective amplification, as would occur subsequent to amplification in cell culture. We have now begun to characterize multiple cloned PCR env fragments from brain and lymph node tissue of these individuals and observe typical quasispecies differences (23) indicating amplification of multiple variants. In addition to depicting the major HIV-1 env genotype (Fig. 3), we observed polymorphisms at a number of restriction endonuclease sites represented by less intense bands and indicating viral subpopulations of increased genome diversity.
FIG. 3.
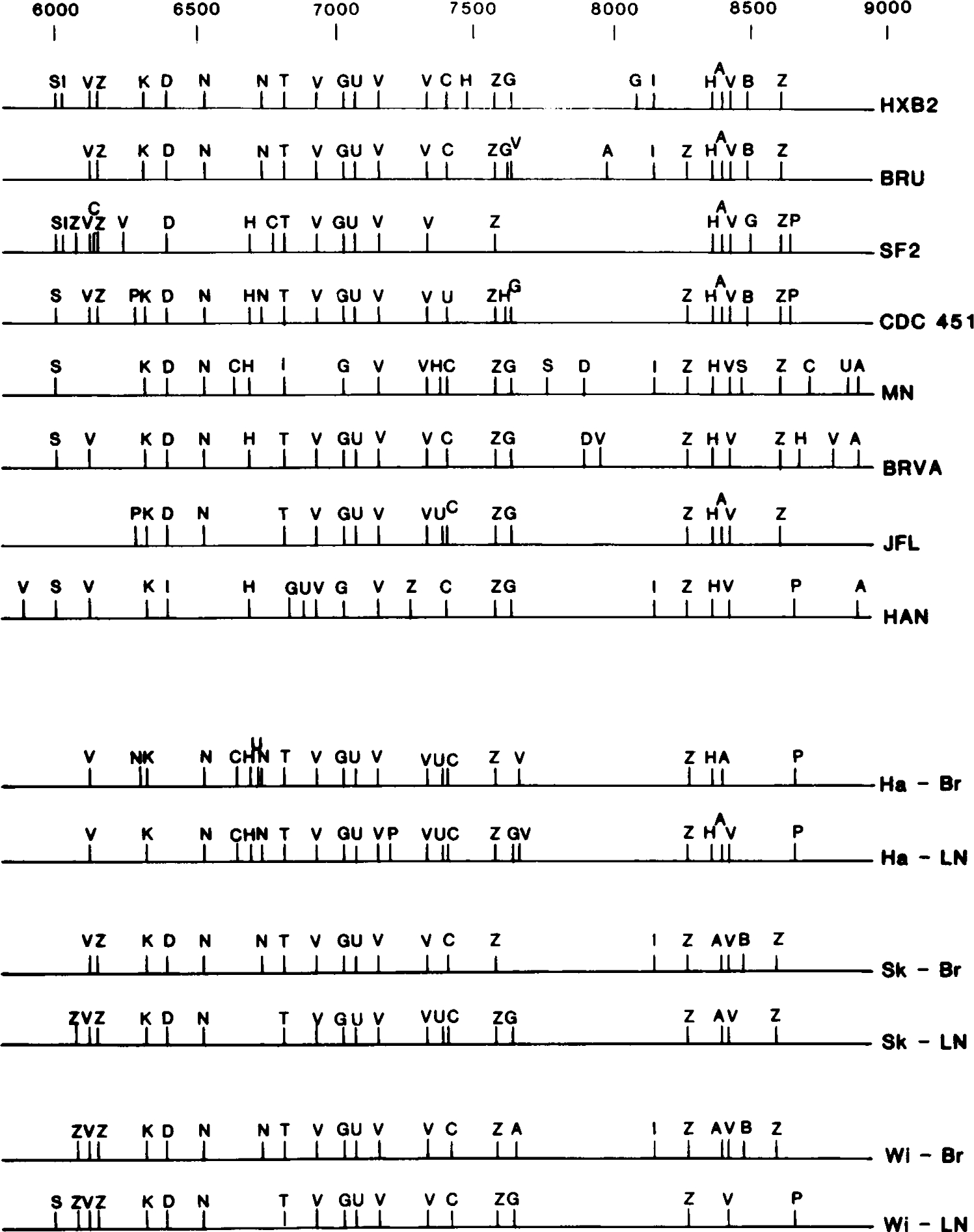
Restriction endonuclease cleavage maps of brain- and lymph node-specific PCR-amplified envelope-coding regions. DNA was prepared and analyzed as described for Figure 2, and the locations of restriction enzyme cleavage sites were deduced from single and double digests and compiled. Only polymorphisms detected exclusively in that particular tissue compartment are indicated on each map. The restriction maps of seven North American isolates (HXB2 [30], BRU [36], SF2 [32], CDC 451 [10], MN [16], BRVA [1], JFL [22]) and one European isolate (HAN [33]) are shown for comparison. Restriction endonuclease cleavage sites are indicated as A, AvaI; V, AvaII; B, BamHI; G, BglII; E EcoRI; H. HphI; I, HindIII; K, KpnI; D, NdeI; N, NsiI; P, PstI; U, PvuII; L, SalI; S, SacI; C, ScaI; Z, SspI; T. StuI; X. XhoI. Derivation of cleavage patterns for all characterized isolates was facilitated by information contained within the Human Retroviruses and AIDS Sequence Database (24).
Several of the HIV-1 env polymorphisms that differentiate brain from lymph node virus populations were consistent between patients; e.g., the lymph node-specific BglII site at position 7600 in all three individuals and brain-specific HindIII site at position 8130 in two individuals. It is possible we are witnessing compartmentalization of quasispecies; however, the small number of subjects studied here limits the significance of such observations. The presence of distinct genotypes within the two tissue compartments did not appear dependent on the neurologic status of the subject, given that patient Wi, who had no AIDS-related neurologic dysfunction, also displayed distinct HIV-1 genotypes in the brain and lymph node compartments.
The results presented here demonstrate that distinct HIV-1 genotypes predominate in two anatomic sites, providing direct evidence for compartmentalization of HIV-1. At present, the basis for this predominance of distinct genotypes is unknown. However, host-induced selective pressure that drives HIV-1 diversity may differ in the two tissue sites. Evidence suggests that HIV-1 replication in macrophages and glial cells in the brain is quite distinct from replication in T cells. In vitro, HIV-1 infection of glial cells results in a minimally replicative and nonproductive persistent infection lacking any of the HIV-1-induced cytopathologic damage evident in a cytocidal T-cell infection. Similarly, infection of macrophages is less efficient than that of T cells. The high levels of unintegrated HIV-1 DNA in terminally differentiated cells of the CNS in AIDS dementia patients (28), the ability of virus to assemble from intracytoplasmic surfaces of monocytes in vitro (14), and the accumulation of self-integrates in this cell type (29) indicate a lifecycle in the CNS that differs from that typical of T-cell infection in lymphoid tissue. By contrast, HIV-1 infection of lymphoid cells in vivo may be characterized by a high-level cytocidal infection leading to the death of the host cell and susceptibility to immune surveillance. This surveillance may favor the rapid evolution of divergent HIV-1 genotypes altered in cytopathogenic property and which are better adapted to persist in the host cell.
We are at present sequencing the primary PCR-amplified DNA and establishing and sequencing multiple env clones from paired brain and lymph node samples from each subject. Future creation of chimeric viruses containing PCR-amplified, authentic tissue-specific envelope-coding regions are under way to determine the significance of specific env alterations.
ACKNOWLEDGMENTS
We thank Jonathan Goldsmith and Steve Suvalsky for clinical material and Steve Dewhurst for suggestions on PCR of large regions of DNA. M.S. was supported by grants A125582 and A130386 from the National Institutes of Health, grant 962–7-RGR from the American Foundation for AIDS Research, and a grant from the Nebraska Research Initiative. S.H. was supported by NIH postdoctoral training grant T32 CA09476.
REFERENCES
- 1.Anand R, Thayer R, Srinivasan A, Nayyar S, Gardner M, Luciw P, and Dandekar S. 1989. Biological and molecular characterization of human immunodeficiency virus (HIV-1 BR) from the brain of a patient with progressive dementia. Virology 168:79–89. [DOI] [PubMed] [Google Scholar]
- 2.Budka H 1989. Human immunodeficiency virus (HlV)-induced disease of the central nervous system: pathology and implications for pathogenesis. Acta Neurol. Pathol. 77:225–236. [DOI] [PubMed] [Google Scholar]
- 3.Cann AJ, Zack JA, Go AS, Arrigo SJ, Koyanagi Y, Green PL, Koyanagi Y, Pang S, and Chen ISY. 1990. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 64:4735–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng–Mayer C, Weiss C, Seto D, and Levy JA. 1989. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc. Natl. Acad. Sci. U.S.A. 86:8575–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiodi F, Valentin A, Keys B. Schwartz S, Asjo B, Gartner S. Popovic M, Albert J, Sundquist V-A, and Fenyo EM. 1989. Biological characterization of paired human immunodeficiency virus type 1 isolates from blood and cerebrospinal fluid. Virology 173:178–187. [DOI] [PubMed] [Google Scholar]
- 6.Chirgwin JM, Pryzbyla AE, MacDonald RJ, and Rutter WJ. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294–5299. [DOI] [PubMed] [Google Scholar]
- 7.Clapham PR, Weber JN, Whitby D. McIntosh K. Dalgleish AG, Maddon PJ, Deen KC, Sweet RW, and Weiss RA. 1989. Soluble CD4 blocks the infectivity of diverse strains of HIV and SIV for T cells and monocytes but not for brain and muscle cells. Nature (London) 337:368–370. [DOI] [PubMed] [Google Scholar]
- 8.Collman R, Godfrey B, Cutilli J. Rhodes A. Hassan NF, Sweet R. Dougals SD, Friedman H, Nathanson N. and Gonzalez-Scarano F. 1990. Macrophage-tropic strains of human immunodeficiency virus type 1 utilize the CD4 receptor. J. Virol. 64:4468–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordonnier A, Montagnier L, and Emerman M. 1989. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature (London) 340:571–574. [DOI] [PubMed] [Google Scholar]
- 10.Desai SM, Kalyanaraman VS, Casey JM. Srinivasan A, Andersen PR, and Devare SG. 1986. Molecular cloning and primary nucleotide sequence analysis of a distinct human immunodeficiency virus isolate reveals significant divergence in its genomic sequences. Proc. Natl Acad. Sci. U.S.A. 83:8380–8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewhurst S, Bresser J, Stevenson M, Sakai K. Evinger-Hodges MJ, and Volsky DJ. 1987. Susceptibility of human glial cells to infection with human immunodeficiency virus. FEBS Lett. 213:138–143. [DOI] [PubMed] [Google Scholar]
- 12.Eilbott DJ, Peress N, Burer H, La Neve D, Orenstein J, Gendelman HE, Seidman R. and Weiser B. 1989. Human immunodeficiency virus type 1 in spinal cords in acquired immune deficiency syndrome patients with myelopathy: expression and replication in macrophages. Proc. Natl. Acad. Sci. U.S.A. 86:3337–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gartner S, Markovitz P, Markovitz DM, Betts RF, and Popovic M. 1986. Virus isolation from and identification of HTLV-III/LAV-producing cells in brain tissue from a patient with AIDS. JAMA 256:2365–2371. [PubMed] [Google Scholar]
- 14.Gendelman HE, Orenstein J, Martin M, Ferrua C, Mitra R, Phipps T, Whal L, and Meltzer M. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomatos PJ, Stamatos NM, Gendelman HE, Tramont EC, and Meltzer MS. 1990. Relative inefficiency of soluble recombinant CD4 for inhibition of infection by monocyte-tropic HIV in monocyte and T cells. J. Immunol. 144:4183–4188. [PubMed] [Google Scholar]
- 16.Gurgo C, Guo H-G, Franchini G, Aldovini A, Collalti E, Farrell K, Wong-Staal F, Gallo RC, and Reitz MS Jr. 1988. Envelope sequences of two new United States HIV-1 isolates. Virology 164:531–536. [DOI] [PubMed] [Google Scholar]
- 17.Harouse JM, Kunsch C, Hartle HT, Laughlin MA, Hoxie JA, Wigdahl B, and Gonzalez–Scarano F. 1989. CD4-independent infection of human neural cells by human immunodeficiency virus type 1. J. Virol. 63:2527–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenig S, Gendelman HE, Orenstein JM, Del Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, and Fauci AS. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089–1093. [DOI] [PubMed] [Google Scholar]
- 19.Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, and Chen ISY. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819–822. [DOI] [PubMed] [Google Scholar]
- 20.Kunsch C, Hartle HT, and Wigdahl B. 1989. Infection of human fetal dorsal root ganglion glial cells with human immunodeficiency virus type 1 involves an entry mechanism independent of the CD4 T4A epitope. J. Virol. 63:5054–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasky LA, Nakamura G, Smith DH, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, and Capon DJ. 1987. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell 50:975–985. [DOI] [PubMed] [Google Scholar]
- 22.McNearney T, Westervelt P, Theilan B, Trowbridge DB, Garcia J, Whittier R, and Ratner L. 1990. Limited sequence heterogeneity among biologically distinct human immunodeficiency virus type 1 isolates from individuals involved in a clustered infectious outbreak. Proc. Natl. Acad. Sci. U.S.A. 87:1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyerhans A, Cheyniér R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt–Manson L, Asjo B, and Wain–Hobson S. 1989. Temporal fluctuations in HIV quasispecies are not reflected by sequential HIV isolations. Cell 58:901–910. [DOI] [PubMed] [Google Scholar]
- 24.Myers G, Rabson AB, Joseph SF, Smith TF, and Wong-Staal F (ed.) 1988. Human retroviruses and AIDS. Los Alamos, N.M., Los Alamos National Laboratory. [Google Scholar]
- 25.Nelson JA, Reynolds–Kohler C, Oldstone MBA, andWiley CA. 1988. HIV and HMCV coinfect brain cells in patients with AIDS. Virology 165:286–290. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson JKA, Cross GD, Callaway CS, and McDougal JS. 1986. In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV). J. Immunol. 137:323–329. [PubMed] [Google Scholar]
- 27.O’Brien WA, Koyanagi Y, Namazie A, Zhao J-Q, Diagne A, Idler K, Zack JA, and Chen ISY. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature (London) 348:69–73. [DOI] [PubMed] [Google Scholar]
- 28.Pang S, Koyanagi Y, Miles S, Wiley C, Vinters HV, and Chen ISY. 1990. High levels of unintegrated HIV-1 DNA in brain tissue of AIDS dementia patients. Nature (London) 343:85–89. [DOI] [PubMed] [Google Scholar]
- 29.Pauza CD, and Galindo J. 1989. Persistent human immunodeficiency virus type 1 infection of monoblastoid cells leads to accumulation of self-integrated viral DNA and to production of defective virions. J. Virol. 63:3700–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich P, Josephs SJ, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, Ivanoff L, Petteway SR Jr., Pearson ML, Lautenberger JA, Papas TS. Ghreyeb J, Chan NT, Gallo RC, and Wong-Staal F. 1985. Complete nucleotide sequence of the AIDS virus, HTLV III. Nature (London) 313:277–284. [DOI] [PubMed] [Google Scholar]
- 31.Rosenblum ML, Levy RM, and Bredesen DE. 1988. In Aids and the nervous system, p. 1–3. Raven Press, New York. [Google Scholar]
- 32.Sanchez–Pescador R, Power MD, Barr PJ, Steimer KS. Stempien MM, Brown–Shimer SL, Gee WW, Renard A, Randolph A, Levy JA, Dina D, and Luciw PA. 1985. Nucleotide sequence and expression of an AIDS-associated retrovirus. (ARV-2). Science 227:484–492. [DOI] [PubMed] [Google Scholar]
- 33.Sauermann U, Schneider J. Mous J, Brunckhorst U, Schedel I, Gensch KD, and Hunsmann G. 1990. Molecular cloning and characterization of a German HIV-1 isolate. AIDS Res. Hum. Retroviruses 6:813–823. [DOI] [PubMed] [Google Scholar]
- 34.Shioda T, Levy JA, and Cheng-Mayer C. 1991. Macrophage and T cell line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature (London) 349:167–169. [DOI] [PubMed] [Google Scholar]
- 35.Stevenson M. Stanwick TL, Dempsey MP, and Lamonica CA. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wain–Hobson S, Sonigo P, Danos O, Cole S, and Alizon M. 1985. Nucleotide sequence of the AIDS virus. LAV. Cell 40:9–17. [DOI] [PubMed] [Google Scholar]
- 37.Watkins BA, Dorn HH, Kelly WB. Armstrong RC, Potts BJ, Michaels F, Kuftka CV, and Dubois–Dalcq M. 1990. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science 249:549–553. [DOI] [PubMed] [Google Scholar]
- 38.Weber J, Clapham PR, McKeating J, Stratton M. Robey E, and Weiss RA. 1989. Infection of brain cells by diverse human immunodeficiency virus isolates: role of CD4 as receptor. J. Gen. Virol. 70:2653–2660. [DOI] [PubMed] [Google Scholar]
- 39.Wiley CA, Schrier RD, Nelson JA, Lampert PW, and Oldstone MBA. 1986. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc. Natl. Acad. Sci. U.S.A. 83:7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.York–Higgins D, Cheng–Mayer C, Baver D, Levy JA, and Dina D. 1990. Human immunodeficiency virus type 1 cellular host range, replication, and cytopathicity are linked to the envelope region of the viral genome. J. Virol. 64:4016–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]


