Highlights
-
•
Carbohydrate metabolic processes and starch and sucrose metabolism play significant roles in β-glucan biosynthesis.
-
•
This study identifies β-glucan synthesis-related DEGs for further investigation and utilization for genetic improvements.
Keywords: Barley (Hordeum vulgare L.), β-Glucan, Molecular mechanism, Quality, Transcriptome
Abstract
The presence of β-glucan in barley grains is one of its important quality traits. Lower β-glucan content is required for the barley used in beer and feed production, while higher β-glucan content is beneficial for food barley. Although intensive research has been carried out on the genotypic and environmental differences in β-glucan content in barley grains, little information is available on the molecular mechanisms underlying their genotypic differences and genetic regulation of β-glucan synthesis and accumulation. In this study, RNA sequencing analysis was conducted to compare the transcriptome profiles of two barley genotypes (BCS192 and BCS297) that greatly differ in grain β-glucan content, in order to identify the key genes responsible for β-glucan synthesis and accumulation during grain development. The results showed that carbohydrate metabolic processes and starch and sucrose metabolism play significant roles in β-glucan synthesis. The identified differently expressed genes (DEGs), which are closely associated with grain β-glucan content, are mainly involved in hydrolase activity and glucan metabolic processes. In addition, β-glucan accumulation in barley grains is predominantly regulated by photosynthesis and carbon metabolism. The DEGs identified in this study and their functions may provide new insights into the molecular mechanisms of β-glucan synthesis and genotypic differences in barley grains.
1. Introduction
Barley (Hordeum vulgare L.) has multiple uses, including feed and brewing in addition to food for some areas. β-glucan content is an important trait affecting the quality and use of barley grains. For example, β-glucan can affect viscosity in animal feed, reducing the absorption of nutrients (McNab & Smithard, 1992) and, in brewing, this viscosity can make filtration difficult, impacting malt and beer quality (Vis & Lorenz, 1998). However, β-glucan has been shown to be beneficial to human health (Wouk et al., 2021, Geng et al., 2022), and regular consumption is associated with a reduced risk of cardiovascular disease and hyperglycemia (Fuse et al., 2020). Thus, controlling β-glucan content in barley grains remains important for its production.
β-glucan is a linear polysaccharide consisting of only β-d-glucan units linked by (1 → 3)- or (1 → 4)-glycosidic bonds (Bin, Maninder, Hongzhi & Baojun, 2019). It is a non-cellulose polysaccharide, commonly found in the cell walls of grass plants. In general, cell walls of cereal endosperms are composed of arabinoxylan and β-glucan, the proportions of which vary widely among plant species. Arabinoxylan is dominant in rye and wheat, while β-glucan is dominant in barley and oats (Fincher & Stone, 1986). In general, β-glucan content in barley grains is around 2 %–8%, depending on genetic and environmental factors (Geng, Li, Xie, Wu, Ye & Zhang, 2021).
The structure and physicochemical properties of β-glucan have been intensively investigated (Giridhar, Paras, Sandeep & Longvah, 2020), along with enzymes and encoding genes responsible for its synthesis. In the early days, some quantitative trait loci (QTLs) associated with β-glucan content were identified (Jingzhao et al., 2008, Kim et al., 2011). In rice, the cellulose synthase-like (Csl) gene family members were considered as the putative genes encoding β-glucan synthase (Burton et al., 2006). This gene family was divided into eight sub-groups, named CslA to CslH, respectively (Farrokhi et al., 2006). However, in grasses, only CslF and CslH groups were identified (Hazen, Scott-Craig & Walton, 2002), and it was confirmed that CslF family genes were the main genes controlling β-glucan biosynthesis (Burton et al., 2006, Burton et al., 2011). Recently, it was demonstrated that only the gene CslF6 had a significant effect on β-glucan content in barley grains (Burton et al., 2011, Garcia-Gimenez et al., 2020). Thus, CslF6 is now known as a major gene controlling barley β-glucan biosynthesis. It is still unknown whether other major genes are associated with β-glucan synthesis in barley grains.
In recent years, studies on transcriptional and biochemical changes in barley during grain development showed that the transcriptional profiles and metabolite levels were dramatically dependent on developmental stages (Tang et al., 2017, Bian et al., 2019). However, little information is available on the expression patterns of genes related to β-glucan synthesis in barley during grain development. As a polysaccharide, β-glucan should be closely associated with assimilation, in particular, glucose metabolism during grain development, although no relevant research has been reported. Accordingly, we aimed to analyze the transcriptional dynamics of two barley genotypes, BCS192 and BCS297, differing in grain β-glucan content during grain development using RNA sequencing (RNA-seq). Then, differentially expressed genes (DEGs) in the two genotypes during grain development were identified and functionally annotated by Gene Ontology (GO) and KEGG pathway analysis. Finally, These DEGs were validated using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). The obtained results explored β-glucan synthesis-related genes for further investigation and utilization in genetic improvements and revealed the regulatory mechanisms of β-glucan synthesis during grain development in barley.
2. Materials and methods
2.1. Plant material and growth conditions
Two barley genotypes, BCS192 and BCS297 obtained from the International Barley Core Selected Collection (BCS), were used in this experiment. They have similar growth and grain characteristics (hulled, two-rowed, and the same heading time) but differ greatly in grain β-glucan content (Geng, Li, Xie, Wu, Ye & Zhang, 2021). The two barley genotypes were grown in a growth chamber in Zhejiang University, with 14 h of light and 10 h of dark, and the temperature of 22 °C during the day and 18 °C during the night, respectively. After heading, developing grains of the two barley genotypes were harvested at 7, 14, 21, and 28 days post-anthesis (DPA) for β-glucan content measurements and RNA extraction. For each genotype, 20 developing grains were obtained from the central part of a spike (shown in Fig. S1) at each developmental stage, and there were three biological replicates for each sampling.
2.2. Quantification of β-glucan content
The sampled barley grains were dried at 65 °C in an oven and thoroughly ground into flour with a grinder, followed by storage in a dryer before β-glucan content measurement. β-glucan content in barley flour was determined using a modified version of the Megazyme mixed-linkage β-glucan assay, according to our previous study (Geng, Li, Xie, Wu, Ye & Zhang, 2021), based on the “Streamlined method” (McCleary method; AOAC Method 995.16, AACC Method 32–23, ICC Standard Method No. 168).
2.3. mRNA library construction
The purified mRNA was fragmented into small pieces with fragment buffer. Then, first-strand cDNA was generated using random hexamer-primed reverse transcription, followed by second-strand cDNA synthesis. Repair was then performed by incubation with A-Tailing Mix and RNA Index Adapters. cDNA fragments obtained from the previous step were amplified by PCR, and products were purified with AMPure XP Beads and then dissolved in EB solution. The product was quality-controlled by the Agilent Technologies 2100 Bioanalyzer. The double-chain PCR product obtained in the previous step was heated and denatured by splinting the oligonucleotide sequence and recycled to obtain the final library. The single-strand circle DNA (ssCir DNA) was formatted as the final library, which was amplified with Phi29 to obtain a molecular DNA nanosphere (DNB) with over 300 copies. DNBs were loaded into the patterned nanoarray and pair-end 100 base reads were generated on the BGIseq500 platform (BGI-Shenzhen, China).
2.4. Read processing and identification of differentially expressed genes (DEGs)
Sequencing data were filtered by SOAPnuke (v1.5.2) (https://github.com/BGI-flexlab/SOAPnukeby) using the following steps: (1) Remove reads containing sequencing adapter; (2) Remove reads whose low-quality base ratio (base quality less than or equal to 5) is higher than 20 %; and (3) Remove reads whose unknown base ('N' base) ratio is higher than 5 %. Clean reads were generated and assessed using the Q20 and Q30, and then stored in FASTQ format. Clean reads were mapped to the reference genome of barley (ftp://ftp.ensemblgenomes.org/pub/plants/release48/fasta/hordeum_vulgare/dna/Hordeum_vulgare.IBSC_v2.dna.toplevel.fa.gz) using HISAT2 (v2.0.4) (https://www.ccb.jhu.edu/software/hisat/index.shtml). Bowtie2 (v2.2.5) (https://bowtiebio.sourceforge.net/%20Bowtie2%20/index.shtml) was applied to align the clean reads to the gene set, a database built by BGI (Beijing Genomic Institute in Shenzhen), including known and novel coding transcripts included. After alignment, normalized gene-level expression values expressed as fragments per kilobase pair of exon model per million fragments mapped (FPKM) were determined using RSEM (v1.2.12) (https://github.com/deweylab/RSEM). The heatmap was drawn by pheatmap (v1.0.8) (https://cran.r-project.org/web/packages/pheatmap/index.html) according to the gene expression in the different samples. Essentially, differential expression analysis was performed using the DESeq2 (v1.4.5) (https://www.bioconductor.org/packages/release/bioc/html/ DESeq2.html) with Q values ≤ 0.05.
2.5. Gene annotation, functional enrichment, and pathway enrichment analysis
To investigate the functions of these putative DEGs in the two genotypes, GO (https://www.geneontology.org/) and KEGG (https://www.kegg.jp/) enrichment analyses of annotated DEGs were performed by Phyper (https://en.wikipedia.org/wiki/Hypergeometric_distribution) based on the hypergeometric test. The significant levels of terms and pathways were corrected by Q values with a rigorous threshold (Q value ≤ 0.05).
2.6. Quantitative real-time PCR validation
To evaluate the accuracy of the RNA-seq data, 10 DEGs were randomly selected for qRT-PCR validation. cDNA synthesis was performed using the PrimeScript™ RT-PCR Master Mix kit (Takara, Japan) according to the manufacturer’s instructions. Absolute mRNA quantification of target genes was performed using a housekeeping gene: glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers used in the qRT-PCR experiment were designed by Genious (v10.22) and listed in Table S1. qRT-PCR was performed on a Light Cycler 480 System using the TB Green™ Premix Ex Taq™ II kit (Takara, Japan). A two-step PCR amplification procedure was conducted according to the standard procedure. The program was set as follows: 95 °C for 30 s, 95 °C for 5 s, and 60 °C for 20 s, with 40 cycles. Melting curves were then generated. Fold changes in target gene expression levels were calculated by the quantitative method (2−△△CT) (Schefe, Lehmann, Buschmann, Unger & Funke-Kaiser, 2006). Three replicates of qRT-PCR reactions were performed for each cDNA sample, including three negative controls with RNAse-free water.
3. Results
3.1. β-glucan accumulation during grain development
Changes in β-glucan content were determined in BCS192 and BCS297 during grain development, i.e., 7, 14, 21, and 28 DPA (Fig. 1). At 7 DPA, the β-glucan content of BCS192 was 1.01 %, which was significantly higher than that of BCS297 (0.7 %). Then, from 7 to 21 DPA, a consistent increase in grain β-glucan content was observed for the two genotypes. Although there were no significant differences between the two genotypes at 14 DPA, the difference became highly significant at 21 DPA, with the β-glucan content of BCS297 being 5.76 % and that of BCS192 being 4.99 %. Moreover, the difference in grain β-glucan content between the two genotypes became larger at 28 DPA, with those of BCS297 and BCS192 being 6.65 % and 4.51 %, respectively (Fig. 1A). Interestingly, BCS192 showed a decline and BCS297 showed a continuous increase in grain β-glucan content from 21 DPA to 28 DPA. The two barley genotypes displayed clear differences in their β-glucan accumulation patterns during grain development.
Fig. 1.
The grain morphology and β-glucan content of BCS192 and BCS297. A. Dynamic β-glucan content of BCS192 and BCS297 during four grain development stages: 7, 14, 21, and 28 days post-anthesis (DPA). The values are represented as mean value. Statistically significant differences were determined by one-way ANOVA (P < 0.05; n = 3; error bars indicate SE). B. The grains of BCS192 and BCS297 in four stages. I, II, III, and IV indicate time points 7, 14, 21, and 28 DPA, respectively. Bar = 1 cm.
3.2. Transcriptome profiles of developing grains
After removal of low-quality reads from the raw reads, a total of 10.49 billion clean reads and 157.39 Gb clean bases were obtained from 24 samples. Among them, clean reads with high-quality scores at Q20 level (a base quality greater than 20 and an error probability of 0.01) and Q30 level (a base quality greater than 30 and an error probability of 0.01) represented 94.84 % and 88.69 % of the total, respectively. Clean reads from BCS192 and BCS297 were mapped to the barley reference genome sequence with the proportion of 50.47 %–69.49 % and 54.83 %–63.4 %, respectively. Moreover, a high ratio of clean reads from BCS192 (79.83 %–83.9 %) and BCS297 (80.19 %–83.57 %) were aligned to the barley transcriptome gene set (Table S2). Finally, 30,849 known transcripts with different sequence sizes and 3621 novel genes were identified from the RNA-seq data. The abundance of expressed genes was determined by FPKM. In order to reflect the correlation of gene expression between different samples, Pearson correlation coefficients of all gene expression levels between every-two samples were calculated. The result indicated a high correlation between sequencing replicates in this study (Fig. S2).
3.3. Functional annotations of DEGs related to grain β-glucan accumulation during grain development
To identify the DEGs related to grain β-glucan accumulation in both genotypes, transcriptional changes were determined by comparing BCS192 and BCS297 with the standard of Fold Change ≥ 2 and Q-value < 0.05. For each barley genotype, gene expression changes in the distinct samples at 14, 21, and 28 DPA were each examined in pair-wise comparisons with those at 7 DPA. As a result, more DEGs were detected at the later stages of grain development in the two genotypes (Fig. S3A). Additionally, genotype-specific co-expressed DEGs were also determined. Overall, 1834 and 4693 co-expressed DEGs related to grain development were identified in BCS192 and BCS297, respectively. Among them, 1160 DEGs were co-modulated in both genotypes during grain development (Fig. S3B). The expression patterns of these co-modulated DEGs were displayed in different colors based on their expression profiles using heatmap software. These co-modulated DEGs could be mainly divided into two classes, each displaying up- and down-regulated expression patterns (Fig. S3C).
To explore the regulatory mechanisms of these co-modulated DEGs, functional enrichment GO term and KEGG pathway analyses were conducted. Overall, 20 GO terms involved in protein dimerization activity, carbohydrate metabolic process, chromosome, DNA packaging complex, protein-DNA complex and protein heterodimerization activity etc. were significantly enriched and commonly over-represented in both genotypes (Fig. S3D). Among them, carbohydrate metabolic process was proposed to be significantly associated with β-glucan biosynthesis, according to previous studies. Moreover, KEGG pathway enrichment analysis of co-modulated DEGs was performed to understand their biological functions. As a result, 7 significantly enriched pathways were identified for the co-modulated DEGs in the two genotypes, including starch and sucrose metabolism, phenylpropanoid biosynthesis, mRNA surveillance pathway, photosynthesis - antenna proteins, glycerolipid metabolism, RNA transport, and cyanoamino acid metabolism (Fig. S3E), indicating that these pathways may play vital roles in grain development. Genotype-specific DEGs were specially identified in this study. GO enrichment analysis showed that DEGs in BCS192 were significantly enriched in almost 20 GO terms, including carbohydrate metabolic process, tetrapyrrole binding, extracellular region, hydrolase activity (hydrolyzing O-glycosyl compounds), hydrolase activity (acting on glycosyl bonds), photosynthesis and thylakoid (Fig. S4A). A large proportion of the DEGs in BCS297 was significantly enriched in the GO terms carbohydrate metabolic process, transferase activity (transferring glycosyl groups), organic acid metabolic process, oxoacid metabolic process, and carboxylic acid metabolic process (Fig. S4B). Moreover, KEGG pathway enrichment analysis identified 10 significantly enriched pathways of the BCS192 genotype-specific DEGs, including phenylpropanoid biosynthesis, mRNA surveillance pathway, starch and sucrose metabolism, carbon metabolism, and photosynthesis (Fig. S4C). Similarly, 5 significantly enriched pathways (metabolic pathways) of the genotype-specific DEGs were identified in BCS297, including phenylpropanoid biosynthesis; biosynthesis of amino acids; glycolysis/ gluconeogenesis; flavonoid biosynthesis; and valine, leucine, and isoleucine biosynthesis (Fig. S4D).
3.4. Identification and classification of DEGs related to β-glucan biosynthesis
To understand the regulatory mechanisms of β-glucan accumulation during grain development, further GO enrichment analyses were performed on the co-modulated DEGs which identified in GO term carbohydrate metabolic process and KEGG pathway starch and sucrose metabolism in the two genotypes. Overall, 85 and 50 co-modulated DEGs were identified in the carbohydrate metabolic process and the starch and sucrose metabolism, respectively. Clustering and heatmap analyses showed that a larger proportion of those DEGs was down-regulated during grain development (Fig. 2A, 2B). In addition, GO enrichment analysis showed that those co-modulated DEGs were significantly enriched in GO terms associated with β-glucan accumulation, especially the GO terms hydrolase activity, hydrolase activity (acting on glycosyl bonds), hydrolase activity (hydrolyzing O-glycosyl compounds), glucan metabolic process, and cellular glucan metabolic process (Fig. 2C, 2D). These identified DEGs associated with β-glucan synthesis were classified based on the significantly enriched GO terms (Table S3). Furthermore, 10 DEGs were randomly selected and validated using qRT-PCR. High consistency (R2 = 0.91) was found between the results of RNA-seq and the qRT-PCR experiments (Fig. 2E), indicating a high reliability of the RNA-seq data.
Fig. 2.
Heatmap and GO enrichment analysis of differentially expressed genes (DEGs) related to grain β-glucan synthesis of BCS192 and BCS297 for grain dynamic development. A. Clustering and heatmap of 85 common DEGs of both BCS192 and BCS297 based on expression profiles. Numbers ranging from −10 to 10 indicate the multiples of difference. B. Clustering and heatmap of 50 common DEGs of both BCS192 and BCS297 based on the expression profiles. Numbers ranging from −10 to 10 indicate the multiples of difference. C. GO enrichment of 50 common DEGs related to grain β-glucan synthesis based on expression profiles. D. GO enrichment of 50 common DEGs related to grain β-glucan synthesis based on the expression profiles. E. Correlation between RNA-seq and qPCR data for DEGs related to β-glucan biosynthesis identified in BCS192 and BCS297. Each RNA-seq expression data point was plotted against that from quantitative real-time PCR and fit into a linear regression. Both x- and y-axes were shown in log2 scale.
3.5. Functional annotation of the DEGs related to β-glucan accumulation in the two genotypes
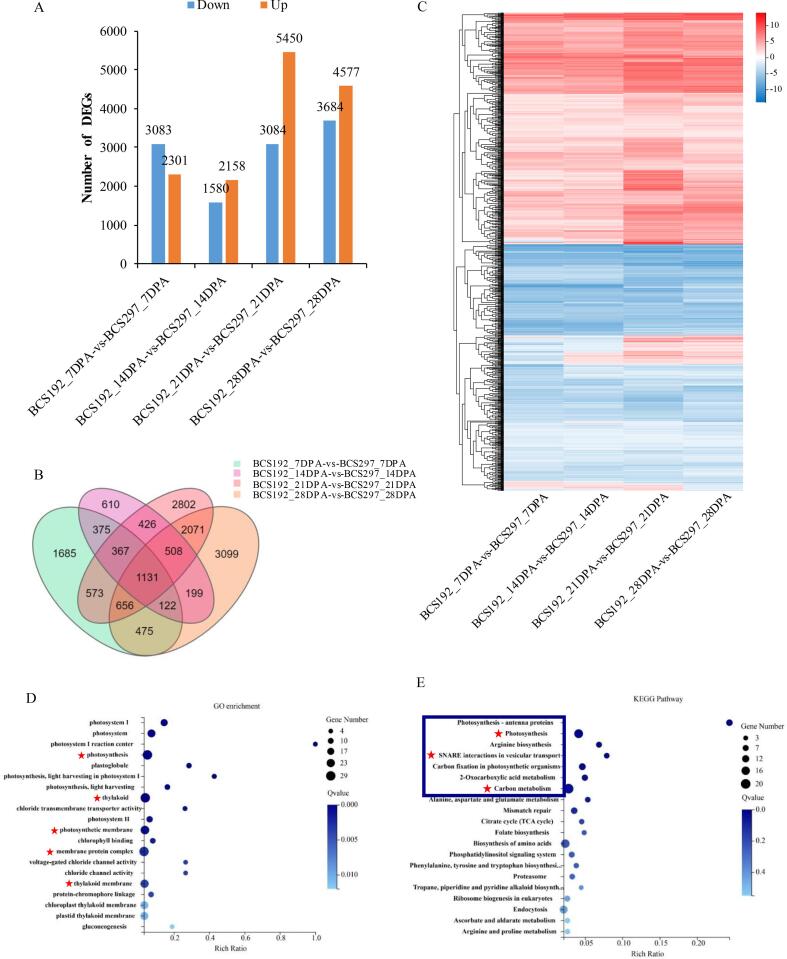
Transcriptional changes of the developing grains were compared between BCS192 and BCS297 with a standard of Fold Change ≥ 2 and Q-value < 0.05. Gene expression in the developing grains at 7, 14, 21, and 28 DPA was compared between BCS297 and BCS192. The expression levels of the up-regulated genes in BCS297 were higher than those in BCS192 at each grain development stage, while the opposite was true for the down-regulated genes. Totally, 15,099 DEGs were identified in BCS192 and BCS297 at four grain development stages. The highest number of DEGs was identified in samples obtained at 21 DPA, with 5,450 genes being up-regulated and 3,684 genes being down-regulated. At a later stage, the number of DEGs showed a slight decline (Fig. 3A). A Venn diagram of 15,099 DEGs in the two genotypes showed that 1131 DEGs were commonly expressed during four stages of grain development (Fig. 3B). Additionally, 11.16 %, 4.04 %, 18.56 %, and 20.52 % of all DEGs were uniquely expressed at 7, 14, 21, and 28 DPA, respectively (Fig. 3B).
Fig. 3.
Transcriptional changes and functional enrichment analysis among differentially expressed genes (DEGs) between BCS192 and BCS297 in four developmental stages. A. Statistic of differentially expressed genes including up-regulated and down-regulated genes in each comparison groups between BCS192 and BCS297. The expression changes of all genes were analyzed by performing pair-wise comparisons of BCS192 and BCS297 in four developmental stages. Samples at 7, 14,21, and 28 DPA group, respectively (7 vs 7, 14vs 14, 21 vs 21, 28 vs 28). Red indicates genes that were up-regulated in BCS297 compared with BCS192, while blue indicates genes that were down-regulated, log2FC = 1. B. Venn diagram analysis of grain development-related DEGs in BCS297 and BCS192 across four time points (7 vs 7, 14 vs 14, 21 vs 21, and 28 vs 28 DPA). log2FC = 1. C. Clustering and heatmap of 1,131 common DEGs based on their expression profiles in BCS192 and BCS297 across four comparison groups (7 vs 7, 14 vs 14, 21 vs 21, and 28 vs 28 DPA). Numbers ranging from −10 to 10 indicate the multiples of difference. D. GO enrichment of 540 commonly up-regulated DEGs during four grain developing stages. Red pentacles represent putative GO terms related to β-glucan content. E. KEGG pathway enrichment of 540 commonly up-regulated DEGs in four grain developing stages. The blue box indicates significantly enriched pathways and red pentacles represent putative KEGG pathways related to β-glucan content. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To explore factors affecting β-glucan accumulation, DEGs that were commonly expressed during the four developmental stages were further analyzed. The expression pattern of 1,131 DEGs in BCS192 and BCS297 could be mainly divided into two classes, up- and down-regulated DEGs (Fig. 3C). The regulatory mechanism of 540 up-regulated DEGs affecting β-glucan accumulation was analyzed to decipher the reason for the significantly higher β-glucan content of BCS297 compared to that of BCS192. GO enrichment analysis showed that the commonly up-regulated DEGs in the two genotypes were significantly enriched in 20 GO terms, especially those related to cellular components, such as photosynthesis, thylakoid, photosynthetic membrane, membrane protein complex, and thylakoid membrane. (Fig. 3D). Furthermore, KEGG pathway enrichment analysis was performed on these DEGs to understand their biological functions. Overall, 7 significantly enriched pathways were identified in the two genotypes, including photosynthesis - antenna proteins, photosynthesis, arginine biosynthesis, SNARE interactions in vesicular transport, carbon fixation in photosynthetic organisms, 2-oxocarboxylic acid metabolism, and carbon metabolism (Fig. 3E). Accordingly, we formulated the hypothesis that β-glucan accumulation is positively regulated by photosynthesis and carbon metabolism during grain development, as higher β-glucan content in BCS297 is consistent with the higher expressions of genes enriched in photosynthesis and carbon metabolism pathways compared to those in BCS192.
3.6. DEGs related to β-glucan biosynthesis at late grain development stages
As mentioned above, BCS192 showed a decline and BCS297 showed a continuous increase from 21 DPA to 28 DPA in grain β-glucan content. Thus, the DEGs of the two genotypes were compared at both 21 and 28 DPA. Overall, 380 DEGs were identified at the two stages. By combining functional annotations of DEGs and the results obtained in the previous studies, 22 DEGs affecting β-glucan accumulation during late developmental stages were selected (Table 1). Most of these DEGs (encoding alpha-amylase inhibitor, glucan endo-1,3-beta-glucosidase, and sugar transporter) showed different expression patterns in the two genotypes, which might explain the genotypic difference in changes in β-glucan content from 21 DPA to 28 DPA. These genes were up-regulated in BCS192 and down-regulated in BCS297, implying that they negatively regulate β-glucan biosynthesis. Furthermore, the genes encoding UDP-glycosyltransferase superfamily protein were up-regulated in both genotypes, suggesting that they positively regulate grain β-glucan accumulation.
Table 1.
Detailed information on the DEGs related to β-glucan biosynthesis in barley during the late developmental stages.
| Gene ID | log2(BCS192_28DPA/BCS192_21DPA) | log2(BCS297_28DPA/BCS297_21DPA) | Description |
|---|---|---|---|
| HORVU6Hr1G005900 | 1.049570541 | −1.458332637 | alpha/beta-Hydrolases superfamily protein |
| HORVU6Hr1G001150 | 1.062971704 | −1.819937234 | Alpha-amylase inhibitor 0.28 |
| HORVU2Hr1G122280 | 1.129132263 | −2.136165107 | Alpha-amylase inhibitor 4 |
| HORVU6Hr1G066330 | −2.105531567 | −1.485381007 | Alpha-amylase/trypsin inhibitor |
| HORVU7Hr1G035020 | 1.059890674 | −1.725241586 | Alpha-amylase/trypsin inhibitor |
| HORVU7Hr1G120960 | −1.24094519 | 1.002412471 | callose synthase 1 |
| HORVU1Hr1G067460 | 1.550872139 | −1.954333224 | Glucan endo-1,3-beta-glucosidase |
| HORVU4Hr1G002360 | 1.907078702 | −1.369124101 | Glucan endo-1,3-beta-glucosidase 8 |
| HORVU3Hr1G105630 | 1.611150579 | −2.065692524 | Glucan endo-1,3-beta-glucosidase GI |
| HORVU7Hr1G027860 | 1.050046245 | −1.678152579 | Glycogen debranching enzyme |
| HORVU1Hr1G021590 | 1.052187379 | −1.414706859 | Glycogen synthase |
| HORVU2Hr1G015720 | 4.837174216 | −3.093722363 | Hexosyltransferase |
| HORVU7Hr1G031650 | 1.882465504 | −1.656510162 | sugar transporter 4 |
| HORVU5Hr1G028030 | 2.588309792 | −1.665160806 | sugar transporter 9 |
| HORVU4Hr1G067450 | 2.234579331 | −2.571257213 | sugar transporter protein 7 |
| HORVU7Hr1G054710 | −2.1605587 | −1.361717672 | Sugar transporter SWEET |
| HORVU1Hr1G012680 | 1.112619697 | 1.045005462 | UDP-Glycosyltransferase superfamily protein |
| HORVU1Hr1G064410 | 3.839332083 | 2.218879372 | UDP-Glycosyltransferase superfamily protein |
| HORVU2Hr1G066780 | 2.009016988 | 1.881924568 | UDP-Glycosyltransferase superfamily protein |
| HORVU5Hr1G096240 | 1.314240367 | 2.826982946 | UDP-Glycosyltransferase superfamily protein |
| HORVU6Hr1G009880 | 1.266635141 | 1.08862923 | UDP-Glycosyltransferase superfamily protein |
| HORVU7Hr1G101710 | 1.509789801 | 1.638485479 | UDP-Glycosyltransferase superfamily protein |
3.7. Cellulose synthase-like genes identified in BCS192 and BCS297
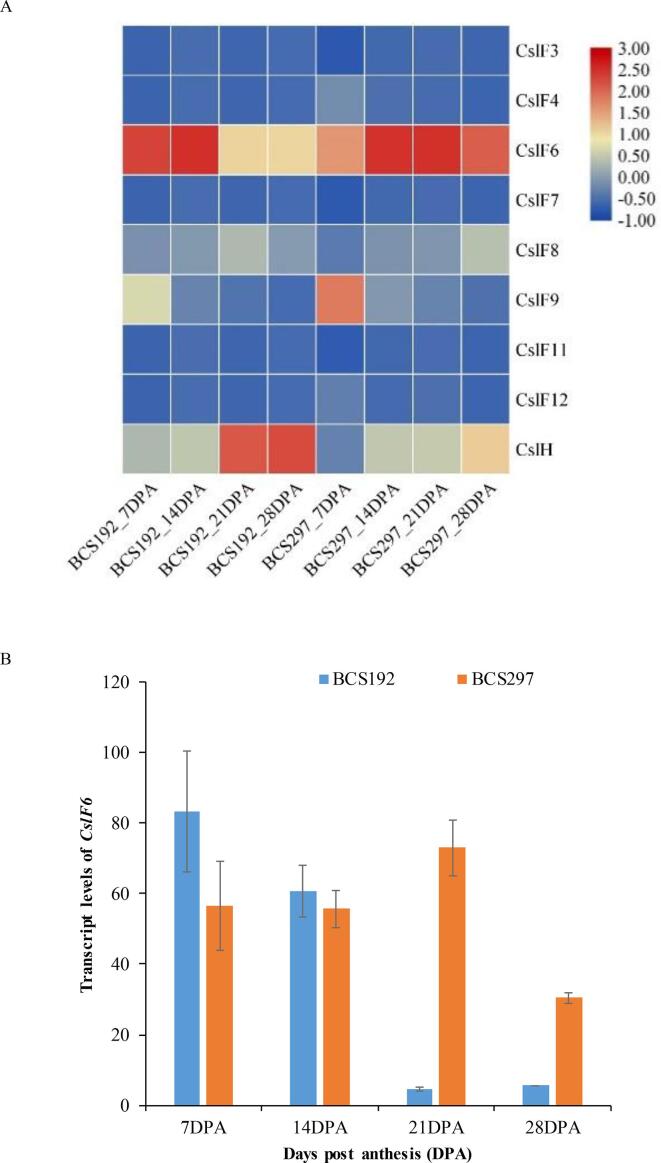
It is well documented that members of two cellulose synthase-like (CslF and CslH) subfamilies are responsible for β-glucan synthesis. In total, 8 CslFs and 1 CslH genes encoding cellulose synthase-like protein were identified in BCS192 and BCS297 during four grain development stages. Among them, only CslF6 and CslH in both genotypes and CslF9 in BCS297 (at 7 DPA) showed a relatively high transcript abundance, while that of others was nearly undetectable during grain development stages in both genotypes (Fig. 4A). CslH showed relatively high expression in BCS192 at 21 and 28 DPA. CslF6 is known as a main gene responsible for β-glucan biosynthesis, and its transcript levels were compared between BCS192 and BCS297 during grain development. As shown in Fig. 4B, the expression of CslF6 was genotype-dependent during the entirety of grain development. During early stages (7 and 14 DPA), the transcript abundance of CslF6 in BCS192 was relatively higher than that in BCS297, which is consistent with the differences in β-glucan content between the two genotypes. During 14 to 28 DPA, a significant decrease in CslF6 transcript abundance was observed in BCS192, but the opposite trend was found in BCS297; i.e., CslF6 expression showed a slight increase from 14 to 21 DPA, and then declined moderately at 28 DPA. Our results indicate that CslF6 expression levels at grain development stages may have a great impact on β-glucan content in barley grains.
Fig. 4.
Transcript levels of HvCslF family genes and HvCslH in developing grains of BCS192 and BCS297. A. Heat map of transcript expression of HvCslF family genes and HvCslH in developing grains of BCS192 and BCS297 at 7, 14, 21, and 28 DPA based on RNA-seq data. Expression data were averaged from three biological replicates per genotype and transcript expression values were calculated as LOG ranging from −1.00 to 300. Red indicates high expression, blue indicates low expression. B. Transcript levels of HvCslF6 in developing grains of BCS192 and BCS297 at 7, 14, 21, and 28 DPA from RNA-seq data. Expression data were averaged from three biological replicates per genotype. Bars indicate standard errors (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
To develop improved barley varieties with desirable traits, it is necessary to exploit elite germplasm and reveal the molecular mechanisms affecting grain quality during development. β-glucan content in barley grains has been especially addressed in recent years, as it is an important trait affecting grain quality for all kinds of uses (Griffey et al., 2010, Villacrés et al., 2019). However, very limited information is available on the biochemical and molecular mechanisms of β-glucan synthesis and accumulation in barley grains. In this study, RNA-seq analysis was conducted to compare the transcriptome profiles of two barley genotypes differing in grain β-glucan content and identify the genes responsible for β-glucan synthesis and accumulation during grain development.
Measurement of β-glucan contents in the grain samples obtained at different developmental stages indicated that β-glucan content varied greatly with development stages and genotypes. Overall, the β-glucan content of the two genotypes increased throughout grain development. However, its accumulation pattern differed greatly between the two genotypes at later developing stages (during 21 to 28 DPA), with BCS297 showing a consistent increase and BCS192 showing a slight decline. In our previous study, the β-glucan contents of BCS192 and BCS297 in mature grains were 2.37 % and 5.02 %, respectively (Geng, Li, Xie, Wu, Ye & Zhang, 2021), being dramatically lower than the values obtained in this study (4.51 % and 6.65 %, respectively). This difference could be attributed to environmental conditions (Geng, Li, Xie, Wu, Ye & Zhang, 2021). In the previous study, the experiment was conducted in the field, while the current study was carried out in a growth chamber. A slight decline in β-glucan content at the late grain-filling (developmental) stage was also found in a previous study (Aman, Graham & Tilly, 1989). Moreover, the greatest change in grain β-glucan content occurred from 7 to 14 DPA, suggesting that this is critical period for β-glucan accumulation in barley grains.
DEGs related to grain β-glucan accumulation were identified in the two barley genotypes. A total of 1,834 and 4,693 DEGs were identified in BCS192 and BCS297, respectively. Among them, 1,160 DEGs were co-modulated in both genotypes during grain development. Functional enrichment analysis of the co-modulated DEGs indicated that the GO terms carbohydrate metabolic process and starch and the sucrose metabolism KEGG pathway could be potentially involved in β-glucan synthesis. The DEGs identified through the above GO terms and KEGG pathways were further classified by GO enrichment analysis. These identified DEGs were primarily associated with hydrolase activity and glucan metabolic process, indicating that they may be directly or indirectly related to β-glucan accumulation during grain development. Especially, two DEGs (HORVU7Hr1G012380 and HORVU7Hr1G038420) encoding starch synthases and a gene (HORVU2Hr1G077120) encoding a starch branching enzyme were identified, revealing the link between β-glucan accumulation and starch biosynthesis. It was reported that mutations to the genes related to starch synthesis occurred in some barley varieties with high β-glucan content, indicating a regulatory link between grain starch and β-glucan contents (Clarke et al., 2008, Munck et al., 2004). The link is also involved in the regulation of sugar nucleotide levels, as ADP-Glc and UDP-Glc are glucose donors for starch and β-glucan synthesis, respectively (Trafford et al., 2013). The over-expression of a CslF gene finally resulted in almost a twofold increase in β-glucan content and a significant decrease in starch content (Burton et al., 2011), also demonstrating the regulatory link between starch and β-glucan synthesis. GWAS revealed that a locus on chromosome 7H, including the GBSS1 gene (granule bound starch synthase I), was significantly associated with grain β-glucan (Chutimanitsakun et al., 2013), again confirming the regulatory link between starch and β-glucan metabolism. However, the precise mechanism of the regulatory link remains unclear. It may be assumed that carbon partitioning between β-glucan and starch could be involved. In this study, we also noticed that a significantly enriched GO term transferase activity (transferring glycosyl groups) was only found in BCS297, which is consistent with our previous findings that the putative candidate genes encoding some enzymes (such as glacosylhydrolase, glycosyltranferase, and glucosidases) in glucose metabolism was closely associated with β-glucan content (Geng, Li, Xie, Wu, Ye & Zhang, 2021). Furthermore, it was once reported that the induction of starch synthase, starch branching enzyme, beta-glucosidases, and glycosyltransferases could enhance β-glucan synthesis (Zhang et al., 2021). Therefore, it may be suggested that a high expression of these genes increases the hydrolysis of polysaccharide glycosidic bonds, thus providing more substrates for β-glucan synthesis during grain development.
To explore the factors affecting β-glucan accumulation, we compared the transcript abundance of the relevant genes in the two genotypes at different grain development stages. Totally, 15,099 DEGs were identified in both BCS192 and BCS297. Of those, 1,131 DEGs were commonly expressed in the two genotypes. Functional enrichment analyses of the up-regulated DEGs showed that β-glucan accumulation is positively regulated by photosynthesis and carbon metabolism during grain development. In fact, previous studies have revealed a close association between light and β-glucan accumulation in cereal grains. Zhang et al. (2021) reported that β-glucan synthesis was regulated by light in oats (Zhang, Yan, Liu, Guo & Wu, 2021), and high light intensities could upregulate the expression of the β-glucan synthase gene AsCslF6. In barley, β-glucan content in seedlings was reduced significantly when transferred to continuous darkness from its naturally growing conditions (Roulin, Buchala & Fincher, 2002). A previous study also found that β-glucan degradation in dark-incubated wheat leaves could be completely reversed by light (Roulin & Feller, 2001). In the current study, GO enrichment analysis showed that a large proportion of up-regulated DEGs was significantly enriched the cellular components, such as photosystem, photosynthesis, thylakoid, photosynthetic membrane, membrane protein complex, and thylakoid membrane. Significantly enriched KEGG pathways of these up-regulated DEGs have been identified in photosynthesis, carbon metabolism, and endocytosis. As higher β-glucan content in BCS297 is consistent with the increased expression of the genes related to photosynthesis, it may be assumed that β-glucan accumulation in barley grains is positively regulated by photosynthesis.
CslF6 is dose-dependent in β-glucan synthesis, with endosperm-specific over-expression leading to significant increases in β-glucan content in barley grains (Burton et al., 2011). Similar roles of CslF6 orthologs were also found in wheat (Nemeth et al., 2010) and rice (Vega-Sanchez et al., 2012). In this study, relatively higher levels of CslF6 expression were found at 7 and 14 DPA. Moreover, BCS192 showed a significantly lower expression of CslF6 at 21 and 28 DPA in comparison with BCS297, which is consistent with its lower β-glucan content. These results support the idea that the expression level of CslF6 during grain development has a great impact on β-glucan accumulation in barley grains (Sie et al., 2015).
5. Conclusion
In this study, we identified DEGs associated with β-glucan biosynthesis and accumulation in barley grains using RNA-seq analysis. Overall, 1,160 DEGs were co-modulated in the two barley genotypes during grain development. The DEGs involved in carbohydrate metabolic process and starch and sucrose metabolism play significant roles in β-glucan synthesis. These DEGs are involved in hydrolase activity and glucan metabolic process. Moreover, there were 1,131 DEGs commonly expressed in the two genotypes during four developmental stages. A functional analysis of the up-regulated DEGs showed that β-glucan accumulation in barley grains is positively regulated by photosynthesis and carbon metabolism. A proposed model for β-glucan synthesis in barley grain was developed (Fig. 5), indicating that photosynthesis is important for providing energy and carbon sources for β-glucan and starch synthesis during grain development. It can be seen from the model that starch and sucrose may compete with β-glucan for glucose. On the other hand, they can also be metabolized to act as glucose donors for β-glucan synthesis. The model also showed some key enzymes and DEGs that are directly involved in β-glucan synthesis during grain development. The identified DEGs and their functions reported in this study may provide new insights into the molecular mechanisms of β-glucan synthesis in barley grains.
Fig. 5.
The proposed biological process of β-glucan synthesis during grain development. A. Proposed model system of barley grain β-glucan biosynthesis. The model shows some key enzymes and DEGs that are involved in β-glucan biosynthesis during grain development.
CRediT authorship contribution statement
La Geng: Investigation, Methodology, Data curation, Formal analysis, Visualization, Writing – original draft. Xinyi He: Data curation, Formal analysis, Visualization. Lingzhen Ye: Investigation, Methodology, Writing – original draft, Writing – review & editing, Project administration, Supervision, Funding acquisition. Guoping Zhang: Conceptualization, Methodology, Data curation, Formal analysis, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This research was funded by the National Key R&D Program of China (2018YFD1000706), Key Research Projects of Zhejiang Province (2021C02057, 2021C02064-3), China Agriculture Research System (CARS-05) and Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2022.100136.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References:
- Aman P., Graham H., Tilly A.C. Content and solubility of mixed-linked (1→3), (1→4)-beta-d-glucan in barley and oats during kernel development and storage. Journal of Cereal Science. 1989;10(1):45–50. [Google Scholar]
- Bian J., Deng P., Zhan H., Wu X., Nishantha M.D.L.C., Yan Z.…Song W. Transcriptional dynamics of grain development in barley (hordeum vulgare l.) International Journal of Molecular Sciences. 2019;20(4) doi: 10.3390/ijms20040962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin D., Maninder M., Hongzhi L., Baojun X. A concise review on the molecular structure and function relationship of beta-glucan. International Journal of Molecular Sciences. 2019;20(16):4032. doi: 10.3390/ijms20164032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R.A., Collins H.M., Kibble N.A.J., Smith J.A., Shirley N.J., Jobling S.A.…Fincher G.B. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-beta-d-glucans and alters their fine structure. Plant Biotechnology Journal. 2011;9(2):117–135. doi: 10.1111/j.1467-7652.2010.00532.x. [DOI] [PubMed] [Google Scholar]
- Burton R.A., Wilson S.M., Hrmova M., Harvey A.J., Shirley N.J., Stone B.A.…Fincher G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-d-glucans. Science. 2006;311(5769):1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Chutimanitsakun Y., Cuesta-Marcos A., Chao S., Corey A., Filichkin T., Fisk S.…Hayes P.M. Application of marker-assisted selection and genome-wide association scanning to the development of winter food barley germplasm resources. Plant Breeding. 2013;132(6):563–570. [Google Scholar]
- Clarke B., Liang R., Morell M.K., Bird A.R., Jenkins C.L.D., Li Z. Gene expression in a starch synthase IIa mutant of barley: Changes in the level of gene transcription and grain composition. Functional & Integrative Genomics. 2008;8(3):211–221. doi: 10.1007/s10142-007-0070-7. [DOI] [PubMed] [Google Scholar]
- Farrokhi N., Burton R.A., Brownfield L., Hrmova M., Wilson S.M., Bacic A., Fincher G.B. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnology Journal. 2006;4(2):145–167. doi: 10.1111/j.1467-7652.2005.00169.x. [DOI] [PubMed] [Google Scholar]
- Fincher G.B., Stone B.A. Cell walls and their components in cereal grain technology., Advances in Cereal. Science and Technology. 1986;8:207–295. [Google Scholar]
- Fuse Y., Higa M., Miyashita N., Fujitani A., Yamashita K., Ichijo T.…Hirose T. Effect of high beta-glucan barley on postprandial blood glucose and insulin levels in type 2 diabetic patients. Clinical Nutrition Research. 2020;9(1):43–51. doi: 10.7762/cnr.2020.9.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gimenez G., Barakate A., Smith P., Stephens J., Khor S.F., Doblin M.S.…Houston K. Targeted mutation of barley (1,3;1,4)-beta-glucan synthases reveals complex relationships between the storage and cell wall polysaccharide content. Plant Journal. 2020;104(4):1009–1022. doi: 10.1111/tpj.14977. [DOI] [PubMed] [Google Scholar]
- Geng L., Li M., Xie S., Wu D., Ye L., Zhang G. Identification of genetic loci and candidate genes related to beta-glucan content in barley grain by genome-wide association study in international barley core selected collection. Molecular Breeding. 2021;41(1) doi: 10.1007/s11032-020-01199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Li M., Zhang G., Ye L. Barley: A potential cereal for producing healthy and functional foods. Food Quality and Safety. 2022;6 [Google Scholar]
- Giridhar G., Paras S., Sandeep J., Longvah T. Effect of processing on barley beta-glucan content, its molecular weight and extractability. International Journal of Biological Macromolecules. 2020;162:1204–1216. doi: 10.1016/j.ijbiomac.2020.06.208. [DOI] [PubMed] [Google Scholar]
- Griffey C., Brooks W., Kurantz M., Thomason W., Taylor F., Obert D.…Hicks K. Grain composition of virginia winter barley and implications for use in feed, food, and biofuels production. Journal of Cereal Science. 2010;51(1):41–49. [Google Scholar]
- Hazen S.P., Scott-Craig J.S., Walton J.D. Cellulose synthase-like genes of rice. Plant Physiology. 2002;128(2):336–340. doi: 10.1104/pp.010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingzhao L., Baga M., Rossnagel B.G., Legge W.G., Chibbar R.N. Identification of quantitative trait loci for beta-glucan concentration in barley grain. Journal of Cereal Science. 2008;48(3):647–655. [Google Scholar]
- Kim H., Park K., Baek S., Kim J. Inheritance of (1–3)(1–4)-beta-d-glucan content in barley (hordeum vulgare l.) Journal of Crop Science and Biotechnology. 2011;14(4):239–245. [Google Scholar]
- McNab J.M., Smithard R.R. Barley beta-glucan: An antinutritional factor in poultry feeding. Nutrition Research Reviews. 1992;5(1):45–60. doi: 10.1079/NRR19920006. [DOI] [PubMed] [Google Scholar]
- Munck L., Moller B., Jacobsen S., Sondergaard I. Near infrared spectra indicate specific mutant endosperm genes and reveal a new mechanism for substituting starch with (1 -> 3,1 -> 4)-beta-glucan in barley. Journal of Cereal Science. 2004;40(3):213–222. [Google Scholar]
- Nemeth C., Freeman J., Jones H.D., Sparks C., Pellny T.K., Wilkinson M.D.…Shewry P.R. Down-regulation of the CSLF6 gene results in decreased (1,3;1,4)-beta-d-glucan in endosperm of wheat. Plant Physiology. 2010;152(3):1209–1218. doi: 10.1104/pp.109.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin S., Buchala A.J., Fincher G.B. Induction of (1 -> 3,1 -> 4)-beta-d-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta. 2002;215(1):51–59. doi: 10.1007/s00425-001-0721-1. [DOI] [PubMed] [Google Scholar]
- Roulin S., Feller U. Reversible accumulation of (1 -> 3,1 -> 4)-beta-glucan endohydrolase in wheat leaves under sugar depletion. Journal of Experimental Botany. 2001;52(365):2323–2332. doi: 10.1093/jexbot/52.365.2323. [DOI] [PubMed] [Google Scholar]
- Schefe J.H., Lehmann K.E., Buschmann I.R., Unger T., Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “gene expression's C-T difference” formula. Journal of Molecular Medicine-Jmm. 2006;84(11):901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- Sie C.W., Shirley N.J., Little A., Khoo K.H.P., Schwerdt J., Fincher G.B.…Mather D.E. Differential expression of the HvCslF6 gene late in grain development may explain quantitative differences in (1,3;1,4)-beta-glucan concentration in barley. Molecular Breeding. 2015;35(1):12–20. doi: 10.1007/s11032-015-0208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Zeng X., Wang Y., Bai L., Xu Q., Wei Z.…Nyima T. Transcriptomics analysis of hulless barley during grain development with a focus on starch biosynthesis. Functional & Integrative Genomics. 2017;17(1):107–117. doi: 10.1007/s10142-016-0537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trafford K., Haleux P., Henderson M., Parker M., Shirley N.J., Tucker M.R.…Burton R.A. Grain development in brachypodium and other grasses: Possible interactions between cell expansion, starch deposition, and cell-wall synthesis. Journal of Experimental Botany. 2013;64(16):5033–5047. doi: 10.1093/jxb/ert292. [DOI] [PubMed] [Google Scholar]
- Vega-Sanchez M.E., Verhertbruggen Y., Christensen U., Chen X., Sharma V., Varanasi P.…Ronald P.C. Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiology. 2012;159(1):56–69. doi: 10.1104/pp.112.195495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villacrés E., Campaña D., Garófalo J., Falconí E., Quelal M., Matanguihan J., Murphy K. Evaluation of β-glucan content, viscosity, soluble dietary fiber and processing effect in grains of ecuadorian barley genotypes. Agronomía Colombiana. 2019;37(3):323–330. [Google Scholar]
- Vis R.B., Lorenz K. Malting and brewing with a high beta-glucan barley. Lebensmittel-Wissenschaft and Technologie. 1998;31(1):20–26. [Google Scholar]
- Wouk J., Dekker R.F.H., Queiroz E.A.I.F., Barbosa-Dekker A.M. Beta-glucans as a panacea for a healthy heart? Their roles in preventing and treating cardiovascular diseases. International Journal of Biological Macromolecules. 2021;177:176–203. doi: 10.1016/j.ijbiomac.2021.02.087. [DOI] [PubMed] [Google Scholar]
- Zhang G., Zhang G., Zeng X., Xu Q., Wang Y., Yuan H.…Nyima T. Quantitative proteome profiling provides insight into the proteins associated with beta-glucan accumulation in hull-less barley grains. Journal of Agricultural and Food Chemistry. 2021;69(1):568–583. doi: 10.1021/acs.jafc.0c05284. [DOI] [PubMed] [Google Scholar]
- Zhang J., Yan L., Liu M., Guo G., Wu B. Analysis of beta-d-glucan biosynthetic genes in oat reveals glucan synthesis regulation by light. Annals of Botany. 2021;127(3):371–380. doi: 10.1093/aob/mcaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.