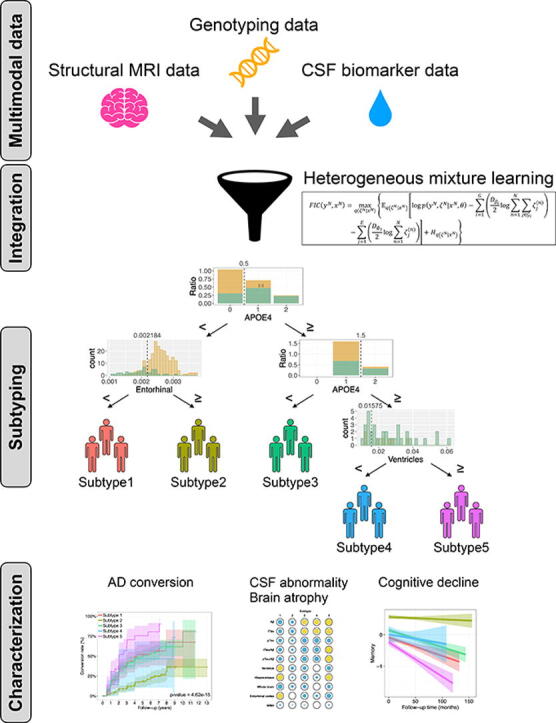
Graphical abstract
Keywords: Alzheimer’s disease, Mild cognitive impairment, Decision trees
Highlights
-
•
There is heterogeneity in the conversion from mild cognitive impairment to dementia.
-
•
Heterogeneous mixture learning (HML) models identified mild cognitive impairment subtypes.
-
•
Multimodal data were used to classify-five subtypes of mild cognitive impairment.
-
•
These subtypes showed low, moderate, or high risk patterns of conversion to AD.
-
•
Moderate risk was associated with brain atrophy or CSF biomarker abnormalities.
Abstract
Mild cognitive impairment (MCI) is a high-risk condition for conversion to Alzheimer's disease (AD) dementia. However, individuals with MCI show heterogeneous patterns of pathology and conversion to AD dementia. Thus, detailed subtyping of MCI subjects and accurate prediction of the patients in whom MCI will convert to AD dementia is critical for identifying at-risk populations and the underlying biological features. To this end, we developed a model that simultaneously subtypes MCI subjects and predicts conversion to AD and performed an analysis of the underlying biological characteristics of each subtype. In particular, a heterogeneous mixture learning (HML) method was used to build a decision tree-based model based on multimodal data, including cerebrospinal fluid (CSF) biomarker data, structural magnetic resonance imaging (MRI) data, APOE genotype data, and age at examination. The HML model showed an average F1 score of 0.721, which was comparable to the random forest method and had significantly more predictive accuracy than the CART method. The HML-generated decision tree was also used to classify-five subtypes of MCI. Each MCI subtype was characterized in terms of the degree of abnormality in CSF biomarkers, brain atrophy, and cognitive decline. The five subtypes of MCI were further categorized into three groups: one subtype with low conversion rates (similar to cognitively normal subjects); three subtypes with moderate conversion rates; and one subtype with high conversion rates (similar to AD dementia patients). The subtypes with moderate conversion rates were subsequently separated into a group with CSF biomarker abnormalities and a group with brain atrophy. The subtypes identified in this study exhibited varying MCI-to-AD conversion rates and differing biological profiles.
1. Background
Worldwide, 55 million people are affected by dementias, including Alzheimer’s disease (AD) dementia, which is characterized by the deposition of amyloid-beta (Aβ) protein and tau protein within the brain, and the number of affected individuals continues to increase [1]. Experimental drugs for AD have failed to prevent or slow cognitive decline in people with AD in clinical trials [2]. Although these drugs do not demonstrate excellent clinical efficacy in patients with late-stage AD, they could potentially be effective for the treatment of patients with early-stage AD or mild cognitive impairment (MCI), which makes it critical to identify the population of patients who will progress to late-stage AD. However, individuals with MCI show heterogeneity in patterns of pathology, and MCI does not always convert to AD dementia. Detailed subtyping of MCI can reduce heterogeneity in the individuals and increase statistical power for accurate prediction of the patients in whom MCI will convert to AD dementia. This may support new trial designs and enable the efficacy of drugs to be evaluated with small numbers of patients in clinical trials.
Petersen and Morris, who focused on the heterogeneity of MCI in early times, classified MCI into four clinical subtypes based on memory impairment [3]. This classification system divides MCI into amnestic and nonamnestic MCI, each further divided into a group with impairment in a single cognitive domain (single-domain MCI) and a group with impairments in multiple cognitive domains (multiple-domain MCI). It has been reported that amnestic MCI, regardless of whether it is single- or multiple-domain MCI, comparatively highly converts to dementia, mainly AD dementia, at a rate of 10 % to 15 % per year [4]. Recent studies based on neuropsychological tests or brain imaging data identified other MCI subtype classification systems in which different subtypes of MCI have different conversion rates to AD [5], [6], [7], [8], [9]. However, these classification systems are based on a single feature. Subtyping using data with multiple features (i.e., multimodal data) may provide a more detailed classification system, which may predict conversion to AD more accurately.
Machine learning approaches help to generate models for prediction and classification (e.g., subtyping) using multimodal data. In particular, nonlinear models such as deep learning have been shown to be useful for prediction and classification with high accuracy in various fields, including the medical field [10], [11]. However, it is difficult to interpret which criteria serve as the basis for predictions made by applied nonlinear models. This is because there is a trade-off between the predictive accuracy and the interpretability of the model [12]. Linear models based on a single decision tree allow the criteria for prediction or classification to be visualized, contributing to interpretability. However, due to the trade-off mentioned above, while linear models are superior in interpretability, they generally have lower prediction accuracy than nonlinear models.
To overcome these problems, we introduced a heterogeneous mixture learning (HML) method. HML is a type of hierarchical mixture of experts [13], [14], [15] that integrates multiple learners using a single decision tree. HML divides individuals into similar groups based on various features of the individuals and generates appropriate predictive models for each group. Using HML has several advantages, including the following: (1) the decision tree facilitates an understanding of how individuals are classified into their subtypes, (2) HML naturally prunes more complex branches of a decision tree, and (3) HML shows predictive accuracy comparable to that of nonlinear models [16], providing a more compact decision tree than those produced by other decision-tree-based methods. In this study, we applied the HML method to construct a versatile classification system of MCI based on multimodal data, namely, five brain region volumes, cerebrospinal fluid (CSF) biomarkers, including Aβ and tau, and genomic data on the apolipoprotein E (APOE) gene. Our HML model produced more interpretable and higher or comparable predictive accuracy than other decision-tree-based methods. We also characterized the subtypes of MCI identified by HML, including their conversion to AD dementia over time.
2. Materials and methods
2.1. ADNI dataset
The data used in this study were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) [17]. The ADNI was launched in 2003 as a public–private partnership led by Principal Investigator Michael W. Weiner, MD. The primary goal of the ADNI is to test whether serial magnetic resonance imaging (MRI) and positron emission tomography (PET) data and the analysis of other biological markers and clinical and neuropsychological assessments can be combined to characterize the progression of MCI and early AD. The ADNI dataset contains data from a large number of cognitively normal (CN), MCI, and AD dementia patients recruited from over 50 different centres in the US and Canada, with follow-up assessments performed every 6 months. Institutional review boards approved the study procedures for the institutions involved in the establishment of the ADNI dataset, and written informed consent was obtained from all participants whose data were used in the ADNI dataset.
This study considered data from 941 subjects (at baseline), comprising 305 CN subjects, 480 MCI subjects, and 156 AD dementia patients, that were included in a publicly available dataset of the ADNI called ADNIMERGE. The dataset included 309 participants (97 CN individuals, 144 MCI subjects, 68 AD dementia patients) from the ADNI 1 project and 632 participants (208 CN individuals, 336 MCI subjects, 88 AD dementia patients) from the ADNI GO/2 project. All subjects had records of CSF biomarker data, structural MRI data, APOE genotype data, and age at examination. The AD dementia patients and the MCI subjects were diagnosed mainly by neuropsychological tests (Mini–Mental State Examination (MMSE), Clinical Dementia Rating–Sum of Boxes (CDR-SB), and Wechsler Memory Scale Logical Memory II). The AD dementia patients who were analysed in this study had AD dementia or Alzheimer’s clinical syndrome diagnosed based on clinical diagnostic and neuropsychological test outcomes without assessments of the levels of pathological markers such as Aβ or tau protein. Table 1 shows a summary of each group.
Table 1.
Summary of sample characteristics at baseline.
| CN | MCI | AD dementia | |
|---|---|---|---|
| N | 305 | 480 | 156 |
| Age in years, mean ± SE | 73.7 ± 0.328 | 71.8 ± 0.339 | 74 ± 0.666 |
| Sex (Female:Male) | 162:143 | 200:280 | 69:87 |
| Education year, mean ± SE | 16.3 ± 0.151 | 16 ± 0.127 | 15.5 ± 0.214 |
| CSF Aβ(1–42) (pg/mL), mean ± SE | 1226 ± 25.27 | 964.2 ± 19.94 | 644.6 ± 22.97 |
| CSF tTau (pg/mL), mean ± SE | 238.4 ± 5.128 | 287.4 ± 6.264 | 373.8 ± 11.1 |
| CSF pTau (pg/mL), mean ± SE | 21.9 ± 0.529 | 27.9 ± 0.693 | 37.3 ± 1.18 |
| tTau / Aβ(1–42), mean ± SE | 2.33e-01 ± 9.01e-03 | 3.86e-01 ± 1.32e-02 | 6.51e-01 ± 2.45e-02 |
| pTau / Aβ(1–42), mean ± SE | 2.19e-02 ± 9.78e-04 | 3.82e-02 ± 1.43e-03 | 6.52e-02 ± 2.55e-03 |
| Whole-brain volume / ICV, mean ± SE | 6.94e-01 ± 2.52e-03 | 6.83e-01 ± 2.28e-03 | 6.45e-01 ± 3.37e-03 |
| Hippocampus volume / ICV, mean ± SE | 5.01e-03 ± 3.37e-05 | 4.5e-03 ± 3.74e-05 | 3.84e-03 ± 5.27e-05 |
| Brain-ventricular volume / ICV, mean ± SE | 2.14e-02 ± 5.9e-04 | 2.45e-02 ± 5.87e-04 | 3.05e-02 ± 9.25e-04 |
| Entorhinal cortex volume / ICV, mean ± SE | 2.58e-03 ± 2.16e-05 | 2.33e-03 ± 2.19e-05 | 1.86e-03 ± 3.48e-05 |
| WMH volume, mean ± SE | 4.03 ± 0.429 | 5.02 ± 0.385 | 4.75 ± 0.539 |
| APOE ε4 carriers (%) | 81 (26.6%) | 239 (49.8%) | 108 (69.2%) |
Abbreviations are as follows: CN, Cognitively normal; MCI, Mild cognitive impairment; AD, Alzheimer's disease; CSF, Cerebrospinal fluid; Aβ, Amyloid-beta; tTau, Total tau; pTau, Phosphorylated tau; ICV, intracranial volume; WMH, White matter hyperintensity; APOE, Apolipoprotein E; SE, Standard error.
2.2. ADNI measures
We generated models using the following five CSF biomarkers, five brain volumes, APOE genotype, and age at examination (sections 2.2.1–3). The other data shown in sections 2.2.4–6 were used to characterize each subtype.
2.2.1. CSF biomarkers
The CSF biomarkers comprised the following five measures: Aβ(1–42) peptide levels, total tau (tTau) protein levels, phosphorylated tau (pTau) protein levels, the tTau/Aβ(1–42) ratio, and the pTau/Aβ(1–42) ratio. The levels of Aβ(1–42), tTau, and pTau were obtained from the ADNI; these data were initially acquired using Roche Elecsys® immunoassays (Roche Diagnostics GmbH, Penzberg, Germany). We calculated the tTau/Aβ(1–42) ratio and pTau/Aβ(1–42) ratio based on the levels of these three CSF biomarkers. The CSF biomarkers were obtained as quantitative variables, but the level of each was often represented by a string containing an inequality sign when the biomarker level reached the upper limit or was below the detection limit of the immunoassays. In these cases, we treated “>1700” for Aβ(1–42) as 1,700 pg/mL and “>1300″ for tTau as 1,300 pg/mL. Similarly, ”<8″ and “>120″ for pTau were treated as 8 pg/mL and 120 pg/mL, respectively.
2.2.2. Structural MRI
Structural MRI data were obtained from the ADNI dataset for the following five markers: whole-brain volume, brain-ventricular volume, hippocampal volume, entorhinal cortex volume, and white matter hyperintensity (WMH) volume. We normalized these volumes as fractions of the intracranial volume (ICV). WMH volumes were calculated based on coregistered T1-, T2-, and proton density-weighted structural MRI images. The ADNIMERGE dataset included results from a 1.5 T MRI scan in the ADNI 1 and a 3.0 T MRI scan in the ADNI GO/2. Cortical reconstruction and volumetric segmentation were performed by the University of California San Francisco with FreeSurfer image analysis suite (version 4.3 for 1.5 T MRI scans and version 5.1 for 3.0 T MRI scans). The scan data were processed cross-sectionally using the 2010 Desikan-Killany atlas. The acquisition parameters were as follows: an inversion time of 1,000 ms (1.5 T MRI) and 853–900 ms (3.0 T MRI); repetition time of 2,400 ms (1.5 T MRI) and 2,300 or 3,000 ms (3.0 T MRI); flip angle of 8°; field of view of 240 × 240 mm2; in-plane resolution of 192 × 192 (1.25 × 1.25 mm2) (1.5 T MRI) or 256 × 256 (0.94 × 0.94 mm2) (3.0 T MRI); and slice thickness of 1.2 mm [18]. We combined these data because the difference between 1.5 T and 3.0 T did not significantly affect the prediction of AD conversion (p = 0.970, likelihood ratio test; Supplemental information).
2.2.3. APOE genotype
APOE genotyping data were obtained from blood DNA samples from each individual using an APOE genotyping kit. APOE has 3 alleles (ε2, ε3, and ε4) and 6 genotypes (ε22, ε23, ε24, ε33, ε34, and ε44). We focused our analysis on the number of ε4 alleles, as the ε4 allele is broadly known to be a risk factor for AD.
2.2.4. Composite scores of cognitive domains
We obtained the composite scores of four cognitive domains (memory, executive function, language, and visuospatial function) from the ADNI dataset. These scores were constructed based on a bifactor model [19], [20]. Detailed protocols for these composite scores are available for download at https://ida.loni.ucla.edu/.
-
-
The composite score for memory was calculated based on the following tests: the word lists from the three forms of the Alzheimer's Disease Assessment Scale–Cognitive Subscale (ADAS-Cog), the word lists from the two forms of the Rey Auditory Verbal Learning Test (RAVLT), the three-word recall items from the MMSE, and Logical Memory scores.
-
-
The composite score for executive function was calculated based on the following tests: the category fluency tests for animals and vegetables, the Trail Making Test (parts A and B), the Digit Span Backwards test, the Wechsler Adult Intelligence Scale–Revised (WAIS-R) Digit Symbol Substitution Test, and five clock-drawing items (circle, symbol, numbers, hands, time).
-
-
The composite score for language was calculated based on the following tests: a neuropsychological battery (including three language-related tests), the MMSE (including eight language tasks), the ADAS-Cog (including three different language tasks), and the Montreal Cognitive Assessment (MoCA) (including six language items).
-
-
The composite score for visuospatial function was calculated based on the following tests: a neuropsychological battery including five tests related to copying a clock, the constructional praxis test from the ADAS-Cog, and the copy design test in the MMSE.
2.2.5. CSF markers of neuronal injury and the inflammatory response
We obtained data on the CSF levels of the following markers from the ADNI dataset: the neuronal injury marker Visinin-like protein-1 (VILIP-1); the synaptic dysfunction markers Synaptosomal-associated protein, 25 kDa (SNAP-25) and Neurogranin (NGRN); and the inflammation marker YKL-40. The levels of VILIP-1, SNAP-25 and NGRN were obtained using the Erenna® immunoassay system (Singulex Inc., Alameda, CA, USA). The levels of YKL-40 were obtained using a MicroVue YKL-40 ELISA (Quidel, San Diego, CA, USA). We analysed the marker levels at baseline in 62 MCI subjects with data on all four markers.
2.3. Prediction model and subtyping
2.3.1. HML model
We applied HML to subtype MCI subjects. HML constructs a decision tree and generates a predictive model at each leaf node, and each leaf node can be regarded as a subgroup (i.e., subtype) with similar characteristics [14], [15]. As described in section 2.3.3, HML simultaneously estimates the parameters for a decision tree and the prediction models using the expectation–maximization (EM) algorithm based on the factorized information criterion (FIC), which is an estimator specific to HML (Supplemental information). A program for HML was provided by NEC Corporation (Tokyo, Japan).
2.3.2. Decision tree in the HML model
We had observation data , where ; N is the number of individuals; and D is the number of dimensions in (variables considered in the study), which, for this study, was equal to 12 (i.e., 5 CSF biomarkers, 5 brain volumes, APOE genotype, and age at examination). HML was used to create a decision tree in which the gating nodes were nonleaf nodes and the expert nodes were leaf nodes (Figure S1). The i-th gating node assigns an individual as input data to an appropriate expert node for prediction based on the rule , where and are the index of a variable and a threshold, respectively, in a gating node . A binary logistic regression model was used in the expert nodes. The prediction model in the j-th expert node is presented in the following equation:
| (1) |
Let us denote the classification target as , where corresponds to and indicates a weight vector of parameters in the j-th expert node.
2.3.3. Estimation of parameters by EM-like iterative optimization
To obtain a decision tree model via HML, we needed the parameters for the gating nodes (i.e., , , and ) and the expert nodes (i.e., ). These parameters were estimated by EM-like iterative optimization (Algorithm 1 in Supplemental information). In the E-step, the variational distribution, which was derived from the FIC, has a regularizing effect and penalizes the expert nodes that contribute to the formation of a complex tree structure that has more variables with small effects (Supplemental information). In this manner, HML automatically selects an optimal decision tree and optimal model parameters to maximize the FIC [14], [15].
2.3.4. Test performance of the HML model
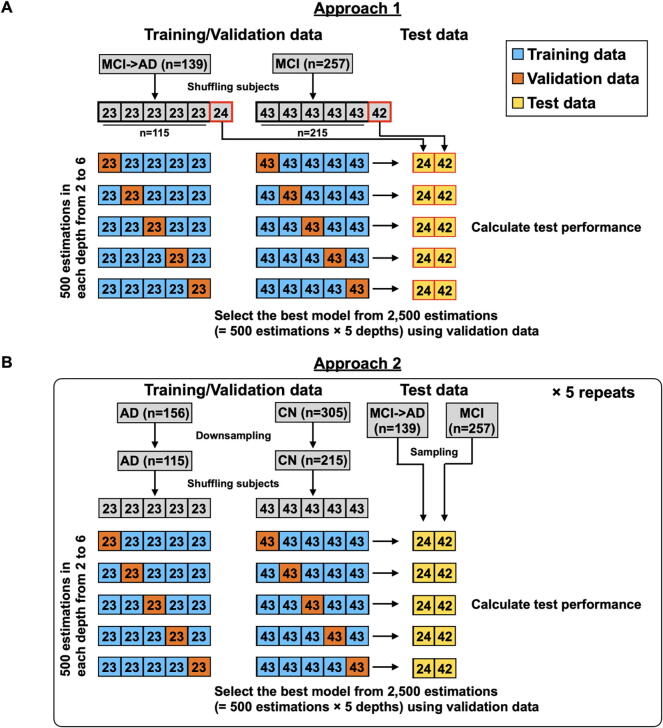
To test the performance of the HML model, we tested two approaches based on the HML method. Approach 1 used the baseline datasets from MCI subjects who converted to AD within 3 years (n = 139) and MCI subjects who did not convert to AD (n = 257) as training, validation, and test datasets (Fig. 1). Approach 2 used the baseline datasets from all 156 AD dementia patients and 305 CN subjects as training and validation data and used the baseline datasets from MCI subjects as test data. The difference between Approach 1 and 2 is whether MCI subjects or AD and CN subjects were used as training and validation data. On the other hand, both approaches used MCI subjects as test data. In Approach 2, we randomly sampled AD and CN subjects to match the sample size of Approach 1 and repeated 5 independent rounds of random sampling to avoid sampling bias. Each of these approaches was used to determine a decision tree and model parameters via HML. In the training and validation data, the classification target was when a subject was an MCI subject who converted to AD dementia within three years (Approach 1) or was an AD dementia patient (Approach 2). On the other hand, the classification target was when a subject was an MCI subject who did not convert to AD dementia within three years (Approach 1) or was a CN individual (Approach 2). Among all MCI subjects, 74.6 % were followed up for more than 3 years (Figure S2). Then, only data from MCI subjects who were followed for more than 3 years or who converted to AD within 3 years were used in this test (396 of 480 MCI subjects). In the test data for both approaches, the classification target was when MCI converted to AD dementia within three years and when there was no conversion to AD dementia. Approach 2 assumed that MCI subjects classified as AD would convert to AD in the future since they had already started to exhibit AD pathology.
Fig. 1.
Procedures for 5-fold cross-validation (CV) to estimate model parameters based on heterogeneous mixture learning (HML).
Using the training dataset, we first set the tree depth d to a value ranging from two to six. Then, we estimated parameters via HML. As mentioned above, HML optimizes the parameters based on EM-like iterative optimization. It is well known that EM-like iterative optimization generally converges to a local optimum depending on an initial value and is not guaranteed to converge to the global optimum. To avoid convergence to a local optimum, we generated 500 models with different initial values at each depth. We next applied the validation dataset to the 2,500 models (= 5 depths × 500 models) generated from the training data and adopted the decision tree model with the highest F1 in the validation dataset as the model with optimal parameters (Fig. 1). We finally calculated the test performance of the model using the test data. These procedures were repeated using a 5-fold cross-validation (CV) approach.
(A) The procedure for Approach 1 used subjects with mild cognitive impairment (MCI) that converted to Alzheimer's disease (AD) within 3 years (n = 139) and subjects with MCI that did not convert to AD (n = 257). (B) The procedure for Approach 2 used AD dementia patients (n = 156) and cognitively normal (CN) subjects (n = 305). Approach 2 was repeated 5 times. Blue, orange, and yellow boxes indicate training, validation, and test data, respectively.
An HML decision tree model generated from the training data classified (positive) or (negative). For test performance, the individuals with who were predicted to be positive were defined as true positives (TPs). The individuals with who were predicted to be negative were defined as false negatives (FNs). In the same way, the individuals with who were predicted to be positive or negative were defined as false positives (FPs) and true negatives (TNs), respectively. We calculated sensitivity, specificity, precision, accuracy, and F1 using these four outcomes as follows:
| (2) |
The conversion from MCI to AD dementia in each subject is presented as a time-to-event value, that is, the number of days from age at baseline to age at onset. In this study, we defined the data for the MCI subjects in whom MCI did not convert to AD dementia during the follow-up period as censored data. The log-rank test was performed to evaluate the differences in conversion among the MCI subtypes. The conversion rate at time t (CRt) was calculated by the following equation:
| (3) |
where nt is the number at risk (the number of MCI subjects) at time t and ct is the number of individuals with MCI who converted to AD dementia during the period from time t-1 to time t.
2.3.5. CART and random forest
We compared the prediction performance of HML with two different methods based on the decision tree: CART [21] and random forest [22] methods. The test performances by each method were calculated using the same training, validation and test datasets used for the HML assessment based on Approach 2 (Fig. 1B). For the CART method, we set the tree depth d to a value ranging from two to six. The GridSearchCV function from the Python scikit-learn package [23] optimized the following parameters in CART: the maximum depth of the tree (2, 3, 4, 5, and 6); the criterion (the “Gini impurity” or the “information gain”); the minimum number of samples required to be at a leaf node (1,…,11); the minimum number of samples required to split an internal node (2,…,11); the random state (0,…,101); and the strategy used to choose the split at each node (“best” or “random”). For the CART method, classification rather than regression was used for binary classification to discriminate between AD or CN. For the random forest method, we used the RandomForestClassifier function from the Python scikit-learn package to implement the random forest method. We adopted the model with the highest F1 score on a validation dataset and calculated the test performance of the adopted model on the test dataset.
2.3.6. MCI subtyping
For MCI subtyping, we focused on a decision tree generated by HML. As will be described in section 3.1, we found that the test performances of Approach 2, which trained AD and CN subjects, were better than Approach 1, which trained MCI subjects, in predicting conversion from MCI to AD. Therefore, a decision tree for MCI subtyping was generated using all AD and CN subjects according to Approach 2. We adopted the decision tree model with the highest F1 among the 2,500 models (= 5 depths × 500 models with different initial values) generated from AD and CN subjects. The F1 score was calculated as the predictive accuracy of AD conversion using all MCI subjects.
2.4. Analyses of composite scores for cognitive domains
Multiple pairwise comparisons were performed with Tukey’s honest significant difference (HSD) test to verify the difference between the baseline scores across different subtypes. We performed linear mixed model (LMM) analyses to compare the associations between MCI subtype and cognitive function with increasing follow-up times. Subtype 2, which is mentioned in the following subsections, was used as the reference. The independent variables included MCI subtype, follow-up time, and the interactions between subtype and follow-up time. The covariates included age, sex, and years of education. The composite scores for cognitive function were used as dependent variables. The random factors included the intercept and follow-up time. Separate models were run for the four domains of cognitive function. We used the false discovery rate (FDR) method to correct for multiple testing.
3. Results
3.1. Test performance of a decision tree model obtained by HML
All test performances of Approach 2, which trained AD and CN subjects, were higher than those of Approach 1, which trained MCI subjects (Table 2) (see 2.3.4 in Materials and methods). These results suggested that a clear discrimination between AD and CN subjects (i.e., Approach 2) led to a more accurate prediction of conversion to AD than the discrimination of a subtle difference between subjects with MCI that converted to AD and those with MCI that did not convert (i.e., Approach 1).
Table 2.
Test performance of each method.
| HML (Approach 1) | HML (Approach 2) | CART | Random forest | |
|---|---|---|---|---|
| Sensitivity | 0.574 ± 0.079 | 0.765 ± 0.011 | 0.682 ± 0.024 (p=0.003)1 |
0.725 ± 0.012 (p=0.231)1 |
| Specificity | 0.848 ± 0.036 | 0.825 ± 0.008 | 0.806 ± 0.011 (p=0.221)1 |
0.834 ± 0.005 (p=0.685)1 |
| Precision | 0.694 ± 0.055 | 0.695 ± 0.010 | 0.658 ± 0.011 (p=0.014)1 |
0.705 ± 0.005 (p=0.710)1 |
| Accuracy | 0.760 ± 0.037 | 0.792 ± 0.006 | 0.760 ± 0.008 (p=0.002)1 |
0.795 ± 0.003 (p=0.956)1 |
| F1 | 0.610 ± 0.039 | 0.721 ± 0.008 | 0.663 ± 0.015 (p<0.001)1 |
0.713 ± 0.006 (p=0.853)1 |
| # of leaf nodes | 3.8 ± 1.095 | 4.2 ± 0.236 | 26.2 ± 2.145 (p<0.001)2 |
– |
Each value shows mean ± SE of performances obtained from 5-fold CV.
1Tukey's HSD tests were performed on three groups: HML (Approach 2), CART, and random forest, and the p-value between HML and the corresponding algorithm is shown.
2The p-value between HML (Approach 2) and CART by Student's t-test is shown.
To compare the prediction performance of HML with the other methods, we next evaluated the test performance of the CART and random forest methods, which are recognized as traditional decision tree methods. All test performances except for the specificity of the HML model (Approach 2) were significantly higher than those of the CART model (Table 2). On the other hand, the HML model showed comparable performance with the random forest method. In addition, the comparison of model complexities showed that the HML method resulted in significantly fewer leaf nodes (expert nodes in HML) than the CART method; thus, the HML method provided a more compact decision tree, improving part of the interpretability. We did not compare the number of leaf nodes based on the HML and random forest methods because random forest is a method that uses multiple decision trees. These analyses showed that HML is able to predict conversion to AD with one decision tree that is more interpretable than conventional methods and with the same or better performance than conventional methods.
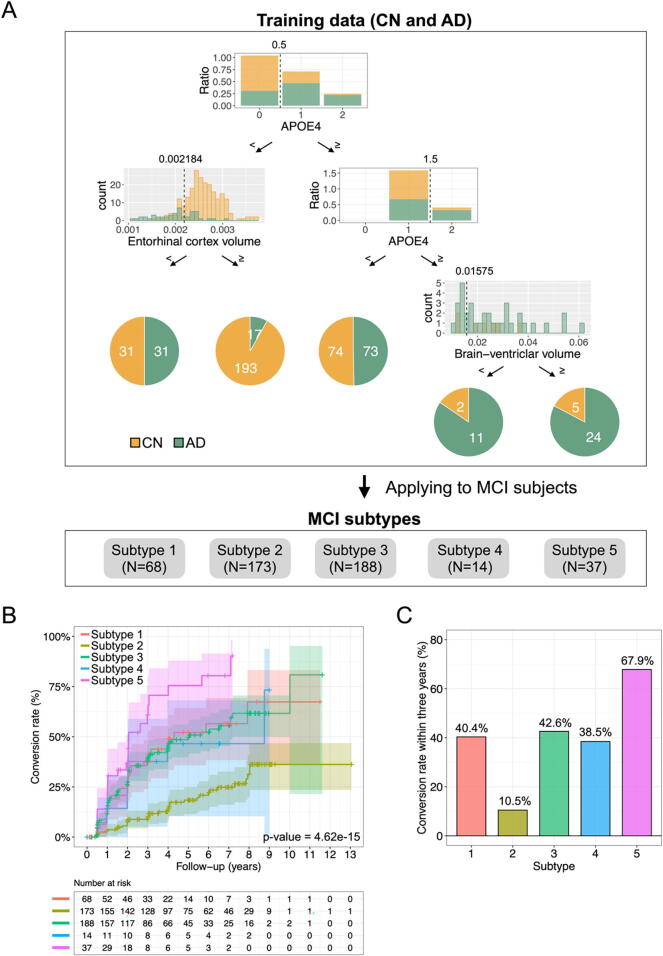
3.2. Characteristics of each subtype
We next examined the subtype of patients with MCI using a decision tree generated by HML and characterized the identified subtypes. The decision tree generated using Approach 2 (with all data from 156 AD dementia patients and 305 CN individuals in this section) had five expert nodes (Fig. 2A). This decision tree divided subjects based on the presence or absence of APOE ε4 alleles, and subjects without APOE ε4 alleles were further divided into two groups based on entorhinal cortex volume. Subjects with APOE ε4 alleles were then divided into two groups: those with one APOE ε4 allele and those with two APOE ε4 alleles. Subjects with two APOE ε4 alleles were subsequently divided into two groups based on brain-ventricular volume. Individuals included in a particular expert node on a decision tree constitute a group of individuals with similar features. We then used this decision tree to classify the MCI individuals in our dataset into one of the five expert nodes. The 480 MCI subjects were divided as follows: 68 subjects were in subtype 1, 173 were in subtype 2, 188 were in subtype 3, 14 were in subtype 4, and 37 were in subtype 5 (Table 3). We compared the conversion rates of MCI to AD dementia in the subjects in each subtype to characterize each subtype (Fig. 2B and 2C). The Kaplan–Meier curves showed different conversion patterns across subtypes. Notably, 67.9 % of the MCI subjects in subtype 5 progressed to AD dementia within three years. On the other hand, the conversion rates in subtypes 1, 3, and 4 were moderate at approximately 40 %. Subtype 2 had a comparatively low conversion rate of 10.5 %.
Fig. 2.
The HML-based decision tree classifies the MCI subjects into five subtypes. (A) Decision tree model generated by HML. The dotted lines in each gating node represent each threshold. The numbers in yellow and green areas of each pie chart indicate the number of subjects with CN and AD, respectively. (B) Conversion rates over time for each MCI subtype (p = 4.62e-15 in the log-rank test). (C) Conversion rates within three years for each MCI subtype. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Summary of each subtype.
| Subtype 1 | Subtype 2 | Subtype 3 | Subtype 4 | Subtype 5 | |
|---|---|---|---|---|---|
| N | 68 | 173 | 188 | 14 | 37 |
| Age in years, mean ± SE | 73.9 ± 0.99 | 71.8 ± 0.568 | 71.4 ± 0.529 | 66 ± 1.62 | 72 ± 0.948 |
| Sex (female:male) | 25:43 | 72:101 | 82:106 | 9:5 | 12:25 |
| Years of education, mean ± SE | 15.8 ± 0.355 | 16.3 ± 0.202 | 15.8 ± 0.206 | 17 ± 0.756 | 16.1 ± 0.452 |
| CSF Aβ(1–42) (pg/mL), mean ± SE | 923.3 ± 46.37 | 1211 ± 32.32 | 852.1 ± 28.32 | 713.9 ± 68.01 | 549.7 ± 30.4 |
| CSF tTau (pg/mL), mean ± SE | 273.2 ± 17.62 | 235.8 ± 7.491 | 323.7 ± 10.54 | 416 ± 56.81 | 321.7 ± 18.28 |
| CSF pTau (pg/mL), mean ± SE | 25.9 ± 1.95 | 22.1 ± 0.843 | 32 ± 1.15 | 42.6 ± 6.5 | 31.6 ± 1.96 |
| tTau / Aβ(1–42), mean ± SE | 3.53e-01 ± 2.85e-02 | 2.37e-01 ± 1.36e-02 | 4.66e-01 ± 2.2e-02 | 6.95e-01 ± 1.47e-01 | 6.25e-01 ± 3.56e-02 |
| pTau / Aβ(1–42), mean ± SE | 3.4e-02 ± 3.07e-03 | 2.29e-02 ± 1.52e-03 | 4.68e-02 ± 2.38e-03 | 7.21e-02 ± 1.66e-02 | 6.13e-02 ± 3.59e-03 |
| Whole-brain volume / ICV, mean ± SE | 6.49e-01 ± 4.88e-03 | 6.99e-01 ± 3.63e-03 | 6.81e-01 ± 3.65e-03 | 7.24e-01 ± 8.36e-03 | 6.72e-01 ± 6.86e-03 |
| Hippocampus volume / ICV, mean ± SE | 3.83e-03 ± 7.68e-05 | 4.87e-03 ± 5.18e-05 | 4.49e-03 ± 6e-05 | 4.91e-03 ± 2.16e-04 | 3.87e-03 ± 8.39e-05 |
| Brain-ventricular volume / ICV, mean ± SE | 3.26e-02 ± 1.69e-03 | 2.21e-02 ± 8.44e-04 | 2.44e-02 ± 9.93e-04 | 1.18e-02 ± 6.66e-04 | 2.61e-02 ± 1.36e-03 |
| Entorhinal cortex volume / ICV, mean ± SE | 1.83e-03 ± 3.46e-05 | 2.59e-03 ± 2.24e-05 | 2.3e-03 ± 3.65e-05 | 2.63e-03 ± 1.42e-04 | 2.05e-03 ± 6.6e-05 |
| WMH volume, mean ± SE | 5.78 ± 1.17 | 5.48 ± 0.624 | 4.29 ± 0.618 | 4.71 ± 1.71 | 5.22 ± 1.25 |
| APOE ε4 carriers (%) | 0 (0%) | 0 (0%) | 188 (100%) | 14 (100%) | 37 (100%) |
Abbreviations are as follows: CN, Cognitively normal; MCI, Mild cognitive impairment; AD, Alzheimer's disease; CSF, Cerebrospinal fluid; Aβ, Amyloid-beta; tTau, Total tau; pTau, Phosphorylated tau; ICV, intracranial volume; WMH, White matter hyperintensity; APOE, Apolipoprotein E; SE, Standard error.
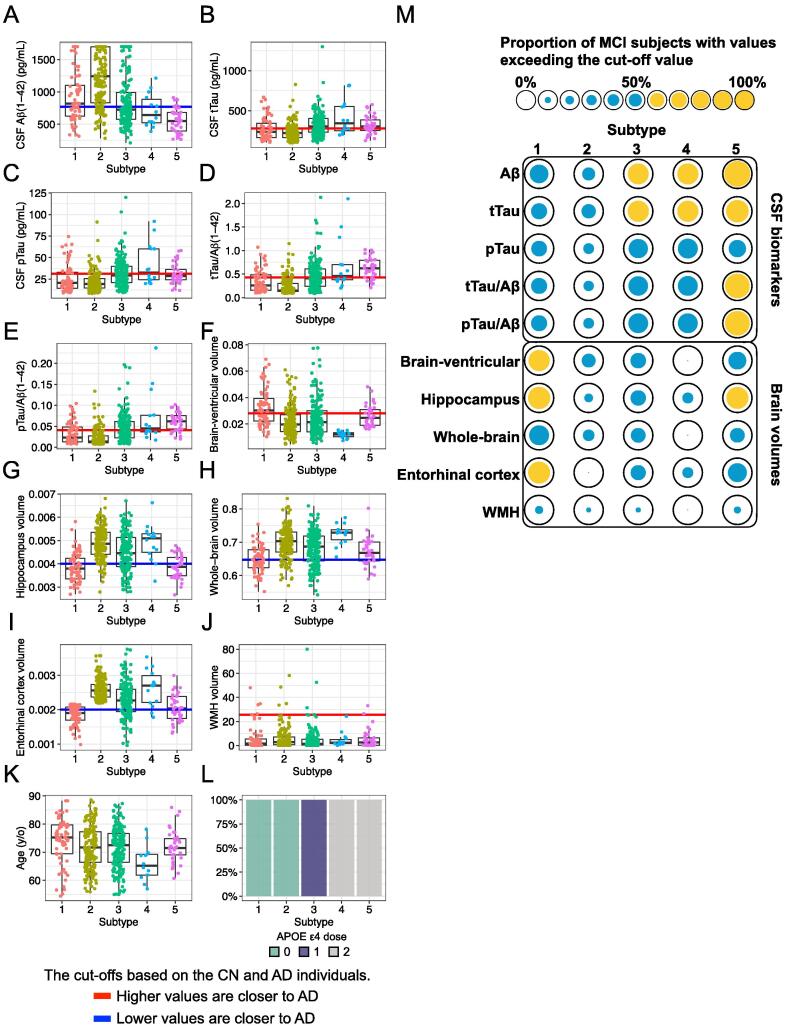
To provide a more detailed characterization of each subtype, we compared the levels of 12 features used in the HML model among the subtypes (Fig. 3). Subtype 2 showed high levels of CSF Aβ(1–42) (Fig. 3A), suggesting low deposition of Aβ in the brain. In general, in AD dementia patients, there is the abnormal accumulation of Aβ produced in the brain resulting in senile plaques, which suppresses the efflux of Aβ from the brain to the CSF, and the amount of Aβ in the CSF decreases. On the other hand, Aβ does not accumulate in the brain in normal subjects; thus, the level of CSF Aβ appears to be higher in normal subjects than in AD dementia patients. The levels of CSF tau (CSF tTau, CSF pTau, tTau/Aβ(1–42) ratio, and pTau/Aβ(1–42) ratio), which indicate the degree of Aβ-dependent neurofibrillary tangles, were high in subtypes 4 and 5 (Fig. 3B-E). These biomarker patterns suggest that individuals classified into subtypes 4 and 5 have AD pathology in the brain. Subtype 1 had a high brain-ventricular volume (Fig. 3F), suggesting brain atrophy. This subtype also had low hippocampal, whole-brain and entorhinal cortex volumes, which is consistent with enlargement of the ventricles (Fig. 3G-H). Low hippocampal and entorhinal cortex volumes were also observed in subtype 5 (Fig. 3G and 3I). Regarding WMH volumes, which reflect white matter lesions caused by cerebral ischaemia, there were no differences across the subtypes (Fig. 3J), implying that most MCI subjects in this study did not present with vascular dementia. A comparison of ages showed that subtypes 1 and 4 included older and younger MCI subjects, respectively (Fig. 3K). Because the decision tree had gating nodes based on APOE ε4 allele numbers, the MCI subjects in subtypes 1 and 2 did not have any APOE ε4 alleles, which is a genetic risk factor, while the individuals in subtype 3 and those in subtypes 4 and 5 had one and two APOE ε4 alleles, respectively (Fig. 3L).
Fig. 3.
Features of each MCI subtype. (A) Cerebrospinal fluid (CSF) Aβ(1–42) level, (B) CSF tTau level, (C) CSF pTau level, (D) tTau/Aβ(1–42) ratio, (E) pTau/Aβ(1–42) ratio, (F) normalized brain-ventricular volume, (G) normalized hippocampal volume, (H) normalized whole-brain volume, (I) normalized entorhinal cortex volume, (J) normalized white matter hyperintensity (WMH) volume, (K) age, and (L) dosage of APOE ε4 alleles. The blue and red lines indicate the cut-off values based on the CN subjects and the AD dementia patients. The adopted cut-off value was that which distinguished the CN subjects and the AD dementia patients with the highest accuracy in the ROC analysis. Points below the blue line or above the red line represent levels similar to those observed in AD dementia patients. (M) The spot matrix shows the proportions of individuals with features similar to those of AD dementia patients. The spot size represents the proportion of MCI subjects with values exceeding the cut-off value. Yellow indicates a value greater than 50% of the proportion. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The overall trend is summarized in the spot matrix in Fig. 3M. Here, we considered features with a majority of MCIs exceeding the cut-off value to be abnormal in that subtype (yellow spots in Fig. 3M). The spot matrix characterized biological features of the subtypes that had the conversion rates shown in Fig. 2B and 2C: subtype 2, with no abnormalities (i.e., no yellow spots in Fig. 3M), had a low conversion rate; subtype 1, with some brain atrophy, and subtypes 3 and 4, with abnormalities in CSF biomarkers, had moderate conversion rates; and subtype 5, with both CSF biomarker abnormalities and brain atrophy, had a high conversion rate.
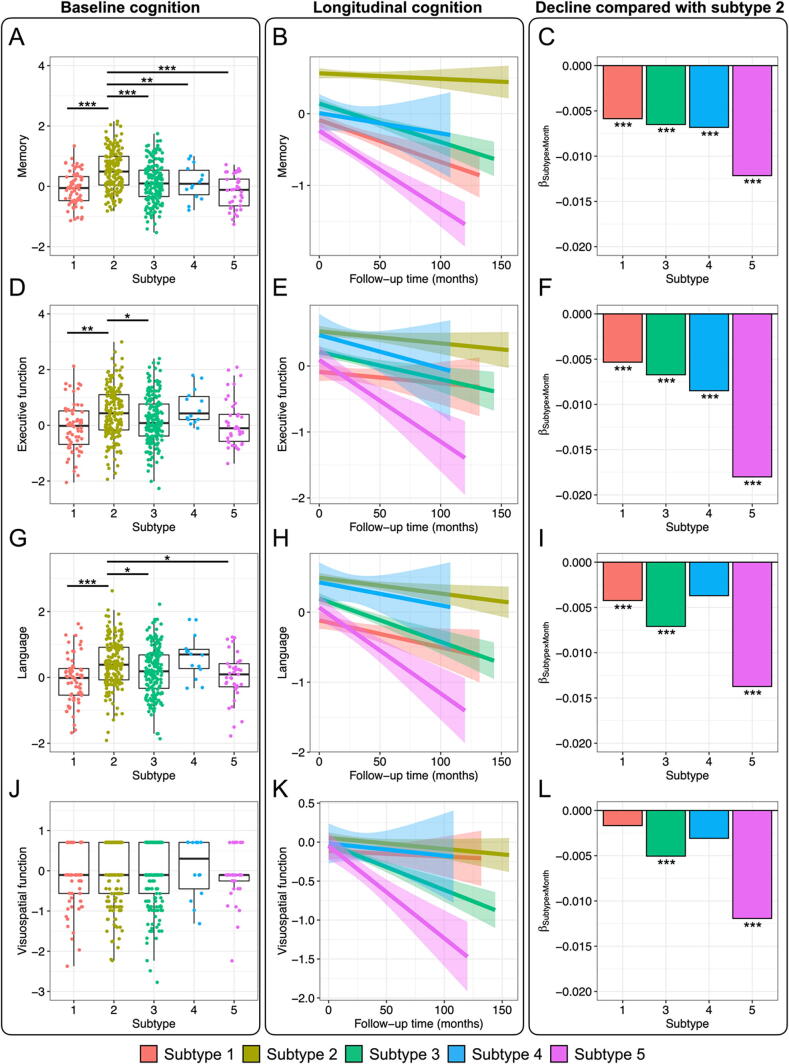
3.3. Cognitive function in each subtype
To examine the differences in cognitive function among the subtypes identified in the previous section and their changes over time, we first compared the four composite scores for memory, executive function, language, and visuospatial function at baseline among the subtypes. Comparisons among the subtypes showed that the three scores (the memory, executive function, and language scores) other than the visuospatial function scores were significantly higher in subjects with subtype 2 than in subjects with the other subtypes (Fig. 4A, 4D, 4G, and 4J). We next examined the trajectories of these scores during the follow-up time. We used subtype 2 (no abnormalities) as the reference to compare the association between the follow-up time and each score. The memory and executive function scores in subtypes 1, 3, 4, and 5 declined significantly more steeply over time than those in subtype 2 (Fig. 4C, 4F, 4I and 4L; Figures S2 and S3). The relative decline in subtype 5 consistently showed the most rapid decreases in all scores. In addition, subtype 1 exhibited slower declines than subtypes 3, 4, and 5, particularly for memory and executive function scores. These results show that the rate of exacerbation of cognitive decline differs depending on the subtype.
Fig. 4.
Cognitive function in each subtype. (A, D, G, J) Comparison of the cognitive function at baseline among subtypes. Higher composite scores in each cognitive domain were indicative of higher cognitive function. Multiple pairwise comparisons were performed with Tukey’s HSD tests to verify the differences in scores between subtypes. (B, E, H, K) Longitudinal changes in cognitive function by subtype. Each trajectory is indicated by a linear regression line. The error bars represent the 95 % confidence intervals. (C, F, I, L) Cognitive decline over time compared with cognitive function in subtype 2. The bar plots represent the interactions between subtypes and follow-up time, as estimated by the linear mixed model (LMM) with subtype 2 as the reference. *p < 0.05, **p < 0.01, ***p < 0.001.
3.4. Neuronal dysfunction and the inflammatory response in each subtype
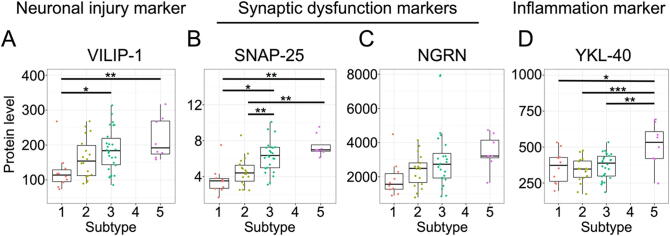
To investigate the phenomena occurring in the brains of individuals belonging to each subtype, we next examined the levels of CSF proteins reflecting neuronal injury, synaptic dysfunction, and inflammation within the brain. CSF markers were measured in the following subjects: 10 subjects in subtype 1, 18 in subtype 2, 26 in subtype 3, and 8 in subtype 5. These CSF markers were not measured in any of the subjects in subtype 4. Levels of the neuronal injury marker VILIP-1 and the synaptic dysfunction markers SNAP-25 and NGRN increased across subtypes in the following order (from lowest to highest levels): 1, 2, 3, and 5 (Fig. 5A-C). VILIP-1 and SNAP-25 levels in subtypes 3 and 5 were significantly higher than those in subtypes 1 or 2 (Fig. 5A and 5B). On the other hand, the level of the inflammation marker YKL-40 was highest in subtype 5 (Fig. 5D). These findings suggest that the accumulation of Aβ and tau proteins within the brain leads to neuronal dysfunction followed by an inflammatory response. Additionally, as we will mention in the Discussion, CSF markers such as VILIP-1 reflect Aβ- and tau-induced neuronal cell death. Therefore, these markers might not have been elevated in subtype 1.
Fig. 5.
Levels of the CSF neuronal and synaptic injury markers VILIP-1 (A), SNAP-25 (B), and NGRN (C) and the inflammatory response marker YKL-40 (D). Multiple pairwise comparisons were performed with Tukey’s HSD tests to verify the differences in CSF levels between subtypes. *p < 0.05, **p < 0.01, ***p < 0.001.
4. Discussion
We classified MCI subjects into subtypes using a highly interpretable decision tree. Our decision tree model predicted the MCI subjects in whom MCI converted to AD dementia with predictive accuracy comparable to that of the random forest method. Furthermore, the decision tree model divided the MCI subjects into five subtypes based on the characteristics reflected in their multimodal data. Detailed analysis showed a relationship between the speed of conversion to AD for each subtype and its biological characteristics, including CSF biomarkers and indicators of brain atrophy and inflammation.
Our final decision tree model was explained by only three features: brain-ventricular volume, entorhinal cortex volume, and dosage of APOE ε4 alleles, even though we used various biological data, including CSF biomarkers, brain imaging, and genomic data (Fig. 2A). This means that we can classify MCI subjects into subtypes by only MRI data and APOE ε4 alleles genotyped from blood without the need for lumbar puncture spinal fluid collection, which is associated with the risk of headache and nausea. This is due to the benefit that HML provides a sophisticated classification system by automatically performing dimensionality reduction from multimodal data. In addition, the decision tree divided the subjects into three groups based on the dosage of APOE ε4 alleles: the APOE ε4 noncarrier group, heterozygous carrier group, and homocarrier group. HML was able to naturally classify subjects according to their genetic background, suggesting that HML can apply to other genetic diseases.
The MCI subjects were categorized into three main groups in terms of AD conversion: MCI subjects with low conversion rates (subtype 2), who appeared similar to CN subjects; MCI subjects with moderate conversion rates (subtypes 1, 3, and 4); and MCI subjects with high conversion rates (subtype 5), who appeared similar to AD dementia patients. Our decision tree classified the MCI subjects with two copies of APOE ε4 alleles and more than 0.01575 of the normalized brain-ventricular volume into subtype 5 and predicted a high AD conversion risk. MCI subjects in subtype 5 may benefit from early intervention.
Furthermore, the group with moderate conversion rates was separated into two groups based on the presence of CSF biomarker abnormalities (subtypes 3 and 4) or brain atrophy (subtype 1). One of the differences among these subtypes was the presence or absence of APOE ε4 alleles (Fig. 3L). APOE ε4 alleles have been found to lead to Aβ and tau accumulation in the brain [24], [25], [26], [27], consistent with our results. Nettiksimmons et al. classified 139 cases of amnestic MCI in the ADNI into four subtypes using MRI data, CSF biomarker data, and serum biomarker data [28]. These subtypes included a CN-like subtype and an AD-like subtype. In addition, the remaining two subtypes showed moderate AD conversion rates. Of these two subtypes with moderate conversion rates, one subtype showed abnormal Aβ and tau levels and had typical AD features. However, the other subtype showed abnormal Aβ levels and brain atrophy but normal tau. This subtype deviates from typical AD and is similar to the subtype 1 we identified. Our results support that MCI has heterogeneity and does not always show consistent ordering to AD.
Recently, the AT(N) system has been proposed to elucidate the heterogeneity of AD by subdividing the pathological condition from the viewpoint of amyloid (A), tau (T), and neurodegeneration (N) [29]. A cohort study in Amsterdam showed that cognitive function varied across different AT(N) profiles [30]. Based on the AT(N) system, the MCI subjects in subtype 1 were A-T-N+, the subjects in subtypes 3 and 4 were A+T+N-, and the subjects in subtype 5 were A+T+N+. A recent study from the Alzheimer’s Biomarkers in Daily Practice (ABIDE) project reported that the A+T+N- classification has a higher percentage of APOE ε4 carriers than the A-T-N+ classification, corresponding with our finding that subtypes 3 and 4 (A+T+N-) include APOE ε4 carriers but subtype 1 (A-T-N+) does not [31].
Although subtypes 3, 4, and 5 are likely to include MCI subjects who develop AD because they display CSF Aβ and tau abnormalities defining the Alzheimer’s continuum, subtype 1 may include MCI subjects with suspected non-AD pathophysiology (SNAP) that is marked by neurodegeneration without Aβ deposition within the brain [32]. Some of the MCI subjects in subtype 1 may develop dementias other than AD in the future. One type of SNAP associated with the A-T-N+ classification that should be considered is limbic-predominant age-related TDP-43 encephalopathy (LATE) [33]. Validation using other biomarkers of neurodegenerative diseases, including the TDP-43 protein, will reveal the pathology of subtype 1 more clearly.
In summary, our results suggest that stratification by APOE ε4 may be helpful in the design of clinical trials for MCI subjects. MCI subjects without the APOE ε4 allele and with progressive brain atrophy should be excluded from clinical trials because they may progress to neurodegenerative diseases other than AD. It is also important to note that APOE ε4 homocarriers include subjects suspected of developing AD.
Subtype 1 showed relatively low levels of CSF markers of neuronal and synaptic injury (VILIP-1 and SNAP-25) despite advanced brain atrophy (Fig. 5A and 5B). Previous studies have shown that CSF levels of VILIP-1 are associated with CSF Aβ and p-tau levels, suggesting that VILIP-1 is a marker of neuronal degeneration related to Aβ and tau pathologies [34], [35]. In addition, a comparison of CSF VILIP-1 levels among CN subjects, MCI subjects, and AD dementia patients showed that VILIP-1 levels increased yearly only in MCI subjects; they did not increase in CN subjects and AD dementia patients [36]. CSF VILIP-1 levels may increase during inflammation and neurodegeneration triggered by Aβ and tau, but they may decrease after neurons have died and brain atrophy has occurred. Based on the findings of these studies, we concluded that subtype 1 did not exhibit increases in the levels of these neuronal degeneration markers because there were no prominent CSF biomarker abnormalities. Additionally, our results suggested that the MCI subjects in subtype 1 might convert to other dementias, as discussed above, because they did not show CSF biomarker abnormalities specific to AD pathologies.
Subtypes 3 and 5 showed high levels of CSF markers for neuronal and synaptic injury, namely, VILIP-1 and SNAP-25 (Fig. 5A and 5B). The levels of these markers gradually increased with the dosages of APOE ε4 alleles, consistent with the findings of recent studies reporting associations between these markers and APOE ε4 [37], [38]. On the other hand, CSF levels of the inflammation marker YKL-40 were increased only in subtype 5 (levels were not assessed in subtype 4) (Fig. 5D).
Our study has several limitations. First, we were not able to analyse all MCI patients in some analyses. For example, the MCI subjects in subtype 4 did not have the neuronal injury, synaptic injury, or inflammation CSF markers assessed. Second, given the correlational nature of some of our results, identification of the underlying mechanisms will require detailed analyses using animal models. Third, our study aimed to subtype MCI using decision trees generated by the HML model, but there is room for improvement in the prediction of AD conversion by our model. To improve accuracy, it may be useful to take into account various neuroimaging modalities rather than brain volumes alone. Gupta et al. combined different neuroimaging modalities, including structural MRI, fluorodeoxyglucose-PET, florbetapir-PET AV45, diffusion tensor imaging, and resting-state functional MRI, with the APOE genotype and showed more than 90 % accuracy discriminating AD from CN and predicting conversion from MCI to AD [39]. Barbará-Morales et al. proposed a new biomarker, three-dimensional brain tortuosity, which yielded improved accuracy when combined with other brain imaging biomarkers, including brain volume and cortical thickness, and CSF biomarkers [40]. The accuracy of the HML model would be improved by incorporating these detailed brain image parameters.
5. Conclusion
In this study, we succeeded in classifying MCI subjects into subtypes using highly interpretable decision trees (i.e., few leaf nodes) generated by the HML approach. Our study identified subtypes with characteristics similar to those of typical AD and identified one subtype in which MCI was likely to convert to other neurodegenerative diseases. These findings imply that the inclusion of additional pathological information can enable a more precise prediction of the onset or progression of a wide variety of neurodegenerative diseases. Moreover, we developed a decision tree model to predict conversion to AD dementia. Although the overall performance of the model can potentially be improved, focusing on specific subtypes in which conversion to AD dementia can be predicted with the most accuracy (e.g., subtype 5, in which the prediction was made with high precision) could enable more efficient clinical trials to be conducted.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (grant number 20K15778 to M.K.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and by grants from the Japan Agency for Medical Research and Development (AMED) (Grant No JP20dk0207045 to M.K. and T.I., JP20ek0109392 to M.K. and T.I., JP20dm0207073 to T.I., JP22wm0525019 to M.K. and T.I., and JP22dk0207060 to M.K., A.M. and T.I.). The funders had no role in the study design, data collection, decision to publish, or preparation of the manuscript.
7. Availability of data and materials
The data used in this study are available from the ADNI dataset (https://ida.loni.ucla.edu/).
8. Ethics approval and consent to participate
This study was approved by the Ethics Committee of Osaka University.
9. Consent for publication
Consent for publication has been granted by ADNI administrators.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The Department of Genome Informatics is a joint research department established with the sponsorship of NEC Corporation. K. Kobayashi., E.Y., Y.K., Y.F., and K. Kamijo are employees of NEC Corporation. The funder (NEC Corporation) provided support to authors in the form of salaries (K. Kobayashi, E.Y., Y.K., Y.F., and K. Kamijo) but did not have any additional roles in the study design, data collection, or decision to publish. The other authors have no competing interests to declare.
Acknowledgement
We thank all the participants and staff of the ADNI.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Alzheimer Report 2021: Journey through the diagnosis of dementia. [Available from: https://www.alzint.org/resource/world-alzheimer-report-2021/.
- 2.Cummings J., Lee G., Ritter A., Sabbagh M., Zhong K. Alzheimer's disease drug development pipeline: 2019. Alzheimers Dement (N Y) 2019;5:272–293. doi: 10.1016/j.trci.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen R.C., Morris J.C. Mild cognitive impairment as a clinical entity and treatment target. Arch Neurol. 2005;62(7):1160–1163. doi: 10.1001/archneur.62.7.1160. discussion 7. [DOI] [PubMed] [Google Scholar]
- 4.Farias S.T., Mungas D., Reed B.R., Harvey D., DeCarli C. Progression of mild cognitive impairment to dementia in clinic- vs community-based cohorts. Arch Neurol. 2009;66(9):1151–1157. doi: 10.1001/archneurol.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanken A.E., Jang J.Y., Ho J.K., Edmonds E.C., Han S.D., Bangen K.J., et al. Distilling heterogeneity of mild cognitive impairment in the national alzheimer coordinating center database using latent profile analysis. JAMA Netw Open. 2020;3(3):e200413. doi: 10.1001/jamanetworkopen.2020.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang J., Kim C.M., Jeon S., Lee J.M., Hong Y.J., Roh J.H., et al. Prediction of Alzheimer's disease pathophysiology based on cortical thickness patterns. Alzheimers Dement (Amst) 2016;2:58–67. doi: 10.1016/j.dadm.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machulda M.M., Lundt E.S., Albertson S.M., Kremers W.K., Mielke M.M., Knopman D.S., et al. Neuropsychological subtypes of incident mild cognitive impairment in the Mayo Clinic Study of Aging. Alzheimers Dement. 2019;15(7):878–887. doi: 10.1016/j.jalz.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulakis K., Pereira J.B., Mecocci P., Vellas B., Tsolaki M., Kloszewska I., et al. Heterogeneous patterns of brain atrophy in Alzheimer's disease. Neurobiol Aging. 2018;65:98–108. doi: 10.1016/j.neurobiolaging.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Whitwell J.L., Graff-Radford J., Tosakulwong N., Weigand S.D., Machulda M., Senjem M.L., et al. [(18) F]AV-1451 clustering of entorhinal and cortical uptake in Alzheimer's disease. Ann Neurol. 2018;83(2):248–257. doi: 10.1002/ana.25142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinney S.M., Sieniek M., Godbole V., Godwin J., Antropova N., Ashrafian H., et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577(7788):89–94. doi: 10.1038/s41586-019-1799-6. [DOI] [PubMed] [Google Scholar]
- 12.Linardatos P., Papastefanopoulos V., Explainable K.S. AI: A review of machine learning interpretability methods. Entropy (Basel) 2020;23(1) doi: 10.3390/e23010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan M.I., Jacobs R.A. Hierarchical mixtures of experts and the Em algorithm. Neural Comput. 1994;6(2):181–214. [Google Scholar]
- 14.Fujimaki R., Morinaga S. In: Proceedings of the Fifteenth International Conference on Artificial Intelligence and Statistics; Proceedings of Machine Learning Research: PMLR. Neil D.L., Mark G., editors. 2012. Factorized asymptotic bayesian inference for mixture modeling; pp. 400–408. [Google Scholar]
- 15.Eto R., Fujimaki R., Morinaga S., Tamano H. Fully-automatic bayesian piecewise sparse linear models. Jmlr Worksh Conf Pro. 2014;33:238–246. [Google Scholar]
- 16.Iwasaki Y., Sawada R., Stanev V., Ishida M., Kirihara A., Omori Y., et al. Identification of advanced spin-driven thermoelectric materials via interpretable machine learning. Npj Comput Mater. 2019;5 [Google Scholar]
- 17.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15(4):869-77, xi-xii. [DOI] [PMC free article] [PubMed]
- 18.Jack C.R., Jr., Bernstein M.A., Fox N.C., Thompson P., Alexander G., Harvey D., et al. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbons L.E., Carle A.C., Mackin R.S., Harvey D., Mukherjee S., Insel P., et al. A composite score for executive functioning, validated in Alzheimer's Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav. 2012;6(4):517–527. doi: 10.1007/s11682-012-9176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crane P.K., Carle A., Gibbons L.E., Insel P., Mackin R.S., Gross A., et al. Development and assessment of a composite score for memory in the Alzheimer's Disease Neuroimaging Initiative (ADNI) Brain Imaging Behav. 2012;6(4):502–516. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breiman L. Classification and regression trees. The Wadsworth and Brooks-Cole statisticsprobability series; 1984.
- 22.Ho T.K. Proceedings of 3rd international conference on document analysis and recognition. 1995. Random decision forests; pp. 278–282. [Google Scholar]
- 23.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 24.Bi X., Yong A.P., Zhou J., Ribak C.E., Lynch G. Rapid induction of intraneuronal neurofibrillary tangles in apolipoprotein E-deficient mice. Proc Natl Acad Sci U S A. 2001;98(15):8832–8837. doi: 10.1073/pnas.151253098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeMattos R.B., Cirrito J.R., Parsadanian M., May P.C., O'Dell M.A., Taylor J.W., et al. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41(2):193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- 26.Tiraboschi P., Hansen L.A., Masliah E., Alford M., Thal L.J., Corey-Bloom J. Impact of APOE genotype on neuropathologic and neurochemical markers of Alzheimer disease. Neurology. 2004;62(11):1977–1983. doi: 10.1212/01.wnl.0000128091.92139.0f. [DOI] [PubMed] [Google Scholar]
- 27.Small S.A., Duff K. Linking Abeta and tau in late-onset Alzheimer's disease: a dual pathway hypothesis. Neuron. 2008;60(4):534–542. doi: 10.1016/j.neuron.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nettiksimmons J., DeCarli C., Landau S., Beckett L. Alzheimer's Disease Neuroimaging I. Biological heterogeneity in ADNI amnestic mild cognitive impairment. Alzheimers Dement. 2014;10(5):511–21 e1. doi: 10.1016/j.jalz.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack C.R., Jr., Bennett D.A., Blennow K., Carrillo M.C., Feldman H.H., Frisoni G.B., et al. A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. 2016;87(5):539–547. doi: 10.1212/WNL.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebenau J.L., Timmers T., Wesselman L.M.P., Verberk I.M.W., Verfaillie S.C.J., Slot R.E.R., et al. ATN classification and clinical progression in subjective cognitive decline: The SCIENCe project. Neurology. 2020;95(1):e46–e58. doi: 10.1212/WNL.0000000000009724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altomare D., de Wilde A., Ossenkoppele R., Pelkmans W., Bouwman F., Groot C., et al. Applying the ATN scheme in a memory clinic population: The ABIDE project. Neurology. 2019;93(17):e1635–e1646. doi: 10.1212/WNL.0000000000008361. [DOI] [PubMed] [Google Scholar]
- 32.Jack C.R., Jr., Knopman D.S., Weigand S.D., Wiste H.J., Vemuri P., Lowe V., et al. An operational approach to National Institute on Aging-Alzheimer's Association criteria for preclinical Alzheimer disease. Ann Neurol. 2012;71(6):765–775. doi: 10.1002/ana.22628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson P.T., Dickson D.W., Trojanowski J.Q., Jack C.R., Boyle P.A., Arfanakis K., et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain. 2019;142(6):1503–1527. doi: 10.1093/brain/awz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutphen C.L., McCue L., Herries E.M., Xiong C., Ladenson J.H., Holtzman D.M., et al. Longitudinal decreases in multiple cerebrospinal fluid biomarkers of neuronal injury in symptomatic late onset Alzheimer's disease. Alzheimers Dement. 2018;14(7):869–879. doi: 10.1016/j.jalz.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H., Ng K.P., Therriault J., Kang M.S., Pascoal T.A., Rosa-Neto P., et al. Cerebrospinal fluid phosphorylated tau, visinin-like protein-1, and chitinase-3-like protein 1 in mild cognitive impairment and Alzheimer's disease. Transl Neurodegener. 2018;7:23. doi: 10.1186/s40035-018-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kester M.I., Teunissen C.E., Sutphen C., Herries E.M., Ladenson J.H., Xiong C., et al. Cerebrospinal fluid VILIP-1 and YKL-40, candidate biomarkers to diagnose, predict and monitor Alzheimer's disease in a memory clinic cohort. Alzheimers Res Ther. 2015;7(1):59. doi: 10.1186/s13195-015-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Zhang J., Pan T. APOE ε4 is associated with higher levels of CSF SNAP-25 in prodromal Alzheimer's disease. Neurosci Lett. 2018;685:109–113. doi: 10.1016/j.neulet.2018.08.029. [DOI] [PubMed] [Google Scholar]
- 38.Wang L., Zhang M., Wang Q., Jiang X., Li K., Liu J. APOE ε4 allele is associated with elevated levels of CSF VILIP-1 in preclinical Alzheimer's disease. Neuropsychiatr Dis Treat. 2020;16:923–931. doi: 10.2147/NDT.S235395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta Y., Kim J.I., Kim B.C., Kwon G.R. Classification and graphical analysis of Alzheimer's disease and its prodromal stage using multimodal features from structural, diffusion, and functional neuroimaging data and the APOE genotype. Front Aging Neurosci. 2020;12:238. doi: 10.3389/fnagi.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbara-Morales E., Perez-Gonzalez J., Rojas-Saavedra K.C., Medina-Banuelos V. Evaluation of brain tortuosity measurement for the automatic multimodal classification of subjects with Alzheimer's disease. Comput Intell Neurosci. 2020;2020:4041832. doi: 10.1155/2020/4041832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study are available from the ADNI dataset (https://ida.loni.ucla.edu/).