Abstract
Rapid, direct and sensitive detection of foodborne bacteria in complex samples is still challenging. Here, we reported a universal surface-enhanced Raman scattering (SERS)-based lateral flow assay (LFA) for highly sensitive detection of foodborne bacteria in food and environmental samples using wheat germ agglutinin (WGA)-modified Fe3O4@Au (Au@MNP-WGA) nanotags. The Au@MNP-WGA tag with numerous intraparticle hotspots was integrated into the LFA system for the first time, which can not only greatly improve the detection sensitivity through the dual amplification effect of magnetic enrichment and SERS enhancement but also achieve the broad-spectrum capture of multiple bacteria. In addition, monoclonal antibodies were separately immobilized onto the test line of different LFA strips to ensure the specific detection of different target pathogens. With this strategy, the proposed assay can achieve the universal and highly sensitive determination of three common foodborne bacteria, namely, Listeria monocytogenes, Campylobacter jejuni, and Staphylococcus aureus, with low detection limit (10 cells mL−1), short testing time (<35 min), and high reproducibility (RSD < 8.14%). Given its good stability and accuracy in complex samples, the Au@MNP-WGA-based SERS-LFA has great potential to be a powerful tool for the universal and on-site detection of different foodborne pathogens.
A universal SERS-based lateral flow assay was proposed for highly sensitive detection of foodborne bacteria in food and environmental samples using wheat germ agglutinin-modified Fe3O4@Au nanotags.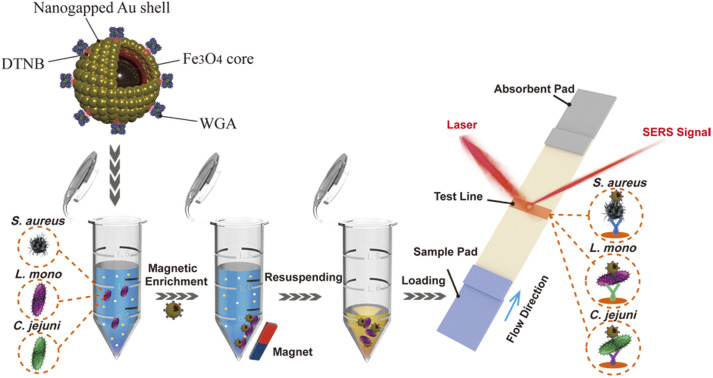
1. Introduction
Infectious diseases caused by foodborne pathogens have always been a major threat to food safety and human public health.1 They infect more than 550 million people and kill millions of people worldwide every year and cause huge economic and social burdens.2 Common foodborne pathogens, such as Staphylococcus aureus (S. aureus), Listeria monocytogenes (L. mono), and Campylobacter jejuni (C. jejuni), are widely distributed in nature and food processing environments and infect humans by contaminating food or water.3 These bacteria are highly contagious and only need very low doses (10–100 cells) to cause infection and lead to similar early symptoms (e.g., nausea, fever, headache, vomiting, abdominal cramps, and persistent diarrhea).4,5 Thus, the early and accurate detection of foodborne pathogens in food and environmental samples is key to curbing the spread of foodborne diseases and guiding effective treatment. Current microbial identification techniques include bacterial isolation and culture methods, polymerase chain reaction (PCR), genome sequencing, and mass spectrometry, which can provide accurate results for pathogen detection under laboratory conditions.6–8 However, these techniques require a clean testing place to avoid contamination, a tedious operation for sample processing, sophisticated instruments, and long testing time (hours to days) to generate results; therefore, they are difficult to use in the on-site detection of bacterial samples. A rapid, sensitive, and reliable point-of-care testing (POCT) method for foodborne bacteria detection is still strongly demanded.
Lateral flow assay (LFA) has developed into the most mature and popular POCT technology with the advantages of portability, real-time analysis, rapidness, and low cost.9–12 The performance of current LFA methods for bacteria detection highly depends on the performance of signal nanotags and biorecognition molecules (mainly antibodies), which cause the poor universality of the LFA system for different pathogens. In general, a pair of antibodies is required for one target bacteria and plays two key roles in the LFA method: (i) it is modified onto the surface of signal nanotags to effectively bind to the target bacterial cell, and (ii) it is immobilized on the test line of the LFA strip to specifically catch the formed bacteria–nanotag complexes. However, high-affinity antibodies are difficult to prepare, easily denatured, and subject to poor reproducibility, which greatly increase the difficulty of LFA construction.13–16 In addition, the complex matrixes (e.g., high sugar, high salt, and low pH) in actual food or environmental samples easily affect the stability of common LFA methods, thus easily causing false-positive results or misdiagnosis.17–20 Until now, a sensitive and universal LFA technique for different bacteria detection that can be directly used in complex samples has not been reported.
Lectins are a class of non-immune proteins that bind specifically to certain carbohydrates.21,22 Some phytolectins have strong affinity to the specific carbohydrates and their derivatives present on the surface of bacteria via multiple forces, such as hydrogen bonding, electrostatic interactions, and hydrophobic interactions. For example, wheat germ agglutinin (WGA) can specifically recognize N-acetylglucosamine on the cell wall of most foodborne pathogens.23,24 Compared with other biorecognition molecules such as antibodies and aptamers, WGA not only have the advantages of good stability and low cost but also has broad-spectrum bacteria binding ability. However, there are still no reports about the introduction of lectin into LFA method for bacteria detection.
In recent years, surface-enhanced Raman scattering (SERS)-based LFA method is considered one of the most sensitive paper-based POCT techniques, because it uses Raman dye-modified SERS tags to provide ultrasensitive (single-molecule level), specific (characteristic peak), and stable (no photobleaching) Raman signal for target determination.25–33 Inspired by these works, we proposed a universal SERS-LFA method for the sensitive and quantitative detection of three different foodborne bacteria in complex samples by utilizing the broad-spectrum recognition ability of WGA and the superior performance of nanogapped Fe3O4@Au tag (Au@MNP). Our assay has two obvious innovations compared with previously reported LFA biosensors for bacteria. First, the WGA-conjugated Au@MNP was used as the universal bacteria capture tool for the rapid and efficient magnetic enrichment of three common pathogens (i.e., L. mono, C. jejuni, and S. aureus) with high capture efficiencies (>90%), which makes the universal detection of bacteria easily realized via LFA. Second, the nanogapped Au@MNP can provide the dual amplification effect of multiple SERS hotspots and magnetic enrichment and thus ensures the highly sensitive detection of foodborne pathogens on the test strips. Given these merits, the proposed Au@MNP-WGA-based SERS-LFA can achieve the universal detection of L. mono, C. jejuni, and S. aureus in various food and environmental samples within 35 min with a limit of detection (LOD) as low as 10 cells mL−1. This work is the first to introduce lectin into the LFA method and the first to achieve the universal detection of foodborne pathogens via LFA. The universal SERS-LFA based on WGA-modified Au@MNP tag has great potential for the rapid, low-cost, and on-site ultrasensitive detection of foodborne bacteria.
2. Experimental section
2.1. Materials, chemicals, and instruments
Lectin from Triticum vulgaris (wheat germ agglutinin, WGA), glycine, polyethylenimine (PEI) branched (MW 25 kDa), 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), polyvinylpyrrolidone (PVP, 40 K), 2-(N-morpholino)ethanesulfonic (MES), phosphate-buffered saline (PBS), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), fetal bovine serum (FBS), N-hydroxysulfosuccinimide sodium salt (sulfo-NHS), and Tween-20 were supplied by Sigma-Aldrich (USA). Anti-WGA antibody (Catalog # ab178444) was purchased from Abcam (Cambridge, UK). Chloroauric acid tetrahydrate (HAuCl4·4H2O), hydroxylamine hydrochloride, and trisodium citrate were purchased from Sinopharm Chemical Reagent Co. (Shanghai, China). Mouse monoclonal antibody to L. mono and Goat anti-mouse IgG antibody were purchased from Fitzgerald (America). Mouse monoclonal anti-S. aureus antibody was obtained from Thermo Fisher (USA). Sample pad, absorbent pad, conjugate pad, PVC bottom plate were bought from Jieyi Biotechnology Co. (Shanghai, China) and Nitrocellulose (NC) membrane (CN95) with 15 μm pore size was obtained from Sartorius (Spain). All the bacteria used in this study were supplied by Pro. Shengqi Wang's group of Beijing Key Laboratory of New Molecular Diagnosis Technologies for Infectious Diseases.
The instruments used in this work and the SERS measurement conditions on LFA strips were shown in ESI S1.1.†
2.2. Synthesis of dual Raman dye-modified nanogapped Au@MNP
AuNPs with 20 nm particle size were synthesized according to the reported literature.34,35 In brief, 1 mL of 1% HAuCl4 (w/v) and 4 mL of 1% trisodium citrate (TSC) were mixed and boiled under stirring (100 °C) for 15 min, and then the mixture was cooled to room temperature and 20 nm AuNPs were obtained. We prepared 3 nm Au seed NPs using the approach reported by Wang et al. with some modifications.36 First, 4 mL of 1 wt% trisodium citrate (TSC) was mixed with 396 mL of HAuCl4 aqueous solutions (0.01 wt%) and then vigorously stirred at room temperature. Second, 12 mL of 0.1 M freshly prepared NaBH4 was quickly injected into the mixture and stirred for 4 h. The AuNPs with 3 nm size were obtained.
The synthesis procedure of the nanogapped Au@MNP with two layers of DTNB loading is shown in Scheme 1a. First, 160 nm superparamagnetic Fe3O4 nanoparticles (Fe3O4 MNPs) were fabricated according to the classical solvothermal reaction.37 Then, 10 mL of PEI solution (0.5 mg mL−1) was reacted with 1 mL of the prepared Fe3O4 MNP solution (10 mg mL−1) under sonication for 30 min. The obtained Fe3O4-PEI MNPs were magnetically separated, and re-dispersed with 40 mL of 20 nm AuNPs, and sonicated for 30 min. Then, 20 mL of 3 nm AuNPs was added into the reaction system, and the mixture was sonicated for another 15 min to form the Fe3O4-20/3 nm Au seed NPs. The fabricated Fe3O4-20/3 nm Au seed NPs were magnetically collected, rinsed with deionized water, dispersed in 10 mL of ethanol, and then reacted with 40 mL of freshly prepared DTNB (10 μM). The mixture of Fe3O4-20/3 nm Au seed NPs and DTNB was vigorously sonicated for 1 h to form DTNB-modified Fe3O4–Au seed NPs.38 Afterward, 1 mL of Fe3O4–Au seed/DTNB was added into 40 mL of deionized water containing 2 mg mL−1 PVP, 0.5 mg mL−1 hydroxylamine hydrochloride, and 200 μL of 1% HAuCl4·4H2O. The mixture was sonicated vigorously for 5 min to form the nanogapped Au shell outside of the Fe3O4 core. Finally, the obtained products were magnetically collected, dispersed in 10 mL of DTNB-ethanol solution (10 μM), and sonicated for 1 h to form dual-layer DTNB-modified Au@MNPs.
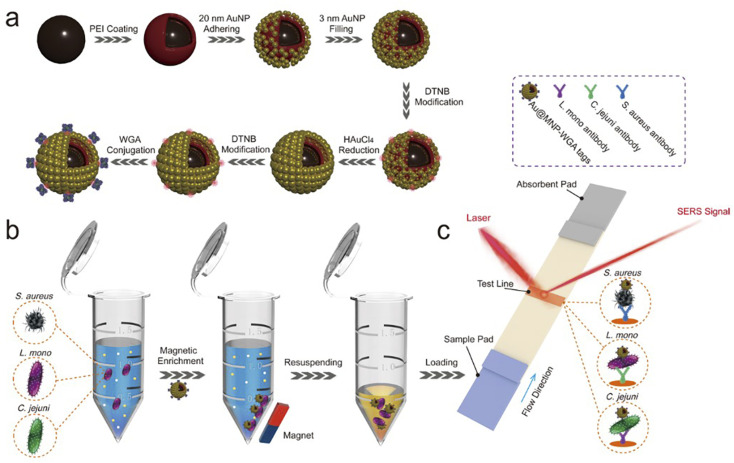
Scheme 1. Principle diagrams of (a) the synthesis procedure of Au@MNP-WGA tag, (b) magnetic enrichment of bacteria via Au@MNP-WGA tags, and (c) the Au@MNP-WGA tag-based universal SERS-LFA for the detection of S. aureus, L. mono, and C. jejuni.
2.3. Preparation of WGA-modified Au@MNPs
WGA was directly conjugated onto the DTNB modified Au@MNPs by EDC/NHS coupling chemistry. Briefly, 1 mL of the prepared Au@MNP-DTNB was magnetically separated and washed twice with ethanol to remove excessive DTNB. Then, the Au@MNP-DTNB was resuspended in 500 μL of MES buffer (0.1 M, pH 5.5) containing 5 μL of EDC (0.1 M) and 10 μL of NHS (0.1 M) and sonicated for 15 min. Then, WGA (20 μg) was added, and the mixture was reacted for 2 h with shaking. After that, 100 μL of glycine solution (1 M) was added, and the mixture was shaken for another 1 h to block the unreacted carboxyl sites. Finally, the formed Au@MNP-WGA SERS tags were magnetically collected, rinsed twice with PBS solution, and re-dispersed in 200 μL of PBST solution (10 mM PBS, 0.05% Tween 20, pH 7.4) for future use.
2.4. Assembly of magnetic SERS tags-based LFA for bacteria detection
The LFA strip with Au@MNP-WGA SERS tag is made up of three parts: a sample pad for magnetic SERS tag loading, a NC membrane with one test zone for antibody modification, and an absorbent pad to provide capillary force (Scheme 1c). C. jejuni detection antibody (1 mg mL−1), L. mono detection antibody (1 mg mL−1), and S. aureus detection antibody (1.4 mg mL−1) were separately dispensed on the NC membranes of different test strips using a spraying apparatus (Biodot Xyz5050) at a dispensing rate of 1 μL cm−1. The antibody-loaded NC membrane was dried at 37 °C for 3 h and then attached onto a plastic backing card for LFA assembly. The final assembled card was cut into independent 3.5 mm-wide strips and stored in a vacuum desiccator.
2.5. Bacteria detection by Au@MNP-WGA-based SERS-LFA
The accurate concentration of the bacteria samples was determined by standard plate counting method.18 Bacterial samples with various concentrations (0–106 cells mL−1) were prepared and spiked into different complex samples (i.e., vegetable juice, fruit juice, and river water). The food samples were purchased from a local supermarket, and the river water was collected from Beijing Yuyuantan Park. The river water samples were sterilized by autoclaving before spiking with target bacteria. As shown in Schemes 1b and c, the detection assays for different pathogens were successively performed in a 1.5 mL Eppendorf tube and on a test strip. First, 5 μL of Au@MNPs and 100 μL of salt ion buffer (10 mM PBS, 10 mM Ca2+, 10 mM Mg2+, 0.05% Tween 20) were added into 1 mL of the sample solution, and the mixture was shaken for 15 min on an oscillator at room temperature. Then, the formed Au@MNP-WGA–bacteria complexes were rapidly separated using an external magnet, resuspended in 100 μL of loading solution (10 mM PBS, pH 7.4, 10% FBS, 0.5% milk, 1% Tween 20),39 and then loaded onto the sample pad of the tested strip to start the chromatographic reaction. After 15 min, the SERS signal on the test line was detected using a portable Raman instrument.
3. Results and discussion
3.1. Bacteria detection principle of the Au@MNP-WGA-based SERS-LFA
The principle of the Au@MNP-WGA-based SERS-LFA for the universal detection of foodborne bacteria is illustrated in Scheme 1. The universal SERS-LFA was designed based on the broad-spectrum bacteria binding capability of WGA and the magnetic enrichment ability and superior SERS activity of Au@MNP, which can achieve the efficient capture of multiple kinds of bacteria from complex samples and the specific and sensitive detection of different foodborne pathogens on the LFA strips. The WGA-modified Au@MNP is the crucial factor for the performance of the universal SERS-LFA. It consists of four components: (i) a big Fe3O4 core ensures the strong magnetic responsiveness, (ii) a nanogapped Au shell outside magnetic core to provide large surface area for Raman dye DTNB modification and multiple high-efficiency hotspots for SERS sensing, (iii) dual-loaded DTNBs on the surface and inside of Au@MNP to provide ultra-strong SERS signals, and (iv) surface-conjugated WGA molecules that can recognize and capture a broad-spectrum of foodborne pathogens.
Three common foodborne pathogens, namely, L. mono, S. aureus, and C. jejuni were used as the model bacteria to demonstrate the feasibility and performance of the Au@MNP-WGA-based SERS-LFA for bacteria universal detection. The operating procedure of the universal SERS-LFA based on Au@MNP-WGA tags can be divided into two parts. First, the Au@MNP-WGA tags were added to the sample solution to capture a broad spectrum of bacteria and enrich them by magnetic force (Scheme 1b). Second, the Au@MNP-WGA–bacteria complexes were redispersed in a running solution and then dropped onto the sample pad of the LFA strip to start the immunochromatographic detection (Scheme 1c). The Au@MNP-WGA tag-matched LFA strip is composed of a sample pad, a NC membrane with a test line for specific antibody loading, and an absorbing pad. The monoclonal antibody-modified test line can ensure the specific capture of Au@MNP-WGA-target bacteria complexes and forms a black test line for target bacteria detection. If no target bacteria is present in the sample solution, the Au@MNP-WGA-nontarget bacteria complexes cannot bind to the test line, and no black band could be observed. Finally, the SERS signal on the test line of the LFA strip was quickly measured by Raman spectroscopy. The SERS intensity of the test zone was proportional to the amount of accumulated Au@MNP-WGA-target bacteria complexes; thus, it can be used for the quantitative determination of target bacteria. In this study, three kinds of LFA strips were prepared by modifying the test line with antibodies specific to L. mono, S. aureus, and C. jejuni to achieve the detection of the three model pathogens.
3.2. Characterization of the WGA-modified Au@MNP
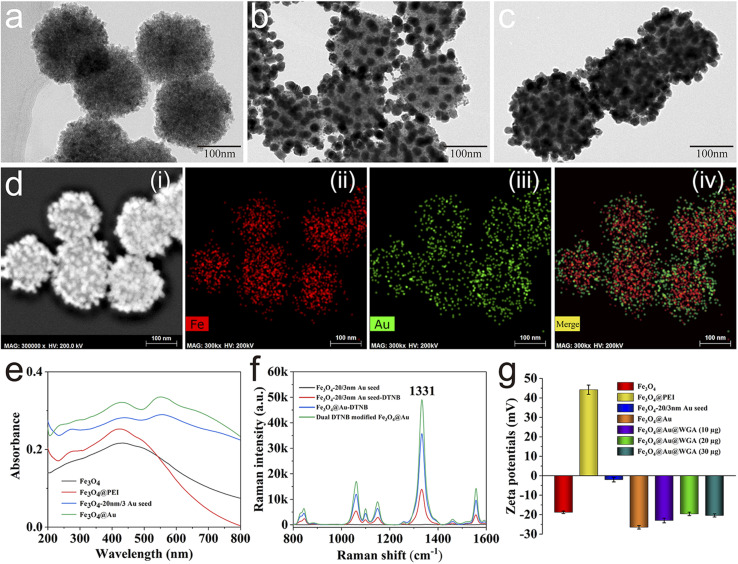
The well-designed Au@MNP was fabricated using an improved seed growth method as illustrated in Scheme 1a. First, Fe3O4 magnetic cores were synthesized through classical solvothermal reaction using PVP as the stabilizer, sodium acetate as the reducing agents, and ethylene glycol as the solvent. As shown in Fig. 1a, the prepared Fe3O4 MNPs had a uniform size and a diameter of ∼160 nm. Second, cationic polymer PEI was used to coat the Fe3O4 core to form a positively charged thin layer, which allows the rapid and efficient adsorption of Au seeds (3 and 20 nm) onto the Fe3O4 surface via electrostatic interaction. The PEI coating was verified by zeta potential. As displayed in Fig. 1g, the zeta potential of the Fe3O4@PEI MNPs remarkably increased from −18.8 mV to 44.42 mV, which indicated the successful coating of the PEI layer. Fig. 1b displays the TEM image of the prepared Fe3O4-20/3 nm Au NPs, which clearly shows the uniform distribution of the 3 and 20 nm AuNPs on the surface of the Fe3O4 core. The zeta potential of the Fe3O4-20/3 nm Au NPs after the adsorption of the Au seeds decreased to −2.1 mV. In our design, 20 nm AuNPs were used to build numerous nanogaps on the Au shell, and 3 nm AuNPs were used to fill the space between two 20 nm AuNPs to ensure the continuous growth of nanogapped Au shells. The 20 and 3 nm AuNPs on the Fe3O4 surface can act as acting sites for DTNB loading and the subsequent fabrication of nanogapped Au shells. The Au shell grew quickly on the 20/3 nm Au seeds after the addition of Au3+ ions and NH2OH·HCl as the reducing agents. As shown in Fig. 1c, the nanogapped Au shell was formed outside the Fe3O4 core with dense nanoscale cracks. The structural composition of the Au@MNPs was further characterized by energy-dispersive spectroscopy (EDS) elemental mapping analysis. As revealed in Fig. 1d, Au element (green) was intensively distributed onto the external surface of the Fe (red) core, which proved that the rough Au shell was successfully coated. The UV-vis absorption spectra of the Au@MNPs are shown in Fig. 1e. A new absorption peak at 558 nm appeared after Au seed adsorption and became remarkably stronger after nanogapped Au shell formation. This phenomenon can be attributed to the strong surface plasmon coupling at the nanogaps between neighboring AuNPs, which can greatly enhance the SERS activity of Au@MNP tags. Compared with previously reported Fe3O4@Au MNPs with complete Au shell,40,41 the nanogapped Au@MNPs can offer a larger surface area for Raman dye and WGA molecule modification and generate sufficient intraparticle hotspots for SERS label detection; thus, it is more suitable for the construction of highly sensitive SERS biosensors.
Fig. 1. Characterization of the nanogapped Au@MNP. TEM images of (a) Fe3O4, (b) Fe3O4-20/3 nm Au seed, and (c) Au@MNP. (d) Elemental mapping images of Au@MNPs. (e) UV-vis spectra of Fe3O4, Fe3O4@PEI, Fe3O4-20/3 nm Au seed, and Au@MNP. (f) Raman intensities of Fe3O4-20/3 nm Au seed, Fe3O4-20/3 nm Au seed-DTNB, Au@MNP-DTNB, and dual DTNB-modified Au@MNP. (g) Zeta potentials in each synthesis stage of Au@MNP-WGA.
In this study, DTNB was employed as the Raman dye molecule to build Au@MNP nanotags due to the following reasons: (i) DTNB has a large Raman cross-section that generates a strong and unique characteristic Raman peak at 1331 cm−1, and (ii) DTNB molecule can be easily labeled onto the Au shell by forming Au–S bond, and its terminal carboxyl group can directly act as a site for WGA modification. As illustrated in Scheme 1a, we used a dual-loading strategy to attach two layers of DTNB inside and on the Au shell of the Au@MNP tags to provide a more stable and stronger SERS signal. The SERS activities of different magnetic tags were compared to assess the advantage of dual DTNB-labeled Au@MNPs. As shown in Fig. 1f, the SERS intensity of the dual DTNB-modified Au@MNPs was 1.4 and 3.5 times higher than those of the single-layer DTNB-modified Au@MNP and DTNB-modified Fe3O4-20/3 Au seed NPs, respectively, at the SERS signal intensity of 1331 cm−1. The results confirm that the dual DTNB-labeled Au@MNPs possess superior SERS activity and are suitable for use in SERS label-based biosensors. WGA is a protein in nature and is therefore easily modified onto the surface of Au@MNP-DTNB via EDC/NHS coupling chemistry. The successful conjugation of WGA onto the Au@MNP tags was confirmed by zeta potential monitoring. As shown in Fig. 1g, the zeta potential of the Au@MNP tags obviously decreased after WGA modification and stabilized at −19.6 mV by conjugation with enough WGA (>20 μg). This result suggested that the amount of WGA modified onto the Au@MNP surface was saturated.
3.3. Verification of the broad-spectrum capture ability of Au@MNP-WGA to foodborne bacteria
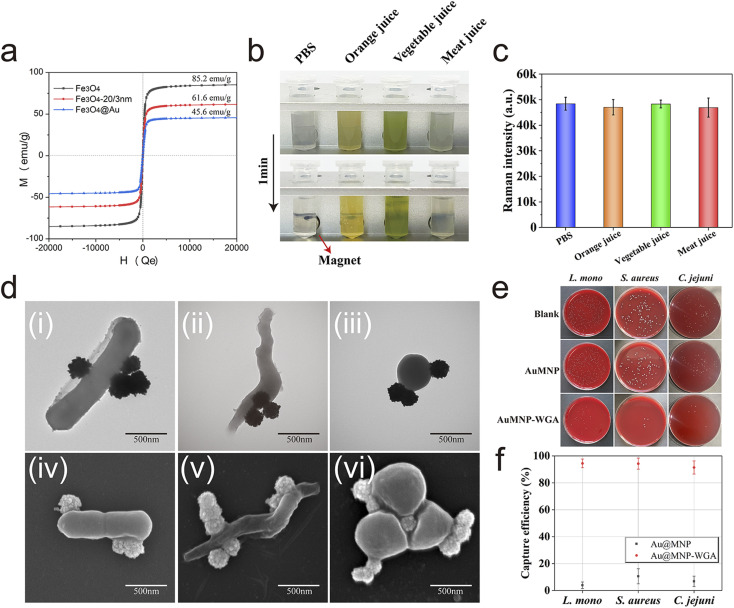
As a broad-spectrum bacterial capture tool for real application, the key performance (magnetic response ability, stability, and capture ability) of Au@MNP-WGA in complex food samples should be investigated. The prepared Au@MNP exhibited superparamagnetism and high saturation magnetization (MS) of 45.6 emu g−1 owing to its big Fe3O4 core (160 nm, Fig. 2a). The high magnetic response property of Au@MNP allows the rapid separation and enrichment of targets from complex samples via an external magnet. Au@MNP-WGA tags (5 μL, 10 mg mL−1) were mixed with 1 mL of different real samples (PBS, orange juice, vegetable juice, and meat juice) and incubated for 10 min to verify its practicability. As revealed in Fig. 2b, the Au@MNP-WGA tags were well dispersed and remained stable in the high-salt solution and complex food samples and can be completely enriched from all the tested samples within 1 min under a magnetic field. In addition, the SERS intensity of the separated Au@MNP-WGA tags from the complex matrixes did not change (Fig. 2c). These results indicated the excellent stability and feasibility of Au@MNP-WGA tags toward real food samples. The broad-spectrum capture ability of Au@MNP-WGA for three common foodborne pathogens was then evaluated. The TEM and SEM images in Fig. 2d clearly demonstrate that Au@MNP-WGA tags can rapidly and efficiently bind to L. mono, C. jejuni, and S. aureus in 5 min. The capture efficiency of Au@MNP-WGA for the three target bacteria was further confirmed by plate counting method (Fig. 2e). The detailed process of capture efficiency verification was shown in ESI S1.2.† The colony counting results verified that the capture efficiencies of Au@MNP-WGA for L. mono, C. jejuni, and S. aureus were 94.5%, 91.4%, and 94.3%, respectively (Fig. 2f). The bare Au@MNP-DTNB without WGA conjugation (control) had no ability to enrich the bacteria. Furthermore, the capture efficiency of Au@MNP-WGA for other common pathogens was also verified (Table S1†).
Fig. 2. (a) Magnetic hysteresis curves of Fe3O4, Fe3O4-20/3 nm Au seed, and Au@MNPs. (b) Magnetic separation behavior and (c) SERS intensity of Au@MNP-WGA tags in various food samples. (d) TEM and SEM images of Au@MNP-WGA-L. mono complexes (i), (iv), Au@MNP-WGA-C. jejuni complexes (ii), (v), and Au@MNP-WGA-S. aureus complexes (iii), (vi). (e) Photographs of the colonies on blood agar plates indicating the amounts of remaining L. mono, S. aureus, and C. jejuni in the supernatant after the magnetic enrichment of Au@MNP and Au@MNP-WGA tags. (f) Corresponding capture efficiencies of Au@MNP-WGA for L. mono, S. aureus, and C. jejuni.
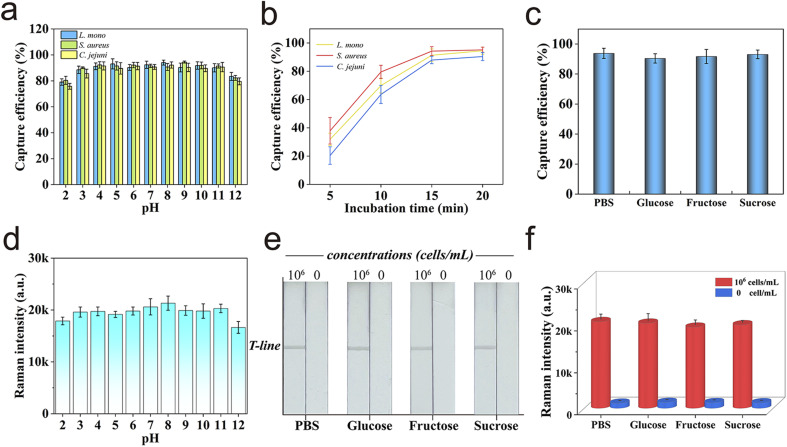
In addition, the high and stable bacteria-capturing ability of Au@MNP-WGA was exhibited in aqueous solution over a wide pH range of 3–11 (Fig. 3a). The relationship between the bacterial capture ability of Au@MNP-WGA and incubation time was further investigated to achieve the highest capture efficiency and suitable capture time. As shown in Fig. 3b, 15 min of incubation was sufficient to capture the three target bacteria. Considering that WGA is a specific protein that binds to the N-acetyl-d-glucosamine and N-acetyl-d-glucosamine derivatives of the bacterial cell wall, the influence of common sugars on the capture ability of WGA was further studied. Glucose, fructose, and sucrose are common sugars in fruits and vegetables, and their concentrations in foods do not exceed 0.2 g mL−1.42 Therefore, 0.2 g mL−1 of the three sugar solutions were prepared, spiked with L. mono (104 cells mL−1), and then captured by Au@MNP-WGA. These low molecular sugars had no obvious effect on the capture of bacteria; thus, WGA has good carbohydrate specificity (Fig. 3c). All the results confirmed that Au@MNP-WGA is an efficient and broad-spectrum tool for bacterial capture in various food samples and a wide pH range. The excellent capture ability of Au@MNP-WGA paved the way to develop a universal SERS-LFA for the detection of different pathogens.
Fig. 3. (a) Capture abilities of Au@MNP-WGA for three target bacteria in aqueous solution with different pH values. (b) Capture efficiency of Au@MNP-WGA versus incubation time. (c) Capture ability of Au@MNP-WGA in PBS and three different sugar solutions. (d) Raman intensities of the Au@MNP-WGA-based LFA strip for L. mono detection at varying pH values. (e) Digital photograph and (f) corresponding Raman intensity at 1331 cm−1 of the proposed LFA for bacteria detection in different sugar solutions. The error bars represent the standard deviations from five measurements.
3.4. Optimization of the Au@MNP-WGA-based SERS-LFA
Important parameters, such as the composition of the running buffer, antibody concentration, and reaction time were optimized to achieve the best bacteria detection performance of SERS-LFA. The optimization results of running buffer, concentration of the detection antibody on the test line, and chromatographic separation time for the SERS-LFA biosensor were demonstrated in ESI S2 and Fig. S1–S3.† The performance of the optimized Au@MNP-WGA-based SERS-LFA was further tested into different aqueous solutions over a wide pH range (pH 2–12) with high sugar content and 106 cells mL−1 of L. mono. As shown in Fig. 3d, the SERS signal values from the test strips remained stable in the solution within the pH range of 3–11. In addition, the photographs of the tested strips in Fig. 3e, and the corresponding SERS signal results in Fig. 3f confirmed that the Au@MNP-WGA-based SERS-LFA can work well in high-sugar samples. The results further demonstrated the good stability of the proposed SERS-LFA biosensor in complex samples.
3.5. Evaluation of universal SERS-LFA system for bacteria detection
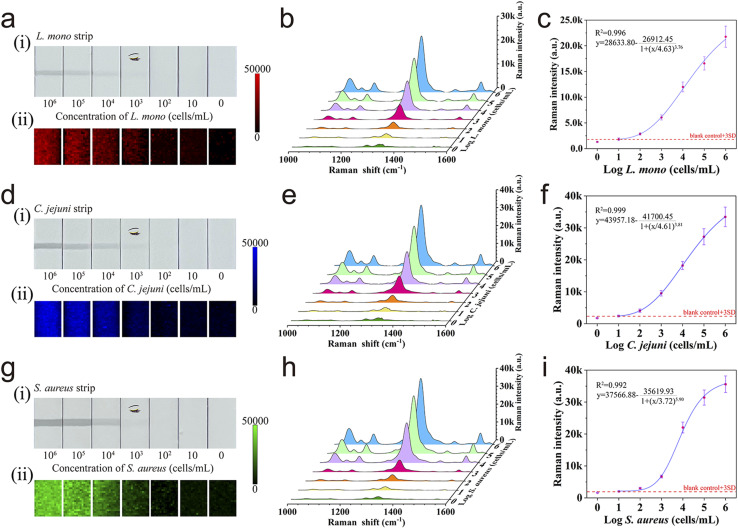
To evaluate the detection performance and universality of Au@MNP-WGA-based SERS-LFA, the established method was validated by detecting three different food-borne pathogens. First, the precise concentrations of the three foodborne pathogens were separately determined by plate colony counting (Fig. S4†). Three different series of L. mono, C. jejuni, and S. aureus samples with various concentrations (106–10 cells mL−1) were prepared by gradient dilution and detected via the proposed SERS-LFA. Fig. 4a(i), 4d(i), and 4g(i) display the photographs of the performance of the Au@MNP-WGA-based SERS-LFA in the detection of L. mono, C. jejuni, and S. aureus samples, respectively. For each group, the black color bands of the test lines generated from the Au@MNP-WGA–bacteria complexes decreased with decreasing concentrations of the target bacteria. The black test line on the corresponding strips became completely invisible to the naked eye when the concentrations of L. mono, C. jejuni, and S. aureus reached 103 cells mL−1. The capture ability of Au@MNP-WGA toward the three target bacteria was similar. The better visual sensitivity of the SERS-LFA to L. mono than the other two bacteria can be attributed to higher activity of anti-L. mono compared with those of the anti-C. jejuni and anti-S. aureus antibodies. Fig. 4a(ii), 4d(ii), and 4g(ii) show the SERS mapping images measured from the test zones of the SERS-LFA strips for L. mono, C. jejuni, and S. aureus, respectively, using the intensity of DTNB at 1331 cm−1 as the source. Each image contains 300-pixel points (1 pixel = 40 μm × 40 μm) over a 600 μm × 800 μm area with the color brightness denoting the SERS intensity, in which red, blue, and green represent the test lines for L. mono, C. jejuni, and S. aureus, respectively. Obviously, the overall SERS intensity of the test lines of the three groups of LFA strips declined with decreasing bacterial concentration. The SERS mapping results show the satisfactory reproducibility of SERS signals at high bacterial concentrations (104–106 cells mL−1) and the nonuniform SERS signal distribution at low bacterial concentrations (10–103 cells mL−1). This phenomenon can be explained by the decreased amount and uneven distribution of Au@MNP-WGA–bacteria complexes on the test zone at low bacterial concentration. A large laser spot (∼105 μm) was employed to randomly measure 20 spots on the test line, and the average of all the obtained SERS spectra was used to generate a reproducible signal to achieve the accurate and rapid measurement of SERS signal. Our previous works demonstrated that this strategy works well in the SERS-LFA system.41,43Fig. 4b, e, and h display the average SERS spectra from the tested SERS-LFA strips for L. mono, C. jejuni, and S. aureus detection, respectively. The main Raman peaks of DTNB (1331 cm−1) from the positive groups were obviously stronger than that of the blank control even when the concentrations of the three pathogens were down to 10 cells mL−1. The four-parameter logistic fitting curves for L. mono, C. jejuni, and S. aureus were drawn according to the sigmoidal function of bacteria concentrations and SERS intensities at 1331 cm−1 on the test lines (Fig. 4c, f, and i). LOD was calculated as the minimal concentration of bacteria corresponding to three times the standard deviation for the blank controls. The LODs of the proposed assay for L. mono, C. jejuni, and S. aureus were as low as 10 cells mL−1. In addition, the SERS intensities of test strips for the three target pathogens exhibited wide dynamic relationships (six orders of magnitude) with bacterial concentrations, with correlation coefficients (R2) of 0.996 for L. mono, 0.999 for C. jejuni, and 0.992 for S. aureus. The excellent detection performance of the proposed assay can be attributed to the synergistic effect of the high capture efficiency and broad-spectrum capture ability of WGA and the superior SERS ability of the Au@MNP-WGA tags. Moreover, the Au@MNP-WGA-based SERS-LFA showed prominent advantages, including universality, higher sensitivity, the need for only one antibody (lower cost and more convenient) than other recently reported LFA biosensors for bacteria detection (Table 1). Notably, only one T line was constructed onto the NC membrane of the proposed SERS-LFA due to WGA is a class of non-immune proteins and its antibody is rather expensive. In theory, using anti-WGA antibody to build the control (C) line is feasible, because the anti-WGA antibody modified C line can catch the superfluous WGA-conjugated Au@MNPs, thus can always generate a visible dark band. To verify this, we sprayed the anti-S. aureus antibody and anti-WGA antibody onto the NC membrane to build a test line for S. aureus and a C line, respectively. Then, a series of S. aureus samples (0–106 cells mL−1) was detected by the prepared LFA strips. As shown in Fig. S5,† the LOD of these strips for S. aureus was determined to be 10 cells mL−1 with R2 of 0.988. Moreover, the SERS intensity, detection range and LOD of LFA strips with C line were consistent with those of SERS-LFA without C line (Fig. 4g and h). This result clearly indicated that the proposed Au@MNP-WGA-based SERS-LFA containing a C line can also work well for bacteria detection.
Fig. 4. Photographs (i) and SERS mapping pictures (ii) of test lines of LFA strips for different L. mono (a), C. jejuni (d), and S. aureus concentrations (g). Corresponding Raman spectra of the test lines for different L. mono (b), C. jejuni (e), and S. aureus concentrations (h). Corresponding calibration lines of L. mono (c), C. jejuni (f), and S. aureus (i).
Comparison of Au@MNP-WGA-LFA with other recently reported LFA-based bacterial detection methods.
| Detection method | Bacteria | LOD (cells mL−1) | Reference |
|---|---|---|---|
| Magnetic-fluorescent LFA | S. typhimurium | 3.75 × 103 | 44 |
| Magnetic-fluorescent LFA | E. coli O157:H7 | 2.39 × 102 | 45 |
| SERS LFA | E. coli O157:H7 | 50 | 46 |
| Colorimetric LFA | S. aureus | 103 | 47 |
| Fluorescent LFA | S. aureus | 6.6 × 102 | 48 |
| Fluorescent LFA | E. coli O157:H7, S. typhi | 50, 50 | 49 |
| Magnetic SERS LFA | L. mono | 10 | This work |
| C. jejuni | 10 | ||
| S. aureus | 10 |
To intuitively evaluate the superiority of WGA than commonly used antibodies on Au@MNP-based LFA system, anti-S. aureus antibodies were paired to separately modify the WGA and test strip to detect the same S. aureus samples. Except for the use of the S. aureus capture antibody-modified Au@MNP instead of the WGA-modified Au@MNP as the magnetic SERS tags, the other parameters of the LFA system were consistent with the established protocol. The preparation and usage of the immuno-Au@MNP-based LFA method are described in ESI S1.3 and S1.4.† Fig. S6a† shows the photographs of the immuno-Au@MNP-based LFA for S. aureus sample detection (0–106 cells mL−1). The visualization limit of the immuno-Au@MNP-based LFA for S. aureus was ∼103 cells mL−1. Fig. S6b and S6c† revealed the corresponding SERS spectra of the test lines and the plotted calibration curve of immuno-Au@MNP-based LFA, respectively. From the results, the LOD of the immuno-Au@MNP-based LFA for S. aureus was determined to be 28 cells mL−1, which was 2.8 times higher than that of the Au@MNP-WGA-based LFA system. The higher sensitivity of the Au@MNP-WGA-based LFA can be attributed to the higher affinity of Au@MNP-WGA tags toward bacteria. This experiment result also indicated WGA has a great potential to act as a more efficient recognition/capture molecule to replace antibody for bacteria detection.
3.6. Specificity and repeatability of the Au@MNP-WGA-based SERS-LFA
To assess the specificity of the proposed assay, seven major foodborne bacteria samples, including L. mono, C. jejuni, S. aureus, Escherichia coli (E. coli), Salmonella typhimurium (S. typhimurium), Shigella flexneri (S. flexneri), and Vibrio parahaemolyticus (V. parahaemolyticus) with the same concentrations (106 cells mL−1) were prepared and separately detected using the Au@MNP-WGA-based SERS-LFA. Moreover, a mixture sample containing 106 cells mL−1L. mono, C. jejuni, and S. aureus simultaneously was used to test the anti-interference ability of the universal SERS-LFA. As shown in Fig. S7,† the target bacteria and mixture groups can generate strong and stable SERS signals on the test lines, whereas all the negative groups containing interfering bacteria exhibited very weak SERS response. The result indicates the good specificity of the proposed assay to the target bacteria and its strong anti-interference ability toward non-target bacteria. The high specificity of the universal SERS-LFA can be attributed to the highly specific monoclonal antibody used in the construction of the test line. Next, the reproducibility of the universal SERS-LFA for the three model pathogens was investigated. Fig. S8† shows the results of 10 independent assays via the Au@MNP-WGA-based SERS-LFA for 104 and 102 cells mL−1L. mono, C. jejuni, and S. aureus. The SERS signals on the test zones of all LFA strips were relatively uniform, and their relative standard deviation (RSD) values were in the range of 4.10–8.14%, which indicated the high reliability of the proposed assay.
3.7. Analysis of real food and environmental samples
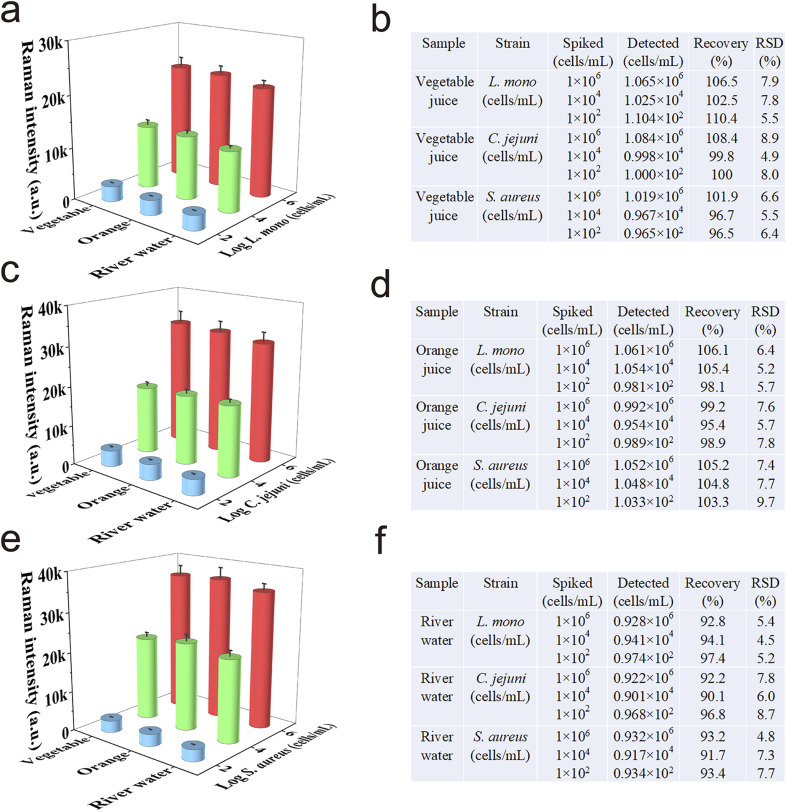
Considering that most foodborne bacteria transmitted through contaminated food or drinking water, the detection performance of Au@MNP-WGA-based SERS-LFA in real food and environmental samples should be assessed. Two common food samples (i.e., vegetable juice and orange juice) and one environmental sample (river water) were spiked with high (1 × 106 cells mL−1), medium (1 × 104 cells mL−1), and low (1 × 102 cells mL−1) levels of pathogens (L. mono, C. jejuni, and S. aureus). Real sample analysis was then performed according to the established protocol described in Experimental section 2.5. Fig. 5a, c, and e show the SERS detection results of the Au@MNP-WGA-based SERS-LFA for three model pathogens in vegetable juice, orange juice, and river water, respectively. The results verified that the SERS signals at 1331 cm−1 of the SERS-LFA strips were stable in detecting different food samples and decreased with decreasing concentrations of the target bacteria. The recoveries of the three bacteria-spiked samples are shown in Fig. 5b, d, and f, respectively. The average recoveries for the three foodborne pathogens in vegetable juice, orange juice, and river water were 96.5–110.4%, 95.4–106.1%, and 90.1–97.4%, respectively, with RSD values of 4.5–9.7%. The results indicated that the proposed assay can work well in complex food and environmental samples and achieve the universal and accurate detection of different pathogens in contaminated foods or water.
Fig. 5. Application of Au@MNP-WGA-based SERS-LFA for the detection of (a) L. mono, (c) C. jejuni, and (e) S. aureus in real complex samples. Recovery results of the three foodborne bacteria detected in (b) vegetable juice, (d) orange juice, and (f) river water via AuMNP-WGA LFA.
4. Conclusion
In this study, we proposed a universal SERS-LFA method for foodborne pathogens detection using WGA-modified Au@MNPs as the broad-spectrum capture tool and monoclonal antibody-modified LFA strip as the specific detection platform. Compared with traditional antibody-modified nanotags, the WGA-conjugated Au@MNPs exhibited great advantages, including higher affinity and broad-spectrum binding ability to pathogens, better stability, lower cost, magnetic enrichment ability, and superior SERS signal for quantitative analysis. The proposed universal SERS-LFA was successfully applied to accurately and specifically detect three common foodborne bacteria (L. mono, C. jejuni, and S. aureus) with low LODs (10 cells mL−1). Moreover, our method showed high reliability and stability in real food and environmental samples. In theory, this strategy can be easily extended for the sensitive detection of more pathogenic bacteria by only changing the detection antibody on the test strip. We believe the WGA-based magnetic SERS-LFA can be developed into a powerful tool for the universal and on-site detection of various pathogens in the future.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant nos. 81830101, 81871734, 82072380, 32200076), and the Natural Science Foundation of Anhui Province (Grant no. 2208085MB29).
Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra04735g
References
- Hameed S. Xie L. Ying Y. Trends Food Sci. Technol. 2018;81:61–73. [Google Scholar]
- Lee W.-I. Park Y. Shrivastava S. Jung T. Meeseepong M. Lee J. Jeon B. Yang S. Lee N.-E. Biosens. Bioelectron. 2020;152:112007. doi: 10.1016/j.bios.2020.112007. [DOI] [PubMed] [Google Scholar]
- Xiao X. Hu S. Lai X. Peng J. Lai W. Trends Food Sci. Technol. 2021;111:68–88. [Google Scholar]
- Wang P. Sun Y. Li X. Wang L. Xu Y. He L. Li G. Anal. Chim. Acta. 2021;1157:338279. doi: 10.1016/j.aca.2021.338279. [DOI] [PubMed] [Google Scholar]
- Dietvorst J. Vilaplana L. Uria N. Marco M.-P. Muñoz-Berbel X. Trends Anal. Chem. 2020;127:115891. [Google Scholar]
- Chen J. Andler S. M. Goddard J. M. Nugen S. R. Rotello V. M. Chem. Soc. Rev. 2017;46:1272–1283. doi: 10.1039/c6cs00313c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. Wu Q. Huang F. Xu Z. Zhou L. Zhao S. ACS Appl. Mater. Interfaces. 2021;13:26782–26789. doi: 10.1021/acsami.1c05522. [DOI] [PubMed] [Google Scholar]
- Chen H. Das A. Bi L. Choi N. Moon J. I. Wu Y. Park S. Choo J. Nanoscale. 2020;12:21560–21570. doi: 10.1039/d0nr06340a. [DOI] [PubMed] [Google Scholar]
- Bu T. Bai F. Zhao S. Cao Y. He K. Sun X. Wang Q. Jia P. Li M. Wang X. Wang L. Biosens. Bioelectron. 2021;192:113538. doi: 10.1016/j.bios.2021.113538. [DOI] [PubMed] [Google Scholar]
- Wang L. Wang X. Cheng L. Ding S. Wang G. Choo J. Chen L. Biosens. Bioelectron. 2021;189:113360. doi: 10.1016/j.bios.2021.113360. [DOI] [PubMed] [Google Scholar]
- Zhou Y. Chen Y. Liu Y. Fang H. Huang X. Leng Y. Liu Z. Hou L. Zhang W. Lai W. Xiong Y. Biosens. Bioelectron. 2021;171:112753. doi: 10.1016/j.bios.2020.112753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X. Zheng S. Wang W. Han H. Yang X. Shen W. Wang C. Wang S. Chem. Eng. J. 2021;426:131836. [Google Scholar]
- Ilhan H. Tayyarcan E. K. Caglayan M. G. Boyaci I. H. Saglam N. Tamer U. Biosens. Bioelectron. 2021;189:113383. doi: 10.1016/j.bios.2021.113383. [DOI] [PubMed] [Google Scholar]
- Zheng S. Wang C. Li J. Wang W. Yu Q. Wang C. Wang S. Chem. Eng. J. 2022;448:137760. [Google Scholar]
- Wang C. Wang C. Li J. Tu Z. Gu B. Wang S. Biosens. Bioelectron. 2022;214:114525. doi: 10.1016/j.bios.2022.114525. [DOI] [PubMed] [Google Scholar]
- Bi L. Wang X. Cao X. Liu L. Bai C. Zheng Q. Choo J. Chen L. Talanta. 2020;220:121397. doi: 10.1016/j.talanta.2020.121397. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T. de la Guardia M. Baradaran B. Trends Anal. Chem. 2020;125:115842. [Google Scholar]
- Shen W. Wang C. Zheng S. Jiang B. Li J. Pang Y. Wang C. Hao R. Xiao R. J. Hazard. Mater. 2022;437:129347. doi: 10.1016/j.jhazmat.2022.129347. [DOI] [PubMed] [Google Scholar]
- Lin D. Qin T. Wang Y. Sun X. Chen L. ACS Appl. Mater. Interfaces. 2014;6:1320–1329. doi: 10.1021/am405396k. [DOI] [PubMed] [Google Scholar]
- Yao L. Xu J. Cheng J. Yao B. Zheng L. Liu G. Chen W. Food Chem. 2022;366:130595. doi: 10.1016/j.foodchem.2021.130595. [DOI] [PubMed] [Google Scholar]
- Mi F. Guan M. Hu C. Peng F. Sun S. Wang X. Analyst. 2021;146:429–443. doi: 10.1039/d0an01459a. [DOI] [PubMed] [Google Scholar]
- Liu X. Lei Z. Liu F. Liu D. Wang Z. Biosens. Bioelectron. 2014;58:92–100. doi: 10.1016/j.bios.2014.02.056. [DOI] [PubMed] [Google Scholar]
- Yang G. Meng X. Wang Y. Yan M. Aguilar Z. P. Xu H. Sens. Actuators, B. 2019;279:87–94. [Google Scholar]
- Cheng S. Tu Z. Zheng S. Cheng X. Han H. Wang C. Xiao R. Gu B. Anal. Chim. Acta. 2021;1187:339155. doi: 10.1016/j.aca.2021.339155. [DOI] [PubMed] [Google Scholar]
- Zhang D. Huang L. Liu B. Su E. Chen H.-Y. Gu Z. Zhao X. Sens. Actuators, B. 2018;277:502–509. [Google Scholar]
- Zhang W. Tang S. Jin Y. Yang C. He L. Wang J. Chen Y. J. Hazard. Mater. 2020;393:122348. doi: 10.1016/j.jhazmat.2020.122348. [DOI] [PubMed] [Google Scholar]
- Wang R. Kim K. Choi N. Wang X. Lee J. Jeon J. H. Rhie G.-e. Choo J. Sens. Actuators, B. 2018;270:72–79. [Google Scholar]
- Sheng E. Lu Y. Xiao Y. Li Z. Wang H. Dai Z. Biosens. Bioelectron. 2021;181:113149. doi: 10.1016/j.bios.2021.113149. [DOI] [PubMed] [Google Scholar]
- Wang C. Wang C. Wang X. Wang K. Zhu Y. Rong Z. Wang W. Xiao R. Wang S. ACS Appl. Mater. Interfaces. 2019;11:19495–19505. doi: 10.1021/acsami.9b03920. [DOI] [PubMed] [Google Scholar]
- Zhang D. Huang L. Liu B. Ni H. Sun L. Su E. Chen H. Gu Z. Zhao X. Biosens. Bioelectron. 2018;106:204–211. doi: 10.1016/j.bios.2018.01.062. [DOI] [PubMed] [Google Scholar]
- Wang Y. Yan B. Chen L. Chem. Rev. 2013;113:1391–1428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- Chen H. Park S.-G. Choi N. Moon J.-I. Dang H. Das A. Lee S. Kim D.-G. Chen L. Choo J. Biosens. Bioelectron. 2020;167:112496. doi: 10.1016/j.bios.2020.112496. [DOI] [PubMed] [Google Scholar]
- Wang X. Choi N. Cheng Z. Ko J. Chen L. Choo J. Anal. Chem. 2017;89:1163–1169. doi: 10.1021/acs.analchem.6b03536. [DOI] [PubMed] [Google Scholar]
- Li J. F. Tian X. D. Li S. B. Anema J. R. Yang Z. L. Ding Y. Wu Y. F. Zeng Y. M. Chen Q. Z. Ren B. Wang Z. L. Tian Z. Q. Nat. Protoc. 2013;8:52–65. doi: 10.1038/nprot.2012.141. [DOI] [PubMed] [Google Scholar]
- Cheng Z. Choi N. Wang R. Lee S. Moon K. C. Yoon S. Y. Chen L. Choo J. ACS Nano. 2017;11:4926–4933. doi: 10.1021/acsnano.7b01536. [DOI] [PubMed] [Google Scholar]
- Wang C. Wang J. Li M. Qu X. Zhang K. Rong Z. Xiao R. Wang S. Analyst. 2016;141:6226–6238. doi: 10.1039/c6an01105e. [DOI] [PubMed] [Google Scholar]
- Wang C. Cheng X. Liu L. Zhang X. Yang X. Zheng S. Rong Z. Wang S. ACS Appl. Mater. Interfaces. 2021;13:40342–40353. doi: 10.1021/acsami.1c11461. [DOI] [PubMed] [Google Scholar]
- Li J. Wu T. Wang C. Tu J. Song X. Shao Y. Wang C. Qi K. Xiao R. ACS Appl. Nano Mater. 2022 doi: 10.1021/acsanm.2c02494. [DOI] [Google Scholar]
- Wang C. Yang X. Gu B. Liu H. Zhou Z. Shi L. Cheng X. Wang S. Anal. Chem. 2020;92:15542–15549. doi: 10.1021/acs.analchem.0c03484. [DOI] [PubMed] [Google Scholar]
- Wang J. Wu X. Wang C. Rong Z. Ding H. Li H. Li S. Shao N. Dong P. Xiao R. Wang S. ACS Appl. Mater. Interfaces. 2016;8:19958–19967. doi: 10.1021/acsami.6b07528. [DOI] [PubMed] [Google Scholar]
- Liu X. Yang X. Li K. Liu H. Xiao R. Wang W. Wang C. Wang S. Sens. Actuators, B. 2020;320:128350. [Google Scholar]
- Boulton J. Hashem K. M. Jenner K. H. Lloyd-Williams F. Bromley H. Capewell S. BMJ Open. 2016;6:e010330. doi: 10.1136/bmjopen-2015-010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Dai E. Xiao R. Zhou Z. Zhang M. Bai Z. Shao Y. Qi K. Tu J. Wang C. Wang S. Sens. Actuators, B. 2021;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. Jiang Y. Z. Tang M. Wu L. L. Xie H. Y. Zhang Z. L. Pang D. W. Anal. Chem. 2019;91:1178–1184. doi: 10.1021/acs.analchem.8b05154. [DOI] [PubMed] [Google Scholar]
- Huang Z. Peng J. Han J. Zhang G. Huang Y. Duan M. Liu D. Xiong Y. Xia S. Lai W. Food Chem. 2019;276:333–341. doi: 10.1016/j.foodchem.2018.09.164. [DOI] [PubMed] [Google Scholar]
- Shi L. Xu L. Xiao R. Zhou Z. Wang C. Wang S. Gu B. Front. Microbiol. 2020;11:596005. doi: 10.3389/fmicb.2020.596005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. Yao X. Liu S. Zhang H. Wang L. Yin X. Su L. Xu B. Wang J. Lan Q. Zhang D. Food Chem. 2021;339:127955. doi: 10.1016/j.foodchem.2020.127955. [DOI] [PubMed] [Google Scholar]
- Yang H. Wang Y. Liu S. Ouyang H. Lu S. Li H. Fu Z. Biosens. Bioelectron. 2021;182:113189. doi: 10.1016/j.bios.2021.113189. [DOI] [PubMed] [Google Scholar]
- Zheng S. Yang X. Zhang B. Cheng S. Han H. Jin Q. Wang C. Xiao R. Food Chem. 2021;363:130400. doi: 10.1016/j.foodchem.2021.130400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.