Abstract
Both of the Saccharomyces cerevisiae 2μm circle-encoded Rep1 and Rep2 proteins are required for efficient distribution of the plasmid to daughter cells during cellular division. In this study two-hybrid and in vitro protein interaction assays demonstrate that the first 129 amino acids of Rep1 are sufficient for self-association and for interaction with Rep2. Deletion of the first 76 amino acids of Rep1 abolished the Rep1-Rep2 interaction but still allowed some self-association, suggesting that different but overlapping domains specify these interactions. Amino- or carboxy-terminally truncated Rep1 fusion proteins were unable to complement defective segregation of a 2μm-based stability vector with rep1 deleted, supporting the idea of the requirement of Rep protein interaction for plasmid segregation but indicating a separate required function for the carboxy-terminal portion of Rep1. The results of in vitro baiting assays suggest that Rep2 contains two nonoverlapping domains, both of which are capable of mediating Rep2 self-association. The amino-terminal domain interacts with Rep1, while the carboxy-terminal domain was shown by Southwestern analysis to have DNA-binding activity. The overlapping Rep1 and Rep2 interaction domains in Rep1, and the ability of Rep2 to interact with Rep1, Rep2, and DNA, suggest a model in which the Rep proteins polymerize along the 2μm circle plasmid stability locus, forming a structure that mediates plasmid segregation. In this model, competition between Rep1 and Rep2 for association with Rep1 determines the formation or disassembly of the segregation complex.
Most strains of the budding yeast Saccharomyces cerevisiae contain an endogenous plasmid, the 2μm circle. This 6,318-bp double-stranded circular DNA plasmid is located in the nucleus at approximately 60 copies per haploid cell and replicates autonomously from, but synchronously with, the chromosomal DNA (for a review, see reference 9). The 2μm circle confers no phenotype or selective advantage on the host yeast; indeed, 2μm plasmid-bearing ([cir+]) cells grow 1% more slowly than isogenic plasmid-free ([cir0]) cells (10). Despite this disadvantage, the 2μm plasmid displays a high level of mitotic stability. This stability results from the presence of a plasmid-encoded copy number amplification system and a partition mechanism which together ensure that the rates of plasmid loss in mitosis and meiosis are very low (4, 10, 16, 18). Partitioning of the 2μm plasmid requires two proteins encoded by the plasmid genes REP1 and REP2 and a cis-acting 2μm locus termed STB (16, 18). The role of these three components has been examined in a variety of studies involving mainly deletion or insertion analysis of 2μm-derived plasmids (17, 18, 23). In the absence of any one of these three components, the 2μm plasmid displays a strong maternal bias in inheritance; most plasmids are retained in the mother cell (22). The 2μm circle partition system overcomes this bias by an as yet unknown mechanism.
The Rep1 and Rep2 proteins mediate 2μm plasmid segregation, but their mode of action is unclear. Lack of either Rep1, Rep2, or the STB locus results in the same degree of mitotic instability (18, 27). Experiments designed to reconstitute efficient 2μm segregation by expressing different amounts of Rep1 and Rep2 in the cell have shown that the relative levels of the two proteins are important for their partitioning function (5, 6). In addition to having a role in plasmid partitioning, the Rep proteins repress transcription of the 2μm plasmid FLP gene (25, 27, 31, 35). The FLP gene encodes a site-specific recombinase required for plasmid amplification, and the control of Flp expression by Rep proteins has been proposed as the mechanism by which 2μm plasmid copy number is regulated (8, 37). Rep protein-mediated transcriptional repression, like Rep protein segregation function, requires the concerted action of both Rep1 and Rep2 (25, 31, 35). Taken together, these data suggest that Rep1 and Rep2 may function as part of a complex. Recently, evidence for interaction between Rep1 and Rep2 and for self-association of the two proteins has been obtained both by two-hybrid genetic assays and in vitro protein interaction assays (1, 30, 36). We have used similar approaches here to delineate the regions of Rep1 and Rep2 that are required for these associations. Amino-terminal truncations of Rep1 abolish Rep1-Rep2 interaction and are unable to complement the segregation defect of a 2μm-based stability vector with rep1 deleted, while Rep2 has two separate domains capable of mediating Rep2 self-association, one of which has DNA-binding activity while the other interacts with Rep1.
MATERIALS AND METHODS
Yeast and bacterial strains.
Escherichia coli strain DH5α (29) was used for propagating plasmids, strain JF1754 (hisB leuB met thi) was used for expression of glutathione S-transferase (GST) fusion proteins, and strain BL21(DE3) (Novagen) was used for expression of pET fusion proteins. S. cerevisiae strains used were isogenic [cir+] and [cir0] AS3 (MATα his3Δ1 ura3-52 leu2-3 leu2-112 trp1-289 ade2Δ::URA3), AS2 (AS3 with an ade2Δ rather than an ade2Δ::URA3 allele), and CTY10/5d [MATa gal4 gal80 his3-200 trp1-901 ade2 ura3-52 leu2-3,112 URA3::(lexAop)8-lacZ] (2).
Media.
Yeast cells were routinely grown at 30°C in liquid or on solid synthetic defined (SD) medium containing 0.67% yeast nitrogen base (without amino acids) and 2.0% dextrose, supplemented with appropriate amino acids as described by Rose et al. (28). Selection for plasmids in CTY10/5d was maintained by omitting either leucine or histidine or both from the SD medium. Yeast was grown in YEPD rich medium (1% yeast extract, 2% Bacto Peptone, 2% dextrose) prior to transformation. SD medium supplemented only with tryptophan and histidine was used for selective growth conditions for plasmid rate loss experiments, whereas SD medium supplemented with tryptophan, histidine, and adenine was used for nonselective growth. E. coli was grown in 2× YT with 50 μg of ampicillin per ml as described by Sambrook et al. (29). Media were solidified with 2.0% agar. All medium reagents were obtained from Difco Laboratories or Sigma Chemical Co.
Generation of REP1 and REP2 BamHI fragments.
BamHI restriction fragments containing REP1 and REP2 open reading frames (ORFs) were generated by PCR using the 2μm-based plasmid pJDB219B (3) as a template and the following oligonucleotide primers: for REP1, REP1-1 (5′GGATCCATATGAATGGCGAGAGACTGC3′) and REP1-2 (5′GGATCCTATATAACCTACCCATCCAC3′) and for REP2, REP2-1 (5′GGATCCAAATGGACGACATTGAAACAGCC3′) and REP2-2 (5′GGATCCTCATACCCTAGAAGTATTACGTG3′). Initiation codons and BamHI sites are underlined. PCR was carried out with Taq DNA polymerase (Boehringer) according to the manufacturer's instructions except that an additional 10 mM Tris, pH 8.8, was added to the commercial buffer, which significantly improved yields. Amplification was carried out for 30 cycles, each consisting of 1 min at 94°C, 2 min at 50°C, and 4 min at 72°C. PCR products were cloned directly into SmaI-digested phagemid pTZ18R (Pharmacia) to which 3′ T overhangs had been added by incubation with Taq DNA polymerase and TTP to give pTZ18-REP1 and pTZ18-REP2. The REP BamHI fragments were sequenced using a Sequenase 2.0 sequencing kit (U.S. Biochemicals) according to the manufacturer's instructions.
Plasmid construction.
REP1 and REP2 BamHI fragments were subcloned into the BamHI site in the HIS3 two-hybrid vector pSH2-1 (2) to give in-frame fusions with LexA, producing pLEX-REP1 and pLEX-REP2, and also at the BamHI site in the LEU2 two-hybrid vector pGAD424 (Clontech) to give in-frame fusions with the Gal4 transcriptional activation domain (Gal4AD), producing pGAD-REP1 and pGAD-REP2. pGADrep1Δ1–76 and pGADrep1Δ1–129 were constructed by ligating an end-filled SpeI/BamHI fragment and an end-filled StuI/BamHI fragment from pTZ18-REP1, respectively, with BamHI-linearized, end-filled pGAD424. In both constructs, amino-terminally truncated Rep1 is fused in frame to the carboxy terminus of the Gal4AD. pGADrep1Δ130–373 was constructed by StuI/BamHI digestion, end filling, and self-ligation of pGAD-REP1. pGADrep2Δ1–57 and pGADrep2Δ59–296 were constructed by EcoRI/NcoI and NcoI/BgIII digestion, respectively, and end filling of pGAD-REP2. pGADrep2Δ1–14 was a gift from J. Sherk and is a MaeII partial digestion product containing the truncated REP2 ORF inserted at the ClaI site in pGAD424. For the expression of GST-Rep fusion proteins in E. coli, the REP BamHI fragments were end-filled with the Klenow fragment and cloned into SmaI-digested pGEX-2T (Pharmacia) in the correct orientation, to give pGEX-REP1 and pGEX-REP2. For the expression of S peptide-tagged, hexahistidine-tagged, thioredoxin-Rep fusion proteins in E. coli (hereinafter termed pET-Rep fusion proteins), EcoRI/SalI fragments containing intact or truncated REP gene ORFs from the appropriate pGAD-REP plasmids were ligated with EcoRI/SalI-restricted pET32 LIC (Novagen).
The construction and mitotic stability of the 2μm stability vector pAS4 will be described elsewhere (A. Sengupta, manuscript in preparation), but in brief, it consists of a complete 2μm plasmid in the B form (15) with insertions at two positions; the FLP gene has been disrupted by end filling the internal EcoRI site with the Klenow fragment and inserting an end-filled 3.5-kb genomic EcoRI/BamHI fragment containing the yeast ADE2 gene (kindly provided by D. Gottschling) (12), and the BamHI-digested phagemid vector pTZ18R was inserted at a unique BamHI site in the inverted repeat in the REP1/REP2 3′ intergenic region, created by a brief BAL 31 digestion at the XbaI site followed by closure on BamHI oligonucleotide linkers (6). The result was an ADE2+ flp− 2μm plasmid which can be propagated in yeast and E. coli. The version of pAS4 with rep1 deleted was created by digestion with PvuII, which excises the entire REP1 coding region, including 126 bp of a 5′-end-flanking sequence and 3′-end-flanking sequence up to the XbaI site, and also removes all but 215 bp of the pTZ18R vector sequence. The large PvuII fragment lacking REP1 sequences was gel purified and self-ligated to create pAS4-Δrep1, which can be propagated only in yeast.
Two-hybrid assays.
Transformants and cotransformants in the two-hybrid yeast host CTY10/5d (2) were assayed for β-galactosidase expression using a permeabilized cell assay (28). Specificity of the interactions was confirmed by expressing all Rep fusion proteins in the absence of other fusion proteins and with either Snf1 or Snf4 fusion proteins (7).
Anti-Rep antibodies.
GST-Rep1 and GST-Rep2 fusion proteins were expressed in, and extracted from, E. coli transformed with pGEX-REP1 and pGEX-REP2, respectively, as described by Koerner et al. (19). Both GST-Rep fusion proteins were found in the insoluble fraction of E. coli extracts and were further purified by electroelution from unfixed sodium dodecyl sulfate (SDS)-polyacrylamide gels in which the insoluble fraction had been electrophoresed (20). Rep1 and Rep2 polyclonal antisera were generated by subcutaneous injection of rabbits with purified GST-Rep fusion proteins monthly and then every 2 weeks until a sufficient titer of anti-Rep antibodies was obtained, as assayed against Western blots of GST-Rep1 and GST-Rep2 fusion proteins and [cir+] and [cir0] yeast whole-cell extracts (14). Anti-GST-Rep1 and anti-GST-Rep2 antibodies were affinity purified from the undiluted crude polyclonal antisera by the method of Pringle et al. (26) using a Trans-Blot 0.2-μm-pore-size polyvinylidene difluoride membrane (Bio-Rad) for immobilization of gel-purified GST-Rep1 and GST-Rep2 and GEG buffer (0.2 M glycine, 1 mM EGTA, pH 2.5) for antibody elution.
Western blotting.
Proteins were separated on SDS–10% polyacrylamide gels (20) and transferred to a polyvinylidene difluoride membrane for Western blotting analysis as described by Towbin et al. (32) except that 0.037% SDS was included in the transfer buffer since this was found to be essential for efficient transfer of Rep2. Rabbit anti-Gal4AD antibodies were purchased from Santa Cruz Biotechnology. The secondary antibody for Western blotting was peroxidase-labeled goat anti-rabbit immunoglobulin G, which was detected by chemiluminescence using a LumiGLO kit (Kirkegaard & Perry Laboratories, Inc.).
In vitro protein interaction assay.
Expression of GST and pET fusion proteins was induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 h at 25°C. GST fusion proteins were isolated and in vitro protein interaction assays were carried out as described by Ahn et al. (1). Equal amounts of GST fusion proteins were incubated with 50 μl of a 50% slurry of glutathione-agarose beads (Sigma) that had been preequilibrated in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3) containing 0.2% Nonidet P40 (NP-40) and a protease inhibitor cocktail (a 1× concentration contains 0.5 μg of leupeptin per ml, 1 μg of aprotinin per ml, and 0.8 μg of pepstatin A per ml). The final volume was increased to 200 μl with buffer A (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 2 mM methionine) containing protease inhibitors, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride, and the beads were incubated for 1 h with gentle rocking at room temperature (RT). Bound GST fusion proteins were collected by centrifugation at 500 × g for 3 min. Beads were washed three times with 10 volumes of cold PBS–0.2% NP-40, with rocking for 5 min between washes. Beads were resuspended in 100 μl of PBS–0.2% NP-40–150 μg of bovine serum albumin per ml. pET fusion proteins were prepared by resuspending cell pellets in ice-cold sonication buffer (50 mM NaH2PO4 [pH 8.0], 10 mM Tris [pH 8.0], 100 mM NaCl) with 15% glycerol, freezing at −20°C, and low-speed centrifugation. The pellet was resuspended in sonication buffer containing 8 M urea. Following sonication at 4°C, the urea-solubilized E. coli extracts were centrifuged at 15,800 × g for 15 min. The pET fusion proteins were purified from the supernatant by affinity chromatography on TALON resin (Clontech) under denaturing conditions according to the manufacturer's directions. The TALON-purified material was diluted to 10 ng/μl in buffer A containing protease inhibitors, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride and dialyzed against three changes of buffer A at 4°C. The purified pET fusion protein extracts were centrifuged at 15,800 × g for 10 min at 4°C to remove any particulate matter. The equivalent of 500 ng of renatured pET fusion protein was added to the bead-bound GST fusion proteins, and the volume was increased to 500 μl with buffer A. Samples were rocked for 1 h at RT, followed by centrifugation at 500 × g for 3 min. Beads were washed five times with an equal volume of PBS–0.2% NP-40 by rocking them for 5 min, followed by centrifugation at 500 × g for 3 min. The beads with any bound proteins were then resuspended in 50 μl of 2× gel loading buffer and one-fifth of each sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on a 12% polyacrylamide gel and Western blot analysis. The presence of pET fusion proteins was detected by incubation with chemiluminescent reagents directed against the S protein tag (Kirkegaard & Perry Laboratories, Inc.).
Southwestern assay.
Southwestern blotting was performed as described by Mellor et al. (21). Western blots of SDS-PAGE-separated TALON-purified pET fusion proteins were incubated with radiolabeled probes generated by using the Klenow fragment; unlabeled dGTP, dATP, TTP; and [α-32P]dCTP to label either a 375-bp EcoRI/BamHI fragment from the E. coli plasmid pBR322 or a 312-bp EcoRI/HindIII fragment containing the 2μm STB-proximal locus [the 2μm STB AvaI/HpaI fragment had been end filled and subcloned at the EcoRV site in the pBluescript II SK(+) (Stratagene) polylinker region, where it is flanked by these two sites]. Specific activities of the probes were 1.4 × 108 for the pBR322 probe and 7.5 × 107 cpm/μg probe for the STB-proximal probe. Duplicate Western blots were preincubated in BBW buffer (3 g of Ficoll per liter, 3 g of polyvinylpyrrolidone per liter, 10 mM NaCl, 20 mM Tris [pH 8.0], 0.25% milk powder) and then placed in 1 ml of this buffer in a sealed bag for a 1-h incubation at RT with 5 μg of unlabeled sonicated salmon sperm DNA and 50 ng of either radiolabeled probe. The membranes were then washed four times for 30 min in the BBW buffer before exposure to X-ray film for 3.5 h at −70°C with an intensifier screen.
Plasmid stability assays.
LEU2 pGAD-REP1 fusion plasmids and the ADE2 pAS4-Δrep1 plasmid were cotransformed into the [cir0] yeast strain AS3 by the LiAc/SS-DNA/PEG protocol of Gietz et al. (11). For the plasmid stability assay, cotransformed yeast cells were initially grown in media lacking adenine and leucine to select for the presence of both plasmids. The cells were then allowed a period of growth where there was no selection for the presence of the ADE2 stability vector. The proportion of cells containing both plasmids, or just the LEU2 pGAD424-based plasmid, was determined both before and after the period of nonselective growth by plating cells on the appropriate selective medium and counting the number of viable colonies formed. The rate of plasmid loss per generation was calculated as previously described (6).
RESULTS
Rep protein interactions in vivo.
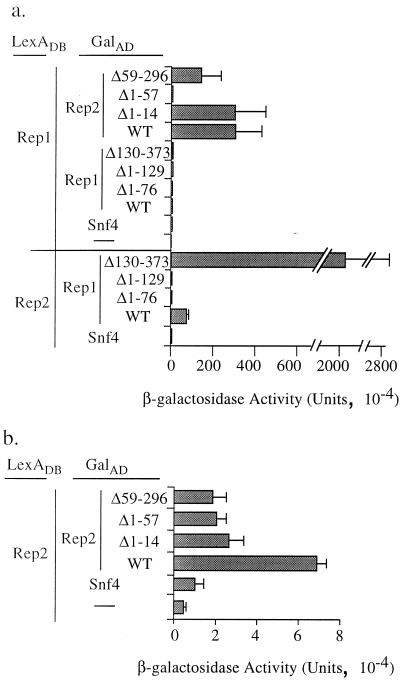
Genetic data have suggested that Rep1 and Rep2 function as part of a complex to regulate FLP gene expression and to mediate 2μm plasmid segregation (5, 6, 35). A two-hybrid genetic assay has previously shown that these two proteins interact in vivo (1, 36). We have used a similar approach here to investigate the ability of truncated versions of Rep1 and Rep2, expressed as in-frame fusions with the Gal4AD, to interact with either Rep1 or Rep2 expressed as in-frame fusion proteins with the bacterial DNA-binding protein LexA. Plasmids were cotransformed into the yeast two-hybrid host, where transcription of a lacZ reporter gene (detected by measuring β-galactosidase activity) requires interaction of the two fusion proteins to recruit RNA polymerase to the LexA binding site upstream of the reporter gene (2). The results of the two-hybrid assay are shown in Fig. 1. None of the Rep fusion proteins or their truncated derivatives activated the lacZ reporter gene when they were singly expressed in the yeast two-hybrid host (data not shown), demonstrating that neither Rep1 nor Rep2 has an intrinsic transcriptional activation function in this assay. In accordance with the results of earlier studies (1, 36), expression in the reporter strain of both Gal4AD-Rep1 and LexA-Rep2, or both Gal4AD-Rep2 and LexA-Rep1, resulted in activation of the lacZ reporter gene, indicating a Rep1-Rep2 interaction. β-Galactosidase production was also observed when the plasmids expressing Gal4AD-Rep2 and LexA-Rep2 were cotransformed into the yeast two-hybrid host, although at a lower level than that observed for the Rep1-Rep2 fusion protein cotransformants. Although Rep1 self-association has previously been observed using a two-hybrid assay (1, 36), no association was detected in our assay. This may reflect interference with the interaction by the LexA moiety fused to the amino terminus of the Rep1 protein or differences between the two-hybrid assay systems used in the two studies. Neither of the pGAD-REP plasmids expressing Rep1 proteins with amino-terminal deletions induced β-galactosidase expression when they were coexpressed with LexA-Rep2, suggesting that loss of Rep1 amino-terminal sequences abolishes interaction with Rep2. In contrast, when LexA-Rep2 was coexpressed with the version of Rep1 with a carboxy-terminal deletion, expression of the reporter gene was significantly enhanced. These results suggest that the first 129 amino acids of Rep1 are sufficient for interaction with Rep2 and that Rep1 carboxy-terminal sequences may influence this association.
FIG. 1.
Two-hybrid assay for in vivo Rep protein interaction. Activation of the lacZ reporter in the two-hybrid yeast strain CTY10/5d transformed with plasmids expressing the LexA and Gal4AD fusion proteins, shown on the left, was measured by a permeabilized cell assay. β-Galactosidase activities represent the averages and standard deviations obtained with four independent transformants. WT, wild type.
In the Rep2 two-hybrid analysis, deletion of either the first 14 amino acids of Rep2 or all but the first 58 amino acids did not significantly reduce the level of expression of β-galactosidase when these fusion proteins were coexpressed with LexA-Rep1, while removal of the first 57 amino acids of Rep2 led to loss of expression of the reporter gene. Taken together, these data suggest that the amino-terminal 57 amino acids of Rep2 are required for Rep1 interaction and that residues 15 to 58 are sufficient for this association. All Rep2 truncations gave lower β-galactosidase levels than full-length Rep2 when they were coexpressed as Gal4AD fusions with LexA-Rep2. However, the level of activation was significantly higher than when LexA-Rep2 was expressed in the absence of a Gal4AD fusion protein, suggesting that none of the truncations completely abolished the ability of Rep2 to interact with the LexA-Rep2 fusion protein.
Rep fusion protein expression.
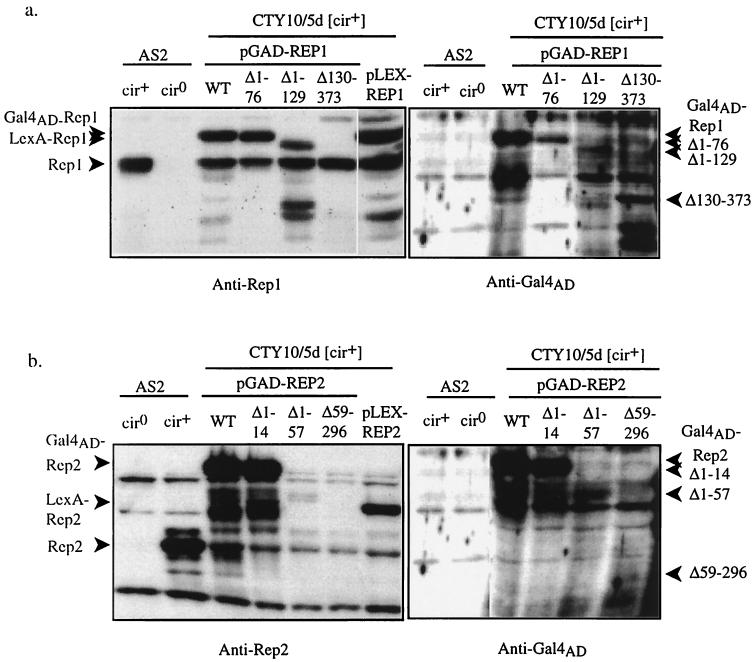
Lack of β-galactosidase expression in the two-hybrid assay may be due to a lack of interaction between the Rep1 and Rep2 portions of the two fusions, or it may be due to a lack of expression or lower abundance of the Rep deletion fusion proteins. To verify expression of the Rep fusion proteins, whole-cell protein extracts from the two-hybrid cotransformants were isolated and analyzed by SDS-PAGE and Western blotting using affinity-purified polyclonal antibodies specific for Rep1, Rep2, and the Gal4AD (Fig. 2). The anti-Rep1 antibody Western blot (Fig. 2a) shows a band migrating with a molecular mass of 50 kDa in all protein extracts made from [cir+] yeast (lanes 1 and 3 to 7) but not from a [cir0] yeast strain (lane 2). The size of this band is slightly larger than the 43-kDa predicted molecular mass for native Rep1 protein (15) but similar to that observed in previous studies (30, 38). A novel Rep1-antigenic band with a mobility of approximately 56 kDa was observed in the extract derived from yeast containing the pLEX-REP1 plasmid. All transformants containing plasmids expressing Gal4AD-Rep1 fusion proteins showed novel Rep1 antigenic bands with mobilities consistent with the amount of the REP1 coding region remaining in each, and these bands were also detected with the Gal4AD-specific antibody. In the case of the Gal4AD-Rep1Δ130–373 fusion protein, the anti-Rep1 antigenic bands were faint, presumably due to loss of epitopes in the deleted carboxy-terminal portion of the protein. Lower-molecular-weight bands were also detected by both antibodies in all extracts derived from yeast expressing Gal4-Rep fusion proteins and probably represent products of premature translational termination or proteolysis.
FIG. 2.
Western blot analysis was used to confirm expression of the Rep fusion proteins in the two-hybrid host. Total protein was extracted and analyzed by SDS-PAGE and Western blotting with anti-Ga14AD antibodies and affinity-purified anti-Rep1 (a) or anti-Rep2 (b) antibodies. Protein was from strain AS2 [cir0] or [cir+] and the two-hybrid host CTY10/5d [cir+] cotransformed with either pLEX-REP2 expressing the LexA-Rep2 fusion protein and pGAD424 containing either full-length or truncated versions of the REP1 ORF, allowing them to be expressed as Ga14AD fusion proteins (a), or pLEX-REP1 and pGAD424 containing either full-length or truncated versions of the REP2 ORF (b). Protein from CTY10/5d transformed with either pLEX-REP1 (a) or pLEX-REP2 (b) (lanes 7) was also included. The positions of native Rep1 and Rep2, as well as those of Rep fusion proteins, are indicated by arrowheads. Autoradiographs were scanned using Molecular Analyst (Bio-Rad) software and prepared for figures using Adobe Photoshop. WT, wild type.
The anti-Rep2 antibody Western blot (Fig. 2b) shows a band migrating with a molecular mass of 35 kDa in all protein extracts made from [cir+] yeast (lanes 2 to 7) but not from the [cir0] yeast strain (lane 1). The size of this band is consistent with the predicted molecular mass of native Rep2 protein, 33 kDa (15). The anti-Rep2 antibody also detected a band of approximately 57 kDa and, more variably, two lower-molecular-mass bands in all protein extracts. These proteins must be non-2μm related since they were present in extracts derived from both [cir+] and [cir0] yeasts. A novel Rep2-antigenic band with a mobility of approximately 43 kDa was observed in the extract derived from yeast containing the pLEX-REP2 plasmid. In addition, novel bands with mobilities of approximately 53, 52, and 45 kDa were detected by both anti-Rep2 and anti-Gal4AD antibodies in extracts derived from yeast containing the plasmids pGAD-REP2, pGADrep2Δ1–14, and pGADrep2Δ1–57, respectively. These molecular masses are consistent with the sizes expected for these Gal4AD-Rep fusion proteins. No novel anti-Rep2 antigenic band was observed in extracts derived from yeast containing the pGADrep2Δ59–296 plasmid. The anti-Gal4AD antibody did detect low levels of a protein with an apparent mobility of 31 kDa in these extracts, close to the expected size for this fusion protein, suggesting that the Rep2 truncation eliminated the epitopes recognized by the polyclonal anti-Rep2 antibodies. Despite the low steady-state level of the Gal4AD-Rep2Δ59–296 fusion protein, the level was sufficient to activate the reporter gene when it was coexpressed with LexA-Rep1. In contrast, the Gal4AD-Rep2Δ1–57 fusion protein, which was expressed at a higher level than the carboxy-terminally truncated Rep2 fusion protein, did not activate the reporter gene when it was coexpressed with LexA-Rep1 and neither did the higher levels of amino-terminally truncated Rep1 when it was coexpressed with LexA-Rep2. In these cases, lack of β-galactosidase expression for the two-hybrid cotransformants must have been due to lack of interaction between the fusion proteins being tested rather than a result of the lack of expression of the truncated protein.
Rep protein interactions in vitro.
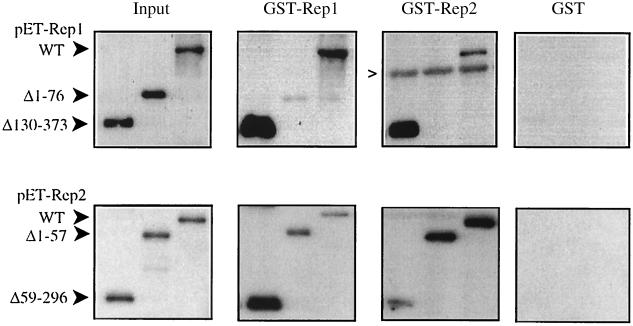
To assess the significance of the two-hybrid assay results, we performed an in vitro protein interaction assay in which GST-Rep fusion proteins and GST, bound to glutathione-agarose beads, were tested for their ability to bind either full-length or truncated Rep proteins expressed as pET fusion proteins (Fig. 3). Equal amounts of the indicated target pET-Rep fusion proteins were incubated with either GST-, GST-Rep1-, or GST-Rep2-bound beads. Neither Rep1, Rep2, nor any of the truncated Rep fusion proteins were brought down by GST. In agreement with previous studies (1, 30), both Rep1 and Rep2 were brought down by either GST-Rep1 or GST-Rep2, although the amount of pET-Rep1 retained on the GST-Rep2 beads was less than that bound by GST-Rep1 beads, while the opposite was observed for pET-Rep2, with more being bound to GST-Rep2 than to GST-Rep1. The pET-Rep1Δ130–373 fusion protein was retained on both the GST-Rep1 and GST-Rep2 beads significantly more efficiently than the nontruncated Rep1 fusion protein. A small amount of the pET-Rep1Δ1–76 fusion protein was bound by the GST-Rep1 beads, but no association of this amino-terminally truncated Rep1 fusion protein with the GST-Rep2 beads was observed, even after prolonged exposure of the membrane to film or when increased amounts of the truncated fusion protein were added to the beads. These results support the findings from the two-hybrid assay indicating that the first 76 amino acids of Rep1 are required for association with Rep2 but that the first 129 amino acids are sufficient for this interaction. The in vitro association of Rep1 supports studies showing Rep1 self-association in vivo (1, 36) and suggests that the region comprising amino acids 77 to 129 is required for this interaction.
FIG. 3.
In vitro baiting assay demonstrating Rep protein interaction. Rep proteins and deletion derivatives were expressed as pET fusions; shown are results with full-length Rep1, Rep1Δ1–76, and Rep1Δ130–373 (top panels) and full-length Rep2, Rep2Δ1–57, and Rep2Δ59–296 (bottom panels). The pET-Rep fusion proteins were incubated with GST-Rep1, GST-Rep2, or GST bound to glutathione-agarose beads. The input (1/10 of that added to the reaction mixtures) and bound pET fusion proteins were analyzed by SDS-PAGE and Western blotting and detected with an S protein probe and chemiluminescence. The positions of the pET-Rep fusion proteins are indicated by filled arrowheads. A caret indicates the position of GST-Rep2, which cross-reacts with the S protein probe. The autoradiograph was scanned using Ofoto (Light Source) software and prepared for figures using Adobe Photoshop. WT, wild type.
The baiting assay with the two truncated pET-Rep2 fusion proteins showed that both parts of the Rep2 protein were capable of independently mediating interaction with either GST-Rep1 or GST-Rep2, although the pET-Rep2Δ59–296 protein was brought down much less efficiently by the GST-Rep2 beads than was the amino-terminally truncated Rep2 fusion. The converse was observed for the interaction with the GST-Rep1 beads; the carboxy-terminally truncated Rep2 was more efficiently retained than the pET-Rep2Δ1–57 fusion. The two-hybrid assay results support an interaction between Rep1 and the amino-terminal portion of Rep2 but did not provide evidence for the in vitro association observed between Rep1 and the carboxy-terminal domain of Rep2.
Rep2 DNA-binding activity.
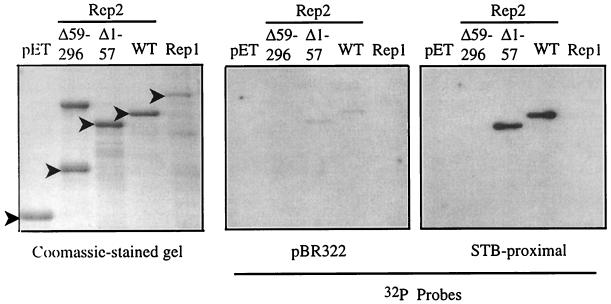
Although the Rep proteins are hypothesized to interact with the 2μm STB locus to mediate plasmid segregation, direct association of either of the Rep proteins with DNA has yet to be demonstrated. Transient association of the Rep proteins with STB DNA has been shown by a biosensor assay but requires the presence of other unidentified host proteins (13). Biochemical studies of Rep1 and Rep2 have been complicated by the inherent insolubility of the native proteins when they were isolated from yeast (13, 38) or when they were expressed as fusion proteins in E. coli. (M. J. Dobson, unpublished results). To circumvent these insolubility problems, a Southwestern assay was used to determine whether either Rep1 or Rep2 might have intrinsic DNA-binding activity (Fig. 4). Rep1 and the carboxy-terminally truncated Rep2 did not demonstrate any DNA-binding activity in this assay. Rep2 and amino-terminally truncated Rep2 both bound a DNA fragment containing the repeat region of STB and with lower efficiency bound a DNA fragment derived from the E. coli plasmid pBR322.
FIG. 4.
Southwestern assay shows Rep2 DNA-binding activity. pET-Rep fusion proteins were purified by affinity chromatography and separated by SDS-PAGE. Triplicate gels were either stained with Coomassie blue or Western blotted and incubated with a radiolabeled DNA (either a 375-bp EcoRI-BamHI fragment from the E. coli plasmid pBR322 or a 312-bp EcoRI-HindIII fragment containing the 2μm STB-proximal locus), and the signal was detected by autoradiography. The positions of the pET-Rep fusion proteins are indicated by filled arrowheads. (A higher-molecular-weight protein that copurifies with the pET-Rep2Δ59–296 fusion protein on Talon resin is observed as a slower-migrating species on the Coomassie blue-stained gel.) WT, wild type.
Rep1 segregation function.
The results of the two-hybrid and baiting assays provide evidence for in vivo association of Rep1 and Rep2. To determine the connection between this association and 2μm plasmid segregation, we decided to test the ability of the truncated forms of Rep1 to mediate plasmid segregation. To assay plasmid segregation, we used a 2μm-based stability vector, pAS4 (A. Sengupta, unpublished data). In addition to containing all 2μm sequences, the vector carries an ADE2 gene, which allows selection for the plasmid in an ade2 host strain. If the REP1 and REP2 genes on the vector are wild type, the plasmid segregates in a [cir0] host almost as efficiently as native 2μm (A. Sengupta, unpublished results). For this study, a rep1 deletion derivative of pAS4, pAS4-Δrep1, was cotransformed into a leu2 ade2 [cir0] yeast host with the LEU2 pGAD424-derived plasmids expressing the Gal4AD-Rep1 fusion proteins to determine whether the fusion proteins could complement the rep1 deletion in the stability vector. pGAD424 is itself a 2μm-based vector but contains only the cis-acting STB locus; its normal mitotic stability relies on Rep1 and Rep2 being supplied in trans from the 2μm plasmids in the [cir+] two-hybrid host. In our [cir0] assay system, the stability vector with rep1 deleted supplies Rep2 while the Gal4AD-Rep1 fusion protein is the only source of Rep1. If the Gal4AD-Rep1 fusion protein is able to supply Rep1 function, most cells grown under conditions selecting for the presence of both plasmids should be able to form colonies on medium lacking adenine and leucine since both contain the STB locus and will be efficiently partitioned at each cell division. Conversely, if the Gal4-Rep1 fusion protein does not restore Rep1 function, a significant proportion of the cells grown in selective medium will not contain both plasmids, reflecting the inability of the plasmids to be segregated to the daughter cells. Similarly, the rate of loss of the ADE2 stability vector with rep1 deleted from cells containing the LEU2 Gal4AD-Rep1 fusion protein-expressing plasmid should be high if the Rep1 fusion protein is nonfunctional but low if Rep1 function is restored. The percentage of Leu+ cells which also contain the ADE2 plasmid was determined for the cotransformants, both during growth selecting for the presence of both plasmids and after a period of growth in which selection for the ADE2 plasmid had been relaxed. The results are shown in Table 1. A comparison of yeast cotransformed with pAS4-Δrep1 and pGAD-REP1 versus those cotransformed with pAS4-Δrep1 and the vector pGAD424 indicates that the full-length Rep1-Gal4AD fusion protein is able to restore mitotic stability to the version of pAS4 with rep1 deleted. The rate of loss of the pAS4-Δrep1 plasmid during nonselective growth was also reduced by expression of the full-length Rep1-Gal4AD fusion protein. In contrast, none of the truncated Rep1-Gal4AD fusion proteins was able to increase the mitotic stability of pAS4-Δrep1 beyond that observed with the pGAD424 cotransformant. The results show that deletion of the amino-terminal 76 amino acids or the carboxy-terminal 244 amino acids of Rep1 destroys its ability to mediate plasmid segregation.
TABLE 1.
Plasmid segregation assaya
| LEU2 plasmid | % of Leu+ cells retaining the ADE2 pAS4-rep1Δ plasmid after growth in medium lacking adenine and leucine | Rate of loss of the ADE2 pAS4-rep1Δ plasmid from Leu+ cells during growth in adenine-containing medium (%/generationb) |
|---|---|---|
| pGAD424 | 66.7 (±14.7) | 13.3 (±8.0) |
| pGAD-REP1 | 96.0 (±7.7) | 0.4 (±1.4) |
| pGAD-rep1Δ1–76 | 61.7 (±12.3) | 13.8 (±6.2) |
| pGAD-rep1Δ1–129 | 72.1 (±5.6) | 13.3 (±4.8) |
| pGAD-rep1Δ130–373 | 44.9 (±12.2) | 15.9 (±5.1) |
The ability of Gal4AD-Rep1 fusion proteins to complement a REP1 deletion was tested by measuring the mitotic stability of the 2μm-based ADE2 plasmid pAS4-Δrep1 when it was cotransformed in an ade2 leu2 [cir0] yeast strain, AS3, with either the LEU2 pGAD424 vector or derivatives of this plasmid expressing Rep1 fusion proteins. Data are averages ± standard deviations from assays of a minimum of four independent yeast transformants. See Materials and Methods for details.
Numbers of generations in adenine-containing medium ranged from 10 to 12 for all cultures.
DISCUSSION
We have used two-hybrid and in vitro baiting assays to determine the regions of the yeast 2μm plasmid-encoded proteins Rep1 and Rep2 that mediate the interactions of these proteins and a Southwestern assay to identify Rep2 DNA-binding activity. These interactions are summarized in Fig. 5. A plasmid segregation assay was used to demonstrate that amino-terminal deletions of Rep1 that abolish Rep1-Rep2 interaction and a carboxy-terminal deletion that does not impair this interaction both result in loss of Rep protein-mediated plasmid partitioning.
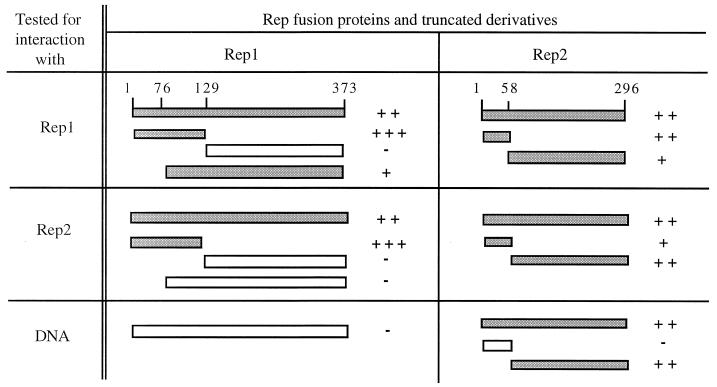
FIG. 5.
Summary of in vivo and in vitro Rep protein interactions. Rep fusion proteins and their truncated derivatives, tested for interaction with Rep1, Rep2, or DNA by two-hybrid, baiting, and Southwestern assays, are shown as rectangles. Gray rectangles indicate interacting fusions, while white rectangles indicate no interaction observed. The degree of interaction is indicated as strongest (+++) to weakest (+). A minus indicates no interaction.
The predicted amino acid sequence of the Rep proteins reveals little about how they mediate plasmid partitioning. Proteins with functions analogous to those of Rep1 and Rep2 are encoded by other 2μm-like plasmids in several closely related yeasts, Zygosaccharomyces rouxii, Zygosaccharomyces bisporus, and Kluyveromyces marxianus (33). Despite the lack of any apparent DNA sequence homology between these yeast plasmids, all share remarkably similar structures; they are similar in size and have a large perfect inverted repeat which divides the plasmid into two unique regions encoding at least three ORFs and a locus required in cis for mitotic stability (33). The encoded proteins include a site-specific recombinase which, like 2μm Flp, promotes amplification of the plasmids by catalyzing recombination between the inverted repeats and two proteins which, like Rep1 and Rep2, are required in trans for mitotic stability of the plasmids. The Rep1-equivalent trans-acting stability factors encoded by the other 2μm-like plasmids share a limited degree of amino acid sequence similarity with 2μm Rep1, whereas the Rep2-like factors are too dissimilar to be aligned (24). The first 129 amino acids of 2μm Rep1, which we have shown to be essential and sufficient for Rep1 self-association and for interaction with Rep2, include a domain which is conserved between Rep1 and the Rep1-like ORFs from the other 2μm-like plasmids (6). The proteins share about 44% amino acid similarity across a 60-amino-acid stretch (residues 30 to 89) (24). The sequence conservation in this amino-terminal domain may reflect its dual functions of mediating Rep1 and Rep2 interaction. However, given the lack of sequence conservation between the Rep2 analogues and their inability to complement the segregation defect of rep2 derivatives of 2μm-like plasmids other than their own, we might not expect the Rep1-Rep2 interaction domain to be as highly conserved as the Rep1 self-association domain (24). In our in vitro association assay, deletion of the first 76 amino acids of Rep1 reduced but did not abolish Rep1 self-association, while data from Velmurugan et al. (36) demonstrated that loss of the first 100 amino acids of Rep1 abolished interaction with both Rep1 and Rep2. Taken together, these data imply that residues critical for Rep1 self-association lie between residues 76 and 100. Almost completely contained within this region is the most significant block of identity shared by the different Rep1 proteins, a region spanning residues 72 to 85. We suggest that this conserved block is at least part of the Rep1 self-association domain. Our data do not allow us to precisely delineate the Rep1 domain responsible for Rep2 interaction, other than by indicating that residues amino terminal to residue 77 and those between positions 77 and 129 must be required.
The inability of the amino-terminally truncated Rep1 fusion proteins analyzed in our study to interact with Rep2 or to provide Rep1 plasmid segregation function indicates that the carboxy-terminal two-thirds of the protein is insufficient for these functions. Interestingly, deletion of this carboxy-terminal portion of Rep1 appeared to enhance both Rep1 self-association and interaction with Rep2, suggesting that this region may play an inhibitory role in Rep protein interactions. Despite the ability of carboxy-terminally truncated Rep1 to associate with both Rep1 and Rep2, the truncated protein was unable to promote segregation of a rep1− 2μm-based plasmid, indicating that this domain is required for the plasmid segregation function of Rep1. Examination of the amino acid sequence of S. cerevisiae Rep1 has revealed that the carboxy-terminal 40% of the protein has about 60% similarity to two coiled-coil proteins: myosin heavy chain and the intermediate filament protein vimentin (38). This carboxy-terminal domain of Rep1 also shows some degree of conservation between Rep1 analogues from other 2μm-like plasmids, although to a lesser extent than that observed for the amino-terminal domain; only 37% similarity is present over an 80-amino-acid stretch (24). Subcellular fractionation experiments have shown that S. cerevisiae Rep1 copurifies with a nuclear karyoskeletal fraction (38). These observations have led to the suggestion that Rep1 intercalates in the nuclear matrix or lamina by means of this carboxy-terminal domain to provide dispersed attachment sites for the 2μm plasmid, thereby ensuring its efficient partitioning when the nucleus divides (38). If Rep1 is normally tethered at these sites through this domain, the Rep1 two-hybrid fusion proteins may be restricted in their ability to participate in the two-hybrid assay. Deletion of this domain may release the sequestered proteins and may account for the enhanced two-hybrid interaction of carboxy-terminally truncated Rep1 with Rep2.
Protein interaction assays with truncated versions of Rep2 showed that the amino-terminal 58 amino acids were sufficient for promoting interaction with Rep1 and, to a lesser extent, self-association but that the carboxy-terminal 239 amino acids promoted self-association. The in vitro baiting assay also indicated that this carboxy-terminal protein of Rep2 might interact with Rep1. These nonoverlapping interaction domains raise the possibility that the Rep proteins can form a multimeric complex in vivo and may explain their inherent insolubility when they are purified from yeast. Further deletion derivatives of both Rep1 and Rep2 are needed to precisely delineate which protein domains determine the ability of the Rep proteins to interact with and to mediate 2μm plasmid segregation.
Most models of 2μm plasmid segregation predict that Rep1 and/or Rep2 will interact with the DNA repeats of the plasmid STB locus. Direct association of either of the Rep proteins with any 2μm DNA sequence has not previously been demonstrated. Evidence for such an association in vivo comes from observations of alterations in the DNase sensitivity of the STB region when Rep1 is not expressed in the cell and in that of the FLP promoter region, if either Rep1 or Rep2 is absent, compared to the pattern observed in [cir+] cells (34). These alterations might also occur if the Rep proteins interact with host proteins that bind to the DNA rather than binding directly to the DNA themselves. Hadfield et al. (13) reported an uncharacterized yeast host protein that interacts with the 2μm segregation locus, STB, in vitro. The binding activity was observed only in urea-solubilized yeast extracts isolated from cells expressing both Rep1 and Rep2 or in [cir0] extracts to which exogenous Rep1 and Rep2 had been added. This result suggests that the host protein may need to interact with the Rep1-Rep2 complex in order to bind STB or that Rep1 and Rep2 may need the host factor to promote formation of a complex that binds STB. The requirement for urea solubilization of the host factor may indicate that this protein(s) is normally associated with a subcellular structure (13). Our Southwestern assay demonstrated that Rep2 can preferentially bind STB DNA, but efficient binding of Rep2 to other AT-rich repeated DNA sequences has also been observed (M. J. Dobson, unpublished results). It is possible that additional sequence specificity is conferred when Rep2 is associated with Rep1 or other host proteins. Recently, Velmurugan et al. (36) isolated a potential candidate for a host protein involved in STB binding when they identified the SHF1 gene on the basis of its ability to activate expression of a reporter gene lying downstream of the STB repeats in a one-hybrid screen.
On the basis of the data presented here and in previous studies of the 2μm plasmid, we propose a model for plasmid partitioning in which there is competition between the Rep proteins for heterodimerization versus self-association. In this model, the Rep1-Rep2 complex is the functional unit for segregation, with Rep2 mediating polymerization of a multimeric Rep protein complex along the STB locus, both through its DNA-binding activity and its potential ability to interact simultaneously with both Rep1 and Rep2. Homodimerization of Rep1 may block the formation or allow the disassembly of this partitioning complex in synchrony with detachment of chromosomal kinetochores from the spindle apparatus during mitosis. The overlap in Rep1 of the regions required for Rep2 interaction and for self-association suggests that these two interactions may be mutually exclusive. Support for this comes from the results of in vitro assays similar to those reported here in which simultaneous addition of Rep1 and Rep2 fusion proteins to GST-Rep1 beads reduced the amounts of bound protein for both compared to when either was added alone, whereas no reduction was observed when they were both added to GST-Rep2 beads (30). In our baiting assays, deletion of the carboxy-terminal portion of Rep1 consistently led to an increase in the amount of self-association of Rep1 while the effect on Rep2 interaction was generally less pronounced. These results suggest that the carboxy-terminal region of Rep1 may inhibit the ability of the amino-terminal domain to interact with either Rep1 or Rep2. In vivo, it is possible that host proteins interacting with the carboxy-terminal domain of Rep1 influence the balance between hetero- and homodimerization and hence between formation and disassembly of the partitioning complex.
There is some experimental support for this model. First, insufficient expression of Rep1 relative to normal Rep2 levels should disrupt segregation of 2μm-based plasmids. Cashmore et al. (5) have shown that a single chromosomally integrated copy of the REP2 gene expressed from its own promoter was able to fully complement the segregation defect of a multicopy rep2− plasmid. In contrast, one integrated copy of the REP1 gene was not sufficient to stabilize a multicopy rep1− plasmid. When the number of integrated copies of the REP1 gene was increased, the stability of the rep1− plasmid increased. These results suggest that Rep1 expression must reach a certain level for efficient plasmid segregation to be established. Further support for this model comes from the experiments of Hadfield et al. (13), in which an observed in vitro STB-binding activity was found to be dependent on the relative levels of expression of Rep1 and Rep2, insufficient Rep1 relative to Rep2 resulting in reduced binding.
Second, if the only requirement for segregation is to maintain a sufficient supply of the Rep1-Rep2 heterodimer, overexpressing Rep1, without altering Rep2 levels, should have no effect on plasmid segregation. Support for this possibility comes from experiments in which an efficient heterologous promoter was used to direct expression of the REP1 ORF on a 2μm-based vector in a [cir+] cell (6). The plasmid displayed high mitotic stability, suggesting that segregation was unperturbed. Concomitant overexpression of Rep1 and Rep2 might have an effect on the cell if the excess Rep1-Rep2 complex can polymerize on other chromosomal DNA sequences, forming a functioning partitioning complex, or is otherwise able to interact with and titrate host proteins, preventing them from performing their normal functions. These host proteins might be required for mediating chromosome segregation. Reynolds et al. (27) reported a reduction in the growth rate of [cir0] cells when Rep1 and Rep2 were simultaneously overexpressed under the control of the GAL1-10 promoter. This reduced growth was not observed when either of the Rep proteins was overexpressed alone, suggesting that the Rep1-Rep2 complex was interfering with some normal cellular process. We are currently continuing our studies to further elucidate the nature of the Rep1-Rep2 complex and to determine the host proteins with which it interacts to ensure the efficient segregation of the 2μm plasmid when the nucleus divides.
ACKNOWLEDGMENTS
We acknowledge that Kristina Blomqvist and Arpita Sengupta contributed equally to this research.
This work was supported by research grant number OGP0155268 from the Natural Science and Engineering Research Council of Canada. K.B. was supported by a postdoctoral fellowship from the Swedish Foundation for International Cooperation in Research and Higher Education. A.P. was supported by an NSERC postgraduate scholarship.
REFERENCES
- 1.Ahn Y-T, Wu X-L, Biswal S, Velmurugan S, Volkert F C, Jayaram M. The 2μm-encoded Rep1 and Rep2 proteins interact with each other and colocalize to the Saccharomyces cerevisiae nucleus. J Bacteriol. 1997;179:7497–7506. doi: 10.1128/jb.179.23.7497-7506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel P, Chien C T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interaction. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: IRL Press; 1993. pp. 153–179. [Google Scholar]
- 3.Beggs J D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978;275:104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- 4.Broach J R, Hicks J B. Replication and recombination functions associated with the yeast plasmid, 2μm circle. Cell. 1980;21:501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- 5.Cashmore A M, Albury M S, Hadfield C, Meacock P A. Genetic analysis of partitioning functions encoded by the 2μm circle of Saccharomyces cerevisiae. Mol Gen Genet. 1986;203:154–162. [Google Scholar]
- 6.Dobson M J, Yull F E, Molina M, Kingsman S M, Kingsman A J. Reconstruction of the yeast 2μm plasmid partitioning mechanism. Nucleic Acids Res. 1988;16:7103–7117. doi: 10.1093/nar/16.14.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durfee T, Becherer K, Chen P, Yeh S, Yang Y, Kilburn A E, Lee W, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 8.Futcher A B. Copy number amplification of the 2μm circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 9.Futcher A B. The 2μm circle plasmid of Saccharomyces cerevisiae. Yeast. 1988;4:27–40. doi: 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- 10.Futcher A B, Cox B S. Maintenance of the 2μm circle plasmid in populations of Saccharomyces cerevisiae. J Bacteriol. 1983;154:612–622. doi: 10.1128/jb.154.2.612-622.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 12.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at Saccharomyces cerevisiae telomeres: reversible repression of polII transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 13.Hadfield C, Mount R C, Cashmore A M. Protein binding interactions at the STB locus of the yeast 2μm plasmid. Nucleic Acids Res. 1995;23:995–1002. doi: 10.1093/nar/23.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 15.Hartley J L, Donelson J E. Nucleotide sequence of the yeast plasmid. Nature. 1980;286:860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- 16.Jayaram M, Li Y-Y, Broach J R. The yeast plasmid 2μm circle encodes components required for its high copy propagation. Cell. 1983;34:95–104. doi: 10.1016/0092-8674(83)90139-3. [DOI] [PubMed] [Google Scholar]
- 17.Jayaram M, Sutton A, Broach J R. Properties of REP3: a cis-acting locus required for stable propagation of the Saccharomyces cerevisiae plasmid 2μm circle. Mol Cell Biol. 1985;5:2466–2475. doi: 10.1128/mcb.5.9.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kikuchi Y. Yeast plasmid requires a cis-acting locus and two plasmid proteins for its stable maintenance. Cell. 1983;35:487–493. doi: 10.1016/0092-8674(83)90182-4. [DOI] [PubMed] [Google Scholar]
- 19.Koerner T J, Hill J E, Myers A M, Tzagoloff A. High expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Mellor J, Fulton A M, Dobson M J, Roberts N A, Wilson W, Kingsman A J, Kingsman S M. The Ty transposon of Saccharomyces cerevisiae determines the synthesis of at least three proteins. Nucleic Acids Res. 1985;13:6249–6263. doi: 10.1093/nar/13.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray A W, Szostak J W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 23.Murray J A H, Cesareni G. Functional analysis of the yeast plasmid partition locus STB. EMBO J. 1986;5:3391–3399. doi: 10.1002/j.1460-2075.1986.tb04655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray J A H, Cesareni G, Argos P. Unexpected divergence and molecular coevolution in yeast plasmids. J Mol Biol. 1988;200:601–607. doi: 10.1016/0022-2836(88)90546-3. [DOI] [PubMed] [Google Scholar]
- 25.Murray J A H, Scarpa M, Rossi N, Cesareni G. Antagonistic controls regulate copy number of the yeast 2μm plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds A E, Murray A W, Szostak J W. Roles of the 2μm gene products in stable maintenance of the 2μm plasmid of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose M D, Winston F, Hieter P. Methods in yeast genetics, a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning, a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Scott-Drew S, Murray J A H. Localization and interaction of the protein components of the yeast 2μm circle plasmid patitioning system suggest a mechanism for plasmid inheritance. J Cell Sci. 1998;111:1779–1789. doi: 10.1242/jcs.111.13.1779. [DOI] [PubMed] [Google Scholar]
- 31.Som T, Armstrong K A, Volkert F C, Broach J R. Autoregulation of 2μm circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 32.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Utatsu I, Sakamoto S, Imura T, Toh-e A. Yeast plasmids resembling 2μm DNA: regional similarities and diversities at the molecular level. J Bacteriol. 1987;169:5537–5545. doi: 10.1128/jb.169.12.5537-5545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veit B E, Fangman W L. Chromatin organization of the Saccharomyces cerevisiae 2μm plasmid depends on plasmid-encoded products. Mol Cell Biol. 1985;5:2190–2196. doi: 10.1128/mcb.5.9.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veit B E, Fangman W L. Copy number and partition of the Saccharomyces cerevisiae 2μm plasmid controlled by transcription regulators. Mol Cell Biol. 1988;8:4949–4957. doi: 10.1128/mcb.8.11.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velmurugan S, Ahn Y, Yang X, Wu X, Jayaram M. The 2μm plasmid stability system: analyses of the interactions among plasmid and host encoded components. Mol Cell Biol. 1998;18:7466–7477. doi: 10.1128/mcb.18.12.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volkert F C, Broach J R. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 38.Wu L C, Fisher P A, Broach J R. A yeast plasmid partitioning protein is a karyoskeletal component. J Biol Chem. 1987;262:883–891. [PubMed] [Google Scholar]