Abstract
The availability of multiple gene sequences, and in particular full genome sequence data, for microbial strains has changed how taxonomists delineate subspecies belonging to the Archaea and Bacteria. Well-defined phylogenetic lineages that share higher genome similarity values compared to the widely used species thresholds are often described as subspecies, despite clear evidence of genetic isolation between them. These well-defined lineages, reflecting notable genetic isolation of the core genome represent more recently evolved, unique and sui generis evolutionary units. Because they bear all of the hallmarks of species, most contemporary subspecies likely represent species in their own right. Although there is considerable value in defining intraspecies variation (e.g., pathovar, serovar and symbiovar), the discriminating properties of such units are mostly encoded on accessory subgenomic compartments. We therefore argue that the taxonomic category of subspecies has become irrelevant and propose that its use should be discontinued. This will minimize inconsistencies related to the subjective nature of species-subspecies distinctions. Formal recognition of biologically relevant variation within species based on the accessory genome information will have practical significance in fields such as clinical, industrial and agricultural microbiology.
Keywords: Accessory genome, core genome, genetic isolation, Prokaryotic taxonomy, species, subspecies, varieties
Introduction
The use of subspecies as taxonomic category is not limited to prokaryotes. In ornithology, for example, it is often applied to geographically or morphologically distinct populations [1,2]. But as with prokaryotes, the nature of species is widely debated and sometimes controversial [2,3]. To resolve the issue in herpetological taxonomy, De Queiroz [3] defined subspecies as incompletely separated lineages where some inter-lineage gene exchange still exists, which is an extension of his general lineage concept of species as portions of “separately evolving metapopulation lineages” [4]. According to De Queiroz's definition, subspecies are units that are not completely isolated in terms of reproduction and overall genetics, a notion that is fully compatible with the semipermeable nature of species boundaries in eukaryotes [5,6]. De Queiroz therefore stressed that in evolutionary terms, subspecies should not be viewed as something less than a species as they have all the hallmarks of species units.
Due to the wide prevalence of horizontal gene transfer (HGT), prokaryotic species boundaries are often many orders of magnitude more permeable than those of most eukaryotes [[7], [8], [9], [10]]. In fact, a large proportion of the genes encoded by a prokaryotic species may be dispensable, the number and identity of which can vary substantially among strains and populations of a species [11]. The flexible, HGT-prone fraction of species' genomes are referred to as the accessory genome or subgenomic compartment, while the stable fraction is denoted as the core genome [12,13]. It is this latter, well-conserved, subgenomic compartment that carries and encodes the properties used by taxonomists to delineate species [[14], [15], [16]]. However, no attempts have been made to reconcile prevailing interpretations of the subspecies category with currently accepted species definitions, especially in light of recent developments in taxonomic practice and the theory underpinning it.
In this article, we consider the nature and value of subspecies as taxonomic category for prokaryotes by making use of contemporary genome-based evidence. To do this, we first describe how species and subspecies are currently defined, and then discuss how genomics data have impacted our view of these taxonomic units. We then present an argument for discontinuing the use of subspecies as a defined taxonomic unit, after which we discuss the value of recognizing varieties to denote intraspecies variation as a viable and functional alternative. We conclude by highlighting how the description of varieties would align with current taxonomic practice and how it would complement and enrich existing taxonomic frameworks, thereby enhancing their value to end-users, while at the same time reflecting their nature more realistically.
What are species and subspecies?
The naming of species and subspecies is governed by the International Code of Nomenclature of Prokaryotes (ICNP). Names of these taxa are recognized as validly published when they meet all the requirements of ICNP Rules 30 to 32 [17]. Based on the original version of the Bacteriological Code [18], phenotypic variants within species could be described as either varieties or subspecies. Subspecies were recommended if the variation was sufficiently distinct and stable. This nomenclature has since changed as Rule 5c of the ICNP clearly states that a variety should be treated as a synonym of subspecies. However, species and subspecies may be delineated using a diverse set of approaches as the ICNP does not oversee the process nor restricts “the freedom of taxonomic thought or action” [17].
Most microbiologists regard an archaeal or bacterial species as a genotypically and phenotypically coherent cluster of isolates with a binomial name that enables unambiguous communication and sharing of information related to the taxon. Accordingly, all of the widely used criteria or species definitions employed for recognizing species revolves around the idea that members of a species are monophyletic and cluster together based on phenotypic and genomic similarities [19]. Of these, the level of phenotypic coherence is often most difficult to determine, but usually includes shared physiological and ecological properties. Phylogenetic coherence is typically determined using sequence information for multiple genes, and most contemporary analyses of genomic coherence are based on the proportion and similarity of shared genes [20,21]. In practice, however, taxonomic decisions are based primarily on genomic coherence where a quantitative threshold or cut-off value of 95% Average Nucleotide Identity (ANI) is used to define species [22]. The latter largely corresponds to 70% DNA/DNA hybridization (DDH), estimated experimentally or using digital DDH (dDDH) [23,24].
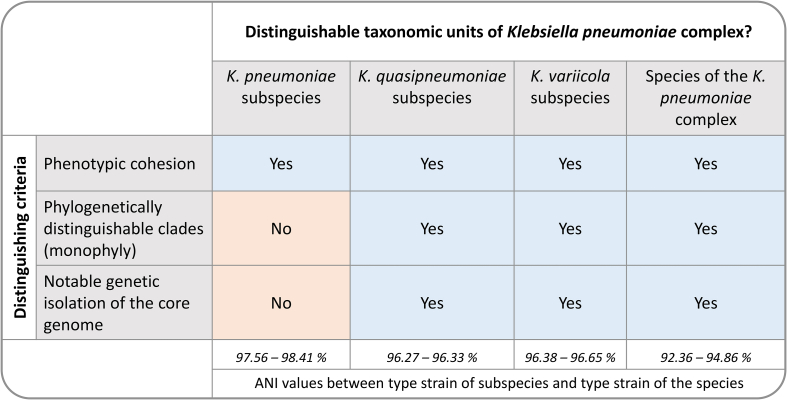
Subspecies are identified by a trinomial name, with Lactobacillus salivarius being one of the earliest species for which varieties or subspecies were formally used to differentiate metabolically distinct strains [25]. Since then numerous traits have been used to recognize subspecies (see Box 1) ranging from clinical, pathological and physiological properties through to phylogenetic monophyly [26]. The latter was introduced in 1987 by an ad hoc committee of the then International Committee on Systematic Bacteriology [24], which subsequently led to the development of numerous multi-locus sequence typing and analysis (MLST and MLSA) schemes for delineating subspecies [[27], [28], [29], [30], [31]]. As a result, phenotype combined with monophyly has become the most important characteristic used for the delineation of subspecies [32]. In July 2022, there were 871 validly named subspecies, of which 454 were considered as “correctly named” based on the most recent taxonomic opinion [33]. An excellent example illustrating the diversity of types of groups being recognized as subspecies is provided by the Klebsiella pneumoniae complex (Box 1).
Box 1.
Subspecies in the Klebsiella pneumoniae complex
The Klebsiella pneumoniae complex is a common cause of nosocomial infections and antimicrobial resistant strains are considered to be a critical health threat [66]. The complex includes a range of populations associated with particular hosts and environmental niches [67]. Through the years, authors have treated its taxonomy differently and the complex thus provides an excellent example of how the delineation of varieties and subspecies changed over time. Based on early work from the 19th century, three subspecies of K. pneumoniae sensu stricto were formally described in the first edition of Bergey's Manual of Systematic Bacteriology [68]. The main methods of differentiation among these taxa were a set of phenotypic characteristics and their clinical manifestations. Subsequent phylogenetic and phylogenomic studies failed to provide sufficient evidence for distinguishing among K. pneumoniae subsp. pneumoniae, K. pneumoniae subsp. ozaenae and K. pneumoniae subsp. rhinoscleromatis [69] and the use of these subspecies has mainly fallen into disuse. By contrast, subspecies of two other species in the complex have been delineated using robust phylogenetic inferences (i.e., K. quasipneumoniae and K. variicola). Despite the limited phenotypic differences among the respective subspecies, they can easily be differentiated based on genetic isolation and limited gene flow of genes associated with the core genome (stable sequence differences) [66,70] and the accessory genomes [71].
Genomics have impacted our understanding of the basic taxonomic unit
The increased availability of whole genome sequences has contributed enormously to the systematics of prokaryotes. It allowed for the development of tools and procedures to streamline species recognition, which is evidenced by the range of overall genome related indexes (OGRI) that have been developed and implemented by taxonomists [15,21,34]. Also, the availability of genome data for species and even populations of species have facilitated an improved understanding of the nature of species in Archaea and Bacteria [7,8,21,[35], [36], [37]]. In other words, wide access to whole genome data for these taxa not only mediated improvements in taxonomic practice, but also stimulated research into the theory underlying prokaryotic species evolution.
From a practical point of view, genomics studies revealed that the distinction between species and subspecies as taxonomic units is not straightforward. For example, extensive genome comparisons showed that species boundaries likely fall in the range of 93-96 % ANI [20], which corresponds well with the previous “gold standard” of 70% DDH [38]. However, this ANI range represents a “fuzzy zone” because classification of strains into separate species depends largely on the interpretation of the taxonomist [20]. For example, certain taxonomists use 95% ANI as species threshold [22,39,40], while others designate strains with > 93% ANI as members of distinct genomovars, i.e., genome-based groups sufficiently distinct to be recognized as separate species, but lacking phenotypic differences for unambiguously differentiating them [20]. The latter could thus also be regarded as subspecies, which are comparable to species, but at lower taxonomic rank due to their strains sharing high genome-based similarity [41]. This would also be in line with proposals that dDDH values between 70% and 80% are used as a quantitative approach for the delineation of subspecies [41]. Indeed, strains belonging to a well-defined phylogenetic lineage, but with ANI/DDH/dDDH values exceeding the suggested species thresholds, are now often described as subspecies [[42], [43], [44]]. In some cases, species have even been lowered in rank to subspecies following the implementation of OGRI measurements [45,46].
A theoretical basis for distinguishing between species and subspecies of prokaryotes has also remained elusive. Current models hold that prokaryotic diversity represent speciation spectra [9] on which gene flow barriers, together with drift and natural selection, lead to the formation and maintenance of discrete and cohesive units [47]. As such, each of these evolutionary units is unique and sui generis (“of its own kind”) in nature [7,8]. In other words, they are produced by distinct evolutionary processes and may differ markedly in terms of intra-unit sequence similarity, population size and evolutionary age, as well as ecological and phenotypic characteristics [[7], [8], [9]]. Whether these units are recognized and described as species or as subspecies depends entirely on the taxonomist's view, because other than convention (e.g., associated with particular taxa or methodologies) a theoretical framework for such decisions are lacking.
The definitions of most contemporary subspecies, especially where genome-based evidence was used, almost exactly match those used for defining species [19]. They represent monophyletic units that are phenotypically and genomically coherent, and potentially only differ from bona fide species in having more recent evolutionary origins, smaller population sizes, and in spanning less phenotypic and genotypic diversity. However, we know that all of these properties are intrinsically variable among the units we recognize as species due to the unique evolutionary trajectories that gave rise to them [8]. How then can one objectively justify why strains belonging to well-defined phylogenetic clusters, with OGRI values higher than the usual species thresholds, should or should not be described as species or subspecies? Hence, we argue that subspecies as a taxonomic category has become irrelevant and of limited value to users of the taxonomic frameworks we establish.
Recognizing varieties within species has practical value
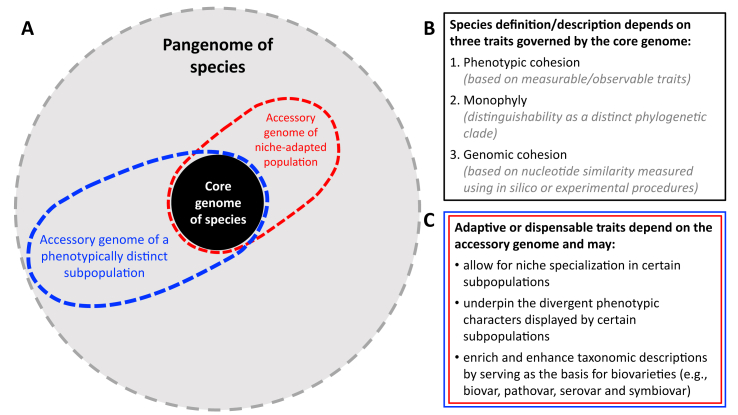
Various aspects of the ecology of prokaryotes depend on functions encoded by their accessory genome. The gene content of this subgenomic compartment largely reflects the response of strains to niche exploration, diversification and their adaptation to ecological changes [48]. This is because accessory genomes are the product of differential gene loss and conservation, together with gene gains from other genomes in the immediate environment via HGT. Although homologous recombination and neutral acquisitions of genes occur, adaptation to various selection pressures is seen as an important driver that shapes the accessory genome of a strain [49]. The accessory genome therefor provides a crucial link to observable intraspecies variation and seldomly exhibits the broad level of genetic isolation associated with species (Fig. 1).
Fig. 1.
The existence of species is governed by properties of core genomes, while varieties within species are mostly dictated by accessory genome features. A: The genomes of prokaryotic species are typically made-up of core and accessory subgenomic compartments, which together form the pangenome of the species [12,13]. While being genetically cohesive with other members of the species based on core genome sequences, individual strains/populations may have substantially diverged accessory genomes, which can cause them to be phenotypically or ecologically distinct (represented by the areas enclosed with red and blue dotted lines). Consequently, species definitions/descriptions are determined by traits governed by the core genome (B), while dispensable traits are determined by the accessory genome (C).
Practically, there is immense importance in recognizing biologically relevant variation within species based on specific operational attributes or phenotypes linked to accessory genomes. For this reason, the pangenomes of human, animals, and plants are a subject of active research [50,51]. Among human and animal pathogens, the focus has been on studying and understanding varieties and variations associated with host specificity, antigenic properties, as well as disease symptoms or clinical presentation, leading to the description of pathovar/pathotype and serovar/serotype, for example (Table 1). Similar accessory genome-derived units are also used within rhizobial species denoting symbiovars associated with particular legume hosts (Table 1). Defining subpopulations or varieties within species using these designations are only important if they convey useful and important biological information for specific user groups such as clinicians or environmental microbiologists with an interest in specific traits linked to the accessory genome. Varieties may therefore have far greater utility and practical relevance compared to many of the currently recognized subspecies, especially those that do not display any biological differences of relevance. Use of such varieties is already well embedded in the scientific literature, where the approach implemented depends on practical needs.
Table 1.
Illustrative examples of species/genera that contain varieties for denoting distinct clusters in which the diverged traits are governed by information encoded on the accessory genome
| Variety recognized | Informative trait | Species or generaa | References |
|---|---|---|---|
| Pathovar | Pathological distinguishability by causing the development of distinctive symptoms on one or more plant or animal host | Escherichia coli, Pseudomonas syringae∗, Xanthomonas hortorum∗ | [[72], [73], [74]] |
| Symbiovar | Nodulation and establishment of the nitrogen-fixing symbiosis with the same legume host, often independently of species affiliation | Bradyrhizobium, Mesorhizobium, Paraburkholderia, Rhizobium | [56,[75], [76], [77]] |
| Serovar | Serological distinguishability due to the presence of similar cell surface antigens (in certain cases, independent of species affiliations) | Leptospira, Listeria monocytogenes, Salmonella enterica | [57,78,79] |
| Morphovar | Morphological characters in culture | Mycobacterium tuberculosis complex∗ | [80] |
| Biovar | Various: | ||
|
Bacillus cereus sensu lato, Lactococcus lactis | [59,81] | |
|
Campylobacter sputorum, Corynebacterium diphtheriae | [82,83] | |
|
Corynebacterium pseudotuberculosis | [84] |
Genome-based evidence showed that determinants for the respective traits are encoded by dispensable/accessory genes. In cases where the molecular basis of traits is yet unknown (indicated with ∗), genome and phylogenetic data together indicated that the traits' genetic determinants are not encoded by the core genome, and subject to HGT, as is typical for accessory genes.
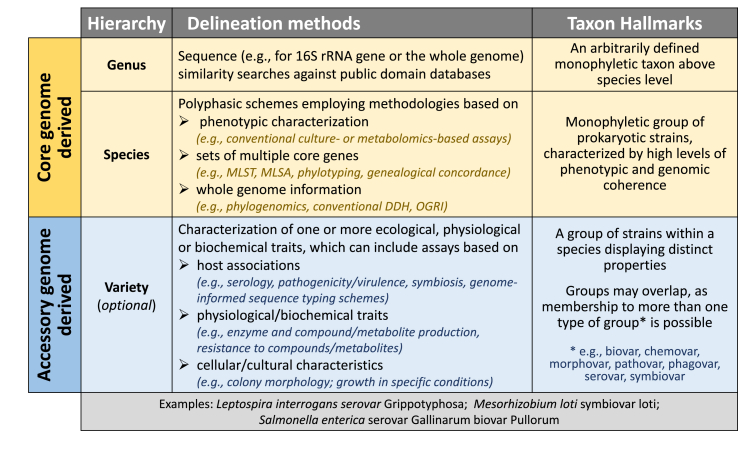
The recognition process for varieties associated with a unique set of accessory genes is flexible (Fig. 2). Also, the same strain may be grouped with a different set of strains depending on the variety of interest and the practical needs of different research fields. For example, the same pathovar could be linked to strains belonging to different serovars [52] or vice versa [53]. It does not require, as in the case of subspecies, that all strains in a species belong to one of the groups defining a variety (e.g. pathogenic vs. commensal E. coli). It also supports the use of typing schemes critical for epidemiological studies of pathogenic bacteria [54,55]. Another practical benefit is that variety-based groupings are transferable across species when their genetic determinants are subject to interspecies HGT [56,57].
Fig. 2.
The core and accessory genomes of prokaryotes inform the demarcation of taxonomic units at different levels. Genus and species units are delineated using properties inherent to the core genome, while varieties are recognized based on traits linked to the accessory genome. Because of the prevalence of HGT, three examples are indicated for cases where variety designations are transferrable across species (serovars of Leptospira and symbiovars of Mesorhizobium) [56,57], and where strains form part of multiple variety forms/types (Salmonella serovars and biovars) [78].
What are the implications of abandoning subspecies as taxonomic category and promoting the use of varieties?
Discontinuing the use of subspecies as category, combined with formal recognition of diagnosable varieties within species, will enhance prokaryote taxonomy among its users (e.g., clinicians and plant pathologists). The reason for this is two-fold: (i) it will reduce taxonomic confusion and instability related to the subjective nature of species-subspecies distinctions; and (ii) the introduction of named varieties within species would have direct practical value. With regards to abandoning the subspecies category, it is well-known that confusion is caused among the users of taxonomy when important groups of organisms are not robustly delineated and/or when their naming conventions are illogical [58,59]. Such issues could also lead to the non-use or disregard of the taxa described by prokaryote taxonomists. Assigning a group of interest to a category (i.e., subspecies) that is essentially indistinguishable from another (i.e., species) would be a good example of such a practice. In terms of the pragmatism related to assigning varieties within species, designations such as pathovar, biovar or symbiovar would complement species descriptions, making their taxonomy more user-friendly and information-rich. These varieties may include any of those listed in Appendix 10 of the ICNP [17], or any other types or forms distinguishing groups within species. Designating varieties in this way further allows clear communication that their distinguishing properties are likely encoded on the species' accessory genomes (which is not the case for “subspecies” whose existence are dictated by core genome information).
Taxonomic implications associated with abandoning the subspecies for prokaryotic species would not be insurmountable and would provide a better reflection of current knowledge of genome organisation. Many of the subspecies described using DNA-based information, especially whole genome sequences, would need to be elevated to species-level as they represent phenotypically and genomically coherent monophyletic units, albeit with more recent evolutionary origins than their contemporaries (Fig. 2). Using this proposal for reviewing the Klebsiella pneumoniae complex (Box 1) for example, will result in the abandonment of K. pneumoniae subspecies. It would only be possible to distinguish the current subspecies as biovars based on phenotypic differences. Their specific clinical manifestations are not conclusive to justify pathovar designations [60]. At the same time the K. quasipneumoniae and K. variicola subspecies would be recognised as distinct species in their own right.
Abandoning the subspecies category also aligns well with the Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org) where the demarcation of subspecies has been discontinued [61]. The GTDB's exclusion of subspecies could potentially have far-reaching significance because its classification system is utilized as the taxonomic framework for the current edition of Bergey's Manual of Systematics of Archaea and Bacteria [62]. Our approach of distinguishing between core and accessory genomes when demarcating taxa and varieties also resonates with the newly proposed SeqCode [63,64] where high quality metagenome assembled genomes (MAGs) will be acknowledged as suitable permanent types for the description of prokaryotic species. However, to ensure consistency with the ICNP and to accommodate community feedback during the SeqCode's development, this new taxonomic initiative also allows for the description of subspecies. Abandonment of the subspecies category would thus require amendments to both Codes.
Genomics provided us with new insights into the evolution and structure of microbial populations, their genomes and the species they belong to. It has also given us the ability to propose a more uniform classification system that will address the needs of end-users, especially those interested in specific traits such as pathogenicity and host specificity which are often shared through HGT. The recognition of intraspecies varieties thus has direct practical value to clinicians, pathologists and industrial microbiologists. From an ecological point of view, delineation of groups within species allows for the contextualization of intraspecies variation relative to the microbial communities within which they appear [65]. Our proposed use of core genome data to define species and the use of accessory derived traits to define intraspecies variation will bring stability to the naming of biologically relevant units and taxa, one of the main principles of the ICNP [17], while at the same time also enhancing taxonomy's value to its end-users.
Conflict of interest
None.
Author contribution
Stephanus Venter: Conceptualization, Investigation, Writing – original draft. preparation, Writing – review & editing, Project administration. Emma Steenkamp: Conceptualization, Writing – original draft. preparation, Writing – review & editing, Visualization. Marike Palmer: Conceptualization, Formal analysis, Writing – review & editing
References
- 1.Mayr E. Of what use are subspecies? The Auk. 1982;99(3):593–595. [Google Scholar]
- 2.O'Neill J.P. The subspecies concept in the 1980's. The Auk. 1982;99(3):609–612. [Google Scholar]
- 3.de Queiroz K. An updated concept of subspecies resolves a dispute about the taxonomy of incompletely separated lineages. Herpetological Review. 2020 [Google Scholar]
- 4.De Queiroz K. Species concepts and species delimitation. Systematic Biology. 2007;56(6):879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- 5.Harrison R.G., Larson E.L. Hybridization, introgression, and the nature of species boundaries. Journal of Heredity. 2014;105(S1):795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- 6.Steenkamp E.T., Wingfield M.J., McTaggart A.R., Wingfield B.D. Fungal species and their boundaries matter–Definitions, mechanisms and practical implications. Fungal Biology Reviews. 2018;32(2):104–116. [Google Scholar]
- 7.Doolittle W.F. Vol. 11. 2018. Speciation without species: a final word. Philosophy, theory, and practice in biology. [Google Scholar]
- 8.Palmer M., Venter S.N., Coetzee M.P., Steenkamp E.T. Prokaryotic species are sui generis evolutionary units. Systematic and Applied Microbiology. 2019;42(2):145–158. doi: 10.1016/j.syapm.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro B.J., Polz M.F. Microbial speciation. Cold Spring Harbor Perspectives in Biology. 2015;7(10) doi: 10.1101/cshperspect.a018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro B.J. Population genomics: microorganisms. Springer; 2018. What microbial population genomics has taught us about speciation; pp. 31–47. [Google Scholar]
- 11.McInerney J.O., McNally A., O'connell M.J. Why prokaryotes have pangenomes. Nature Microbiology. 2017;2(4):1–5. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 12.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome. Proceedings of the National Academy of Sciences. 2005;102(39):13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young J.P.W., Crossman L.C., Johnston A.W., Thomson N.R., Ghazoui Z.F., Hull K.H., et al. Genome Biology. 2006;7(4):R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schleifer K.H. Classification of Bacteria and Archaea: past, present and future. Systematic and Applied Microbiology. 2009;32(8):533–542. doi: 10.1016/j.syapm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Chun J., Oren A., Ventosa A., Christensen H., Arahal D.R., da Costa M.S., et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. International Journal of Systematic and Evolutionary Microbiology. 2018;68(1):461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 16.Chung M., Munro J.B., Tettelin H., Dunning Hotopp J.C. Using core genome alignments to assign bacterial species. MSystems. 2018;3(6):e00236. doi: 10.1128/mSystems.00236-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker C.T., Garrity G.M., Tindall B.J. International code of nomenclature of prokaryotes. International Journal of Systematic and Evolutionary Microbiology. 2019;69(1A):S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 18.Buchanan R.E., John-Brooks R.S., Breed R.S. International bacteriological code of nomenclature. Journal of Bacteriology. 1948;55(3):287–306. doi: 10.1128/jb.55.3.287-306.1948. [DOI] [PubMed] [Google Scholar]
- 19.Rossello-Mora R., Amann R. The species concept for prokaryotes. FEMS Microbiology Reviews. 2001;25(1):39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 20.Rossello-Mora R., Amann R. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol. 2015;38(4):209–216. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Hugenholtz P., Chuvochina M., Oren A., Parks D.H., Soo R.M. Prokaryotic taxonomy and nomenclature in the age of big sequence data. The ISME Journal. 2021;15(7):1879–1892. doi: 10.1038/s41396-021-00941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auch A.F., von Jan M., Klenk H.-P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Standards in Genomic Sciences. 2010;2(1):117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wayne L., Brenner D., Colwell R., Grimont P., Kandler O., Krichevsky M., et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. International Journal of Systematic and Evolutionary Microbiology. 1987;37(4):463–464. [Google Scholar]
- 25.Rogosa M., Wiseman R., Mitchell J.A., Disraely M., Beaman A. Species differentiation of oral lactobacilli from man including descriptions of Lactobacillus salivarius nov spec and Lactobacillus cellobiosus nov spec. Journal of Bacteriology. 1953;65(6):681–699. doi: 10.1128/jb.65.6.681-699.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen K.D., Christensen H., Bisgaard M., Olsen J.E. Genetic diversity of Pasteurella multocida fowl cholera isolates as demonstrated by ribotyping and 16S rRNA and partial atpD sequence comparisons the GenBank accession numbers for the 16S rRNA sequences of strain HIM 830-7T (NCTC 10204T) and 77179 of P. m. Microbiology. 2001;147(10):2739–2748. doi: 10.1099/00221287-147-10-2739. [DOI] [PubMed] [Google Scholar]
- 27.Maiden M.C. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 28.Brady C., Cleenwerck I., Venter S., Coutinho T., De Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov as Lelliottia nimipressuralis comb. nov and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov as Pluralibacter gergoviae comb. nov and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol. 2013;36(5):309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Jolley K.A., Bliss C.M., Bennett J.S., Bratcher H.B., Brehony C., Colles F.M., et al. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology. 2012;158(4):1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiden M.C., Van Rensburg Mjj, Bray J.E., Earle S.G., Ford S.A., Jolley K.A., et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nature Reviews Microbiology. 2013;11(10):728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaeser S.P., Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Systematic and Applied Microbiology. 2015;38(4):237–245. doi: 10.1016/j.syapm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Christensen H., Kuhnert P., Busse H.-J., Frederiksen W.C., Bisgaard M. Proposed minimal standards for the description of genera, species and subspecies of the Pasteurellaceae. International Journal of Systematic and Evolutionary Microbiology. 2007;57(1):166–178. doi: 10.1099/ijs.0.64838-0. [DOI] [PubMed] [Google Scholar]
- 33.Parte A.C., Carbasse J.S., Meier-Kolthoff J.P., Reimer L.C., Göker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. International Journal of Systematic and Evolutionary Microbiology. 2020;70(11):5607. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi Sant’Anna F., Bach E., Porto R.Z., Guella F., Hayashi Sant’Anna E., Passaglia L.M. Genomic metrics made easy: what to do and where to go in the new era of bacterial taxonomy. Critical Reviews in Microbiology. 2019;45(2):182–200. doi: 10.1080/1040841X.2019.1569587. [DOI] [PubMed] [Google Scholar]
- 35.VanInsberghe D., Arevalo P., Chien D., Polz M.F. How can microbial population genomics inform community ecology? Philosophical Transactions of the Royal Society B. 2020;375(1798) doi: 10.1098/rstb.2019.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achtman M., Wagner M. Microbial diversity and the genetic nature of microbial species. Nature Reviews Microbiology. 2008;6(6):431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 37.Bobay L.-M., Ochman H. Factors driving effective population size and pan-genome evolution in bacteria. BMC Evolutionary Biology. 2018;18(1):1–12. doi: 10.1186/s12862-018-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goris J., Konstantinidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 39.Chan J.Z., Halachev M.R., Loman N.J., Constantinidou C., Pallen M.J. Defining bacterial species in the genomic era: insights from the genus Acinetobacter. BMC Microbiology. 2012;12(1):1–11. doi: 10.1186/1471-2180-12-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer M., Steenkamp E.T., Blom J., Hedlund B.P., Venter S.N. All ANIs are not created equal: implications for prokaryotic species boundaries and integration of ANIs into polyphasic taxonomy. International Journal of Systematic and Evolutionary Microbiology. 2020;70(4):2937–2948. doi: 10.1099/ijsem.0.004124. [DOI] [PubMed] [Google Scholar]
- 41.Meier-Kolthoff J.P., Hahnke R.L., Petersen J., Scheuner C., Michael V., Fiebig A., et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Standards in Genomic Sciences. 2014;9(1):2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brady C., Hunter G., Kirk S., Arnold D., Denman S. Description of Brenneria roseae sp. nov. and two subspecies, Brenneria roseae subspecies roseae ssp. nov. and Brenneria roseae subspecies americana ssp. nov. isolated from symptomatic oak. Systematic and Applied Microbiology. 2014;37(6):396–401. doi: 10.1016/j.syapm.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Jin H., Wang H., Zhang Y., Hu T., Lin Z., Liu B., et al. Description of Azotobacter chroococcum subsp. isscasi subsp. nov. isolated from paddy soil and establishment of Azotobacter chroococcum subsp. chroococcum subsp. nov. International Journal of Systematic and Evolutionary Microbiology. 2020;70(3):2124–2131. doi: 10.1099/ijsem.0.004026. [DOI] [PubMed] [Google Scholar]
- 44.Garcia C., Mesnil A., Tourbiez D., Moussa M., Dubreuil C., Gonçalves De Sa A., et al. Vibrio aestuarianus subsp. cardii subsp. nov., pathogenic to the edible cockles Cerastoderma edule in France, and establishment of Vibrio aestuarianus subsp. aestuarianus subsp. nov. and Vibrio aestuarianus subsp. francensis subsp. nov. International Journal of Systematic and Evolutionary Microbiology. 2021;71(2) doi: 10.1099/ijsem.0.004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.García-López M., Meier-Kolthoff J.P., Tindall B.J., Gronow S., Woyke T., Kyrpides N.C., et al. Analysis of 1,000 type-strain genomes improves taxonomic classification of Bacteroidetes. Frontiers in Microbiology. 2019;10:2083. doi: 10.3389/fmicb.2019.02083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behrendt U., Wende S., Kolb S., Ulrich A. Genome-based phylogeny of the genera Proteus and Cosenzaea and description of Proteus terrae subsp. terrae subsp. nov. and Proteus terrae subsp. cibarius subsp. nov. International Journal of Systematic and Evolutionary Microbiology. 2021;71(3) doi: 10.1099/ijsem.0.004651. [DOI] [PubMed] [Google Scholar]
- 47.Bobay L.-M. The prokaryotic species concept and challenges. The Pangenome. 2020:21–49. [PubMed] [Google Scholar]
- 48.Azarian T., Huang I.-T., Hanage W.P. Structure and dynamics of bacterial populations: pangenome ecology. The Pangenome. 2020:115–128. [PubMed] [Google Scholar]
- 49.Brockhurst M.A., Harrison E., Hall J.P.J., Richards T., McNally A., Maclean C. The ecology and evolution of pangenomes. Current Biology. 2019;29(20):R1094–R1103. doi: 10.1016/j.cub.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Arnold D.L., Jackson R.W. Bacterial genomes: evolution of pathogenicity. Current Opinion in Plant Biology. 2011;14(4):385–391. doi: 10.1016/j.pbi.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Kim Y., Gu C., Kim H.U., Lee S.Y. Current status of pan-genome analysis for pathogenic bacteria. Current Opinion in Biotechnology. 2020;63:54–62. doi: 10.1016/j.copbio.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Rasko D.A., Rosovitz M., Myers G.S., Mongodin E.F., Fricke W.F., Gajer P., et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. Journal of Bacteriology. 2008;190(20):6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanner J.R., Kingsley R.A. Evolution of Salmonella within hosts. Trends in Microbiology. 2018;26(12):986–998. doi: 10.1016/j.tim.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolley K.A., Maiden M.C. Using MLST to study bacterial variation: prospects in the genomic era. Future Microbiology. 2014;9(5):623–630. doi: 10.2217/fmb.14.24. [DOI] [PubMed] [Google Scholar]
- 55.Schürch A.C., Arredondo-Alonso S., Willems R.J.L., Goering R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clinical Microbiology and Infection. 2018;24(4):350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 56.Paulitsch F., Delamuta J.R.M., Ribeiro R.A., da Silva Batista J.S., Hungria M. Phylogeny of symbiotic genes reveals symbiovars within legume-nodulating Paraburkholderia species. Systematic and Applied Microbiology. 2020;43(6) doi: 10.1016/j.syapm.2020.126151. [DOI] [PubMed] [Google Scholar]
- 57.Guglielmini J., Bourhy P., Schiettekatte O., Zinini F., Brisse S., Picardeau M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLOS Neglected Tropical Diseases. 2019;13(4) doi: 10.1371/journal.pntd.0007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garnett S.T., Christidis L., Conix S., Costello M.J., Zachos F.E., Bánki O.S., et al. Principles for creating a single authoritative list of the world’s species. PLOS Biology. 2020;18(7) doi: 10.1371/journal.pbio.3000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carroll L.M., Wiedmann M., Kovac J. Proposal of a taxonomic nomenclature for the Bacillus cereus group which reconciles genomic definitions of bacterial species with clinical and industrial phenotypes. MBio. 2020;11(1):e00034. doi: 10.1128/mBio.00034-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldstein E.J., Lewis R.P., Martin W.J., Edelstein P.H. Infections caused by Klebsiella ozaenae: a changing disease spectrum. Journal of Clinical Microbiology. 1978;8(4):413–418. doi: 10.1128/jcm.8.4.413-418.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks D.H., Chuvochina M., Rinke C., Mussig A.J., Chaumeil P.-A., Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Research. 2021 doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosselló-Móra R., Stackebrandt E. 2021. 1 bridging 200 Years of bacterial classification. [Google Scholar]
- 63.Murray A.E., Freudenstein J., Gribaldo S., Hatzenpichler R., Hugenholtz P., Kämpfer P., et al. Roadmap for naming uncultivated Archaea and bacteria. Nature Microbiology. 2020;5(8):987–994. doi: 10.1038/s41564-020-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitman W.B., Chuvochina M., Hedlund B.P., Hugenholtz P., Konstantinidis K.T., Murray A., et al. Development of the SeqCode: a proposed nomenclatural code for uncultivated prokaryotes with DNA sequences as type. Systematic and Applied Microbiology. 2022:126305. doi: 10.1016/j.syapm.2022.126305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Rossum T., Ferretti P., Maistrenko O.M., Bork P. Diversity within species: interpreting strains in microbiomes. Nature Reviews Microbiology. 2020;18(9):491–506. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyres K.L., Lam M.M., Holt K.E. Population genomics of Klebsiella pneumoniae. Nature Reviews Microbiology. 2020;18(6):344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 67.Bagley S.T. Habitat association of Klebsiella species. Infection Control & Hospital Epidemiology. 1985;6(2):52–58. doi: 10.1017/s0195941700062603. [DOI] [PubMed] [Google Scholar]
- 68.Orskov I. Genus V. Klebsiella trevisan 1885, 105AL. Bergey's Manual of Systematic Bacteriology. 1984;1:461–464. [Google Scholar]
- 69.Brisse S., Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. International Journal of Systematic and Evolutionary Microbiology. 2001;51(3):915–924. doi: 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 70.Rodrigues C., Passet V., Rakotondrasoa A., Diallo T.A., Criscuolo A., Brisse S. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Research in Microbiology. 2019;170(3):165–170. doi: 10.1016/j.resmic.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Long S.W., Linson S.E., Ojeda Saavedra M., Cantu C., Davis J.J., Brettin T., et al. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. Msphere. 2017;2(4):e00290. doi: 10.1128/mSphereDirect.00290-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Denamur E., Clermont O., Bonacorsi S., Gordon D. The population genetics of pathogenic Escherichia coli. Nature Reviews Microbiology. 2021;19(1):37–54. doi: 10.1038/s41579-020-0416-x. [DOI] [PubMed] [Google Scholar]
- 73.Gomila M., Busquets A., Mulet M., García-Valdés E., Lalucat J. Clarification of taxonomic status within the Pseudomonas syringae species group based on a phylogenomic analysis. Frontiers in Microbiology. 2017;8:2422. doi: 10.3389/fmicb.2017.02422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morinière L., Burlet A., Rosenthal E.R., Nesme X., Portier P., Bull C.T., et al. Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynarae Trébaol et al. 2000 emend. Timilsina et al. 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al. 1995. Systematic and Applied Microbiology. 2020;43(4):126087. doi: 10.1016/j.syapm.2020.126087. [DOI] [PubMed] [Google Scholar]
- 75.Laranjo M., Alexandre A., Oliveira S. Legume growth-promoting rhizobia: an overview on the Mesorhizobium genus. Microbiological Research. 2014;169(1):2–17. doi: 10.1016/j.micres.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 76.Ramírez-Bahena M.H., Flores-Félix J.D., Velázquez E., Peix Á. The Mimosoid tree Leucaena leucocephala can be nodulated by the symbiovar genistearum of Bradyrhizobium canariense. Systematic and Applied Microbiology. 2020;43(1) doi: 10.1016/j.syapm.2019.126041. [DOI] [PubMed] [Google Scholar]
- 77.Young J.P.W., Moeskjær S., Afonin A., Rahi P., Maluk M., James E.K., et al. Defining the Rhizobium leguminosarum species complex. Genes. 2021;12(1):111. doi: 10.3390/genes12010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Banerji S., Simon S., Tille A., Fruth A., Flieger A. Genome-based Salmonella serotyping as the new gold standard. Scientific Reports. 2020;10(1) doi: 10.1038/s41598-020-61254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henri C., Leekitcharoenphon P., Carleton H.A., Radomski N., Kaas R.S., Mariet J.-F., et al. An assessment of different genomic approaches for inferring phylogeny of Listeria monocytogenes. Frontiers in Microbiology. 2017;8:2351. doi: 10.3389/fmicb.2017.02351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riojas M.A., McGough K.J., Rider-Riojas C.J., Rastogi N., Hazbón M.H. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis. International Journal of Systematic and Evolutionary Microbiology. 2018;68(1):324–332. doi: 10.1099/ijsem.0.002507. [DOI] [PubMed] [Google Scholar]
- 81.Wels M., Siezen R., Van Hijum S., Kelly W.J., Bachmann H. Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Frontiers in Microbiology. 2019;10:4. doi: 10.3389/fmicb.2019.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hennart M., Panunzi L.G., Rodrigues C., Gaday Q., Baines S.L., Barros-Pinkelnig M., et al. Population genomics and antimicrobial resistance in Corynebacterium diphtheriae. Genome Medicine. 2020;12(1):1–18. doi: 10.1186/s13073-020-00805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miller W.G., Yee E., Chapman M.H., Bono J.L. Comparative genomics of all three Campylobacter sputorum biovars and a novel cattle-associated C. sputorum clade. Genome Biology and Evolution. 2017;9(6):1513–1518. doi: 10.1093/gbe/evx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soares S.C., Silva A., Trost E., Blom J., Ramos R., Carneiro A., et al. The pan-genome of the animal pathogen Corynebacterium pseudotuberculosis reveals differences in genome plasticity between the biovar ovis and equi strains. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0053818. [DOI] [PMC free article] [PubMed] [Google Scholar]