Abstract
ςB, the general stress response sigma factor of Bacillus subtilis, is activated when the cell's energy levels decline or the bacterium is exposed to environmental stress (e.g., heat shock, ethanol). Physical stress activates ςB through a collection of regulatory kinases and phosphatases (the Rsb proteins) which catalyze the release of ςB from an anti-ςB factor inhibitor. The means by which diverse stresses communicate with the Rsb proteins is unknown; however, a role for the ribosome in this process was suggested when several of the upstream members of the ςB stress activation cascade (RsbR, -S, and -T) were found to cofractionate with ribosomes in crude B. subtilis extracts. We now present evidence for the involvement of a ribosome-mediated process in the stress activation of ςB. B. subtilis strains resistant to the antibiotic thiostrepton, due to the loss of ribosomal protein L11 (RplK), were found to be blocked in the stress activation of ςB. Neither the energy-responsive activation of ςB nor stress-dependent chaperone gene induction (a ςB-independent stress response) was inhibited by the loss of L11. The Rsb proteins required for stress activation of ςB are shown to be active in the RplK− strain but fail to be triggered by stress. The data demonstrate that the B. subtilis ribosomes provide an essential input for the stress activation of ςB and suggest that the ribosomes may themselves be the sensors for stress in this system.
The ςB transcription factor controls the general stress regulon of Bacillus subtilis, a collection of at least 22 operons whose products confer multiple stress resistances on the bacterium (13, 19, 31, 34). Induction of this regulon occurs by the activation of ςB itself, a process that is triggered by exposure to an environmental insult (e.g., heat, salt, acid, or ethanol) or a drop in energy charge (e.g., entry into stationary phase, glucose limitation, or azide treatment) (7, 13, 15, 31, 35, 36). ςB is present in unstressed B. subtilis but is inactive due to an association with the anti-ςB protein RsbW (regulator of sigma B-W) (5, 10). A model of how ςB and its regulators are likely to interact is illustrated in Fig. 1. ςB is released from RsbW when an additional protein, RsbV, binds to RsbW in lieu of ςB (10). In the absence of stress, RsbV is unable to bind to RsbW due to an RsbW-dependent phosphorylation (2, 10, 35). The abundance of active RsbV determines the level of free ςB (35). Exposure to physical stress or a drop in energy charge induces stress- or energy-dependent phosphatases to dephosphorylate and reactivate RsbV-P (30, 35). The mechanism by which the energy-dependent phosphatase (RsbP) is activated is unknown; however, a number of the components which control the stress-induced phosphatase (RsbU) have been identified. The best-characterized members of the stress activation pathway are the products of five genes (rsbR, -S, -T, -U, and -X) that are cotranscribed with the ςB structural gene (sigB) and its two principal regulators (rsbV and -W) (1, 7, 11, 14, 15, 32, 39). As can be seen in Fig. 1, RsbT is the pivotal component of the stress activation pathway. When B. subtilis is exposed to stress, RsbT, previously held inactive by its negative regulator RsbS, is triggered to inactivate RsbS by phosphorylation and then activate RsbU, the stress pathway's RsbV-P phosphatase (40). The exact role of RsbR is unclear, but it is thought to mediate RsbT-RsbS interactions (1, 11). RsbX limits the stress induction process by dephosphorylating RsbS-P and reestablishing the RsbS-dependent inhibition of RsbT (33, 40). An additional component of the stress pathway is Obg, an essential GTP binding protein that is required for stress to activate ςB (22). It is unknown whether stress communicates directly with RsbT through Obg or whether an Obg-dependent process functions as a cofactor for stress to activate ςB.
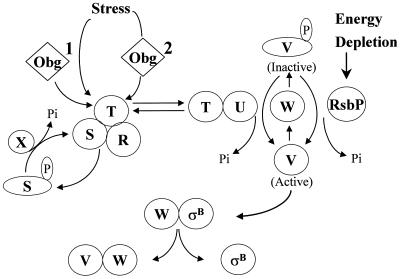
FIG. 1.
Activation of ςB. ςB is held inactive in unstressed B. subtilis as a complex with an anti-ςB protein, RsbW (W). ςB is freed from RsbW when a release factor, RsbV (V), binds to RsbW. In unstressed B. subtilis, RsbV is inactive due to an RsbW-catalyzed phosphorylation (V-P). Environmental stress activates an RsbV-P phosphatase, RsbU (U), which reactivates RsbV. RsbT (T) is the RsbU activator. RsbT is normally bound to a negative regulator, RsbS (S), which inhibits its activity. RsbR (R) also binds to RsbS and -T and is believed to facilitate their interactions. Upon exposure to stress, RsbT phosphorylates and inactivates RsbS and then activates the RsbU phosphatase. Obg, an essential GTPase that can also bind to RsbT, is required for stress to trigger the activation of RsbT. It is unknown whether an Obg-dependent process serves as a coinductant for stress to activate RsbT (process 1) or as the vehicle through which stress directly communicates to RsbT (process 2). RsbS-P is dephosphorylated and reactivated by a phosphatase, RsbX (X), that is encoded by one of the genes downstream of the sigB operon's ςB-dependent promoter. RsbX levels become elevated when ςB is active, which may facilitate a return of RsbT to an inactive complex with RsbS. Energy depletion activates a separate pathway in which a novel RsbV-P phosphatase (RsbP) is triggered, by unknown means, to reactivate RsbV. This model is based on references 1, 3, 5, 7, 10, 15, 22, 30, 32, and 40.
A key unanswered question is how diverse physical stresses are sensed and communicated to RsbT. In other bacterial systems, protein denaturation and chaperone activation play important roles in sensing and communicating stress to responsive transcription factors (reviewed in reference 42); however, no correlation has been found between chaperone activity and B. subtilis ςB induction (17, 23). In addition, the known Rsb proteins are insufficient to detect stress and activate ςB when they are expressed with ςB in Escherichia coli (23). Thus, a bacillus-specific process is needed to communicate stress to the Rsb cascade.
A clue to Bacillus stress signaling was obtained when both Obg and a portion of the cell's RsbR, -S, and -T were found to cofractionate with B. subtilis ribosomes during gel filtration chromatography (24). Obg was subsequently found to bind specifically to a protein (L13) from the 50S ribosomal subunit in an affinity blot assay (24). These results suggested that a ribosome-mediated process might be involved in the stress activation of ςB. To explore this notion, we examined the effects of known ribosome mutations on the stress activation of ςB and discovered that a thiostrepton-resistant mutant of B. subtilis is unable to activate ςB following exposure to environmental stress. The energy-dependent activation of ςB continues to occur in the mutant strain, as does stress-triggered chaperone gene induction, a ςB-independent process (17, 18). The results argue that the B. subtilis ribosomes are part of the apparatus that communicates environmental stress to the ςB regulon.
MATERIALS AND METHODS
Bacterial strains.
All of the strains and plasmids used in this study are listed in Table 1. The BSA and BSZ strains are derivatives of PY22. BSA46 and BSA419 carry a specialized SPβ prophage encoding a translational fusion of the ςB-dependent gene ctc to the E. coli lacZ gene (SPβ ctc::lacZ). This fusion allows β-galactosidase activity to be monitored as a measure of ςB activity (22). BSA419 and BSZ10 have an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, PSPAC, placed upstream of rsbT in the sigB operon. Induction of PSPAC at this location artificially activates the ςB stress pathway by upregulating the expression of RsbT (27). IS169 is a B. subtilis 168 strain carrying a mutation (tsp-6) that confers resistance to thiostrepton (26). The allele, originally isolated as a spontaneous bryamycin resistance mutation and called bry-2 (12), was renamed tsp-6 for uniformity of nomenclature (26). B. subtilis strains carrying the tsp-6 allele are missing ribosomal protein L11 (38). We PCR amplified and sequenced the L11-encoding gene (rplK) of a tsp-6 strain. The amplified rplK allele has a frameshift mutation at codon 57 of the 141-codon gene (S.Z., unpublished data). With this new information, we redesignated the allele rplK57. BSZ5, an RelA− strain, is BSA46 transformed to Spcr with a plasmid (pUS-RE1) that is incapable of autonomous replication in B. subtilis but carries an internal fragment (nucleotides 3 to 542 of the 2,202-nucleotide relA gene) to target its Campbell-like integration into relA. The internal relA fragment was generated by PCR and cloned into pUS-19 (4). BSZ9 is BSA46 transformed to thiostrepton resistance (1 μg/ml) using chromosomal DNA from IS169.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Construction or source |
|---|---|---|
| Bacillus strains | ||
| PY22 | trpC2 | Laboratory strain |
| BSA46 | trpC2 SPβ ctc::lacZ | 3 |
| BSA419 | trpC2 SPβ ctc::lacZ PSPAC::rsbT | 22 |
| IS1 | trpC2 thr-5 | 27 |
| IS169 | trpC2 thr-5 tsp-6 (rplK57) | 27 |
| BSZ5 | trpC2 SPβ ctc::lacZ relA::Spcr | pUS-RE1→BSA46 |
| BSZ9 | trpC2 SPβ ctc::lacZ rplK57 | IS169→BSA46 |
| BSZ10 | trpC2 SPβ ctc::lacZ PSPAC::rsbT rplK57 | BSZ9→BSA419 |
| Plasmids | ||
| pUS-19 | Apr Spcr | 4 |
| pUS-RE1 | Apr SpcrrelA | This study |
Culture conditions and stress induction.
Strains were grown in LB (21) and stressed during exponential growth by exposure to ethanol or sodium azide at a final concentration of 4% or 2 mM, respectively. Cultures to be pulse-labeled during heat shock were grown in Difco Methionine Assay Medium. Following growth to an A540 of approximately 0.3, a portion of the culture was pulse-labeled with EXPRE35S35S [35S] (New England Nuclear/Life Science Products, Boston, Mass.) protein labeling mix (0.5 μCi/ml, 1,175 Ci/mmol) for 5 min. A second portion was transferred to 48°C and similarly pulse-labeled at different times after transfer. Samples were lysed, fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by fluorography as previously described (6).
General methods.
SDS-PAGE, Western blot analyses, β-galactosidase assays, and DNA sequencing were performed as previously described (22, 24). Isoelectric focusing (IEF) was performed in the horizontal Multiphor II electrophoresis system (LKB) using 5% acrylamide gels containing 8 M urea and a 1:1 mixture of ampholytes with pH ranges of 2.5 to 5 and 4 to 6.5 (Pharmacia) at a final ampholyte concentration of 3%. The gel was prerun for 10 min at 10 W, 5-μl samples were loaded, and electrophoresis was conducted at 25 and 35 W for 60 and 30 min, respectively, at a temperature of 15°C. The proteins were transferred by capillary action onto a nitrocellulose membrane and probed with antibodies as previously described (35). B. subtilis transformation was carried out as described by Yasbin et al. (41).
RESULTS
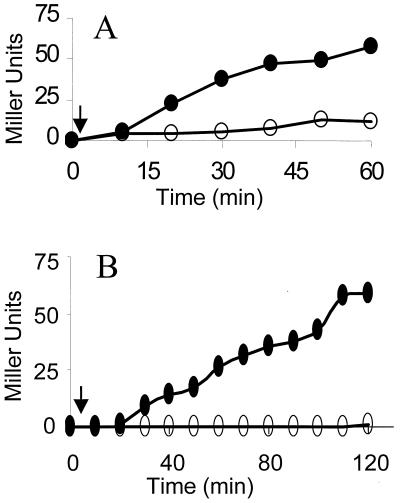
The mechanism by which B. subtilis “senses” environmental stress and channels this to the ςB regulatory cascade is unknown. It is therefore intriguing that the three principal upstream components (RsbR, -S, and -T) of this cascade, as well as a GTP binding protein (Obg) essential for stress signaling to the cascade, cofractionate with ribosomes during Sephacryl chromatography of crude B. subtilis extracts (24). This observation suggested that the ribosome might be involved in the stress activation of ςB. We sought to investigate this possibility by examining B. subtilis strains with characterized ribosome mutations for defects in ςB induction. We focused on a particularly interesting class of mutations which confer resistance to the antibiotic thiostrepton. Thiostrepton resistance mutations map to the rplK gene, which encodes ribosomal protein L11 (25, 26, 38). L11 is located within a region of the ribosome that includes its GTPase center (20). Null mutations in rplK are not lethal but reduce the bacterium's growth rate threefold. In addition, RplK− strains lack the stringent response to amino acid starvation; i.e., they are unable to undertake ribosome-mediated synthesis of ppGpp and thus cannot communicate translational arrest to the bacterium's transcriptional machinery (26). Although ςB activation is not triggered by amino acid starvation, it seemed possible that regions of the ribosome which normally interact with GTPases might also influence the Obg GTPase and, through that, the activity of ςB. This prompted us to test L11's potential involvement in ςB activation. A B. subtilis rplK null allele (rplK57) was transformed into a laboratory strain that carries a ςB-dependent reporter gene system (i.e., ctc::lacZ). The resulting RplK− strain and its wild-type parent were grown in LB and exposed to ethanol stress. ςB activity quickly increased in the wild-type strain but failed to be induced in the RplK− strain (Fig. 2A). It therefore appears that loss of L11 not only eliminates the stringent response but also blocks the stress activation of ςB.
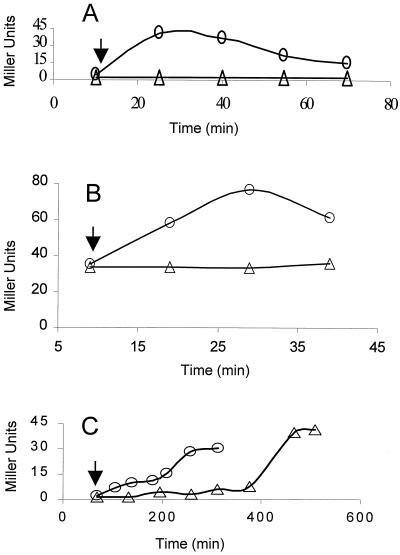
FIG. 2.
Activation of ςB by ethanol stress or sodium azide. B. subtilis strains growing in LB were treated with either 4% ethanol or 2 mM sodium azide. Culture samples were taken at the indicated times and analyzed for ςB-dependent β-galactosidase activity. The arrows indicate the times at which either ethanol or azide was added to the cultures. (A) BSA46 (○) wild-type and BSZ9 (▵) RplK− strains exposed to ethanol at an A540 of approximately 0.2. (B) Strain BSZ5 (RelA−) treated with ethanol at an A540 of 0.1 (○) or untreated (▵). (C) BSA46 (○) wild-type and BSZ9 (▵) RplK− strains treated with azide at an A540 of 0.35.
In order to determine if the effect of the rplK mutation on ςB activation was mediated by its block on the stringent response and RelA-dependent ppGpp synthesis, we repeated the stress induction experiment with a B. subtilis strain with a disruption of the relA gene. This was accomplished by integrating a plasmid within the relA coding sequence of our reporter gene-containing strain (see Materials and Methods). Disruption of B. subtilis relA generates a (p)ppGpp0 phenotype and a strain that is incapable of inducing the stringent response (37). When the RelA− strain was exposed to ethanol, it still displayed stress induction of ςB (Fig. 2B). Thus, the block in stress activation of ςB in the RplK− mutant is not due to the failure of this mutant to activate RelA.
ςB activity can be enhanced by either a stress-dependent or an energy-responsive pathway. The energy-responsive pathway requires neither Obg nor the Rsb components of the stress pathway cascade (Fig. 1). Instead, it employs an alternative RsbV-P phosphatase (RsbP) to reactive RsbV and activate ςB (30). To test whether the block in ςB activation caused by the loss of RplK was limited to the stress pathway, ATP levels were lowered in wild-type and mutant B. subtilis by exposing cultures to Na azide (22). Unlike ethanol treatment, the azide treatment led to enhanced ςB activity in both the wild-type and mutant strains (Fig. 2C). The azide-induced activation of ςB did, however, take twice as long to develop in the RplK− strain as it did in the wild-type parent. Although this could indicate an involvement of RplK in the energy-responsive pathway, it is more likely that it reflects the diminished growth rate of the mutant strain. The data argue that the loss of ribosome protein L11 blocks the stress-induced but not the energy-responsive pathway for ςB activation and that the L11-dependent process in ςB activation does not rely on the RelA ppGpp synthetase.
RplK loss does not block ςB-independent heat shock responses.
Although the ςB regulon is induced by heat shock, the classic heat shock genes (i.e., groEL and dnaK) of B. subtilis are not under ςB control (13, 18, 42). Instead, they are expressed from promoters recognized by RNA polymerase carrying the B. subtilis housekeeping ς factor ςA and controlled by the HrcA repressor protein (13, 17). If the effects of the rplK mutation are directed toward the stress activation of ςB and not the ability of B. subtilis to respond to stress in general, we would expect heat shock induction of chaperone gene expression to persist in the rplK mutant strain. This proved to be true. When wild-type and RplK− B. subtilis strains were heat shocked and pulse-labeled with [35S]methionine, proteins the size of characteristic heat shock proteins Lon, DnaK, and GroEL rapidly accumulated in both the wild-type and mutant strains (Fig. 3). The level of induction of the heat shock proteins was somewhat reduced in the RplK− strain, but given its threefold slower growth rate, this was not unexpected.
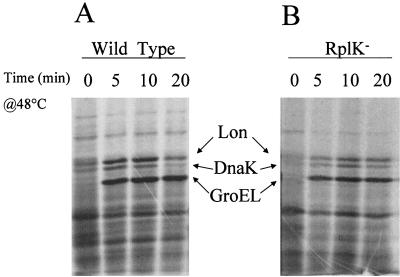
FIG. 3.
Heat shock induction of chaperone proteins in RplK+ and RplK− B. subtilis strains. B. subtilis strains BSA46 (A) and BSZ9 rplK57 (B) were grown to an A540 of 0.3 and pulse-labeled for 5 min with 35[S]Met-Cys (1 μCi/ml) either at 37°C (0) or at 5, 10, or 20 min after transfer to 48°C (33). Cell lysates were fractionated by SDS-PAGE, and labeled protein bands were visualized by fluorography. The positions to which B. subtilis proteins with the molecular weights of Lon, DnaK, and GroEL would migrate in our gel system are indicated (6).
Loss of RplK blocks RsbV-P dephosphorylation.
Our assay for ςB activation was based on the induction of reporter gene activity. Given that RplK is part of the cell's translation machinery, it is formally possible that the apparent block on stress activation of reporter gene activity in the RplK− mutant is due to an unforeseen effect of impaired translation in the stressed rplK57 strain rather than a direct effect of the loss of L11 on ςB activation. To eliminate this possibility, we examined a more upstream event in the activation process. The seminal reaction in the activation of ςB is the dephosphorylation of RsbV-P, leading to a pool of unphosphorylated RsbV that can drive the release of ςB from RsbW (Fig. 1). The activation of the stress-dependent phosphatase that is responsible for this dephosphorylation does not require new protein synthesis and still occurs when translation is blocked by chloramphenicol treatment (35). We therefore examined the effects of the RplK− mutation on the stress-dependent dephosphorylation of RsbV-P. Cultures of RplK+ and RplK− B. subtilis strains were grown, treated with chloramphenicol, and then stressed by the addition of ethanol. The chloramphenicol treatment blocked the synthesis of new RsbV, so as to ensure that any RsbV that was detected would come from the dephosphorylation of preexisting RsbV-P. Crude extracts were prepared from culture samples that had been taken before and at various times after ethanol addition. The extracts were fractionated by Multiphor II IEF and probed by Western blot assay using an anti-RsbV monoclonal antibody (35). Although this technique is only semiquantitative, it clearly showed a difference between the RplK− mutant and the parental strain. As illustrated in Fig. 4, ethanol treatment rapidly converted a portion of the wild-type B. subtilis strain's RsbV-P to RsbV but failed to trigger the appearance of an RsbV pool in the RplK− strain. Thus, the mutant strain's failure to activate ςB can be attributed to its inability to respond to ethanol stress and catalyze the dephosphorylation of RsbV-P.
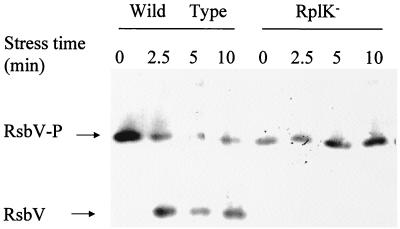
FIG. 4.
Stress-dependent dephosphorylation of RsbV-P. BSA46 (wild-type) and BSZ9 (RplK−) cultures were grown in LB and exposed to ethanol (4% final concentration) during exponential growth at an A540 of 0.4. Bacteria were harvested before (0) and at various intervals after ethanol addition (2.5 to 10 min). Crude extracts were subjected to IEF and transferred to nitrocellulose, and the membrane was probed with an anti-RsbV monoclonal antibody (35). The positions to which phosphorylated (RsbV-P) and unphosphorylated RsbV migrate in this system are indicated.
RsbT is capable of activating the ςB stress pathway in the absence of RplK.
Stress triggers a process in which RsbT frees itself from the inhibitory effects of RsbS and then activates RsbU, the RsbV-P phosphatase (Fig. 1). The RplK-dependent step might occur at any of a number of points in this process. To determine whether RplK is needed for the activation of RsbT or a downstream event, we took advantage of the observation that the induced expression of additional RsbT, in the absence of a corresponding increase in the synthesis of its RsbS inhibitor, can trigger the activation of ςB in the absence of stress (15, 24). If the RplK-dependent function is involved in stress activation of RsbT, then the induced synthesis of RsbT should activate ςB in the absence of RplK. Conversely, if RplK has a critical role in RsbT's ability to activate RsbU or RsbU's capacity to dephosphorylate RsbV-P, ςB should not become active in RplK− cells. In order to test these possibilities, we used strains in which an IPTG-inducible promoter was placed within the sigB operon, downstream of rsbS and immediately upstream of rsbT. When this promoter is activated, there is enhanced rsbT expression and a readily detectable increase in ςB activity (24). Using this inducible system, elevation of ςB activity was seen in both the wild-type and RplK− strains following IPTG addition (Fig. 5). Activation of ςB by RsbT expression in the absence of RplK reveals that RplK's essential role in the stress induction of ςB is upstream of RsbT activation of RsbU, likely in the activation of RsbT itself.
FIG. 5.
Activation of ςB by RsbT overexpression. Strains BSA419 (PSPAC::rsbT) (A) and BSZ10 (PSPAC::rsbT rplK57) (B) were grown in LB. At the times indicated by the arrows (A540, approximately 0.1), IPTG (1 mM) was added to half of each culture. Samples of the IPTG-induced (●) and control (○) cultures were analyzed for ςB-dependent β-galactosidase activity.
DISCUSSION
The mechanism by which environmental stress is detected and communicated to the regulators of the ςB transcription factor is unknown; however, there is evidence of a ribosome-mediated event in this process. The possibility of ribosome involvement was initially suggested by the cofractionation of Obg, a GTP binding protein essential for stress to activate ςB, as well as at least a portion of the upstream components of the ςB stress activation cascade, with ribosomes (24). Our current results support this idea. The inability of a B. subtilis strain lacking ribosome protein L11 to activate ςB in response to environmental stress, while still maintaining energy-dependent activation of this transcription factor, argues that the ribosome plays a role in the stress activation pathway. Furthermore, the observation that the stress pathway can still be activated in an RplK− mutant strain by the induced synthesis of RsbT supports the notion that the stage at which the ribosome acts in the pathway is upstream of RsbT. This is a point at which stress signaling would be expected to interface with the ςB activation cascade.
The idea that ribosomes could be sensitive to stress and direct changes in transcription is not unprecedented. The best-characterized example of this phenomenon is the stringent response, in which amino acid starvation triggers the ribosome to induce the synthesis of ppGpp and alter the cell's transcription pattern (reviewed in reference 8). In addition to the stringent response, there is evidence that the E. coli ribosome is a sensor for heat and cold shock networks. It has been found that partially blocking E. coli translation with ribosome-specific antibiotics elevates the synthesis of proteins associated with heat and cold shock responses (29). It is not implausible that a device with which to detect environmental stress and convey this to the ςB regulators is incorporated in the B. subtilis ribosome.
How might stress alter the ribosome so as to generate a signal that could activate ςB? Simple stress-induced arrest of translation is unlikely to be sufficient. Inhibiting translation with chloramphenicol neither activates ςB nor blocks the stress-dependent dephosphorylation of RsbV-P, the critical step in ςB activation (35). If stress activates ςB by disrupting translation, it will likely involve an event that alters the ribosome in a particular way so as to activate a specific process. One possibility is that stress physically alters the structure of the ribosome in a way that modulates the activities of the associated Rsb proteins. In E. coli, heat shock appears to be able to abort translation and dissociate 70S ribosomes (16). This leads to the formation of separated 30S and 50S subunits, with the nascent polypeptide chain still attached to the 50S particle. The 50S ribosome subunit released by heat shock appears to be in a unique state. It has been shown that a particular E. coli heat shock protein (Hsp15) can bind to the heat shock-disrupted 50S subunit but not to 50S subunits generated by in vitro dissociation of 70S ribosomes (16). A similar dissociation of B. subtilis ribosomes might lead to the structure that could trigger ςB activation. A hypothetical stress-induced change in the ribosome need not be as drastic as one which could dissociate the ribosome itself. More subtle alterations might be sufficient to alter the activities of the Rsb proteins either directly or via ribosome-associated chaperones (9, 28).
Protein denaturation plays a significant role in the activation of other stress-induced systems (42). Thus, models which envision ςB activation that is triggered by stress-induced protein misfolding to alter the structure of the ribosome, or the state of the nascent peptide associated with it, are attractive. However, the fact that loss of ribosome protein L11 blocks ςB activation makes models based solely on protein denaturation less likely. One would expect that any stress-induced misfolding of nascent peptides, or dissociation of the ribosome, would still occur in the mutant strain, and yet ςB fails to be activated.
L11 is believed to be within a part of the ribosome which modulates the activities of the GTP-dependent factors that promote translation (20). Loss of L11 not only blocks ςB activation by stress but also significantly reduces the cell's growth rate and eliminates the stringent response. Although induction of ςB by stress and triggering of the stringent response have distinct aspects (i.e., they are each induced by unique stresses, and ςB activation does not depend on the RelA ppGpp synthetase), both of these inductions are dependent on the same ribosome protein (L11) and rely on processes in which guanine nucleotides are implicated (i.e., the synthesis of ppGpp in the stringent response and the requirement for the Obg/GTPase in ςB stress activation). It is possible that particular stresses induce unique changes in the ribosome's GTPase center, thereby communicating a distinct signal to either RelA, to induce the stringent response, or Obg, to modulate the ςB activation response. In the case of ςB activation, this might be a linear pathway (2 in Fig. 1) by which stress communicates with the RsbR, -S, and -T proteins using the ribosome to induce changes in Obg. Alternatively, it may reflect converging pathways (1 in Fig. 1) in which stress-induced ribosome changes signal the Rsb proteins directly, with Obg providing an essential secondary input for activation of the cascade. Although the specific details of how the ribosome is involved in ςB activation remain highly speculative, the discovery that a ribosome function is needed for a ςB activation process promotes the idea that bacterial ribosomes may have additional, unappreciated, capacities to sense and communicate diverse forms of stress to the cell's transcriptional apparatus.
ACKNOWLEDGMENTS
We thank I. Smith for providing strains IS1 and IS169.
This work was supported by NIH grant GM-48220.
REFERENCES
- 1.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 2.Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 3.Benson A K, Haldenwang W G. Characterization of a regulatory network that controls ςB expression in Bacillus subtilis. J Bacteriol. 1992;174:749–757. doi: 10.1128/jb.174.3.749-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson A K, Haldenwang W G. Regulation of ςB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson A K, Haldenwang W G. The ςB-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993;175:1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 9.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 10.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-ς factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaidenko T A, Yang X, Lee Y M, Price C W. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the stress signaling pathway of Bacillus subtilis. J Mol Biol. 1999;288:29–39. doi: 10.1006/jmbi.1999.2665. [DOI] [PubMed] [Google Scholar]
- 12.Goldthwaite C, Dubnau D, Smith I. Genetic mapping of antibiotic resistance in markers in Bacillus subtilis. Proc Natl Acad Sci USA. 1970;65:96–103. doi: 10.1073/pnas.65.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecker M, Schumann W, Voelker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 14.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korber P, Stahl J M, Nierhaus K H, Bardwell J C A. Hsp15: a ribosome-associated heat shock protein. EMBO J. 2000;19:741–748. doi: 10.1093/emboj/19.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mogk A, Voelker A, Engelmann S, Hecker M, Schumann W, Voelker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersohn A, Bernhardt J, Gerth U, Hoper D, Koburger T, Voelker U, Hecker M. Identification of ςB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J Bacteriol. 1999;181:5718–5724. doi: 10.1128/jb.181.18.5718-5724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poise B T, Cundliffe E, Garrett R A. The antibiotic micrococcin acts on protein L11 at the ribosomal GTPase centre. J Mol Biol. 1999;287:33–45. doi: 10.1006/jmbi.1999.2600. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Scott J M, Haldenwang W G. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor ςB. J Bacteriol. 1999;181:4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott J M, Smirnova N, Haldenwang W G. A Bacillus-specific factor is needed to trigger the stress-activated phosphatase/kinase cascade of ςB induction. Biochem Biophys Res Commun. 1999;257:106–110. doi: 10.1006/bbrc.1999.0418. [DOI] [PubMed] [Google Scholar]
- 24.Scott J M, Ju J, Mitchell T, Haldenwang W G. The Bacillus subtilis GTP binding protein Obg and regulators of the ςB stress response transcription factor cofractionate with ribosomes. J Bacteriol. 2000;182:2771–2777. doi: 10.1128/jb.182.10.2771-2777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith I, Weiss D, Pestka S. A micrococcin-resistant mutant of Bacillus subtilis: localization of resistance to the 50S subunit. Mol Gen Genet. 1976;144:231–233. doi: 10.1007/BF00341720. [DOI] [PubMed] [Google Scholar]
- 26.Smith I, Paress P, Pestka S. Thiostrepton-resistant mutants exhibit relaxed synthesis of RNA. Proc Natl Acad Sci USA. 1978;75:5993–5997. doi: 10.1073/pnas.75.12.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith I, Paress P, Cabane K, Dubnau E. Genetics and physiology of the rel system of Bacillus subtilis. Mol Gen Genet. 1980;178:271–279. doi: 10.1007/BF00270472. [DOI] [PubMed] [Google Scholar]
- 28.Valent Q A, de Gier J-W L, von Heijne G, Kendall D A, ten Hagen-Jongman C M, Oudega B, Luirink J. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol Microbiol. 1997;25:53–64. doi: 10.1046/j.1365-2958.1997.4431808.x. [DOI] [PubMed] [Google Scholar]
- 29.Van Bogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PA5 domain is required to convey signals of energy stress to the ςB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 31.Voelker U, Engelmann S, Maul B, Riethdorf S, Voelker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 32.Voelker U, Dufour A, Haldenwang W G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of ςB. J Bacteriol. 1995;177:114–122. doi: 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voelker U, Luo T, Smirnova N, Haldenwang W G. Stress activation of Bacillus subtilis ςB can occur in the absence of the ςB negative regulator RsbX. J Bacteriol. 1997;179:1980–1984. doi: 10.1128/jb.179.6.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voelker U, Maul B, Hecker M. Expression of the ςB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J Bacteriol. 1999;181:3942–3948. doi: 10.1128/jb.181.13.3942-3948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voelker U, Voelker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wendrich T M, Marahiel M A. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol. 1997;26:65–79. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]
- 38.Wienen B, Ehrlich R, Stoffler-Meilicke M, Stoffler G, Smith I, Weiss D, Vince R, Pestka S. Ribosomal protein alterations in thiostrepton-and micrococcin-resistant mutants of Bacillus subtilis. J Biol Chem. 1979;254:8031–8041. [PubMed] [Google Scholar]
- 39.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor ςB in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 41.Yasbin R E, Wilson G A, Young F E. Transformation and transfection of lysogenic strains of Bacillus subtilis 168. J Bacteriol. 1973;113:540–548. doi: 10.1128/jb.113.2.540-548.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]