Abstract
The 2,3-dihydroxybiphenyl 1,2-dioxygenase from Sphingomonas xenophaga strain BN6 (BphC1) oxidizes 3-chlorocatechol by a rather unique distal ring cleavage mechanism. In an effort to improve the efficiency of this reaction, bphC1 was randomly mutated by error-prone PCR. Mutants which showed increased activities for 3-chlorocatechol were obtained, and the mutant forms of the enzyme were shown to contain two or three amino acid substitutions. Variant enzymes containing single substitutions were constructed, and the amino acid substitutions responsible for altered enzyme properties were identified. One variant enzyme, which contained an exchanged amino acid in the C-terminal part, revealed a higher level of stability during conversion of 3-chlorocatechol than the wild-type enzyme. Two other variant enzymes contained amino acid substitutions in a region of the enzyme that is considered to be involved in substrate binding. These two variant enzymes exhibited a significantly altered substrate specificity and an about fivefold-higher reaction rate for 3-chlorocatechol conversion than the wild-type enzyme. Furthermore, these variant enzymes showed the novel capability to oxidize 3-methylcatechol and 2,3-dihydroxybiphenyl by a distal cleavage mechanism.
Many chlorinated aromatic compounds are of environmental concern because of their potential toxicity and persistence. Nevertheless, several microorganisms with the ability to degrade chlorinated aromatics have been described (9, 27, 28). The aerobic bacterial degradation of chlorinated aromatic compounds usually proceeds via the intermediate formation of chlorinated catechols. The further metabolism of these chlorocatechols is generally catalyzed by dioxygenases which cleave the aromatic ring between the two hydroxy groups (intradiol, ortho, or 1,2-cleavage) (35). In contrast, only a few dioxygenases are known which cleave chlorocatechols at a bond adjacent to the two hydroxy groups (extradiol, meta, or 2,3-cleavage) (14, 24, 30, 32, 40). Moreover, the degradation of certain chlorinated aromatics (e.g., 3-chlorobenzoate) results in the intermediate formation of (substituted) 3-chlorocatechols, and most extradiol dioxygenases are inactivated very rapidly during the conversion of 3-chlorinated catechols (13, 21, 33). For the catechol 2,3-dioxygenase from Pseudomonas putida strain mt-2 (C23O-mt2), it was demonstrated that the inactivation of the enzyme by 3-chlorocatechol was irreversible (2). The inactivation of the extradiol dioxygenases by 3-chlorinated catechols appears to be one major reason why many strains which are able to degrade nonchlorinated (methyl-) aromatics are unable to convert mixtures of chlorinated and methylated aromatics (31, 39).
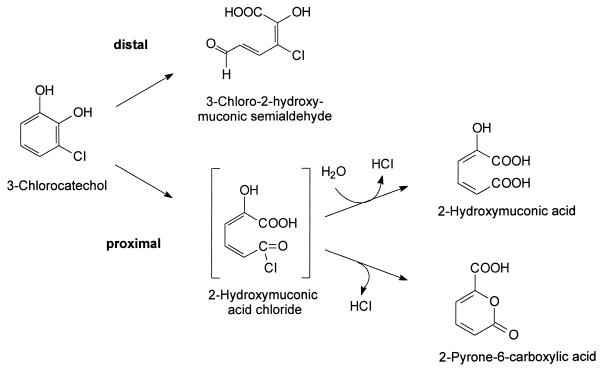
There are very few extradiol dioxygenases which are able to efficiently transform (substituted) 3-chlorocatechols. Thus, the catechol 2,3-dioxygenase from P. putida strain GJ31 transforms 3-chlorocatechol by a proximal cleavage to 2-hydroxymuconic acid (17, 24) (Fig. 1).
FIG. 1.
Distal and proximal extradiol ring cleavage of 3-chlorocatechol and formation of possible reaction products.
A distal extradiol ring cleavage of 3-chlorocatechol resulting in the formation of 3-chloro-2-hydroxymuconic semialdehyde was demonstrated for the 2,3-dihydroxybiphenyl (2,3-DHBP) 1,2-dioxygenase BphC1 from Sphingomonas xenophaga BN6 (29, 30) (Fig. 1). However, during the conversion of 3-chlorocatechol and all other substrates tested, the activity of BphC1 decreased rapidly, presumably due to the loss of the catalytically active Fe(II) ions. This resulted in a relatively low turnover capacity of the enzyme for the oxidation of 3-chlorocatechol. In a previous study (29), an attempt was made to improve the efficiency of this reaction by site-directed mutagenesis of bphC1, but no significant improvement was achieved, basically due to the lack of structural information about BphC1.
In contrast to site-directed mutagenesis, directed evolution offers the possibility of altering an enzyme feature significantly, also in the absence of structural information about the corresponding enzyme (11, 22, 38). In the present study, an attempt was made to generate variant forms of BphC1 with improved efficiency of 3-chlorocatechol conversion. Therefore, bphC1 was randomly mutated using error-prone PCR (6, 23). The catalytic properties of the variants obtained were analyzed in order to obtain a better understanding of the interrelation between the substrate specificity, enzyme stability, and the rather unique ability of BphC1 to cleave 3-chlorocatechol by a distal cleavage mechanism.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The 2,3-DHBP 1,2-dioxygenase from strain BN6 (BphC1) was produced in Escherichia coli DH5α(pGHS118). Plasmid pGHS118 is a derivative of pJOE 2783.1. In pGHS118, bphC1 is under the control of the lac promoter (30). The source of catechol 2,3-dioxygenase from P. putida mt-2 (C23O-mt2) was E. coli BL21(DE3)(pJF150) (30). All E. coli strains were routinely cultured in Luria-Bertani (LB) medium with the appropriate antibiotics.
Random mutagenesis.
Plasmid pGHS118 harboring bphC1 was used as a template for the error-prone PCR. In this plasmid, bphC1 (489 bp) is flanked by an Ndel restriction site 3 bp upstream of the start codon and a BamHI restriction site 4 bp downstream from the stop codon. Using the two oligonucleotide primers 5′-CCC CAG GCT TTA CAC TTT-3′ and 5′-GCT TCT GCG TTC TGA TTT-3′, a DNA fragment was amplified which contained the entire bphC1 gene, 95 bp upstream of the NdeI restriction site and 86 bp downstream of the BamHI site. The following PCR program was used for the amplification of the 677-bp DNA fragment. After 3 min of denaturing at 95°C, 40 cycles of annealing (1.5 min at 48°C), polymerization (3 min at 72°C), and denaturing (40 s at 95°C) followed. The program was completed by an additional annealing (1.5 min at 48°C) and polymerization (10 min at 72°C) step.
In a final volume of 50 μl, the reaction mixtures contained 0.1 ng of template DNA, 10 pmol of each primer (MWG Biotech, Ebersberg, Germany), 1× PCR buffer, 2 U of Taq DNA polymerase (both from Gibco BRL, Life Technologies, Karlsruhe, Germany), MgCl2 (4.7 mM), 0.25 μg of bovine serum albumin (New England Biolabs, Schwalbach/Taunus, Germany), and 3 μl of dimethyl sulfoxide. To obtain mutations during the PCR process, the reaction mixtures were supplemented with MnCl2 (0.05 mM), and dCTP was added in excess (3.5 mM) compared to the other three deoxynucleoside triphosphates (0.1 mM each) (8).
Cloning of the mutated forms of bphC1.
The success of the PCR amplification reaction was verified by agarose gel electrophoresis. Amplified DNA fragments were cut out from the gel and extracted using an Easy-Pure kit (Biozym, Hessisch Oldendorf, Germany). The DNA fragments containing the mutated forms of bphC1 were digested with NdeI and BamHI. After purification by gel electrophoresis, these DNA fragments were ligated with the linearized vector DNA generated by digestion of pJOE 2783.1 with NdeI and BamHI. E. coli DH5α was transformed with the ligation mixture and plated onto LB plates containing 100 μg of ampicillin ml−1.
Screening for clones with an improved activity for 3-chlorocatechol.
The transformants obtained on LB plates with ampicillin were replicated to sterile filter discs (Schleicher und Schuell, Dassel, Germany), which were subsequently sprayed with a solution of 3-chlorocatechol (5 mM concentration in 100 mM Na- K phosphate buffer [pH 7.5]). After this treatment, the clones which were able to convert 3-chlorocatechol by a distal extradiol ring cleavage mechanism turned pale yellow within about 5 min due to the formation of the distal ring cleavage product (λmax = 378 nm). Unfortunately, it was not possible by this method to detect differences in the ring cleavage activity of different clones. In order to make the formation of the distal ring cleavage product more visible, the filter discs were placed under a UV lamp (λ = 366 nm; Desaga, Heidelberg, Germany). Under these conditions, the conversion of 3-chlorocatechol to the distal ring cleavage product resulted in a dark coloration of the respective colonies. To further intensify the UV and visible (UV/Vis) absorption of the distal ring cleavage product, polyethyleneglycol 4000 (10% in methanol) was sprayed on these filters (15) immediately after the treatment with 3-chlorocatechol. The colonies that showed the highest efficiency for 3-chlorocatechol conversion were restreaked on LB plates containing 100 μg of ampicillin ml−1 to obtain single colonies for further investigations.
DNA sequencing and sequence analysis.
The DNA sequences were determined by the method of Sanger et al. (34). Cycle sequencing was performed using the Pharmacia ALFexpress DNA sequencer and a Thermo Sequenase Cy 5 Dye Terminator kit (both from Amersham Pharmacia Biotech, Uppsala, Sweden). Computer-based sequence analysis was performed with the PC/GENE program package (Intelligenetics Inc., Mountain View, Calif.).
Recombination and site-directed mutagenesis.
The wild-type gene (bphC1) and the mutated forms harboring two or three nucleotide exchanges were digested at the unique AluI or MslI restriction sites at positions 163 and 261 of bphC1, respectively. After ligation of mutated and unmutated gene fragments, variant enzymes with single amino acid exchanges were generated.
In some experiments, mutations were introduced into bphC1 by site-directed mutagenesis using a QuikChange kit from Stratagene (29). All mutations were verified by sequencing.
Preparation of cell extracts.
Single colonies were used to inoculate 200 ml of LB medium containing ampicillin (100 μg ml−1). These cultures were grown overnight at 37°C. The cells were harvested by centrifugation (7,000 × g; 15 min; 4°C) and washed with Na-K phosphate buffer (100 mM; pH 7.5). The cells were finally suspended in 2 ml of the same buffer and disrupted by using a French press (Aminco, Silver Spring, Md.) at 80 MPa. Cell debris was removed by centrifugation (100,000 × g; 45 min; 4°C). The protein concentration was determined by the method of Bradford (3), with bovine serum albumin as the standard.
Enzyme assays and analysis of enzyme kinetics.
In general, enzyme assays were performed in 1 ml of Na-K phosphate buffer (100 mM; pH 7.5) with the respective substrate, started by the addition of 1 μl of cell extract (with a protein concentration of about 50 mg ml−1) containing the corresponding enzyme, and measured for 30 s.
The relative activities of the mutant and variant enzymes with 3-chlorocatechol (0.1, 0.5, or 1.0 mM) were determined spectrophotometrically by measuring the change of absorbance at 378 nm to monitor the formation of the distal ring cleavage product.
For the determination of kinetic parameters, 0.05 to 2.0 mM concentrations of catechol, 2,3-DHBP, 3-methyl-, or 3-chlorocatechol were converted by cell extracts containing the wild-type enzyme or the variant enzymes R3, R6, or R9. The wavelengths and extinction coefficients used for the determination of the enzyme activities with the wild-type enzyme were reported previously (12). The variant enzymes converted the substrates to different ratios of distal and proximal ring cleavage products. Therefore, the reaction coefficients for the oxidation of 2,3-DHBP, 3-chloro-, and 3-methylcatechol by the variant enzymes had to be determined experimentally. Thus, each of the substrates (0.05 mM concentration in 1 ml of 100 mM Na-K phosphate buffer [pH 7.5]) was completely converted by the repeated addition of the respective variant enzyme. The resulting absorption of the reaction products was determined spectrophotometrically at those wavelengths at which the enzyme assays were performed (Table 1).
TABLE 1.
Reaction coefficients of ring cleavage products from conversion of different substrates by wild-type enzyme and the variant enzymes R3, R6, and R9
| Enzyme | Reaction coefficient ɛ (M−1 cm−1) of ring cleavage products froma:
|
|||
|---|---|---|---|---|
| Catechol | 2,3-DHBP | 3-Methylcatechol | 3-Chlorocatechol | |
| Wild type | 36,000b | 13,200b | 13,800b | 33,000b |
| Variant R6 | 36,000 | 13,200 | 13,800 | 36,000 |
| Variant R3 | 36,000 | 6,000 | 21,500 | 54,000 |
| Variant R9 | 36,000 | 6,600 | 23,000 | 54,000 |
Values were determined at the following wavelengths: for catechol, 375 nm; for 2,3-DHBP, 434 nm; for 3-methylcatechol, 388 nm; and for 3-chlorocatechol, 378 nm.
Taken from the work of Heiss et al. (12).
The kinetic analyses of substrate turnover and enzyme inactivation during substrate conversion were performed as described previously (29).
One unit of enzyme activity was defined as the amount of enzyme that converts 1 μmol of substrate per min.
Determination of the different ratios of distal to proximal ring cleavage products.
Cell extracts containing the wild type or a variant enzyme (R3, R6, or R9) were repeatedly added to different concentrations of 2,3-DHBP, 3-methyl-, or 3-chlorocatechol (0.01, 0.05, 0.1, and 0.3 mM concentrations, in 1 ml of 100 mM Na-K phosphate buffer [pH 7.5]) until no further formation of the ring cleavage products could be detected spectrophotometrically. The concentrations of the ring cleavage products were determined by high-performance liquid chromatography (HPLC) (see below). The solvent system A was used for the analysis of the ring cleavage products of 2,3-DHBP, and the solvent system B was used for the products of 3-methylcatechol. For the analysis of the distal and proximal products of 3-chlorocatechol, the solvent systems B and C (see below), respectively, were used.
Preparation of substituted picolinic acids from the ring cleavage products of 3-methylcatechol.
3-Methylcatechol (1 mM) was dissolved in 1 ml of Na-K phosphate buffer (50 mM; pH 7.5) and completely converted by the repeated addition of cell extracts with extradiol dioxygenase activities (C23O-mt2 or the variant enzyme R9). The reaction mixtures were filtered through a 10,000-molecular-weight cutoff membrane (Centricon; Millipore, Bedford, Mass.) in order to separate macromolecular contaminants. The filtrates containing the ring cleavage products were incubated with NH4Cl (1.2 M) and shaken overnight at 30°C (25, 30). Subsequently, these solutions were analyzed by HPLC using solvent system A (see below).
HPLC.
All ring cleavage products and the picolinic acids were monitored by HPLC (HPLC Millenium Chromatography Manager 2.0, equipped with a photodiode array detector, model 486, and HPLC pumps, model 510; Waters Associates, Milford, Mass.). The following solvent systems were used.
Solvent system A consisted of water-methanol (60/40 [vol/vol]) containing Na carbonate buffer (50 mM; pH 10) and tetrabutylammonium hydrogen sulfate as an ion pair reagent (5 mM). Solvent system B consisted of water-methanol (90/10 [vol/vol]) containing Na carbonate buffer (50 mM; pH 10) and tetrabutylammonium hydrogen sulfate as an ion pair reagent (5 mM). Solvent system C consisted of water-methanol (90/9.8 [vol/vol]) plus 0.2 [vol/vol] H3PO4.
For the solvent systems A and B, a reversed-phase column (150 by 4 mm; Grom-Polymer C18; 5-μm particles; Grom, Herrenberg, Germany) was used with a flow rate of 0.7 ml min−1. For the solvent system C, a reversed-phase column (125 by 4 mm; Grom-Sil 100 Octyl-4; 5-μm particles; Grom) was used and a flow rate of 0.8 ml min−1 was applied.
Chemicals.
3-Methylpicolinic acid was purchased from TCI (Tokyo, Japan). 2-Hydroxymuconic acid and 2-pyrone-6-carboxylic acid were kindly donated by S. Kaschabek and W. Reineke (Universität-Gesamtschule Wuppertal, Wuppertal, Germany). The sources of all other chemicals have been described previously (29, 30).
RESULTS
Random mutagenesis for improved conversion of 3-chlorocatechol.
To improve the catalytic efficiency of BphC1 from strain BN6 for the extradiol cleavage of 3-chlorocatechol, bphC1 was randomly mutated using error-prone PCR (see Materials and Methods). The amplification products obtained were cloned into the expression vector pJOE 2783.1 (30), and cells of E. coli DH5α were transformed with these plasmids. Using a spray test, about 5,000 colonies were screened on agar plates for the improved formation of the distal ring cleavage product of 3-chlorocatechol (see Materials and Methods). For 15 colonies an improvement in this reaction in comparison to results for the wild-type enzyme was supposed. These clones were cultivated in liquid medium, cell extracts were prepared, and the formation of the distal ring cleavage product from 3-chlorocatechol (λmax = 378 nm) was analyzed spectrophotometrically for different substrate concentrations (0.1, 0.5, and 1.0 mM). Only three cell extracts showed increased activities compared to the wild-type enzyme (Table 2). The most significant improvement for the formation of the distal ring cleavage product was observed with the mutant enzyme 1, which revealed about fivefold-higher relative activities than the wild-type enzyme for all substrate concentrations tested. The mutant enzyme 3 showed higher relative activities than the wild-type enzyme only for substrate concentrations of ≥0.5 mM.
TABLE 2.
Relative activities for conversion of 3-chlorocatechol by wild-type enzyme and mutant forms of BphC1
| Concn of 3-chlorocatechol (mM) | Relative activitya (%)
|
|||
|---|---|---|---|---|
| Wild type | Mutant 1 | Mutant 2 | Mutant 3 | |
| 0.1 | 100 | 530 | 540 | 70 |
| 0.5 | 180 | 700 | 500 | 180 |
| 1.0 | 140 | 630 | 260 | 180 |
The assays were performed with cell extracts of recombinant E. coli strains. The relative activities were determined spectrophotometrically by measuring the change of absorbance at 378 nm. They are given in comparison to the activity of the wild-type enzyme at a 0.1 mM substrate concentration (0.1 U of protein mg−1 = 100%).
From these three clones the mutated forms of bphC1 were isolated and sequenced, and the proteins encoded were found to contain the following amino acid exchanges: M37T and F66L (mutant 1); F66L, N96T, and K106R (mutant 2); and K22E and D155Y (mutant 3).
Recombination and site-directed mutagenesis.
In order to identify the mutations responsible for the changed catalytic properties, the following variant enzymes were constructed by combination of mutated and unmutated gene fragments (see Materials and Methods): R1 (M37T, F66L, N96T, and K106R); R2 (M37T); R3 (F66L); R4 (N96T and K106R); R5 (K22E); and R6 (D155Y).
Cell extracts from cells producing the variant enzyme R3, which carried the common mutation of the mutant enzymes 1 and 2 (F66L), showed almost the same increased relative activities with 3-chlorocatechol as the mutant enzyme 1 (M37T and F66L). For the variant enzyme R6 (D155Y), the relative activities were very similar to those of the mutant enzyme 3 (K22E and D155Y). The other variant enzymes displayed no improved conversion of 3-chlorocatechol in comparison to the wild-type enzyme. This demonstrated that single amino acid exchanges in the mutant enzymes were responsible for the altered catalytic properties.
The substitution for the amino acid phenylalanine at position 66 of BphC1 by the smaller amino acid leucine altered the catalytic properties of the enzyme significantly. This region is probably involved in the binding of the substrate in the catalytic center, since in the crystal structure of the 2,3-DHBP 1,2-dioxygenase from Pseudomonas sp. KKS102 the corresponding region is considered to be involved in substrate binding (36).
In order to further characterize the importance of this region in BphC1 for the conversion of 3-chlorocatechol, further variant enzymes were constructed by site-directed mutagenesis. An acidic (aspartic acid) or a basic (histidine) amino acid was introduced at position 66 of BphC1 to generate the new variant enzymes R7 (F66D) and R8 (F66H). Also, the variant enzyme R3 (F66L) was further modified by replacing the adjacent proline residue in position 67 by a smaller alanine residue, giving the variant enzyme R9 (F66L and P67A).
From these variant enzymes, R7 (F66D) and R8 (F66H) showed no improved formation of the distal ring cleavage product from 3-chlorocatechol (0.1 to 1.0 mM). In contrast, the double mutant R9 (F66L and P67A) demonstrated about sixfold-higher relative activities than the wild-type enzyme for all substrate concentrations tested. This corresponded to an increase of the activity of about 20% in comparison to the variant enzyme R3 (F66L).
Distal and proximal cleavage of 3-chlorocatechol by the different forms of BphC1-BN6.
3-Chlorocatechol (0.05 mM concentration in 100 mM Na-K phosphate buffer [pH 7.5]) was completely converted by the repeated addition of cell extracts containing the wild-type enzyme or the variant enzymes R3 (F66L), R6 (D155Y), or R9 (F66L and P67A) until no further formation of the distal ring cleavage product could be detected spectrophotometrically at 378 nm. Surprisingsly, the reaction mixtures containing the variant enzyme R3 or R9 showed an about 1.6-fold-higher rate of final absorption at 378 nm than the assay mixtures containing the wild-type enzyme or the variant enzyme R6. This indicated that the different forms of BphC1 produced different amounts of the distal ring cleavage product during the conversion of identical amounts of 3-chlorocatechol. This suggested that some of the enzymes oxidized 3-chlorocatechol not only to the distal ring cleavage product but also to another product(s), presumably by a proximal ring cleavage mechanism.
In previous experiments with the wild-type enzyme (29), a proximal cleavage of 3-chlorocatechol by BphC1 was excluded because there were no indications for the formation of 2-hydroxymuconic acid, which had been identified as the product of the proximal cleavage of 3-chlorocatechol by the catechol 2,3-dioxygenase from P. putida GJ31 (17, 24). For those experiments the reaction mixtures had been analyzed after a time-consuming separation of the proteins by ultrafiltration. Because of our new observations, the experiments were repeated and the reaction mixtures were analyzed without further treatment of the sample by HPLC (after about 30 s). In this way, the formation of 2-hydroxymuconic acid during the conversion of 3-chlorocatechol by the wild-type enzyme was detected. A further incubation of the reaction mixture suggested a half-life of about 10 min for 2-hydroxymuconic acid under the reaction conditions used. The reaction analysis by UV/Vis spectroscopy demonstrated the conversion of 2-hydroxymuconic acid (λmax = 290 nm) to a new product with an absorption maximum at λmax = 235 nm, as previously described (16, 17).
In contrast to the catechol 2,3-dioxygenase from P. putida GJ31, which formed 2-hydroxymuconic acid as the single product during the proximal cleavage of 3-chlorocatechol (17), with BphC1 the formation of 2-pyrone-6-carboxylic acid also was observed (Fig. 1).
After the complete conversion of 3-chlorocatechol (0.01, 0.05, 0.1, or 0.3 mM) by the different forms of BphC1, the reaction mixtures were analyzed by HPLC in order to quantify the formation of the distal and proximal ring cleavage products (see Materials and Methods). The concentrations of 2-hydroxymuconic acid and 2-pyrone-6-carboxylic acid were determined using authentic standards. It was found that the ratio by which these products were formed during the conversion of 3-chlorocatechol was independent of the substrate concentrations used. The HPLC analysis demonstrated that the wild-type enzyme oxidized 3-chlorocatechol to 40% by a proximal cleavage. In contrast, the variant enzymes R3 and R9 converted 3-chlorocatechol to only insignificant amounts of the proximal ring cleavage products (Table 3). The quantification of the proximal ring cleavage products was in agreement with the quantification of the distal ring cleavage product, showing that the wild-type enzyme formed the distal cleavage product in an amount up to 60% of that obtained with the variant enzymes R3 and R9.
TABLE 3.
Formation of proximal and distal ring cleavage products during oxidation of 3-chlorocatechol by wild-type enzyme BphC1 and variant enzymes R3, R6, and R9a
| Enzyme | Ratio (%) of ring cleavage products formed
|
||
|---|---|---|---|
| Proximal products
|
Distal product
|
||
| 2-Hydroxy-muconic acid | 2-Pyron-6-carboxylic acid | 3-Chloro-2-hydroxymuconic semialdehyde | |
| Wild type | 26 | 14 | 60 |
| Variant R6 (D155Y) | 22 | 12 | 66 |
| Variant R3 (F66L) | 1 | 0 | 99 |
| Variant R9 (F66L and P67A) | 1 | 0 | 99 |
Cell extracts of recombinant E. coli strains producing the different forms of BphC1 were repeatedly added to the assays containing 3-chlorocatechol (0.05 mM concentration in 1 ml of 100 mM Na-K phosphate buffer [pH 7.5]) to obtain a complete conversion of the substrate. The products were rapidly quantified by HPLC (see Materials and Methods).
Conversion of 3-methylcatechol and 2,3-DHBP by the variant enzymes R3, R6, and R9.
The experiments with 3-chlorocatechol demonstrated that the mutations in the variant forms R3 and R9 of BphC1 resulted in significant changes in the regioselectivity of the enzyme. These variant enzymes showed an enhanced tendency to perform a distal ring cleavage reaction. Therefore, the conversion of 3-methylcatechol and 2,3-DHBP by the wild-type enzyme and the variant enzymes R3, R6, and R9 also was investigated with regard to the regioselectivity.
The conversion of 3-methylcatechol or 2,3-DHBP (0.1 mM, in a total volume of 1 ml containing 100 mM Na-K phosphate buffer [pH 7.5]) by the wild-type enzyme or the variant enzyme R6 resulted in UV/Vis spectra almost identical to that obtained after the conversion of these substrates by the catechol 2,3-dioxygenase from P. putida mt-2 (C230-mt2) (λmax = 388 nm for the oxidation of 3-methylcatechol; λmax = 434 nm for 2,3-DHBP). Since C230-mt2 performs exclusively a proximal ring cleavage (26), the UV/Vis spectra obtained indicated that the wild-type enzyme and the variant enzyme R6 also converted the two substrates by a proximal ring cleavage mechanism.
After conversion of 3-methylcatechol by the variant enzymes R3 or R9, both reaction mixtures showed a UV/Vis spectrum with an absorption maximum at 382 nm which was significantly different from the UV/Vis spectrum observed for the proximal ring cleavage product (λmax = 388 nm).
The difference between the variant enzymes R3 and R9 and the wild-type form of BphC1 was even more obvious during the oxidation of 2,3-DHBP. The spectrophotometrical analysis of the reactions catalyzed by the variant enzymes R3 or R9 showed, besides the known absorption maximum of the proximal ring cleavage product (λmax = 434 nm), an additional absorption maximum at 381 nm with about double intensity, indicating the formation of two reaction products.
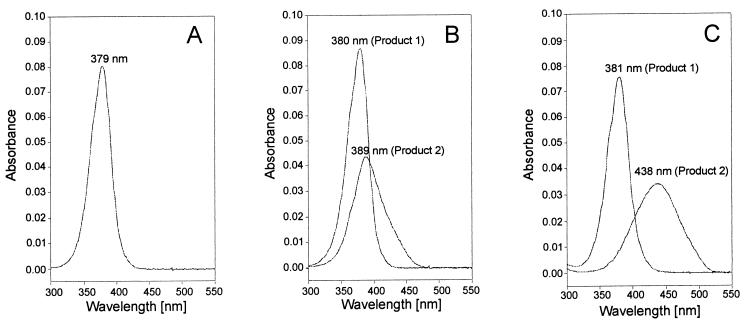
An attempt was therefore made to analyze the reaction mixtures after the conversion of 3-methylcatechol and 2,3-DHBP by the different enzymes using HPLC. In order to obtain a clear separation and detection of the reaction products, a reversed-phase column with an alkaline solvent system and a photodiode array detector was used (see Materials and Methods). The HPLC analyses demonstrated that the variant enzymes R3 and R9 oxidized 3-methylcatechol and 2,3-DHBP each to two products with different UV/Vis spectra (Fig. 2).
FIG. 2.
UV/Vis spectra (at pH 10) of the reaction products formed during the conversion of 3-chlorocatechol (A), 3-methylcatechol (B), or 2,3-DHBP (C). The substrates (0.1 mM concentrations in 100 mM Na-K phosphate buffer [pH 7.5]) were converted with cell extracts of recombinant E. coli strains containing the variant form R9 of BphC1. The products were analyzed by HPLC using a photodiode array detector (see Materials and Methods). The analysis of the product formed from 3-chlorocatechol (Rt = 10.8 min) and also of the products from 3-methylcatechol (for product 1, Rt = 4.9 min; for product 2, Rt = 5.9 min) was performed using solvent system B. The oxidation products of 2,3-DHBP were analyzed with solvent system A (for product 1, Rt = 1.8 min; for product 2, Rt = 3.1 min).
The products with retention times (Rt) of 5.9 min (λmax = 389 nm; from 3-methylcatechol) and 3.1 min (λmax = 438 nm; from 2,3-DHBP) were also formed during the conversion of 3-methylcatechol and 2,3-DHBP by C23O-mt2 and were therefore assigned as the proximal ring cleavage products. The UV/Vis spectra recorded in situ for the respective second products formed from 3-methylcatechol (Rt = 4.9 min; λmax = 380 nm) and 2,3-DHBP (Rt = 1.8 min; λmax = 381 nm) were very similar to the UV/Vis spectrum of the distal ring cleavage product of 3-chlorocatechol (λmax = 379 nm). This strongly indicated a distal cleavage of 3-methylcatechol and 2,3-DHBP by the variant enzymes R3 and R9.
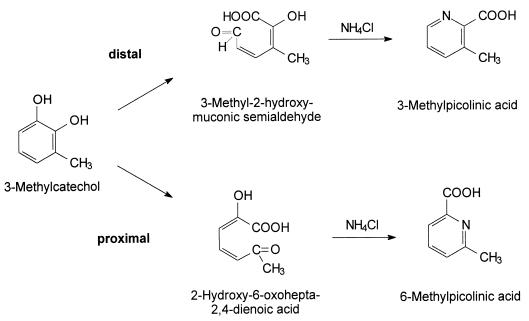
To prove the distal cleavage of 3-methylcatechol, the reaction products formed by the variant enzyme R9 were incubated with NH4Cl in order to produce substituted picolinic acids (25). The HPLC analysis demonstrated that the two ring cleavage products from 3-methylcatechol (Rt = 4.9 and λmax = 380 nm; Rt = 5.9 min and λmax = 389 nm) were transformed by this treatment and that two new products (Rt = 5.7 min and λmax = 266 nm; Rt = 9.0 min and λmax = 270 nm) were formed.
One of these products (Rt = 5.7 min) was also obtained after the conversion of 3-methylcatechol by C23O-mt2 and subsequent incubation of the reaction mixture with NH4Cl. Since C23O-mt2 cleaves 3-methylcatechol only by a proximal ring cleavage mechanism (26), this product was tentatively identified as 6-methylpicolinic acid (1).
The other product (Rt = 9.0 min) was identified as 3-methylpicolinic acid by using an authentic standard. This finding proved the assignment of the ring cleavage product with an Rt of 4.9 min and a λmax of 380 nm from the conversion of 3-methylcatechol as 3-methyl-2-hydroxymuconic semialdehyde and thus the distal ring cleavage product of 3-methylcatechol (Fig. 3).
FIG. 3.
Possible directions for the extradiol ring cleavage of 3-methylcatechol and transformation of the ring cleavage products into different substituted picolinic acids.
3-Methylcatechol and 2,3-DHBP (0.01, 0.05, 0.1, and 0.3 mM) were completely converted by C23O-mt2, the wild-type enzyme, and the variant enzymes R3, R6, and R9, and the ring cleavage products were analyzed by HPLC (see Materials and Methods) in order to determine the ratio in which the proximal and distal ring cleavage products were formed. The ratios in which the proximal and the distal ring cleavage products were formed did not depend on the substrate concentration converted. As expected from the literature, C23O-mt2 converted 3-methylcatechol and 2,3-DHBP exclusively to the proximal ring cleavage products. This enabled the quantification of the proximal products formed by the different forms of BphC1 from 3-methylecatechol and 2,3-DHBP. Thus it was shown that the variant enzymes R3 and R9 converted 3-methylcatechol and 2,3-DHBP to about equal proportions of the distal and proximal ring cleavage products (Table 4).
TABLE 4.
Formation of distal and proximal ring cleavage products during conversion of 3-methylcatechol and 2,3-DHBP by wild-type enzyme and variant enzymes R3, R6, and R9a
| Enzyme | Ratio (%) of proximal/distal ring cleavage products obtained during conversion of:
|
|
|---|---|---|
| 3-Methylcatechol | 2,3-DHBP | |
| C23O-mt2 | 100/0 | 100/0 |
| Wild type | 95/5 | 100/0 |
| R6 (D155Y) | 95/5 | 100/0 |
| R3 (F66L) | 60/40 | 45/55 |
| R9 (F66L and P67A) | 50/50 | 50/50 |
Cell extracts of recombinant E. coli strains producing the different forms of BphC1 were repeatedly added to the assays containing 3-methylcatechol or 2,3-DHBP (0.05 mM concentration in 1 ml of 100 mM Na-K phosphate buffer [pH 7.5]) in order to obtain a complete conversion of the substrates. The products were rapidly quantified by HPLC (see Materials and Methods).
Kinetic data for the conversion of different substrates by the variant enzymes and the wild-type enzyme.
Various concentrations of 2,3-DHBP, catechol, 3-methyl-, or 3-chlorocatechol were converted by cell extracts containing the wild-type enzyme or the variant enzymes R3, R6, or R9. The different forms of BphC1 produced different ratios of proximal and distal ring cleavage products during conversion of the same substrate. Therefore, reaction coefficients had to be determined for the respective reactions (see Materials and Methods).
The kinetic parameters obtained for the conversion of different substrates demonstrated that the variant enzymes R3 and R9 displayed substrate affinities similar to those of the wild-type enzyme. Furthermore, the maximal reaction rates with 3-methylcatechol were almost identical with those for cell extracts containing the wild-type enzyme and the variant enzymes (Table 5). The variant enzymes converted catechol and 3-chlorocatechol with about twofold- and sixfold-higher reaction rates, respectively, than for 3-methylcatechol. This demonstrated, in comparison to the relative activities of the wild-type enzyme with 3-methylcatechol, catechol, and 3-chlorocatechol (1:0.16:1.3), a significant increase in the ability of the mutant enzymes to oxidize catechol and 3-chlorocatechol. In contrast, both variant enzymes converted 2,3-DHBP at a much lower rate than the wild-type enzyme (Table 5). This resulted in a completely altered substrate specificity of the variant enzymes R3 and R9 compared to that for BphC1.
TABLE 5.
Kinetic parameters of the wild-type enzyme and variant enzymes R3, R6, and R9a
| Enzyme | Kinetic parameters for conversion ofb:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 3-Methylcatechol
|
3-Chlorocatechol
|
2,3-DHBP
|
Catechol
|
|||||
| Km (mM) | V (U mg−1) | Km (mM) | V (U mg−1) | Km (mM) | V (U mg−1) | Km (mM) | V (U mg−1) | |
| Wild type | 0.55 ± 0.13 | 0.5 ± 0.08 (100) | 0.23 ± 0.03 | 0.67 ± 0.04 (134) | 0.36 ± 0.06 | 6.5 ± 0.5 (1300) | 3.1 ± 0.8 | 0.085 ± 0.01 (17) |
| R6 (D155Y) | 0.77 ± 0.13 | 0.46 ± 0.06 (100) | 0.43 ± 0.03 | 0.59 ± 0.02 (128) | 0.91 ± 0.33 | 7.0 ± 1.2 (1628) | 9.6 ± 3.7 | 0.19 ± 0.06 (41) |
| R3 (F66L) | 0.61± 0.20 | 0.41 ± 0.07 (100) | 0.24 ± 0.04 | 2.6 ± 0.2 (634) | 0.43 ± 0.14 | 0.87 ± 0.10 (212) | 2.4 ± 0.4 | 0.70 ± 0.08 (171) |
| R9 (F66L and P67A) | 0.63 ± 0.20 | 0.53 ± 0.09 (100) | 0.21 ± 0.03 | 3.2 ± 0.2 (604) | 0.49 ± 0.14 | 1.4 ± 0.16 (264) | 2.4 ± 0.3 | 1.0 ± 0.08 (189) |
The enzyme assays were performed in 1 ml of Na-K phosphate buffer (100 mM [pH 7.5]) and started by the addition of 1 μl of cell extract from recombinant E. coli strains (with a protein concentration of about 50 mg ml−1) containing the corresponding enzyme. The relative activities were determined spectrophotometrically by measuring the change of absorbance at the indicated wavelengths (see footnote b). The extinction and reaction coefficients used are reported in Materials and Methods. The kinetic parameters were determined using different substrate concentrations as described previously (29), and the specific activities indicated in the table were the maximal activities observed with the cell extracts for the respective substrates. The relative activities given in parentheses are expressed for each cell extract as percentages of that for 3-methylcatechol. Results are means ± the standard errors of the means.
Values were determined at the following wavelengths: for 3-methylcatechol, 388 nm; for 3-chlorocatechol, 378 nm; for 2,3-DHBP, 434 nm; and for catechol, 378 nm.
The variant enzyme R6 showed, for all substrates tested, a lower affinity than the wild-type enzyme but reached almost the same maximal velocities with these substrates. Moreover, it was obvious that this enzyme was more stable during substrate conversion than BphC1.
For the wild-type enzyme, a decrease in the reaction rate during the conversion of different substrates was observed. This time-dependent enzyme inactivation was probably caused by the loss or oxidation of the catalytically active ferrous iron. Since this kind of inactivation was most pronounced at higher substrate concentrations, it resulted in apparent substrate inhibition kinetics (29). This time-dependent inactivation was also observed with the variant enzymes. It was most pronounced for all forms of BphC1 with 3-chlorocatechol as a substrate. Rate constants of enzyme inactivation (kinact) for the conversion of 3-chlorocatechol were determined for the different forms of BphC1 in order to compare their stability during substrate turnover (29). A higher kinact value corresponded to a more rapid inactivation of the enzyme during substrate oxidation (4, 5). The kinact value obtained for the variant form R6 (kinact = 0.08 ± 0.02 s−1) demonstrated the higher stability of this enzyme during substrate conversion in comparison to the wild-type enzyme (kinact = 0.13 ± 0.02 s−1) and the variant enzymes R3 (kinact = 0.14 ± 0.03 s−1) and R9 (kinact = 0.13 ± 0.03 s−1).
DISCUSSION
Until now, the crystal structures of three extradiol dioxygenases have been resolved. These are the 2,3-dihydroxybiphenyl 1,2-dioxygenases originating from Burkholderia (Pseudomonas) cepacia LB400 and Pseudomonas sp. KKS102 and the catechol 2,3-dioxygenase from P. putida mt-2 (C23O-mt2). These crystal structures showed very similar active centers and indicated a similar reaction mechanism for this class of dioxygenases (10, 20, 36).
However, sequence alignments of various extradiol dioxygenases showed that these enzymes can be separated into different groups with very low total sequence homology (7). This was very evident for BphC1, which showed less than 20% sequence identity to the most related enzymes, two 2,3-DHBP 1,2-dioxygenases from Rhodococcus globerulus P6 (12). Furthermore, BphC1 from strain BN6 and the 2,3-DHBP dioxygenases from strain P6 belong to a specific group of small extradiol dioxygenases which show significantly lower molecular weights of the single subunits compared to almost all other extradiol dioxygenases. For this group of extradiol dioxygenases, no crystal structure is currently available. Thus, although the catalytic mechanism of BphC1 is probably similar to that of most other extradiol dioxygenases, it is difficult to rationalize the connection between the amino acid sequence and the catalytic function of this enzyme.
In the present study, basically two different types of variant enzymes were obtained. The variant enzyme R6 (D155Y) demonstrated a lower substrate affinity and a higher stability during substrate turnover. In contrast, the variant enzymes R3 (F66L) and R9 (F66L and P67A) showed a changed regioselectivity of ring cleavage and a higher catalytic activity for 3-chlorocatechol compared to the wild-type enzyme.
In the variant enzyme R6, an aspartate residue at the C-terminal part was replaced with a tyrosine residue. From the crystal structure of the C23O-mt2, it was proposed that the catechols must enter the active site of the enzyme through a channel (barrel structure) and that some of the amino acid residues at the C terminus narrow the substrate entrance (20). Furthermore, it may be deduced from the structures obtained for the C23O-mt2 that the carboxy-terminal region of the enzyme is located rather near to the catalytic center. Thus it is possible that the reduced substrate affinity of the variant enzyme R6 is caused by a reduced accessibility of the active center to the organic substrate or by subtle changes in the active center caused by the mutation in the C-terminal region.
The major aim of this study was to obtain variant forms of BphC1 with improved efficiency for 3-chlorocatechol oxidation. This was achieved by the isolation of the variant enzymes R3 and R9, which showed about fivefold-higher reaction rates for the conversion of 3-chlorocatechol than the wild-type enzyme. This is the first example showing that the catalytic activity of an extradiol dioxygenase could be increased by random mutagenesis, and this demonstrated that error-prone PCR combined with an appropriate screening method is a simple and effective method for altering the catalytic properties of an enzyme in the desired way. A different strategy of directed evolution of extradiol dioxygenases was performed by Kikuchi et al. (18, 19) using family shuffling in order to construct a catechol 2,3-dioxygenase with increased thermostability.
The variant enzymes R3 and R9 were selected for the improved oxidation of 3-chlorocatechol. Therefore, the observation that these variant enzymes also showed an altered regioselectivity for the ring cleavage of 3-methylcatechol was rather astonishing. The variant enzymes R3 and R9 oxidized about half of the 3-methylcatechol by a distal cleavage mechanism. This finding was very unexpected, because until now only a single extradiol dioxygenase has been known (from an Achromobacter sp.), which was claimed to convert 3-methylcatechol by a distal ring cleavage mechanism (14). Furthermore, the HPLC analysis and the UV/Vis spectra obtained for the conversion of 2,3-DHBP by these variant enzymes also suggested that this substrate was oxidized to significant amounts of a distal cleavage product. This indicates that the variant enzymes R3 and R9 have an enhanced tendency for a distal cleavage of all 3-substituted catechols. This demonstrates that the substitution of only a single amino acid can change the catalytic properties of the enzyme significantly. Moreover, this is the first time that a change in the direction of ring cleavage was described for an extradiol dioxygenase.
The altered catalytic properties of the variant enzymes R3 and R9 were caused by the replacement of the amino acids at positions 66 and 67 of BphC1 by smaller ones. These residues probably belong to a region of the enzyme which is involved in substrate binding. A possible explanation for the changed regioselectivity in the mutant enzymes may be deduced from the reaction mechanism of extradiol dioxygenases. For the catechol 2,3-dioxygenase from P. putida mt-2, the catalytic cycle is supposed to involve a complexation of the catalytically active Fe(II) ion by a monoanionic catecholate as a bidentate ligand. The ring cleavage reaction is proposed to proceed via an attack of the iron-bound activated oxygen species on the nonhydroxylated position vicinal to the carbon atom bearing the phenolate anion (20, 36, 37). A substituted catechol can in principle bind in two different modes as a bidentate ligand to the catalytically active ferrous iron. This would finally result in a different regioselectivity of the substrate cleavage. The mutation F66L may therefore result in an altered sterical situation in the catalytic center which could allow binding of 3-methylcatechol and 2,3-DHBP to the catalytically active ferrous iron in a way which finally results in a distal cleavage of the substrates. However, preliminary experiments which were performed in order to detect differences in the kinetic parameters for the formation of the proximal and distal ring fission products from 2,3-DHBP by the variant enzymes did not show significant differences in the substrate affinities for the formation of the proximal and distal ring fission products. Thus, probably only the crystal structure of BphC1 and its variant forms could completely explain the unique ability of the mutant enzymes to perform a distal ring cleavage of different substituted catechols.
The variant forms of BphC1 may be used for the production of novel picolinic acid derivatives or for the construction of strains with an improved capacity to degrade chlorinated aromatic compounds.
REFERENCES
- 1.Asano Y, Yamamoto Y, Yamada H. Catechol 2,3-dioxygenase-catalyzed synthesis of picolinic acids from catechols. Biosci Biotechnol Biochem. 1994;58:2054–2056. [Google Scholar]
- 2.Bartels I, Knackmuss H-J, Reineke W. Suicide inactivation of catechol 2,3-dioxygenase from P. putida mt-2 by 3-halocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cerdan P, Rekik M, Harayama S. Substrate specificity differences between two catechol 2,3-dioxygenases encoded by the TOL and NAH plasmids from Pseudomonas putida. Eur J Biochem. 1995;229:113–118. doi: 10.1111/j.1432-1033.1995.tb20445.x. [DOI] [PubMed] [Google Scholar]
- 5.Cerdan P, Wasserfallen A, Rekik M, Timmis K N, Harayama S. Substrate specificity of catechol 2,3-dioxygenase encoded by TOL plasmid pWWO of Pseudomonas putida and its relationship to cell growth. J Bacteriol. 1994;176:6074–6081. doi: 10.1128/jb.176.19.6074-6081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen K, Arnold F H. Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc Natl Acad Sci USA. 1993;90:5618–5622. doi: 10.1073/pnas.90.12.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eltis L D, Bolin J T. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromant M, Blanquet S, Plateau P. Directed random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 9.Häggblom M. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev. 1992;103:29–72. doi: 10.1111/j.1574-6968.1992.tb05823.x. [DOI] [PubMed] [Google Scholar]
- 10.Han S, Eltis L D, Timmis K N, Muchmore S W, Bolin J T. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 11.Harayama S. Artificial evolution by DNA shuffling. Trends Biotechnol. 1998;16:76–82. doi: 10.1016/s0167-7799(97)01158-x. [DOI] [PubMed] [Google Scholar]
- 12.Heiss G, Stolz A, Kuhm A E, Müller C, Klein J, Altenbuchner J, Knackmuss H-J. Characterization of a 2,3-dihydroxybiphenyl dioxygenase from the naphthalenesulfonate-degrading bacterium strain BN6. J Bacteriol. 1995;177:5865–5871. doi: 10.1128/jb.177.20.5865-5871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollender J, Dott W, Hopp J. Regulation of chloro- and methylphenol degradation in Comamonas testosteroni JH5. Appl Environ Microbiol. 1994;60:2330–2338. doi: 10.1128/aem.60.7.2330-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath R S. Co-metabolism of methyl- and chloro-substituted catechols by an Achromobacter sp. possessing a new meta-cleaving oxygenase. Biochem J. 1970;119:871–876. doi: 10.1042/bj1190871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jork H, Funk W, Fischer W, Wimmer H. Dünnschichtchromatographie: Reagenzien und Nachweismethoden. Weinheim, Germany: VCH Verlagsgesellschaft; 1989. [Google Scholar]
- 16.Junker F, Field J A, Bangerter F, Ramsteiner K, Kohler H-P, Joannou C L, Mason J R, Leisinger T, Cook A M. Oxygenation and spontaneous deamination of 2-aminobenzenesulphonic acid in Alcaligenes sp. strain O-1 with subsequent meta ring cleavage and spontaneous desulphonation to 2-hydroxymuconic acid. Biochem J. 1994;300:429–436. doi: 10.1042/bj3000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaschabek S R, Kasberg T, Müller D, Mars A E, Janssen D B, Reineke W. Degradation of chloroaromatics: purification and characterization of a novel type of chlorocatechol 2,3-dioxygenase of Pseudomonas putida GJ31. J Bacteriol. 1998;180:296–302. doi: 10.1128/jb.180.2.296-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kikuchi M, Ohnishi K, Harayama S. Novel family shuffling methods for in vitro evolution of enzymes. Gene. 1999;236:159–167. doi: 10.1016/s0378-1119(99)00240-1. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi M, Ohnishi K, Harayama S. An effective family shuffling method using single-stranded DNA. Gene. 2000;243:133–137. doi: 10.1016/s0378-1119(99)00547-8. [DOI] [PubMed] [Google Scholar]
- 20.Kita A, Kita S, Fujisawa I, Inaka K, Ishida T, Horiike K, Nozaki M, Miki K. An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Pseudomonas putida mt-2. Struct Fold Des. 1999;7:25–34. doi: 10.1016/s0969-2126(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 21.Klecka G M, Gibson D T. Inhibition of catechol 2,3-dioxygenase from Pseudomona putida mt-2 by 3-chlorocatechol. Appl Environ Microbiol. 1981;41:1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchner O, Arnold F H. Directed evolution of enzyme catalysts. Trends Biotechnol. 1997;15:523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]
- 23.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 24.Mars A E, Kasberg T, Kaschabek S R, van Agteren M, Janssen D B, Reineke W. Microbial degradation of chloroaromatics. Use of the meta-cleavage pathway for the mineralization of chlorobenzene. J Bacteriol. 1997;179:4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews C, Rossiter J T, Ribbons D W. Production of pyridine synthons by biotransformations of benzene precursors and their cyclization with nitrogen nucleophiles. Biocatal Biotransform. 1995;12:241–254. [Google Scholar]
- 26.Nozaki M, Kotani S, Ono K, Senoh S. Metapyrocatechase. III. Substrate specificity and mode of ring fission. Biochim Biophys Acta. 1970;220:213–223. doi: 10.1016/0005-2744(70)90007-0. [DOI] [PubMed] [Google Scholar]
- 27.Reineke W. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu Rev Microbiol. 1998;52:287–331. doi: 10.1146/annurev.micro.52.1.287. [DOI] [PubMed] [Google Scholar]
- 28.Reineke W, Knackmuss H-J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- 29.Riegert U, Heiss G, Kuhm A E, Müller C, Contzen M, Knackmuss H-J, Stolz A. Catalytical properties of the 3-chlorocatechol-oxidizing 2,3-dihydroxybiphenyl 1,2-dioxygenase from Sphingomonas sp. strain BN6. J Bacteriol. 1999;181:4812–4817. doi: 10.1128/jb.181.16.4812-4817.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riegert U, Heiss G, Fischer P, Stolz A. Distal cleavage of 3-chlorocatechol to 3-chloro-2-hydroxymuconic semialdehyde by an extradiol dioxygenase. J Bacteriol. 1998;180:2849–2853. doi: 10.1128/jb.180.11.2849-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojo F, Pieper D H, Engesser K-H, Knackmuss H-J, Timmis K N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987;238:1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- 32.Sala-Trepat J M, Evans W C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971;20:400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 33.Sandossi M, Sylvestre M, Ahmad D. Effects of chlorobenzoate transformation on the Pseudomonas testosteroni biphenyl and chlorobiphenyl degradation pathway. Appl Environ Microbiol. 1992;58:485–495. doi: 10.1128/aem.58.2.485-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlömann M. Evolution of chlorocatechol catabolic pathways. Biodegradation. 1994;5:301–321. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 36.Senda T, Sugiyama K, Narita H, Yamamoto T, Kimbara K, Fukuda M, Sato M, Yano K, Mitsui Y. Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. strain KKS102. J Mol Biol. 1996;255:735–752. doi: 10.1006/jmbi.1996.0060. [DOI] [PubMed] [Google Scholar]
- 37.Shu L, Chiou Y-M, Orville A M, Miller M A, Lipscomb J D, Que L., Jr X-ray absorption spectroscopic studies of the Fe(II) active site of catechol 2,3-dioxygenase. Implications for the extradiol cleavage mechanism. Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 38.Stemmer W P. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taeger K, Knackmuss H-J, Schmidt E. Biodegradability of mixtures of chloro- and methylsubstituted aromatics: simultaneous degradation of 3-chlorobenzoate and 3-methylbenzoate. Appl Microbiol Biotechnol. 1988;28:603–608. [Google Scholar]
- 40.Wieser M, Eberspächer J, Vogler B, Lingens F. Metabolism of 4-chlorophenol by Azotobacter sp. GP1: structure of the meta cleavage product of 4-chlorocatechol. FEMS Microbiol Lett. 1994;116:73–78. doi: 10.1111/j.1574-6968.1994.tb06678.x. [DOI] [PubMed] [Google Scholar]