Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a recently emerged pathogenic human coronavirus that belongs to the sarbecovirus lineage of the genus Betacoronavirus. The ancestor strain has evolved into a number of variants of concern, with the Omicron variant of concern now having many distinct sublineages. The ongoing COVID-19 pandemic caused by SARS-CoV-2 has caused serious damage to public health and the global economy, and one strategy to combat COVID-19 has been the development of broadly neutralizing antibodies for prophylactic and therapeutic use. Many are in preclinical and clinical development, and a few have been approved for emergency use. Here we summarize neutralizing antibodies that target four key regions within the SARS-CoV-2 spike (S) protein, namely the N-terminal domain and the receptor-binding domain in the S1 subunit, and the stem helix region and the fusion peptide region in the S2 subunit. Understanding the characteristics of these broadly neutralizing antibodies will accelerate the development of new antibody therapeutics and provide guidance for the rational design of next-generation vaccines.
Subject terms: SARS-CoV-2, Viral immune evasion, Antibodies
The ancestral strain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has evolved into a number of variants of concern. In this Review, Wang and colleagues discuss progress in the development and characterization of broadly neutralizing antibodies to SARS-CoV-2, which may lead to new antibody therapeutics and inform the design of next-generation vaccines.
Introduction
Following the emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, a novel pathogenic human coronavirus (HCoV) emerged in 2019 that soon spread around the world, resulting in the COVID-19 pandemic1,2. This novel virus was named ‘severe acute respiratory syndrome coronavirus 2’ (SARS-CoV-2) owing to its close sequence homology (~79.6%) with SARS-CoV3–6. Compared with SARS-CoV and MERS-CoV, SARS-CoV-2 has a much lower case–fatality ratio. However, the high proportions of asymptomatic or mildly symptomatic infections caused by the original strain of SARS-CoV-2 and its ensuing variants have led to higher and more rapid transmissibility of this virus, which has resulted in serious complications for all populations of the world7–11.
Coronaviruses belong to the subfamily Coronavirinae from the family Coronaviridae, and they are genotypically and serologically diversified into four major genera: alphacoronaviruses (alpha-CoVs), betacoronaviruses (beta-CoVs), gammacoronaviruses (gamma-CoVs) and deltacoronaviruses (delta-CoVs)5,7. HCoVs are those coronaviruses that can infect humans. Taxonomically, historically occurring HCoV-229E and HCoV-NL63 are classified as alpha-CoVs, whereas HCoV-HKU1, HCoV-OC43, SARS-CoV, SARS-CoV-2 and MERS-CoV are beta-CoVs. Alpha-CoVs and beta-CoVs mainly infect mammals, whereas gamma-CoVs and delta-CoVs primarily infect birds. Both SARS-CoV-2 and SARS-CoV belong to Sarbecovirus, which is a subgenus of Betacoronavirus. By contrast, MERS-CoV belongs to Merbecovirus, another subgenus of Betacoronavirus. Two other HCoVs of note, HCoV-HKU1 and HCoV-OC43, which can cause common cold-like illnesses, belong to the subgenus Embecovirus of Betacoronavirus7,12–14.
HCoVs contain phosphorylated nucleocapsid (N) protein with a single-stranded genomic RNA as a core. The viral core is encapsulated by phospholipid bilayers to form spherical or pleomorphic particles 80–120 nm in size, and is characterized by the presence of the outer surface spike (S) protein7,8. The S protein is composed of two subunits, S1 and S2. S1 contains an important receptor-binding domain (RBD), which is responsible for the recognition of host cell surface receptors that enable virus entry. Both SARS-CoV and SARS-CoV-2 engage angiotensin-converting enzyme 2 (ACE2), which is widely expressed by a variety of human cells, as the primary entry receptor15–17. Dipeptidyl peptidase 4 (DPP4; also known as CD26) is the corresponding entry receptor for MERS-CoV17,18. The S2 subunit is mainly responsible for subsequent viral fusion with and entry into the host cell. The junction of S1 and S2 contains a specific furin cleavage site, which is cleaved by host cell furin to facilitate virus entry into cells19. ACE2 engagement by the virus exposes the S2′ cleavage site, and S2 is further cleaved into two parts at this site by transmembrane serine protease 2 (TMPRSS2) at the cell membrane surface, facilitating the process of membrane fusion between the host cell and the virus20. ACE2-bound virus can also be internalized via endocytosis, and in this case, cleavage of the S2′ site is mediated by cathepsins, especially cathepsin L in endosomes21 (Fig. 1a).
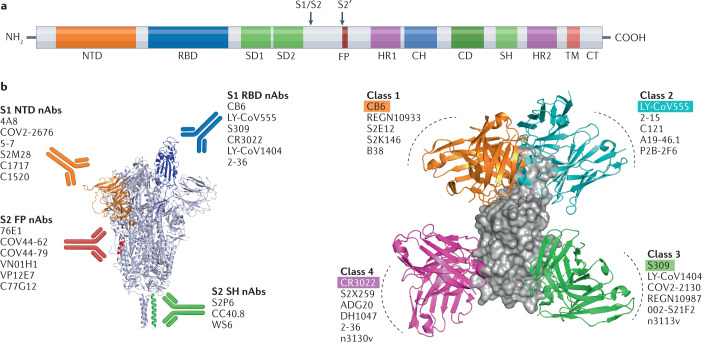
Fig. 1. Neutralizing antibodies directed against the SARS-CoV-2 spike protein.
a | Schematic representation of the main domains of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein. Arrows denote S1/S2 and S2′ protease cleavage sites. b | Different groups of neutralizing antibodies (nAbs) that target the S protein. Representative nAbs targeting the S1 N-terminal domain (NTD), S1 receptor-binding domain (RBD), and S2 stem helix (SH) and S2 fusion peptide (FP) regions are shown with the S protein depicted in the RBD ‘up’ conformation. c | RBD-directed nAbs can be divided into four main classes depending on the epitopes they target in the RBD of the S protein. For each class, one representative nAb bound to the RBD monomer is shown: class 1, CB6; class 2, LY-CoV555; class 3, S309; class 4, CR3022. CD, connector domain; CH, central helix; CT, cytoplasmic tail; HR, heptad repeat; SD, subdomain; TM, transmembrane domain.
Antibodies that recognize pathogens can be categorized as neutralizing antibodies (nAbs) or non-neutralizing antibodies (non-nAbs). The difference between the two generally depends on whether the antibody binding to a specific pathogen can block cell invasion or inhibit membrane fusion after recognition (in the case of nAbs), or not block cell invasion or inhibit membrane fusion after recognition (in the case of non-nAbs)22. In general, nAbs are effective in neutralizing pathogens, reducing pathogen titres and protecting tissues or cells from infection. The neutralization activity of non-nAbs is usually undetectable23–25, but they can exert their protective effects through Fc-mediated effector functions, such as antibody-dependent cell-mediated cytotoxicity, antibody-dependent cellular phagocytosis and complement-dependent cytotoxicity25,26. In this Review, we focus solely on the broadly neutralizing antibodies (bnAbs) that target the neutralizing epitopes in the N-terminal domain (NTD) and the RBD of the S1 subunit, and those that target the stem helix (SH) and fusion peptide (FP) regions in the S2 subunit (Fig. 1b,c).
The NTD
The S1 subunit of the SARS-CoV-2 S protein has two important domains that are targets of monoclonal antibodies (mAbs), namely the NTD and RBD. 4A8 is one of the earliest identified nAbs targeting the NTD, with its heavy chain complementarity-determining regions (HCDRs) — HCDR1, HCDR2 and HCDR3 — interacting with NTD residues27. Chi et al. defined five structural loops (N1–N5) in the NTD, with N3 and N5 mediating interaction with 4A8 (ref.27). Similarly, other NTD-targeting mAbs, such as COV2-2676, COV2-2489, 4-8 and 5-24, can also recognize the epitope comprising the N1, N3 and N5 loops28,29. This strongly positively charged epitope in the NTD was therefore dubbed ‘the NTD supersite’30,31. However, many naturally circulating SARS-CoV-2 variants carry mutations within the NTD supersite, which could dampen the neutralization activities of these NTD supersite-recognizing mAbs. For example, a deletion of NTD amino acid residues 242–244 made 4A8, 4-8 and 5-24 almost completely lose their ability to neutralize the SARS-CoV-2 Beta variant of concern (VOC)32. Similarly, deletion of Y144 in the NTD abolished the neutralization activities of the S2M28 (Fig. 2a), S2X28 and S2X333 nAbs33. Of note, mAbs targeting the non-supersite on the NTD have broad neutralizing potential. For example, mAb 5-7 (Fig. 2a), isolated from a patient recovering from COVID-19 (refs.29,34), retained its neutralizing potency partially against the SARS-CoV-2 Alpha and Beta VOC, as well as against the Omicron BA.1, BA.1.1 and BA.3 sublineages32,35,36. Although some residues in the NTD supersite are highly mutatable under selection pressure, one feasible strategy for developing NTD-targeting bnAbs is to identify 5-7-like mAbs, which bind the non-supersite on the NTD. More importantly, as the bulk of NTD-targeting antibodies did not compete with antibodies targeting other regions of the S protein, such as the RBD27,28,30, combining NTD-targeting antibodies with antibodies binding non-NTD regions may be an ideal way to combat COVID-19 (see Table 1, Supplementary Table S1 and Fig. 2a for a summary of more NTD-targeting nAbs37–40).
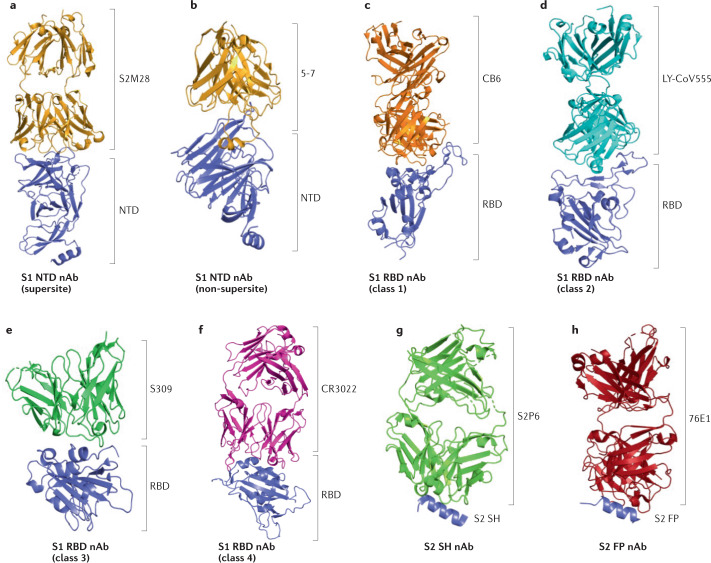
Fig. 2. Structures of neutralizing antibodies bound to the SARS-CoV-2 spike protein.
Three-dimensional modelling is used here to depict the complexes of representative neutralizing antibodies (nAbs) interacting with their targets in the S1 and S2 subunits of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein. a | S1 N-terminal domain (NTD) nAbs, supersite (S2M28, Protein Data Bank (PDB) ID 7LY3). b | S1 NTD nAbs, non-supersite (5-7, PDB ID 7RW2). c | S1 receptor-binding domain (RBD) nAbs, class 1 (CB6, PDB ID 7C01). d | S1 RBD nAbs, class 2 (LY-CoV555, PDB ID 7KMG). e | S1 RBD nAbs, class 3 (S309, PDB ID 7TLY). f | S1 RBD nAbs, class 4 (CR3022, PDB ID 7JN5). g | S2 stem helix (SH) nAb (S2P6, PDB ID 7RNJ). h | S2 fusion peptide (FP) nAb (76E1, PDB ID 7X9E).
Table 1.
Neutralizing antibodies targeting the N-terminal domain of the spike protein
| Antibodies | Binding epitope in NTD | Mechanism of neutralization | Viruses neutralized | Refs. |
|---|---|---|---|---|
| 4A8 | Supersite | Restrains the conformational changes of the S protein | SARS-CoV-2 | 27 |
| COV2-2676, COV2-2489, 5-24, BLN12 | Supersite | Unknown | SARS-CoV-2 | 28,29,34,37,38 |
| 4-8, BLN14 | Supersite | Unknown | SARS-CoV-2; Alpha VOC | 29,34,37,38 |
| 5-7 | Non-supersite | Restrains the conformational changes of the S protein | SARS-CoV-2; Alpha and Beta VOC; BA.1, BA.3 | 29,34 |
| S2M28, S2X28, S2X333 | Supersite | Prevents interaction with an auxiliary receptor, proteolytic activation or membrane fusion | SARS-CoV-2 | 33 |
| C1717 | Non-supersite | Prevents access to the S2′ cleavage site or destabilizes S1 | SARS-CoV-2; Alpha, Beta and Gamma VOC; BA.1 | 40 |
| C1520, C1791 | Non-supersite | Unknown | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1 | 40 |
| ADI-56479 | Supersite | Inhibits the attachment of ACE2 | SARS-CoV-2 | 39 |
The table provides an overview of neutralizing antibodies targeting the N-terminal domain (NTD) of the S1 subunit of the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and related coronaviruses. SARS-CoV-2 indicates the wild type strain. See Supplementary Table S1 for a more detailed description of each antibody. ACE2, angiotensin-converting enzyme 2; VOC, variant of concern.
The RBD
Most of the anti-SARS-CoV-2 antibodies discovered to date target the RBD, and can be further divided into different classes on the basis of their targeted epitopes. Although several different classification systems have been proposed, the most commonly referenced is that proposed by Barnes et al.41, who grouped RBD-targeting antibodies into four classes on the basis of their mode of binding to the S protein (Fig. 1c).
Class 1 RBD-targeting antibodies
The antibody-binding epitope targeted by class 1 RBD-targeting antibodies overlaps with the receptor-binding motif (RBM) in the RBD, and antibodies in this class are mostly encoded by VH3-53 and VH3-66 germ lines and recognize only the ‘up’ RBD conformation41. The substantial neutralization mechanism of class 1 antibodies is achieved by their preventing ACE2 from binding to the S protein. During the early phase of the SARS-CoV-2 outbreak, many class 1 RBD-targeting antibodies were identified, such as CB6 and CT-P59 (refs.42,43), and some were discovered by our own group, including 1-20, 4-20 and 910-30 (refs.29,44). Many of these antibodies lost their neutralizing activities as a result of the E484K mutation seen in the Beta VOC, Gamma VOC and other SARS-CoV-2 variants32,45,46. While the neutralization potency of 1-20 is not apparently affected by the E484K mutation, another common mutation, N501Y, decreases the neutralizing potency of 1-20 almost 10-fold. It is possible that the synergy of various mutants in the Beta VOC resulted in the 600-fold reduction in the neutralizing potency of 1-20 against Beta compared with wild type SARS-CoV-2 (ref.32). CB6 (Fig. 2b) engages in polar and hydrophobic interactions with wild type SARS-CoV-2 S protein primarily through HCDR1, HCDR2 and HCDR3 (ref.42), but its binding epitope is heavily mutated in the S protein of VOC such as Beta and Omicron. In particular, the critical mutation K417N eliminates the neutralizing potency of CB6 and limits its neutralizing breadth32. As a result, the presence of common mutations in the RBD, such as K417N, E484K and N501Y, to which most class 1 mAbs bind, causes most of these antibodies that show great neutralizing potency against wild type SARS-CoV-2 to lose their neutralizing abilities for variants such as Beta, Gamma and Omicron.
Nevertheless, S2E12 is one of the few class 1 mAbs that retains broad-spectrum neutralizing activity47. Although its binding region is similar to that of the other class 1 mAbs mentioned earlier, S2E12 can still neutralize all current VOC, showing exceptional neutralization breadth47,48. The cryogenic electron microscopy structure revealed that S2E12 binds the receptor-binding ridge, contacting 18 residues in the RBD. The S protein F486 residue fills in a buried cavity lined by aromatic residues formed by its interface of heavy and light chains, while residue N487 initiates a polar interaction with S2E12 (ref.47). Although common mutation sites, such as E484 and S477, do exist in S2E12-binding variants, they are not functional residues that interact with S2E12. Moreover, they are located at the edge of the S2E12–RBD interface, conferring considerable neutralizing activity of S2E12 against variants containing these highly frequent mutation sites47. Another ACE2-mimic antibody, S2K146, also demonstrates sizable neutralizing breadth against SARS-CoV-2 variants and other sarbecoviruses49,50. As such, comparison of S2E12 and S2K146 with other class 1 antibodies also highlights the importance of microstructure for an antibody’s neutralizing performance.
Class 2 RBD-targeting antibodies
Antibodies in class 2 are similar to those in class 1 on the basis of their binding to the RBM where the RBD interacts with ACE2. Consistency also derives from the neutralizing mechanism of these antibodies, which prevents the reciprocal binding of the RBD to ACE2. Notably, mAbs in class 2 can bind both ‘up’ and ‘down’ conformations of the S ptotein41. For example, LY-CoV555 (Fig. 2b), which was isolated from a patient recovering from COVID-19, both bound and neutralized SARS-CoV-2 and displayed protective efficacy against SARS-CoV-2 in clinical trials51. Although the antibody neutralized SARS-CoV-2 pseudovirus very effectively (with a half-maximal inhibitory concentration (IC50) of 0.004 μg ml−1 (ref.48)), LY-CoV555 lost most of its neutralization activity against the subsequently discovered VOC, which can be attributed to critical single viral mutations such E484K and Q493R32,36. As a result, the broadly neutralizing capacity of LY-CoV555 was constrained as these mutations were found to be prevalent in many SARS-CoV-2 variants. Similarly, 2-15 (ref.29), another mAb isolated from a patient with COVID-19, is also evaded as a result of the E484K and Q493R mutations, which cause an antigenic structure change32,36. Although the RBD region targeted by class 1 and class 2 antibodies covers the RBM, the amino acid identity of SARS-CoV and SARS-CoV-2 is only 59% shared in the RBM and 94% shared for the rest of the RBD excluding the RBM52. Hence, most antibodies in class 1 and class 2 targeting this region do not have a superior broad spectrum for the inhibition of SARS-CoV and other SARS-like coronaviruses.
Class 3 RBD-targeting antibodies
Class 3 antibodies bind the outside the ACE2-binding region, and they can also bind to RBDs regardless of their ‘up’ and ‘down’ conformations41. Most class 3 antibodies, including REGN10987, COV2-2130, 2-7,1-57, A19-61.1, P2G3, S309 and LY-CoV1404, have demonstrated potent neutralizing activities against SARS-CoV-2 variants29,48,53–60. S309 and LY-CoV1404 are two representative antibodies with great neutralization breadth (refs.56,57). S309 (Fig. 2b) was isolated from a patient recovering from SARS and can efficiently bind to the SARS-CoV-2 S protein without blocking the binding between ACE2 and the RBD56. Structurally, S309 HCDR3 was shown by cryogenic electron microscopy to interact mainly with eight residues (337–344) of the RBD helix, as well as with six residues of the RBD β-sheet (356–361). In addition, the S309 light chain complementarity-determining region 1 (LCDR1) and LCDR2 directly interact with the epitope of the S protein that spans residues 440–444 (ref.56). The epitopes recognized by S309 are the most highly conserved residues in the SARS-CoV and SARS-CoV-2 RBDs, conferring broad cross-reactivity on the S309 antibody56. Moreover, S309 can broadly neutralize sarbecoviruses, including all currently identified SARS-CoV-2 VOC, as the epitope sequences recognized by S309 are highly conserved among sarbecoviruses48,56. Interestingly, although the single mutation S371L in SARS-CoV-2 causes the loss of S309’s neutralizing potency, the detrimental effect of S371L on S309 was counteracted by the combined effect of amino acids near the point of mutation in Omicron such that S309 can still effectively neutralize Omicron (BA.1), with IC50 = 0.28 μg ml−1 (refs.48,56).
Similarly, LY-CoV1404 can also bind to the viral RBDs, irrespective of their ‘up’ and ‘down’ conformations. However, LY-CoV1404 differs from S309 in that it binds a part of the RBD epitope that overlaps with the ACE2-binding domain57. As the epitope bound by LY-CoV1404 is structurally closer to that recognized by class 3 antibodies, LY-CoV1404 also belongs to the class 3 antibody group57. The neutralization mechanism of LY-CoV1404 is achieved by its preventing ACE2 from binding to the RBD57. Of note, the epitope bound by LY-CoV1404 is also conserved in SARS-CoV-2 and its variants. Although the epitope targeted by LY-CoV1404 was associated with two high-frequency mutations at positions 439 and 501, these two mutations did not affect the binding of LY-CoV1404 to and its neutralizing potency against SARS-CoV-2 and its variants containing these mutations57. Compared with S309, LY-CoV1404 was able to neutralize all SARS-CoV-2 VOC without much change in neutralization potency, especially against the Omicron variants48. Among the antibodies approved for clinical trials, LY-CoV1404 was the only antibody that retained its neutralization potency against Omicron sublineages35,36,48,57,61,62. Taken together, these findings suggest LY-CoV1404 is a specific and effective mAb for the treatment of COVID-19.
Recently, another class 3 mAb, named ‘SP1-77’, was obtained from a humanized mouse model (the Vh1–2/Vκ1–33-rearranging mouse model). SP1-77 showed potent neutralization activity against all currently known SARS-CoV-2 variants, including the recently emerging Omicron variant BA.1 (IC50 = 6.5 ng ml−1), and its sublineages BA.2 (IC50 = 33 ng ml−1), BA.3 (IC50 = 7 ng ml−1), BA.4/BA.5 (IC50 = 16 ng ml−1) and BA.2.12.1 (IC50 = 8 ng ml−1)63. Interestingly, SP1-77 does not block the RBD–ACE2 binding interaction or viral endocytosis, but instead mediates virus neutralization by preventing shedding of S1, which blocks membrane fusion63. This study provides insight into how a non-ACE2 blocking antibody can also potently neutralize SARS-CoV-2 infection.
Class 4 RBD-targeting antibodies
The epitope recognized by class 4 antibodies is highly conserved in the RBD, and these antibodies bind to the RBD, but do not directly block ACE2–RBD binding. This epitope has also been described as a cryptic region, which is consistent with the well-described cryptic epitope recognized by the CR3022 antibody (Fig. 2b), an antibody isolated from a patient who had recovered from SARS-CoV infection41,64. The epitope targeted by class 4 antibodies is conserved by up to 86% in SARS-CoV and SARS-CoV-2, and CR3022 is thus able to effectively bind to both coronaviruses64. Owing to a glycosylation site on N370 within the targeted epitope in SARS-CoV, CR3022 binds to SARS-CoV with greater affinity than to SARS-CoV-2. Of note, CR3022 can bind to SARS-CoV-2 only when at least two RBDs are in the ‘up’ conformation, which partly explains its lower binding affinity for SARS-CoV-2 and poor neutralizing potency against this virus (IC50 > 400 μg ml−1)64.
The 2-36 antibody identified by our group competes with CR3022 for binding to the SARS-CoV-2 RBD, and is therefore classified as a class 4 antibody29,65. Similarly to CR3022, 2-36 also recognizes and binds the RBD in the ‘up’ conformation. Cryogenic electron microscopy data revealed that HCDR3 of 2-36 forms the majority of interactions by recognizing loops on the RBD spanning residues 369–385, whereas HCDR1 and LCDR2 of the antibody interact to a lesser extent with the RBD65. The interaction between 2-36 and the SARS-CoV-2 RBD is dependent mainly on hydrophobic effects, as HCDR3 of 2-36 contains a large number of hydrophobic amino acids. Compared with CR3022, 2-36 can more effectively neutralize SARS-CoV, SARS-CoV-2 and related sarbecoviruses that use ACE2 as the entry receptor, with IC50 < 0.1 μg ml−1 in both authentic virus and pseudovirus assays65. 2-36 retained its neutralization potency against the SARS-CoV-2 Alpha, Beta, Gamma and Delta VOC65, and retained partial activity against Omicron BA.1, with IC50 ~ 1 μg ml−1 (refs.35,36). Taken together, these findings indicate that 2-36, as a bnAb to SARS-CoV-2 and related sarbecoviruses, could be a specific drug candidate for the treatment or prevention of COVID-19 after proper engineering, and its binding epitope is a promising target for the development of pan-sarbecovirus vaccines.
Another class 4 antibody, S2X259, can also broadly neutralize SARS-CoV-2 and related sarbecoviruses66. A very recent study demonstrated that S2X259 lost its neutralizing activity against SARS-CoV-2 strains containing the G504D mutant66. Fortunately, the G504D substitution is rarely observed in SARS-CoV-2 strains, with a mutant rate below 0.002%66. This antibody can neutralize not only many currently circulating SARS-CoV-2 variants and SARS-CoV but also a panel of SARS-related sarbecoviruses. Other known class 4 antibodies, such as ADG20, DH1047, COVA1-16 and H014, also demonstrate neutralizing breadth against SARS-CoV-2, SARS-CoV and other relevant sarbecoviruses67–71. This suggests that the epitope recognized by class 4 antibodies is an ideal target for the development of bnAbs.
In summary, RBD-targeting antibodies in class 1 and class 2 will probably lose their neutralizing abilities with the emergence of the next major SARS-CoV-2 variant that carries new mutations in the RBM; thus, their neutralizing breadth is limited. By contrast, antibodies in class 3 and class 4 that bind highly conserved epitopes hold promise as candidates for neutralizing SARS-CoV-2 variants and other SARS-like coronaviruses. This suggests that selecting such conserved epitopes for vaccine design may elicit potent broad-spectrum antibodies that could help to overcome the current COVID-19 pandemic. In addition, it is clear that epitopes of RBD-targeting antibodies in class 1 and class 2 overlap with the ACE2 footprint on the RBD, and these antibodies achieve neutralizing activities by directly blocking the interaction between the RBD and ACE2. However, the major class 3 and class 4 antibodies do not show such explicit neutralization mechanisms, and their mechanisms of neutralization need to be further explored. Studies of antibodies such as SP1-77 may provide insight into the neutralizing mechanisms of other, similar RBD-binding (but non-RBM-targeting) antibodies (See Table 2, Supplementary Table S2 and Fig. 2b for a summary of more RBD-targeting nAbs58–62,72–81).
Table 2.
Neutralizing antibodies targeting the receptor-binding domain of the spike protein
| Antibodies | Binding epitope in RBD | Mechanism of neutralization | Viruses neutralized | Refs. |
|---|---|---|---|---|
| 1-20, 4-20, 910-30, CB1 | Class 1 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2 | 29,42,44 |
| CB6, CC12.3 | Class 1 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha and Delta VOC | 42,48,67,72 |
| REGN10933 | Class 1 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha and Delta VOC; BA.2.75 | 48,53,62 |
| CT-P59 | Class 1 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC | 43,48 |
| A23-58.1, S2E12 | Class 1 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.2.75 | 47–49,58,81 |
| S2K146 | Class 1 | Blocks the interaction between the RBD and ACE2 | Sarbecoviruses (SARS-CoV; SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.3, BA.4/5, BA.2.75) | 49,50,80,81 |
| B38 | Class 1 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Beta VOC | 73 |
| 2-15, LY-CoV555, C121, C144 | Class 2 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha VOC | 29,32,48,51,74 |
| COV2-2196 | Class 2 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.3, BA.2.75 | 48,54,61,62,80 |
| A19-46.1 | Class 2 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta and Gamma VOC | 48,58 |
| P2B-2F6 | Class 2 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2 | 77 |
| S309 | Class 3 | Leads to trimeric S protein crosslinking, causes steric hindrance or aggregation of virions | Sarbecoviruses (SARS-CoV; SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.3, BA.4/5, BA.2.75) | 36,48,56,61,62 |
| LY-CoV1404 | Class 3 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.3, BA.4/5, BA.2.75 | 36,48,57,61,62 |
| COV2-2130 | Class 3 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.3, BA.4/5, BA.2.75 | 48,54,61,62,80 |
| REGN10987 | Class 3 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.4/5 | 48,53,61,62 |
| SP1-77 | Class 3 | Prevents the shedding of S1 and blocks membrane fusion | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.3, BA.4/5 | 63 |
| A19-61.1 | Class 3 | Causes steric hindrance between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1 | 48,58 |
| 1-57 | Class 3 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta and Gamma VOC | 29,32,46,55 |
| 2-7 | Class 3 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta and Gamma VOC; BA.1, BA.2, BA.4/5 | 29,32,46,55,61 |
| 002-S21F2 | Class 3 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2 | 76 |
| P2G3 | Class 3 | Blocks the interaction between the RBD and ACE2 | SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1, BA.2, BA.4/5 | 59,60 |
| n3113v | Class 3 | Inhibits SARS-CoV-2 S protein-mediated membrane fusion | SARS-CoV-2; Alpha, Beta, Gamma, Delta and Omicron VOC | 78,79 |
| CR3022 | Class 4 | Unknown | SARS-CoV | 64 |
| 2-36 | Class 4 | Causes steric hindrance between the RBD and ACE2 | Sarbecoviruses (SARS-CoV; SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1) | 29,36,65 |
| S2X259 | Class 4 | Blocks the interaction between the RBD and ACE2 | Sarbecoviruses (SARS-CoV; SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1) | 50,66 |
| ADG20 | Class 4 | Competes with ACE2 for RBD binding | Sarbecoviruses (SARS-CoV; SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1) | 67,68 |
| DH1047 | Class 4 | Unknown | Sarbecoviruses (SARS-CoV; SARS-CoV-2; Alpha, Beta, Gamma and Delta VOC; BA.1) | 61,69 |
| COVA1-16 | Class 4 | Causes steric hindrance between the RBD and ACE2 | Sarbecoviruses (SARS-CoV; SARS-CoV-2) | 70 |
| H014 | Class 4 | Prevents attachment of SARS-CoV-2 to ACE2 | Sarbecoviruses (SARS-CoV; SARS-CoV-2; Beta VOC) | 71 |
| EY6A | Class 4 | Interferes with ACE2 attachment | SARS-CoV-2 | 75 |
| n3130v | Class 4 | Induces S protein trimer to adopt unstable ‘up’ states | SARS-CoV-2; Alpha, Beta, Gamma, Delta and Omicron VOC | 78 |
The table provides an overview of neutralizing antibodies targeting the receptor-binding domain (RBD) of the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and related coronaviruses. ‘SARS-CoV-2’ indicates the wild-type strain. See Supplementary Table S2 for a more detailed description of each antibody. ACE2, angiotensin-converting enzyme 2; VOC, variant of concern.
The S2 SH region
The SARS-CoV-2 S protein is composed of S1 and S2 subunits. Most SARS-CoV-2 nAbs target the neutralizing epitopes in the NTD and RBD in the S1 subunit. However, such epitopes are likely to succumb to selective pressure, increasing the likelihood of immune escape by virus mutants. By contrast, the neutralizing epitopes in the S2 subunit are more conserved than those in the S1 subunit82. Therefore, nAbs targeting the S2 epitopes would have a greater probability of being broad-spectrum nAbs to SARS-CoV-2 and other HCoVs. For example, S2P6 (Fig. 2c), which was isolated from a patient recovering from COVID-19, could broadly neutralize all beta-CoVs by targeting the S2 subunit83. Further study revealed that the epitope bound by this antibody is located in the S2 SH region that spans 14 residues (1146–1159), which is conserved across beta-CoVs. Moreover, the S2 SH region interacts with the S2P6 antibody’s HCDR1, HCDR2 and HCDR3, as well as with its LCDR1 and LCDR3, mainly through the formation of hydrophobic interactions and hydrogen bonds. Single-substitution analysis revealed that mutations at positions 1148, 1151–1153 and 1155–1156 abolished S2P6 binding affinity, suggesting that these are critical residues for S2P6 binding83. Unlike the neutralization mechanism of antibodies targeting S1, S2P6 can inhibit SARS-CoV-2 infection by preventing S protein-mediated fusion of viral and cellular membrane83. The Fc effector functions of S2P6 also play a critical role in fighting coronavirus infection in vivo83. Several studies reported that S2P6 can broadly neutralize beta-CoVs, including SARS-CoV-2 and SARS-like viruses, which belong to the subgenus Sarbecovirus, MERS-CoV, which belongs to subgenus Merbecovirus, and HCoV-HKU1 and HCoV-OC43, which belong to the subgenus Embecovirus. S2P6 shows variable IC50 of 1.4 μg ml−1 for SARS-CoV-2, 2.4 μg ml−1 for SARS-CoV, 17.1 μg ml−1 for MERS-CoV and 1.3 µg ml−1 for HCoV-OC43 (ref.83). Although S2P6 does not show as great neutralizing potency as some RBD-targeting antibodies, it still has the potential to become a very effective antibody drug for the treatment of COVID-19 or diseases caused by other coronaviruses owing to its broad-spectrum neutralizing properties.
Another antibody targeting the S2 SH region, CC40.8, was isolated from a patient with COVID-19 (ref.84). It binds to residues from 1140 to 1164, and was found to broadly bind and neutralize beta-CoVs84. An in vivo study in hamsters confirmed that CC40.8 mediates effective protection against SARS-CoV-2 infection84. WS6, an antibody isolated from an mRNA-immunized mouse, also binds the SARS-CoV-2 S2 SH region that spans residues 1143–1159 (ref.85). Mechanistically, WS6 can neutralize SARS-CoV-2 by inhibiting the membrane fusion process following virus contact with ACE2 (ref.85). As expected, WS6 can also broadly neutralize beta-CoVs, and pseudovirus neutralization experiments showed its neutralizing potency against all SARS-CoV-2 VOC, with IC50 ranging from 2.46 to 26.52 μg ml−1 (ref.85). The three bnAbs mentioned here all target the S2 SH epitope, highlighting the importance of finding and developing bnAbs that recognize these conserved epitopes.
The S2 FPs
Apart from the SH region mentioned earlier, S2 FPs are also highly conserved among all coronavirus genera, suggesting that broad-spectrum antibodies could be found by targeting this epitope86–88. Some recently identified antibodies to this epitope have excellent broadly neutralizing activity against alpha-CoVs, beta-CoVs and even some gamma-CoVs and delta-CoVs86,87. For example, COV44-62 and COV44-79, which were both isolated from patients recovering from COVID-19, can bind the S2 FP region through recognition of the ‘RSFIEDLLF’ motif. Interestingly, these antibodies do not compete with S2P6, the aforementioned S2 SH-targeting antibody, for binding to the SARS-CoV-2 S protein86, suggesting the possibility of combining S2 SH and S2 FP recognition in a bispecific antibody. Crystal structure analysis revealed that COV44-62 interacts with S2, that spans residues 814–824, mainly through hydrogen bonds, salt bridges and hydrophobic interactions within HCDR1, HCDR2, HCDR3, LCDR1 and LCDR3. Similarly, COV44-79 utilizes HCDR1, HCDR2, HCDR3 and LCDR3 to bind SARS-CoV-2 and interacts with S2 residues 812–823 (ref.86). Of note, both antibodies broadly neutralize beta-CoVs, but show some differences in activity against MERS-CoV. The neutralizing activity of COV44-62 against MERS-CoV could be detected, whereas that of COV44-79 could not. These antibodies can also act against the more distantly evolved alpha-CoVs, including HCoV-NL63 and HCoV-229E, although they are less potent at neutralizing these viruses, with IC50 ~ 10 μg ml−1 (ref.86). Peptide alanine scanning revealed that E819, D820, L822 and F823 in the S2 FP region are crucial for COV44-62 binding, whereas R815, E819, D820 and F823 are crucial for COV44-79 binding86. More importantly, the entire FP sequence is highly conserved among the four coronavirus genera86, giving antibodies targeting this region an extremely broad ability to neutralize SARS-CoV-2 and numerous other coronaviruses.
Another panel of nAbs targeting S2 FP — VN01H1, VP12E7 and C77G12 — were recently identified87. Among them, VN01H1 and VP12E7 can neutralize all alpha-CoV and beta-CoV pseudoviruses, including HCoV-NL63, HCoV-229E, SARS-CoV, SARS-CoV-2 and MERS-CoV; C77G12 exhibited higher neutralization potency than VN01H1 and VP12E7, but only against beta-CoVs87. Crystal structures showed that all three antibodies concealed the R815 residue at the interface when interacting with the S protein. Furthermore, R815 is not only a conserved residue in the S protein but is also the site of cleavage of S2′ by TMPRSS2, suggesting that these three antibodies neutralize SARS-CoV-2 by preventing S2′ cleavage, thereby inhibiting subsequent membrane fusion87. Intriguingly, a potent synergistic neutralization activity was found when C77G12 was combined with S2E12, a nAb in RBD class 1, implying that the FP is most likely a cryptic epitope that is normally inaccessible, but when the RBD interacts with ACE2 or ACE2-mimic antibodies, the exposed epitope is more accessibly bound by FP-targeting antibodies87.
More recently, 76E1 (Fig. 2d) was identified as another S2 FP-targeting nAb that shows extraordinary neutralizing breadth, including for alpha-CoVs, beta-CoVs and a few gamma-CoVs and delta-CoVs88. It has been proved that 76E1 can potently neutralize SARS-CoV-2 both in vitro and in vivo88. Like C77G12, a synergistic effect of 76E1 can be observed when the S protein contacts some RBD-targeting nAbs as well as ACE2 (ref.88). Taken together, these findings show that S2-directed antibodies target prominently conserved epitopes and exhibit the broadest neutralizing spectrum to date. Therefore, they can guide the design of bnAbs to highly variable SARS-CoV-2 variants and even other HCoVs (see Table 3, Supplementary Table S3 and Figs. 2c,d for a summary of the S2-targeting nAbs mentioned above).
Table 3.
Neutralizing antibodies targeting the S2 subunit of the spike protein
| Antibodies | Binding epitope in S2 subunit | Mechanism of neutralization | Viruses neutralized | Refs. |
|---|---|---|---|---|
| S2P6 | S2 stem helix | Inhibits membrane fusion | Beta-CoVs (sarbecoviruses, merbecoviruses and embecoviruses) | 83 |
| CC40.8 | S2 stem helix | Inhibits membrane fusion | Beta-CoVs (sarbecoviruses, HCoV-HKU1) | 84 |
| WS6 | S2 stem helix | Inhibits membrane fusion | Beta-CoVs (sarbecoviruses) | 85 |
| COV44-79 | S2 fusion peptide | Inhibits membrane fusion | Alpha-CoVs and beta-CoVs (except MERS-CoV) | 86 |
| COV44-62, VN01H1, VP12E7, 76E1 | S2 fusion peptide | Inhibits membrane fusion | Alpha-CoVs and beta-CoVs | 86–88 |
| C77G12 | S2 fusion peptide | Inhibits membrane fusion | Beta-CoVs | 87 |
The table provides an overview of neutralizing antibodies targeting the S2 subunit of the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and related coronaviruses. See Supplementary Table S3 for a more detailed description of each antibody. alpha-CoV, alphacoronavirus; beta-CoV, betacoronavirus; HCoV, human coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus.
Conclusion
The engineering of already discovered antibodies can be used to improve their performance, including their neutralizing potency and breadth. One approach is to construct libraries containing a large number of different Vh/Vl sequences by phage or yeast display methods to yield antibodies with better performance89–92. Another approach is to try different formats of antibodies, such as nanobodies. In contrast to human IgG, camelid antibodies (also known as nanobodies or Vhhs), lack a light chain and are composed of two identical heavy chains. The nanobody is the smallest antigen-binding fragment (~15 kDa) discovered to date93–95. Owing to their smaller size, nanobodies can bind more cryptic epitopes that are not easily accessible. Together with their high tissue penetration ability, they can also be formulated as an inhalable atomized powder78,96.
Other effective approaches to improve antibody function include engineering them into bispecific or multispecific antibodies97–100, which can bind to multiple epitopes simultaneously and synergistically. Combining two or more bnAbs targeting different epitopes as therapeutics (for example, different RBD class antibodies) or combining RBD-targeting antibodies with NTD-targeting and/or S2-targeting antibodies might be a feasible strategy against COVID-19. Some antibody cocktails, such as S2E12 and C77G12, 76E1 and CB6, and COV2-2196 and COV2-2130, have already been explored54,87,88, but more combinations need to be tested in clinical trials. Furthermore, some ACE2-targeting antibodies have been reported to confer protection in animal models against infection by SARS-CoV-2 and other SARS-like coronaviruses by competing with the S protein for receptor binding101,102, and these antibodies could be used in combination with the S protein-targeting antibodies. Therefore, we can take advantage of the synergistic enhancement between antibodies to make them exert better neutralization and protective effects in vivo.
The emergence of numerous SARS-CoV-2 variants calls for the generation of bnAbs as a therapy for COVID-19. In this Review, we have described some representative nAbs that bind the NTD and RBD in the S1 subunit and the SH and FP regions in the S2 subunit of SARS-CoV-2 and have summarized more known nAbs in Tables 1–3. Overall, antibodies in RBD class 1 and class 2, as well as those targeting the NTD supersite, are more likely to lose their neutralizing activities as the viral epitopes they target are more prone to mutate. By contrast, antibodies targeting the more conserved S2 epitopes are able to exert an incredibly broad neutralization spectrum against HCoVs, including alpha-CoVs and beta-CoVs. However, despite their having a broader neutralization spectrum, it is important to point out that the S2 antibodies are much less potent than RBD-targeting antibodies. Therefore, these findings put great emphasis on balancing the breadth and potency of nAbs when one is selecting nAbs as candidates for antibody therapy for COVID-19. On the other hand, comprehending the characteristics of these bnAbs could provide guidance for devising more effective vaccines. Consistent with others103–105, we stress the importance of focusing on conserved viral epitopes for the development of broad-spectrum antibody therapies, as well as for vaccine design. In general, the immunogens with conserved epitopes — such as the FP and SH regions in the S2 subunit83,86–88 — are unable to elicit potent nAb responses, possibly owing to their inappropriate conformation and/or low immunogenicity. Therefore, vaccines containing different conformations of these immunogens should be tested in combination with a highly potent adjuvant, such as the STING agonist-based adjuvant CF501 (ref.106). Another feasible strategy for the design of next-generation vaccines would be heterologous or multivalent immunization with S proteins from different HCoVs, which might induce the host immune system to generate bnAbs to highly conserved viral epitopes present in these coronaviruses.
Supplementary information
Acknowledgements
This work was sponsored by the National Natural Science Foundation of China (32270142) and the Shanghai Rising-Star Program (22QA1408800) to P.W. X.Z. acknowledges support from the International Postdoctoral Exchange Fellowship Program (Talent-Introduction Program, YJ20220071).
Glossary
- Furin
A protease belonging to the proprotein convertase family that processes latent precursor proteins into biologically active products.
- Transmembrane serine protease 2
(TMPRSS2). A plasma membrane-anchored serine protease that proteolytically cleaves and activates the spike (S) glycoproteins of human coronaviruses and some other viruses.
- Cathepsins
A family of proteases that are responsible for recycling cellular proteins inside the lysosomes, comprising serine, aspartate and cysteine peptidases, and that exhibit endopeptidase or exopeptidase activities.
- Stem helix
(SH). A helix structure in the S2 stem region that forms part of the spike fusion machinery and is conserved among multiple betacoronaviruses.
- Fusion peptide
(FP). A conserved hydrophobic domain of a fusion protein that inserts itself into membranes during membrane fusion, which is required for the fusogenic activity of glycoproteins from divergent virus families.
- STING agonist
A modulator of stimulator of interferon genes (STING) that can facilitate the phosphorylation of the transcription factor interferon regulatory factor 3 (IRF3), resulting in an increase in the expression of type I interferon genes, through the binding of STING to cyclic GMP–AMP (cGAMP).
Author contributions
P.W. and S.J. conceived ideas. Y.C. X.Z. and P.W. wrote the article. H. Zhou, H. Zhu and S.J. reviewed and edited the manuscript. Y.C. created the tables and H. Zhou prepared the figures with suggestions from P.W. and S.J. All authors reviewed and approved the manuscript before submission.
Peer review
Peer review information
Nature Reviews Immunology thanks S. Liu, Y. Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
P.W. has filed patent applications for antibodies 4-8,5-24, 5-7,1-20, 4-20, 910-30, 2-15, 2-7,1-57 and 2-36. The other authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanjia Chen, Xiaoyu Zhao, Hao Zhou.
Contributor Information
Shibo Jiang, Email: shibojiang@fudan.edu.cn.
Pengfei Wang, Email: pengfei_wang@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41577-022-00784-3.
References
- 1.Mackay IM, Arden KE. MERS coronavirus: diagnostics, epidemiology and transmission. Virol. J. 2015;12:222. doi: 10.1186/s12985-015-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stadler K, et al. SARS–beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Umakanthan S, et al. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19) Postgrad. Med. J. 2020;96:753–758. doi: 10.1136/postgradmedj-2020-138234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirtipal N, Bharadwaj S, Kang SG. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020;85:104502. doi: 10.1016/j.meegid.2020.104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang MY, et al. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell Infect. Microbiol. 2020;10:587269. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaworski JP. Neutralizing monoclonal antibodies for COVID-19 treatment and prevention. Biomed. J. 2021;44:7–17. doi: 10.1016/j.bj.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oran DP, Topol EJ. The proportion of SARS-CoV-2 infections that are asymptomatic: a systematic review. Ann. Intern. Med. 2021;174:655–662. doi: 10.7326/M20-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma A, Ahmad Farouk I, Lal SK. COVID-19: a review on the novel coronavirus disease evolution, transmission, detection, control and prevention. Viruses. 2021;13:202. doi: 10.3390/v13020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo PC, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier RA, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl Acad. Sci. USA. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llanes A, et al. Betacoronavirus genomes: how genomic information has been used to deal with past outbreaks and the COVID-19 pandemic. Int. J. Mol. Sci. 2020;21:4546. doi: 10.3390/ijms21124546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W. Delving deep into the structural aspects of a furin cleavage site inserted into the spike protein of SARS-CoV-2: a structural biophysical perspective. Biophys. Chem. 2020;264:106420. doi: 10.1016/j.bpc.2020.106420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bestle D, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3:e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayati A, Kumar R, Francis V, McPherson PS. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021;296:100306. doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moog C, et al. Protective effect of vaginal application of neutralizing and nonneutralizing inhibitory antibodies against vaginal SHIV challenge in macaques. Mucosal Immunol. 2014;7:46–56. doi: 10.1038/mi.2013.23. [DOI] [PubMed] [Google Scholar]
- 23.Cheeseman HM, et al. Broadly neutralizing antibodies display potential for prevention of HIV-1 infection of mucosal tissue superior to that of nonneutralizing antibodies. J. Virol. 2017;91:e01762-16. doi: 10.1128/JVI.01762-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan GS, et al. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 2016;12:e1005578. doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F, et al. Generation of neutralizing and non-neutralizing monoclonal antibodies against H7N9 influenza virus. Emerg. Microbes Infect. 2020;9:664–675. doi: 10.1080/22221751.2020.1742076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alter G, Moody MA. The humoral response to HIV-1: new insights, renewed focus. J. Infect. Dis. 2010;202:S315–S322. doi: 10.1086/655654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi X, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suryadevara N, et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 30.Cerutti G, et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lok SM. An NTD supersite of attack. Cell Host Microbe. 2021;29:744–746. doi: 10.1016/j.chom.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 33.McCallum M, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerutti G, et al. Neutralizing antibody 5-7 defines a distinct site of vulnerability in SARS-CoV-2 spike N-terminal domain. Cell Rep. 2021;37:109928. doi: 10.1016/j.celrep.2021.109928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, et al. Homologous or heterologous booster of inactivated vaccine reduces SARS-CoV-2 Omicron variant escape from neutralizing antibodies. Emerg. Microbes Infect. 2022;11:477–481. doi: 10.1080/22221751.2022.2030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ai J, et al. Antibody evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell Host Microbe. 2022;30:1077–1083. doi: 10.1016/j.chom.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makdasi E, et al. The neutralization potency of anti-SARS-CoV-2 therapeutic human monoclonal antibodies is retained against viral variants. Cell Rep. 2021;36:109679. doi: 10.1016/j.celrep.2021.109679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noy-Porat T, et al. Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice. iScience. 2021;24:102479. doi: 10.1016/j.isci.2021.102479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haslwanter D, et al. A combination of receptor-binding domain and N-terminal domain neutralizing antibodies limits the generation of SARS-CoV-2 spike neutralization-escape mutants. mBio. 2021;12:e0247321. doi: 10.1128/mBio.02473-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, et al. Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins. Immunity. 2022;55:998–1012. doi: 10.1016/j.immuni.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes CO, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi R, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 43.Kim C, et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banach BB, et al. Paired heavy- and light-chain signatures contribute to potent SARS-CoV-2 neutralization in public antibody responses. Cell Rep. 2021;37:109771. doi: 10.1016/j.celrep.2021.109771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Annavajhala MK, et al. Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York. Nature. 2021;597:703–708. doi: 10.1038/s41586-021-03908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starr TN, et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou T, et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science. 2022;376:eabn8897. doi: 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park YJ, et al. Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science. 2022;375:449–454. doi: 10.1126/science.abm8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cameroni E, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen P, et al. SARS-CoV-2 Neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen WH, Hotez PJ, Bottazzi ME. Potential for developing a SARS-CoV receptor-binding domain (RBD) recombinant protein as a heterologous human vaccine against coronavirus infectious disease (COVID)-19. Hum. Vaccin. Immunother. 2020;16:1239–1242. doi: 10.1080/21645515.2020.1740560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen J, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zost SJ, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cerutti G, et al. Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies. Structure. 2021;29:655–663. doi: 10.1016/j.str.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinto D, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 57.Westendorf K, et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022 doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 2021;373:eabh1766. doi: 10.1126/science.abh1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fenwick C, et al. Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys. Nat. Microbiol. 2022 doi: 10.1016/10.1038/s41564-022-01198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turelli P, et al. P2G3 human monoclonal antibody neutralizes SARS-CoV-2 Omicron subvariants including BA.4 and BA.5 and Bebtelovimab escape mutants. bioRxiv. 2022 doi: 10.1101/2022.07.28.501852. [DOI] [Google Scholar]
- 61.Wang Q, et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao Y, et al. Characterizations of enhanced infectivity and antibody evasion of Omicron BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.18.500332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo S, et al. An Antibody from Single Human VH-rearranging Mouse Neutralizes All SARS-CoV-2 Variants Through BA.5 by Inhibiting Membrane Fusion. Sci. Immuno. 2022 doi: 10.1126/sciimmunol.add5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan M, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang P, et al. A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses. Emerg. Microbes Infect. 2022;11:147–157. doi: 10.1080/22221751.2021.2011623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tortorici MA, et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yuan M, et al. A broad and potent neutralization epitope in SARS-related coronaviruses. Proc. Natl Acad. Sci. USA. 2022;119:e2205784119. doi: 10.1073/pnas.2205784119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rappazzo CG, et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez DR, et al. A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. Med. 2022;14:eabj7125. doi: 10.1126/scitranslmed.abj7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu H, et al. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020;53:1272–1280. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lv Z, et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan M, et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Y, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robbiani DF, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou D, et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 76.Kumar S, et al. Structural insights for neutralization of BA.1 and BA.2 Omicron variants by a broadly neutralizing SARS-CoV-2 antibody. bioRxiv. 2022 doi: 10.1101/2022.05.13.491770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ju B, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 78.Li C, et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell. 2022;185:1389–1401. doi: 10.1016/j.cell.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Z, et al. A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants. Signal. Transduct. Target. Ther. 2021;6:378. doi: 10.1038/s41392-021-00810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao Y, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheward DJ, et al. Evasion of neutralizing antibodies by Omicron sublineage BA.2.75. Lancet Infect. Dis. 2022 doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shrestha LB, Tedla N, Bull RA. Broadly-neutralizing antibodies against emerging SARS-CoV-2 variants. Front. Immunol. 2021;12:752003. doi: 10.3389/fimmu.2021.752003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pinto D, et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou P, et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci. Transl. Med. 2022;14:eabi9215. doi: 10.1126/scitranslmed.abi9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shi W, et al. Vaccine-elicited murine antibody WS6 neutralizes diverse beta-coronaviruses by recognizing a helical stem supersite of vulnerability. Structure. 2022 doi: 10.1016/j.str.2022.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dacon C, et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science. 2022;377:728–735. doi: 10.1126/science.abq3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Low JS, et al. ACE2 engagement exposes the fusion peptide to pan-coronavirus neutralizing antibodies. Science. 2022;377:735–742. doi: 10.1126/science.abq2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun X, et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat. Microb. 2022;7:1063–1074. doi: 10.1038/s41564-022-01155-3. [DOI] [PubMed] [Google Scholar]
- 89.Zhao F, et al. Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape. iScience. 2022 doi: 10.1016/j.isci.2022.104914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Z, et al. Extremely potent monoclonal antibodies neutralize Omicron and other SARS-CoV-2 variants. medRxiv. 2022 doi: 10.1101/2022.01.12.22269023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wellner A, et al. Rapid generation of potent antibodies by autonomous hypermutation in yeast. Nat. Chem. Biol. 2021;17:1057–1064. doi: 10.1038/s41589-021-00832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rouet R, et al. Potent SARS-CoV-2 binding and neutralization through maturation of iconic SARS-CoV-1 antibodies. MAbs. 2021;13:1922134. doi: 10.1080/19420862.2021.1922134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu M, Li L, Jin D, Liu Y. Nanobody-A versatile tool for cancer diagnosis and therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021;13:e1697. doi: 10.1002/wnan.1697. [DOI] [PubMed] [Google Scholar]
- 94.Mitchell LS, Colwell LJ. Comparative analysis of nanobody sequence and structure data. Proteins. 2018;86:697–706. doi: 10.1002/prot.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stanfield RL, Wilson IA. Antibody structure. Microbiol. Spectr. 2014 doi: 10.1128/microbiolspec.AID-0012-2013. [DOI] [PubMed] [Google Scholar]
- 96.Schoof M, et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.De Gasparo R, et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature. 2021;593:424–428. doi: 10.1038/s41586-021-03461-y. [DOI] [PubMed] [Google Scholar]
- 98.Cho H, et al. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021;13:eabj5413. doi: 10.1126/scitranslmed.abj5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ku Z, et al. Engineering SARS-CoV-2 cocktail antibodies into a bispecific format improves neutralizing potency and breadth. bioRxiv. 2022 doi: 10.1101/2022.02.01.478504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, et al. An engineered bispecific human monoclonal antibody against SARS-CoV-2. Nat. Immunol. 2022;23:423–430. doi: 10.1038/s41590-022-01138-w. [DOI] [PubMed] [Google Scholar]
- 101.Du Y, et al. A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants. Nat. Commun. 2021;12:5000. doi: 10.1038/s41467-021-25331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Y, et al. ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker. Signal. Transduct. Target. Ther. 2022;6:315. doi: 10.1038/s41392-021-00740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qi H, Liu B, Wang X, Zhang L. The humoral response and antibodies against SARS-CoV-2 infection. Nat. Immunol. 2022;23:1008–1020. doi: 10.1038/s41590-022-01248-5. [DOI] [PubMed] [Google Scholar]
- 104.Zhou H, et al. Sensitivity to vaccines, therapeutic antibodies, and viral entry inhibitors and advances to counter the SARS-CoV-2 Omicron variant. Clin. Microbiol. Rev. 2022 doi: 10.1128/cmr.00014-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Z, et al. A novel STING agonist-adjuvanted pan-sarbecovirus vaccine elicits potent and durable neutralizing antibody and T cell responses in mice, rabbits and NHPs. Cell Res. 2022;32:269–287. doi: 10.1038/s41422-022-00612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.