Abstract
SARS-CoV-2 infection, which is the cause of the COVID-19 pandemic, has expanded across various animal hosts, and the virus can be transmitted particularly efficiently in minks. It is still not clear how SARS-CoV-2 is selected and evolves in its hosts, or how mutations affect viral fitness. In this report, sequences of SARS-CoV-2 isolated from human and animal hosts were analyzed, and the binding energy and capacity of the spike protein to bind human ACE2 and the mink receptor were compared. Codon adaptation index (CAI) analysis indicated the optimization of viral codons in some animals such as bats and minks, and a neutrality plot demonstrated that natural selection had a greater influence on some SARS-CoV-2 sequences than mutational pressure. Molecular dynamics simulation results showed that the mutations Y453F and N501T in mink SARS-CoV-2 could enhance the binding of the viral spike to the mink receptor, indicating the involvement of these mutations in natural selection and viral fitness. Receptor binding analysis revealed that the mink SARS-CoV-2 spike interacted more strongly with the mink receptor than the human receptor. Tracking the variations and codon bias of SARS-CoV-2 is helpful for understanding the fitness of the virus in virus transmission, pathogenesis, and immune evasion.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-022-05612-6.
Introduction
SARS-CoV-2 is a betacoronavirus that emerged in 2019 and spread worldwide, leading to an ongoing global pandemic [1, 2]. By May 27, 2022, the number of human infections had reached 527 million, resulting in 6.28 million deaths (Johns Hopkins University statistics; https://coronavirus.jhu.edu/map.html). SARS-CoV-2 has a single-stranded positive-sense RNA genome with 29,903 nucleotides that contains 14 open reading frames (ORFs) encoding 29 proteins [3]. One of these viral proteins, the spike glycoprotein (S protein), is an envelope protein that is responsible for the recognition of the host receptor ACE2 and fusion of the viral and host cell membranes [4].
SARS-CoV-2 has a broad host spectrum, probably because ACE2, the receptor used by the virus for cell entry, is a relatively conserved protein in mammals [5]. Many animal species, including cats, dogs, tigers, lions, ferrets, minks, and white-tailed deer, have been found to be susceptible to SARS-CoV-2 infection [6–11]. The earliest animal infections of SARS-CoV-2 in the COVID-19 pandemic were pet cats and dogs [6, 7, 12]. Later, in a report on infection of SARS-CoV-2 in tigers, lions, and human keepers at a New York zoo, the epidemiological and genomic data indicated human-to-animal transmission [8]. Other nondomestic animals, including snow leopards and gorillas, have also tested positive for SARS-CoV-2 after showing signs of illness [13, 14]. Notably, a study from the Netherlands reported the transmission of SARS-CoV-2 both from humans to minks and back from minks to humans on mink farms [15]. Eighty-eight minks and 18 staff members from sixteen mink farms were confirmed to be infected with SARS-CoV-2, as determined by high-throughput sequence analysis. The adaptation of SARS-CoV-2 to the mink receptor and viral evolution in the mink host are thus worthy of further study.
Codon usage bias refers to differences in the frequency of occurrence of synonymous codons for protein translation. For a certain virus, the codon usage pattern may vary when it is adapted to a different host cell [16]. Codon usage bias in some viruses is mainly driven by natural selection pressure [17], while in other viruses such as Ebola virus, mutational bias is a major force determining codon usage [18]. Viruses differ significantly in their host specificity, and analysis of the viral genome structure and composition can contribute to the understanding of virus evolution and adaptation in their hosts [15, 19].
For SARS-CoV-2, exploration of its codon usage pattern in different hosts, especially that of the gene coding for the spike protein, will help to reveal adaptations related to cross-species transmission. Surveillance of nucleotide substitutions and selection in SARS-CoV-2 genomes is important for studying viral evolution and tracking viral transmission. In particular, studying the S gene is important for predicting the efficacy of vaccines and adjusting vaccine design in a timely manner. To investigate the natural selection of SARS-CoV-2 that might play a role in virus evolution, fitness, and transmission, we analyzed the base composition and codon usage of viral genomes isolated from human and animal hosts.
Materials and methods
SARS-CoV-2 sequences and data collection
A total of 258 SARS-CoV-2 genome sequences from humans, cats, dogs, tigers, lions, hamsters, minks, and white-tailed deer were used for genetic analysis. Information about these isolates is given in Supplementary Table S1. All genome sequences were obtained from the GISAID database (https://www.gisaid.org/). Isolate Wuhan/WIV04 was used as the reference strain.
Evolutionary analysis
All 258 SARS-CoV-2 genome sequences were used for phylogenetic analysis. The evolutionary history was inferred using the maximum-likelihood method and the Tamura-Nei model [20]. The tree with the highest log likelihood (-1129076.99) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 1.7203]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. There were 29,899 positions in the final dataset. Evolutionary analysis were performed using MEGA-X [21].
Identification of mutations
Sequences were aligned using MEGA X, and single-nucleotide polymorphisms were analyzed using the SNiPlay pipeline by uploading an aligned Fasta format file (https://sniplay.southgreen.fr/cgi-bin/analysis_v3.cgi) [22]. Complete sequences, including the coding regions, 5' UTR, and 3' UTR, were used for the analysis.
Calculation of nonsynonymous and synonymous substitution rates
The number of nonsynonymous substitutions per synonymous site (dN) and the number of synonymous substitutions per nonsynonymous site (dS) for each coding site were calculated using the Nei–Gojobori method (Jukes–Cantor) in MEGA X. The Datamonkey adaptive evolution server (http://www.datamonkey.org) was used to identify sites where only some branches had undergone selective pressure. The mixed-effects model of evolution (MEME) and fixed-effects likelihood (FEL) approaches were used to determine the nonsynonymous and synonymous substitution rates.
Codon usage analysis
The codon adaptation index (CAI) of coding sequences was calculated using R script [23]. CAI analysis of the coding sequences from different hosts was performed using DAMBE 5.0 and CAIcal [24, 25]. The codon usage patterns of different hosts were obtained from the codon usage database (http://www.kazusa.or.jp/codon/), and the relative synonymous codon usage (RSCU) values were determined using MEGA X software. The accession numbers for mink, cat, dog, tiger, and lion SARS-CoV-2 are MT457401.1, MT747438.1, MT215193.1, MT704316.1, and MT704312.1. Bat-CoV refers to isolate RaTG13 (GenBank no. MN996532.2), obtained from a bat, Pangolin-CoV refers to pangolin coronavirus (GenBank no. QLR06867.1).
Neutral evolution analysis
Neutrality plot analysis was performed to investigate the influence of natural selection and mutation pressure on the codon usage bias [26]. The GC12 values were plotted against GC3 values with a regression line. The slope of the regression line represents the evolutionary speed of the mutation pressure and natural selection pressure. For points lying close to the regression line, there are no significant differences at the three codon positions. If the point is located above the regression line, it means that mutation pressure dominates evolution, whereas, for the points below the line, natural selection plays a more important role.
Spike protein structures and their docking with ACE2
The crystal structure of the receptor-binding domain (RBD) of the SARS-CoV-2 S protein in complex with human ACE2 (PBD ID: 6M0J) was used for structural analysis. Structures of ACE2 and the mink SARS-CoV-2 S protein were predicted using the SWISS Model server (https://swissmodel.expasy.org/). The stability of RBD-ACE2 complexes was calculated using mCSM-PPI2 (http://biosig.unimelb.edu.au/mcsm_ppi2/). The predicted protein structures and pairwise comparisons were performed using PyMOL software.
Molecular dynamics simulation
The binding free energy (E) and the minimized annealing energy were predicted by molecular dynamics (MD) simulation using YASARA [27]. We performed three iterations for the energy minimization of each complex structure of wild-type or mutant mink SARS-CoV-2 RBDs bound to human or mink ACE2. The relative binding energy (∆E) is reported as the mean and standard deviation values from three replicates.
Selection coefficient index
The selection coefficient index (S) of all SARS-CoV-2 codons was calculated using the FMutSel0 model in the program CODEML (PAML package) [28]. The fitness parameter of the most common residue at each position was fixed at 0, while the other fitness parameters were limited to the range of −20 < F < 20.
Plasmid construction and cell culture
Gene fragments encoding the RBD and N-terminal domain (NTD) of the SARS-CoV-2 S protein (NCBI ID no. MN996528.1) were inserted into the plasmid pCAGGS (donated by Prof. Jianguo Wu). Gene fragments encoding the mink RBD and mink ACE2 (NCBI ID no. MW269526.1) were synthesized by Genscript Inc. Mutations were introduced into the RBD gene using a Mut Express II Fast Mutagenesis Kit (Vazyme, C214). All of the wild-type and mutant gene fragments were cloned into the pCAGGS vector with a 6× histidine tag, using EcoRI and XhoI restriction sites. A gene fragment encoding the mink ACE2 was inserted into the vector pcDNA3.1-eGFP to generate pcDNA3.1-mACE2-eGFP. The ligation products were introduced by transformation into competent E. coli Top10 cells. Recombinant plasmids were verified by DNA sequencing. Plasmid pcDNA3.1-hACE2-eGFP expressing hACE2 fused to eGFP was purchased from Fubio Ltd. (MC_0101086). HEK293T and BHK-21 cells were cultured in high-glucose DMEM medium in a 5% CO2 atmosphere in a 37℃ incubator.
Protein expression and purification
Recombinant pCAGGS plasmids for expression of the RBD or NTD variants were introduced by transfection into CHO cells in 245-mm dishes using Lipofectamine 6000 according to the manufacturer's recommendations. The supernatants were collected 5 days after transfection and centrifuged. The soluble proteins were purified using HisPur™ Ni-NTA Resin (Thermo Scientific, 88221) and eluted in 20 mM Tris-HCl (pH 8.0) buffer containing 150 mM NaCl.
Flow cytometry
BHK-21 cells were transfected with pcDNA3.1-hACE2-GFP and pcDNA3.1-mACE2-eGFP using Lipofectamine 6000 (Beyotime, C0526) according to the manufacturer's instructions. The cells expressing hACE2-GFP and mACE2-GFP were collected at 24 h post-transfection, resuspended in phosphate-buffered saline (PBS), and then incubated with the purified His-tagged RBDs at a final concentration of 30 μg/mL at 37ºC for 30 min. The NTD was used as a negative control. After being washed twice with PBS, the cells were incubated with anti-His/APC antibodies (1:5000) and then examined using a BeckMan CytoFLEX Flow Cytometer. The data were analyzed using FlowJo V10 software.
Statistical analysis and mapping
Statistical analysis was performed using ANOVA followed by Turkey's post-hoc test (Fig. 2C and F) or Student's t-test (Fig 3B). The data were considered significantly different if the P-value was less than 0.05. SPSS 20.0 software was used to perform regression curve fitting. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. The figures were made using GraphPad PRISM 5.0.
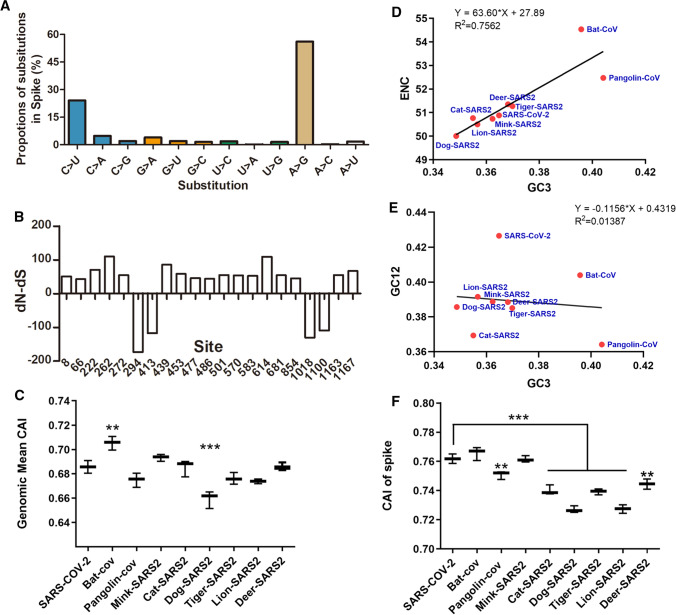
Fig. 2.
The mutation spectrum of the spike protein and selection pressure analysis. (A) Substitutions in the animal-derived SARS-CoV-2 S gene. (B) dN-dS value for S gene sequences. dN = nonsynonymous changes/nonsynonymous site. dS = synonymous changes/synonymous site. (C) CAI values for SARS-CoV-2 sequences from humans and animals. "Bat-CoV" refers to RaTG13 from bat, "Pangolin-COV" refers to pangolin coronavirus (GenBank no. QLR06867.1), "other animal-SARS2" means SARS-CoV-2 isolated from the indicted animals, "the first SARS-CoV-2" refers to the virus isolated from a human host. (D) ENC plot analysis of the coronaviruses from animals and humans. (E) Neutrality plot analysis of the coronaviruses from animals and humans. (F) CAI values of the S sequences of SARS-CoV-2 from humans and animals.
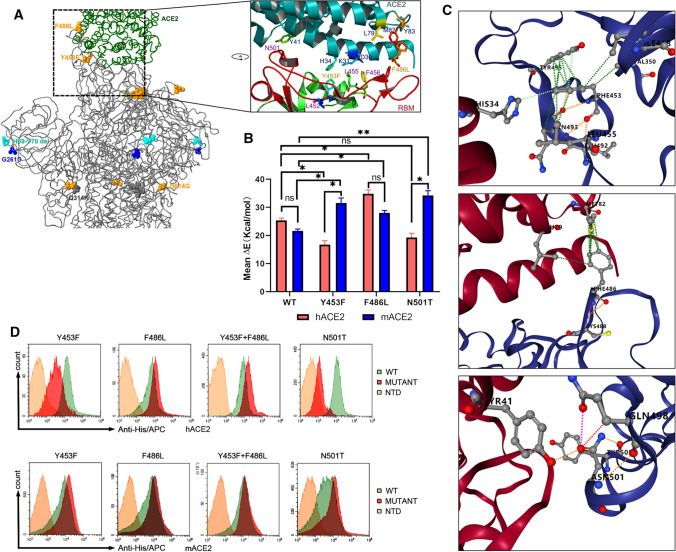
Fig. 3.
Analysis of binding of the SARS-CoV-2 spike protein with human and mink receptors. (A) Comparison of the spike structure of mink SARS-CoV-2 with that of reference strain WIV04. The changed residues within mink SARS-CoV-2 are highlighted as yellow balls. (B) The free energy of binding of the wild-type RBD or mutants from mink SARS-CoV-2 to the human and mink receptor. (C) Amino acid changes involved in the stability of the RBD-ACE2 complex. Detailed structures for Y453, F486, and N501 are arranged from top to bottom. The green lines represent hydrophobic interactions, the orange lines indicate polar H-bonds, the red lines represent hydrogen bonds, and the pink-purple lines represent clashes. (D) Measurement of the binding of RBD mutants to human ACE2 (hACE2, upper panel) and mink ACE2 (mACE2, lower panel) by FACS. His-tagged wild-type RBD, RBD mutants, and NTD were incubated with cells expressing eGFP-fused ACE2. NTD was used as a negative control.
Results
Sequence and analysis of SARS-CoV-2 isolated from animals
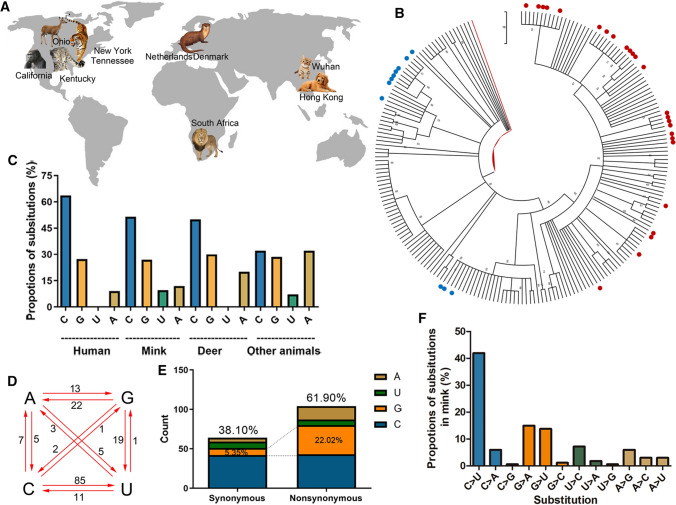
As of June 20, 2021, more than 2.53 million SARS-CoV-2 genome sequences had been uploaded to the GISAID database. It is important to study the mutation rates and selective pressures on the SARS-CoV-2 genome during the spread of the epidemic. In addition to humans, SARS-CoV-2 infects other animals (Fig. 1A) and evolves in these animals. A phylogenetic tree was constructed based on animal-derived whole-genome consensus sequences, using the SARS-CoV-2 human isolate WIV04 as the outgroup (Fig. 1B). Most SARS-CoV-2 sequences isolated from animals in the same geographic region were clustered together, so a single clade could contain sequences from different animals (See Supplementary Figure S1 for details). Previous investigations found evidence of human-to-animal spillover and further transmission of SARS-CoV-2 in minks and white-tailed deer [12, 15], and the available sequences suggesting mink-to-human transmission were mainly from regions where mink infection had been reported (Netherlands and Denmark).
Fig. 1.
Composition and substitution analysis of SARS-CoV-2 isolated from animals. (A) The reported animals infected with SARS-CoV-2 with the defined transmission route from humans to animals. (B) Phylogenetic tree constructed by the maximum-likelihood method with the Tamura-Nei model in MEGA X with 500 bootstrap replicates. Red dots represent human sequences from infected animals, and blue dots represent sequences from infected white-tailed deer. (C) The proportions of uracil, guanine, thymine, and cytidine substitutions (nonsynonymous) in SARS-CoV-2 isolated from human or animals. (D) Base pair changes observed in the SARS-CoV-2 genomes. All of the transitions and transversions are listed in Supplementary Table S2. (E) The synonymous and nonsynonymous substitutions in mink SARS-Cov-2. (F) The relative proportion of each nucleotide substitution in the mink SARS-CoV-2 genome.
In the cluster of SARS-CoV-2 from minks, the sequences had more substitutions than the human SARS-CoV-2 isolates transmitted from infected animals, when compared to the reference sequence WIV04 (Supplementary Table S2). Cytidine substitutions in mink SARS-CoV-2 accounted for nearly 50% of the total substitutions, whereas the replacement of nucleotides with cytidine in isolates from animals other than minks and deer accounted for only 30% of the substitutions (Fig. 1C). Adenine substitution in SARS-CoV-2 in other animals was threefold higher than in mink SARS-CoV-2 (Fig. 1C). To track how the substitutions in the mink SARS-CoV-2 genome occurred, we analyzed all of the mutations in the mink SARS-CoV-2 genome, using the WIV04 genome as a reference sequence. As shown in Fig. 1D and F, cytidine-to-uracil transitions occurred in more than 40% of cases, which was eightfold higher than the rate of uracil-to-cytidine substitution. Notably, guanine and adenine substitutions were more than threefold higher in nonsynonymous mutations than in synonymous mutations (Fig. 1E).
Mutational spectra of spike proteins in human and animal samples
Comparison of S gene sequences from humans and animals revealed considerable variation and allowed the identification of several highly variable residues. C-to-U substitutions were scattered throughout the SARS-CoV-2 genome and accounted for 24.06% of the substitutions in the S gene in all epidemic strains analyzed in this study (Fig. 2A). The dN-dS ratios indicated that natural selection had occurred at most of the mutated sites in the S gene (dN-dS>0 indicates positive selection, and dN-dS<0 indicates purification selection). The data also suggested that positions 222, 262, 439, and 614 were exposed to strong positive selection pressure, while positions 294, 413, 1018, and 1100 had undergone purification selection (Fig. 2B).
CAI analysis was used to quantify the codon usage similarities between different coding sequences based on a reference set of highly expressed genes [29]. To evaluate the adaptation of SARS-CoV-2 in different hosts, we calculated the average CAI for the viral genome (Fig. 2C). Interestingly, the CAI value for SARS-CoV-2 from bats was significantly higher than that from humans, while dog-SARS2 had a much lower CAI value than human SARS-CoV-2 (Fig. 2C). ??Effective number of codons?? (ENC) plot analysis was used to investigate factors influencing SARS-CoV-2 codon usage bias. The results (Fig. 2D) showed that most of the SARS-CoV-2 isolates gave values slightly below the standard curve (R2 = 0.7562, P = 0.0023), indicating that the codon usage bias was affected by both mutation pressure and evolutionary pressure, consistent with previous reports [30, 31]. Neutrality plot analysis showed that the linear regression coefficient of SARS-CoV-2 sequences was -0.1156 (Fig. 2E), indicating that the first and second positions of the codons were mainly affected by mutation pressure and that the third positions were mainly influenced by selection pressure. Codon usage analysis showed that SARS-CoV-2 S genes from pangolins, cats, dogs, tigers, lions, and deer had significantly lower CAI values than those from humans (Fig. 2F), implying that the viruses were less adapted in these animal hosts. All of the nucleotide substitutions in the codons of the S genes are listed in Supplementary Table S3.
Stronger binding to the mink receptor by SARS-CoV-2 spike mutants
Structure analysis of the SARS-CoV-2 and ACE2 complex and sequence alignment of the spike proteins showed that the spike protein interacts with amino acids 34, 41, 79, 82, and 354 of human and mink ACE2 (Supplementary Fig. S2A and B), which form electrostatic and hydrophobic interactions with residues Asn439, Tyr453, Phe486, and Asn501 of the spike protein. An alignment of ACE2 amino acid sequences of humans, minks, ferrets, tigers, cats, and dogs showed that the critical changes H34Y, L79H, and G354R had occurred in minks and ferret ACE2 (Supplementary Fig. S2B). On the other hand, virus mutation is another important factor that should be considered for transmission of viruses between animals and humans. Sequence alignment of the RBD of the spike proteins revealed that most of the residues binding to the receptor were conserved in these virus strains (Supplementary Fig. S2C). However, residue 453 of the mink SARS-CoV-2 spike protein, which was predicted to interact with residue 34 on ACE2 (Fig. 3A), had changed from Y to F. An MD simulation suggested that the binding interaction of F453-Y34 in minks was stronger than that of Y453-H34 in humans (Fig. 3B). The N501T substitution in the spike protein of mink isolates also resulted in stronger binding to the mink receptor (Fig. 3B). Overall, the change in receptor binding affinity was mainly due to the mutations that altered hydrophobic interactions (Y453-H34, F486-M82) and a polar H-bond (N501-Y41) at the binding interface (Fig. 3C). Flow cytometry results showed that the mutations Y453F, F486L, and N501T all enhanced the binding of the spike RBD to the mink ACE2, whereas only F486L and the double mutant Y453F&F486L showed increased binding to the human receptor (Fig. 3D). These data indicate that key point mutations in the spike protein contribute to the adaptation of SARS-CoV-2 to minks.
Codon usage and fitness analysis of SARS-CoV-2-encoded proteins
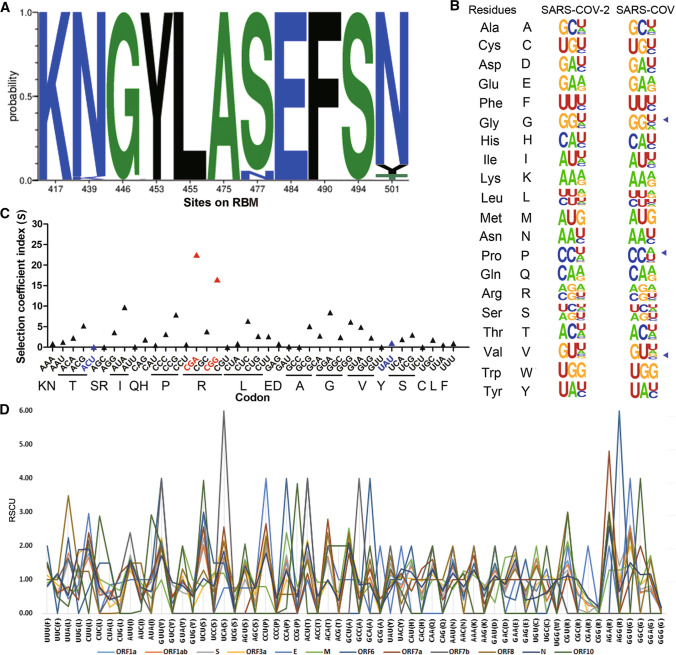
Amino acid substitutions within the SARS-CoV-2 spike receptor-binding motif (RBM) may contribute to host adaption and cross-species transmission. N439K, S477N, and N501Y were found to be the most common variations in the RBM region of the SARS-CoV-2 spike protein (Fig. 4A). Amino acid 439 in the spike does not bind directly to ACE2, but it acts to stabilize the 498–505 loop [32], and the N439K substitution was not found in the animal CoVs analyzed in this study (Supplementary Fig. S2C). A previous computational analysis combined with entropy analysis of the spike showed that the S477N mutant may be less stable than the wild-type protein [33]. For the mutations N501T (AAU>ACU) in minks and N501Y (AAU>UAU) in humans, nonsynonymous nucleotide substitutions were in the first and second codon positions, and interestingly, statistics data showed that both A>C and A>U substitutions occurred at very low frequency in the SARS-CoV-2 genome (Fig. 1F and 2A). Comparison of the synonymous codon usage of mink SARS-CoV-2 and SARS-CoV demonstrated similar codon usage patterns for these two viruses strains (Fig. 4B), consistent with their adaptability in ferrets, which served as hosts for both viruses. The selective coefficient indices shown in Fig. 4C reveal that the relative fitness of SARS-CoV-2 codons differs, with CGA and CGG showing higher fitness scores than the others. The codons for the N501 mutants, ACU for T and UAU for Y, had low fitness scores, indicating that receptor binding ability rather than codon usage bias was the main determinant of selection for these two mutations. In addition, when comparing the 12 ORFs in the SARS-CoV-2 genome, diverse RSCU values were obtained for the different ORFs. UCA-encoded Ser in ORF7b and AGG-encoded Arg in ORF6 were notably preferred (Fig. 4D and Supplementary Table S4).
Fig. 4.
Codon usage and fitness analysis of the SARS-CoV-2-encoded proteins. (A) Probability of mutations in the receptor-binding motif (RBM). The data were analyzed using MEGA X software. The frequency was calculated using the Datamonkey server, and the figure was produced using WebLogo (https://weblogo.berkeley.edu/logo.cgi). (B) The synonymous codon usage of SARS-CoV-2. The figure was produced using WebLogo. The mink SARS-CoV-2 sequence (GenBank no. MT396266) was compared with that of SARS-CoV strain Tor2 (GenBank no. NC_004718.3). (C) Selective coefficient index for SARS-CoV-2 codons. The codons for the N501 mutants are shown in blue and the codons with the highest fitness are highlighted in red. (D) Analysis of relative synonymous codon usage in SARS-CoV-2-encoded proteins.
Discussion
Tracking of viral variants transmitted among animal hosts or transmitted to animals by human contact could be helpful for understanding the evolution and host adaptation of SARS-CoV-2. The correlation between codon optimization of viral genomes and their host adaptation process has been observed in some viruses such as rotaviruses, ??cyprinid herpesvirus 3??, and Marburg virus [26, 34, 35]. Mink was the first extensively farmed species affected by COVID-19, and epidemiological investigation has suggested that mustelids, including minks and ferrets, are more susceptible to SARS-CoV-2 than other animals [36]. Mink-to-mink and mink-to-human transmission of SARS-CoV-2 have been reported on several mink farms in the Netherlands, Denmark, the USA, and Spain [37–39]. Some mutations have accumulated in the viral genomes during transmission of the virus between humans and minks. However, it is challenging to pinpoint whether mutations happened before or after the virus spillover to mink, because it is difficult to distinguish the sequences circulating in the human and mink populations from those involved in cross-species transmission.
In this study, we compared the sequences of mink-derived SARS-CoV-2 isolates and sequences from humans who had contact with infected minks, using human SARS-CoV-2 as a reference sequence. The substitutions are listed in Supplementary Table S2. Some nonsynonymous substitutions, such as C1380U in ORF1a and C14408U in ORF1b, were found in both mink isolates and those from humans who had contact with infected minks. Some unique mutations were found only in the mink isolates and not in those from humans who had contact with infected minks. For example, the nonsynonymous substitutions G520A, G1599U, and A2280C in ORF1a and U23018C in the spike protein (F486L) were present only in the mink isolates and not in the human isolates, which is consistent with the data reported in a previous study [38]. Taking into consideration the high mutation numbers and frequencies, these unique substitutions in mink SARS-CoV-2 are more likely to have appeared after the spillover of infection from human to minks and accumulated during virus spread on mink farms with a large animal population.
Other animals, including tigers, lions, and white tail-deer, have also been found to be susceptible to SARS-CoV-2 infection [12, 40]. Adaptive mutations have also been reported in isolates from deer, and it appears that minimal adaptation is required for onward transmission in minks and deer following human-to-animal spillover [41]. Here, we compared the sequences from deer with those from humans who had contact with infected minks and identified some new nonsynonymous substitutions in the deer sequences, e.g., G8083A and C10319U in ORF1a and G25563U and G25907U in ORF3a. No unique mutations were found in the S gene of the deer sequences.
Given that the usage of synonymous codons in viral genomes varies with the host [26], adaptation to different hosts may affect the codon usage bias of the virus. A previous study revealed that synonymous mutations in SARS-CoV-2 may boost the adaptation of the virus to human codon usage and positively affect viral evolution [42]. In our study, we compared the codon bias of SARS-CoV-2 in minks with that of SARS-CoV in ferrets, both of which can infect both ferrets and minks. Threonine (T) and tyrosine (Y) residues shared comparable codon biases in SARS-CoV-2 and SARS-CoV (Fig. 4B). Notably, the N501Y mutation occurred exclusively in humans, while N501T was found more frequently in minks, suggesting that amino acid 501 played a key role in the virus adaptation in humans and minks that was not related to codon bias. Our data here also show that Y453F could enhance the binding ability of the spike to mink ACE2 (Fig. 3B and F), and we speculate that this mutation was beneficial for virus adaptation and transmission in minks and thus resulted in the extensive spread of SARS-CoV-2 among minks.
The WebLogo diagram in Fig. 4B shows that SARS coronaviruses preferentially have U- or A-ending codons, as has been shown previously [43]. The biased use of purine nucleotide at the third codon position could lead to an imbalance in the tRNA pool and a decrease in host protein synthesis in infected cells. The C-to-U substitution is the most frequent mutation in most of the reported SARS-CoV-2 sequences isolated from animals, with an 8-fold higher level of C-to-U substitutions in the mink sequences than U-to-C substitutions (Fig. 1C and F). This is higher than the previously reported 3.5-fold higher level in minks [15], suggesting that SARS-CoV-2 has evolved over time in minks. Our CAI data suggest that the virus may have adapted with optimized codons better in bats than in dogs and other animals, which is consistent with a previous finding that humans are more favored hosts for SARS-CoV-2 adaptation than dogs [30]. A similar substitution level was found across the whole genome of mink SARS-CoV-2 when compared to human SARS-CoV-2, indicating that the virus could still be in the process of adaptation to its new host species. Further studies are needed to investigate whether the CAI values will increase when the virus has had more time to adapt.
The spike protein is essential for both host adaptability and virus infection. We discovered that three nonsynonymous changes in the RBM domain – Y453F, F486L, and N501T – emerged independently in the mink lineage. These residues are directly involved in receptor binding at the interface of the S-ACE2 complex and are thus important for adaptation of the virus to new hosts. In addition to the mutations in the S protein, amino acid substitutions in ORF1a, ORF9b, E, N, and M have also been identified as significantly associated with increased fitness [44], suggesting that the RBD of the spike protein is not the only region that affects SARS-CoV-2 fitness.
Other mutations within the RBM domain should be monitored for viral transmission. Some residues in this region have been reported to be involved in evading host humoral immunity. For example, the B.1.351 (Beta) SARS-CoV-2 variant carrying the E484K and N501Y mutations, the B.1.617.2 (Delta) variant carrying the L452R mutation, and the BA.2 (Omicron) variant containing the E484A mutation, have improved ability to enter cells, and they can re-infect recovered or vaccinated individuals [45]. The currently available vaccines are less protective against the Delta and Omicron variants, but they can still prevent severe cases [46]. The fitness of these SARS-CoV-2 mutants should be evaluated further to optimize vaccine design and block virus transmission.
In conclusion, we have shown in this study that spike proteins with the mutations Y453F and N501T in mink SARS-CoV-2 recognize the mink receptor better than the human receptor. Our findings may provide a new perspective for the understanding of natural selection and viral fitness of SARS-CoV-2.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Prof. Jianguo Wu and Prof. Hongying Chen for their technical support in the experiments.
Author contributions
YNF, FZ, YPH, JJR, and LL contributed to the design of experiments. YNF, YPH, JJR, FZ, RPY, YNF, and ZXL contributed to the performance of experiments. JJR, RPY, and FZ contributed reagents. HBT, WXD, and ZXL contributed to the analyses of the data. LL, WXD, and ZXL contributed to the writing of the manuscript. WXD and LL contributed to the revision of the paper.
Funding
This work was supported by the Natural Science Foundation of China (82002149 and 81902066), the Principal Investigator Program at Hubei University of Medicine (HBMUPI202102), the Foundation of Health Commission of Hubei Province (WJ2021M059), and the Scientific and Technological Project of Shiyan City (2021K65). The funders had no role in the study design, data collection and analysis, or manuscript writing and submission.
Data availability statement
All datasets presented in this study are included in the article or supplementary material.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanan Fu and Yanping Huang contributed equally.
Contributor Information
Zhixin Liu, Email: lzx20022456@126.com.
Weixing Du, Email: duwei-080@163.com.
Long Liu, Email: liulong2015@outlook.com.
References
- 1.Laha S, Chakraborty J, Das S, Manna SK, Biswas S, Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol J Mol Epidemiol Evolut Genet Infect Dis. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK, Rodriguez-Morales AJ. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med. 2020;28:174–184. [PubMed] [Google Scholar]
- 3.Yang H, Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat Rev Microbiol. 2021;19:685–700. doi: 10.1038/s41579-021-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli K-P, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, Chen Q, Liu K, Wang J, Han P, Zhang Y, Hu Y, Meng Y, Pan X, Qiao C, Tian S, Du P, Song H, Shi W, Qi J, Wang H-W, Yan J, Gao GF, Wang Q. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAloose D, Laverack M, Wang L, Killian ML, Caserta LC, Yuan F, Mitchell PK, Queen K, Mauldin MR, Cronk BD, Bartlett SL, Sykes JM, Zec S, Stokol T, Ingerman K, Delaney MA, Fredrickson R, Ivančić M, Jenkins-Moore M, Mozingo K, Franzen K, Bergeson NH, Goodman L, Wang H, Fang Y, Olmstead C, McCann C, Thomas P, Goodrich E, Elvinger F, Smith DC, Tong S, Slavinski S, Calle PP, Terio K, Torchetti MK, Diel DG. From people to panthera: natural SARS-CoV-2 infection in tigers and lions at the bronx zoo. MBio. 2020 doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW, Yu VYT, Sims LD, Tsang DNC, Chu DKW, Perera R, Poon LLM, Peiris M. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y-I, Kim S-G, Kim S-M, Kim E-H, Park S-J, Yu K-M, Chang J-H, Kim EJ, Lee S, Casel MAB, Um J, Song M-S, Jeong HW, Lai VD, Kim Y, Chin BS, Park J-S, Chung K-H, Foo S-S, Poo H, Mo I-P, Lee O-J, Webby RJ, Jung JU, Choi YK. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e702. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutter JS, de Meulder D, Bestebroer TM, Lexmond P, Mulders A, Richard M, Fouchier RAM, Herfst S. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat Commun. 2021;12:1653. doi: 10.1038/s41467-021-21918-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(894–904):e899. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale VL, Dennis PM, McBride DS, Nolting JM, Madden C, Huey D, Ehrlich M, Grieser J, Winston J, Lombardi D, Gibson S, Saif L, Killian ML, Lantz K, Tell RM, Torchetti M, Robbe-Austerman S, Nelson MI, Faith SA, Bowman AS. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2022;602:481–486. doi: 10.1038/s41586-021-04353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daly N (2021) Several gorillas test positive for COVID-19 at California zoo—first in the world. National Geographic. https://www.nationalgeographic.com/animals/article/gorillas-san-diego-zoo-positive-coronavirus. Accessed 1 Feb 2021
- 14.Jo WK, de Oliveira-Filho EF, Rasche A, Greenwood AD, Osterrieder K, Drexler JF. Potential zoonotic sources of SARS-CoV-2 infections. Transbound Emerg DisDoi. 2020 doi: 10.1111/tbed.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munnink BBO, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester-Vincken N, Harders F, Hakze-van der Honing R, Wegdam-Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM, Koopmans MPG. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belalov IS, Lukashev AN. Causes and implications of codon usage bias in RNA viruses. PLoS ONE. 2013;8:e56642. doi: 10.1371/journal.pone.0056642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Shi Y, Deng H, Gu T, Xu J, Ou J, Jiang Z, Jiao Y, Zou T, Wang C. Characterization of the porcine epidemic diarrhea virus codon usage bias. Infect Genet Evol. 2014;28:95–100. doi: 10.1016/j.meegid.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristina J, Moreno P, Moratorio G, Musto H. Genome-wide analysis of codon usage bias in Ebolavirus. Virus Res. 2015;196:87–93. doi: 10.1016/j.virusres.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Di Giallonardo F, Schlub Timothy E, Shi M, Holmes EC. Dinucleotide composition in animal RNA viruses is shaped more by virus family than by host species. J Virol. 2017;91:e02381–02316. doi: 10.1128/JVI.02381-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dereeper A, Nicolas S, Le Cunff L, Bacilieri R, Doligez A, Peros JP, Ruiz M, This P. SNiPlay: a web-based tool for detection, management and analysis of SNPs. Application to grapevine diversity projects. BMC Bioinform. 2011;12:134. doi: 10.1186/1471-2105-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp PM, Li WH. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia X. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 2013;30:1720–1728. doi: 10.1093/molbev/mst064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puigbo P, Bravo IG, Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct. 2008;3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasrullah I, Butt AM, Tahir S, Idrees M, Tong Y. Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on Marburg virus evolution. BMC Evol Biol. 2015;15:174. doi: 10.1186/s12862-015-0456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Land H, Humble MS. YASARA: a tool to obtain structural guidance in biocatalytic investigations. Methods Mol Biol. 2018;1685:43–67. doi: 10.1007/978-1-4939-7366-8_4. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, Nielsen R. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol Biol Evol. 2008;25:568–579. doi: 10.1093/molbev/msm284. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Weon S, Lee S, Kang C. Relative codon adaptation index, a sensitive measure of codon usage bias. Evolut Bioinform. 2010;6:EBO.S4608. doi: 10.4137/EBO.S4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutta R, Buragohain L, Borah P. Analysis of codon usage of severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) and its adaptability in dog. Virus Res. 2020;288:198113. doi: 10.1016/j.virusres.2020.198113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyagi N, Sardar R, Gupta D. Natural selection plays a significant role in governing the codon usage bias in the novel SARS-CoV-2 variants of concern (VOC) PeerJ. 2022;10:e13562. doi: 10.7717/peerj.13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verkhivker G. Coevolution, dynamics and allostery conspire in shaping cooperative binding and signal transmission of the SARS-CoV-2 spike protein with human angiotensin-converting enzyme 2. Int J Mol Sci. 2020 doi: 10.3390/ijms21218268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alaofi AL, Shahid M. Mutations of SARS-CoV-2 RBD may alter its molecular structure to improve its infection efficiency. Biomolecules. 2021 doi: 10.3390/biom11091273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kattoor JJ, Malik YS, Sasidharan A, Rajan VM, Dhama K, Ghosh S, Bányai K, Kobayashi N, Singh RK. Analysis of codon usage pattern evolution in avian rotaviruses and their preferred host. Infect Genet Evol. 2015;34:17–25. doi: 10.1016/j.meegid.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Ma YP, Liu ZX, Hao L, Ma JY, Liang ZL, Li YG, Ke H. Analysing codon usage bias of cyprinid herpesvirus 3 and adaptation of this virus to the hosts. J Fish Dis. 2015;38:665–673. doi: 10.1111/jfd.12316. [DOI] [PubMed] [Google Scholar]
- 36.Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Hakze-van der Honing RW, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MG, de Rooij MM, Weesendorp E, Engelsma MY, Bruschke CJ, Smit LA, Koopmans M, van der Poel WH, Stegeman A. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020 doi: 10.2807/1560-7917.Es.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahdy MAA, Younis W, Ewaida Z. An overview of SARS-CoV-2 and animal infection. Front Vet Sci. 2020;7:596391. doi: 10.3389/fvets.2020.596391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester-Vincken N, Harders F, Hakze-van der Honing R, Wegdam-Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM, Koopmans MPG. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manes C, Gollakner R, Capua I. Could mustelids spur COVID-19 into a panzootic? Vet Ital. 2020;56:65–66. doi: 10.12834/VetIt.2375.13627.1. [DOI] [PubMed] [Google Scholar]
- 40.McAloose D, Laverack M, Wang L, Killian ML, Caserta LC, Yuan F, Mitchell PK, Queen K, Mauldin MR, Cronk BD, Bartlett SL, Sykes JM, Zec S, Stokol T, Ingerman K, Delaney MA, Fredrickson R, Ivančić M, Jenkins-Moore M, Mozingo K, Franzen K, Bergeson NH, Goodman L, Wang H, Fang Y, Olmstead C, McCann C, Thomas P, Goodrich E, Elvinger F, Smith DC, Tong S, Slavinski S, Calle PP, Terio K, Torchetti MK, Diel DG, Meng X-J. From people to panthera: natural SARS-CoV-2 infection in tigers and lions at the bronx zoo. MBio. 2020;11:e02220–02220. doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan CCS, Lam SD, Richard D, Owen CJ, Berchtold D, Orengo C, Nair MS, Kuchipudi SV, Kapur V, van Dorp L, Balloux F. Transmission of SARS-CoV-2 from humans to animals and potential host adaptation. Nat Commun. 2022;13:2988. doi: 10.1038/s41467-022-30698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramazzotti D, Angaroni F, Maspero D, Mauri M, D'Aliberti D, Fontana D, Antoniotti M, Elli EM, Graudenzi A, Piazza R. Large-scale analysis of SARS-CoV-2 synonymous mutations reveals the adaptation to the human codon usage during the virus evolution. Virus Evol. 2022;8:veac26. doi: 10.1093/ve/veac026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou W. Characterization of codon usage pattern in SARS-CoV-2. Virol J. 2020;17:138. doi: 10.1186/s12985-020-01395-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obermeyer F, Jankowiak M, Barkas N, Schaffner SF, Pyle JD, Yurkovetskiy L, Bosso M, Park DJ, Babadi M, MacInnis BL, Luban J, Sabeti PC, Lemieux JE. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science. 2022;376:1327–1332. doi: 10.1126/science.abm1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez CE, Perdiguero B, Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines. 2021 doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, Stowe J, Tessier E, Groves N, Dabrera G, Myers R, Campbell CNJ, Amirthalingam G, Edmunds M, Zambon M, Brown KE, Hopkins S, Chand M, Ramsay M. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets presented in this study are included in the article or supplementary material.