Abstract
The Streptomyces glaucescens β-ketoacyl-acyl carrier protein (ACP) synthase III (KASIII) initiates straight- and branched-chain fatty acid biosynthesis by catalyzing the decarboxylative condensation of malonyl-ACP with different acyl-coenzyme A (CoA) primers. This KASIII has one cysteine residue, which is critical for forming an acyl-enzyme intermediate in the first step of the process. Three mutants (Cys122Ala, Cys122Ser, Cys122Gln) were created by site-directed mutagenesis. Plasmid-based expression of these mutants in S. glaucescens resulted in strains which generated 75 (Cys122Ala) to 500% (Cys122Gln) more straight-chain fatty acids (SCFA) than the corresponding wild-type strain. In contrast, plasmid-based expression of wild-type KASIII had no effect on fatty acid profiles. These observations are attributed to an uncoupling of the condensation and decarboxylation activities in these mutants (malonyl-ACP is thus converted to acetyl-ACP, a SCFA precursor). Incorporation experiments with perdeuterated acetic acid demonstrated that 9% of the palmitate pool of the wild-type strain was generated from an intact D3 acetyl-CoA starter unit, compared to 3% in a strain expressing the Cys122Gln KASIII. These observations support the intermediacy of malonyl-ACP in generating the SCFA precursor in a strain expressing this mutant. To study malonyl-ACP decarboxylase activity in vitro, the KASIII mutants were expressed and purified as His-tagged proteins in Escherichia coli and assayed. In the absence of the acyl-CoA substrate the Cys122Gln mutant and wild-type KASIII were shown to have comparable decarboxylase activities in vitro. The Cys122Ala mutant exhibited higher activity. This activity was inhibited for all enzymes by the presence of high concentrations of isobutyryl-CoA (>100 μM), a branched-chain fatty acid biosynthetic precursor. Under these conditions the mutant enzymes had no activity, while the wild-type enzyme functioned as a ketoacyl synthase. These observations indicate the likely upper and lower limits of isobutyryl-CoA and related acyl-CoA concentrations within S. glaucescens.
One of the conserved features of fatty acid and polyketide biosynthesis is that the elongation steps that generate the carbon chains in both processes are accomplished by a ketoacyl synthase (KAS) (30). These enzymes typically catalyze a Claisen condensation between an acyl-acyl carrier protein (ACP) substrate and malonyl-ACP (alternative substrates such as methylmalonyl- and ethylmalonyl-ACP can be used in some processes) to generate a 3-ketoacyl-ACP product (5). These enzymes all operate via a ping-pong mechanism in which an active-site cysteine is essential for formation of the acyl enzyme intermediate. In contrast to this unified mechanism of elongation, at least three different processes are used for initiation of fatty acid and polyketide biosynthesis from simple acyl thioester precursors.
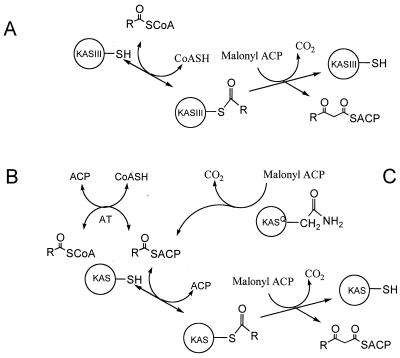
For a type II dissociable fatty acid synthase (FAS) of plants and bacteria a KAS isozyme, KASIII, plays a critical role in initiating fatty acid biosynthesis (Fig. 1A) (17, 23). Elongation steps in a type II FAS are catalyzed by KAS isozymes (KASI and KASII) which have broad overlapping substrate specificities for a range of acyl-ACPs (more than four carbons) (17). KASIII uses an acyl-coenzyme A (CoA) thioester as a substrate to directly generate a 3-ketoacyl-ACP intermediate. This enzyme is thought to play an important role in both regulating fatty acid biosynthesis and determining the type of fatty acids made by an organism. Thus the Escherichia coli KASIII has a strong preference for acetyl-CoA and produces only straight-chain fatty acids (SCFAs) (23). Organisms such as Streptomyces glaucescens and Bacillus subtilis produce a mixture of SCFAs and branched-chain fatty acids (BCFAs) and contain KASIII enzymes with relaxed specificity, processing both BCFA precursors (isobutyryl-CoA and methylbutyryl-CoA) and acetyl-CoA (4, 7). In these organisms the KASIII specificity and the availability of the various acyl-CoA precursors contribute to determining the type of fatty acids made.
FIG. 1.
Pathways for initiation of fatty acid and polyketide biosynthesis using simple acyl-CoA precursors. (A) Direct elongation of an acyl-CoA starter unit by KASIII in a type II dissociable FAS. (B) A transacylation of acyl-CoA to acyl-ACP precedes the first KAS-catalyzed elongation step in type I FASs and in some modular type I PKSs. (C) The acyl-ACP used for the first elongation step is generated by a KASQ-catalyzed decarboxylation of malonyl-ACP in some modular type I PKSs. A similar process is catalyzed by the KSα and KSβ subunits of some type II dissociable PKSs. AT, AT domain. For clarity purposes only processes using a malonyl-CoA-derived malonyl-ACP are shown (methylmalonyl-ACP and other related substrates can also be used). In the type I systems the AT, ACP, KAS, and KASQ catalytic domains are typically contained on one polypeptide chain.
In contrast, initiation in a type I multifunctional FAS involves a malonyl/acetyl transferase domain-catalyzed transfer of an acetyl group from acetyl-CoA to ACP (Fig. 1B) (27, 30). Elongation of the resulting acetyl-ACP and all subsequent acyl-ACPs is catalyzed by the one KAS present in a type I FAS. A similar method for initiation is seen in some modular type I polyketide synthases (PKSs) (15). For the erythromycin PKS an acyltransferase (AT) domain catalyzes the transfer of propionyl-CoA onto an ACP in the loading module (6). Similarly an AT domain in the avermectin PKS loads branched-chain starters isobutyryl-CoA and 2-methylbutyryl-CoA (12). In both cases a KAS domain in the first extension module catalyzes the first elongation step using the resulting acyl-ACP substrate.
A third mechanism for initiation has been observed in numerous modular type I PKSs (Fig. 1C). Analysis of the loading domains of PKSs involved in the biosynthesis of polyketides such as niddamycin, pikromycin, and monensin has revealed a KAS domain (termed KASQ or KSQ) in which the active-site cysteine is replaced with a glutamine (2, 13, 31). These domains do not have KAS activity but are able to catalyze the decarboxylation of malonyl- or methylmalonyl-ACP residues to generate acetyl- and propionyl-ACP, respectively. The AT domains in the loading modules of these PKSs thus transfer malonyl-CoA or methylmalonyl-CoA rather than acetyl-CoA or propionyl-CoA, respectively, onto the cognate ACP domain. Evidence indicates that a similar decarboxylation of malonyl-ACP is used to initiate polyketide biosynthesis in certain type II dissociable PKS systems (1–3). In this context it is interesting that the KAS used to catalyze all extension steps in this iterative process is a heterodimer (KSα and KSβ). The second subunit, KSβ, is similar to KSα but has the essential conserved cysteine replaced with a glutamine (2).
Using S. glaucescens as a host, we describe in this paper how a similar pathway for initiation can be engineered into a type II FAS using a KASIIIQ mutant. This mutated enzyme has no KAS activity and cannot initiate fatty acid biosynthesis in the normal manner but can catalyze the decarboxylation of malonyl-ACP. Thus an extender unit used for both BCFA and SCFA biosynthesis is converted to acetyl-ACP, which is used only to initiate SCFA biosynthesis.
MATERIALS AND METHODS
Materials.
The following chemicals and reagents were purchased from Sigma (St. Louis, Mo.): acetyl-CoA, isobutyryl-CoA, butyryl-CoA, E. coli ACP, and malonyl-CoA. Oligonucleotides were synthesized at Gibco BRL (Life Technologies). Restriction endonucleases and other enzymes were purchased from New England Biolabs (Beverly, Mass.), Perkin-Elmer (Branchburg, N.J.), and Boehringer Mannheim (Bedford, Mass.). Malonyl-ACP was synthesized and purified as previously described (7, 8).
Bacterial strains, plasmids, and cultivation of bacteria.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli TG-2 was used as a host for all plasmid construction. E. coli cells carrying plasmid constructs were grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) at 37°C. E. coli BL21(DE3)/pLysS was used for expression of N-terminal polyhistidyl (His)-tagged KASIII upon isopropyl-β-d-thiogalactopyranoside (IPTG) induction. IPTG (1 mM) was added when the optical density of the culture at 540 nm reached 0.5, and the culture was incubated at 30°C for an additional 3 h. Streptomyces cells carrying plasmids were grown in YEME medium (5) at 30°C in the presence of thiostrepton (5 μg/ml) for 3 days.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterium or plasmid | Relevant genotype or features | Source |
|---|---|---|
| Bacteria | ||
| E. coli TG-2 | supE hsdΔ5 thi Δ(lac-proAB) Δ(srl-recA)306::Tn10(Tetr) | Novagen |
| E. coli BL21 (DE3)/pLysS | F− ompT hsdSB(rB−mB−) gal dcm(DE3) pLysS(Cmr) | Novagen |
| S. glaucescens | Wild type | C. R. Hutchinson |
| Plasmids | ||
| pET15b | Apr; expression vector with His tag | Novagen |
| pLH14 | pET15b with 1.0-kb S. glaucescens fabH | L. Han and K. A. Reynolds (7) |
| pES2 | pET15b with 1.0-kb S. glaucescens fabH122CS | This study |
| pEA23 | pET15b with 1.0-kb S. glaucescens fabH122CA | This study |
| pEG4 | pET15b with 1.0-kb S. glaucescens fabH122CQ | This study |
| pSE34 | Apr Tsr PermE∗; expression vector for Streptomyces | Pfizer Inc. |
| pSW7 | Apr Tsr PermE∗::fabH; from S. glaucescens | This study |
| pSS4 | Apr Tsr PermE∗::fabH122CS; from S. glaucescens | This study |
| pSA11 | Apr Tsr PermE∗::fabH122CA; from S. glaucescens | This study |
| pSG14 | Apr Tsr PermE∗::fabH122CQ; from S. glaucescens | This study |
Site-directed mutagenesis of S. glaucescens KASIII.
A four-primer site-directed mutagenesis system was used to create point mutations within the fabH gene (4, 10). Primers used for site-directed mutagenesis and cloning of mutant fabH are listed in Table 2. The Cys122Ser amino acid substitution was obtained by using two pairs of primers, FORBGLII-REVSER and REVBGLII-FORSER (Table 2), to amplify two overlapping fragments from chromosomal DNA. The resulting two fragments were mixed and further amplified using primer pair FORBGLII-REVBGLII (Table 2). The resulting 1.08-kb PCR fragment contained the desired mutant fabH gene flanked with BglII sites. The Cys122Ala and Cys122Gln amino acid substitutions were obtained by following the same procedure and using overlapping primer pairs REVBGLII-FORALA/FORBGLII-REVALA and REVBGLII-FORGLN/FORBGLII-REVGLN correspondingly (Table 2). The presence of the appropriate mutation was confirmed by DNA sequence analysis. The wild-type fabH flanked with BglII sites was obtained by PCR amplification of a chromosomal template with FORBLGII and REVBGLII primers.
TABLE 2.
Primers used in this study
| Primer | Sequence (5-OH–3-OH) |
|---|---|
| FORBGLII | CTCTTCGCAGATCTGGCCTGAGT |
| REVBGLII | TCCGCAGCAGATCTCGTGATCCG |
| FORSER | CGACATCTCGGCGGGATCCGCCGGCTTCGGCTACG |
| REVSER | CGTAGCCGAAGCCGGCGGATCCCGCCGAGATGTCG |
| FORALA | CGACATCTCGGCGGGCGCCGCCGGCTTCGGCTACG |
| REVALA | CGTAGCCGAAGCCGGCGGCGCCCGCCGAGATGTCG |
| FORGLN | CGACATCTCGGCGGGCCAGGCCGGCTTCGG |
| REVGLN | CGTAGCCGAAGCCGGCCTGGCCCGCCGAGA |
| FABHNDEI | GCCGACCGACATATGTCGAAGATCAAGCC |
Construction of plasmids for expression of mutant and wild-type fabH in E. coli and S. glaucescens.
The DNA fragments encoding wild-type KASIII and KASIII with Cys122Ser, Cys122Ala, and Cys122Gln mutations were inserted into the BamHI site of pSE34 (a streptomycete expression plasmid) downstream of PermE∗ to create pSW7, pSS4, pSA11, and pSG14, respectively (Table 1). Transformation of S. glaucescens with pSW7, pSS4, pSA11, and pSG14 led to strains that produced either elevated levels of the wild-type KASIII or a mutant KASIII (Cys122Ser, Cys122Ala, or Cys122Gln). Plasmids pSS4, pSA11, and pSG14 were also used as templates for PCR with FabHNDEI and REVBGLII primers to create an NdeI site at the codon for the N-terminal methionine and a BglII site downstream of the fabH stop codon. The resulting PCR fragments containing the mutated fabH genes were cloned into the corresponding sites of pET15b to create pES2 (Cys122Ser mutation), pEA23 (Cys122Ala mutation), and pEG4 (Cys122Gln mutation). Transformation of E. coli with pES2, pEA23, and pEG4 gave strains that have high-level expression of S. glaucescens fabH122CS, fabH122CA, and fabH122CQ upon IPTG induction. E. coli transformed with pLH14 (7) was used for overexpression of wild-type S. glaucescens fabH.
PCR and DNA sequencing.
PCR was performed with the Gene AMPXL-PCR kit from Perkin-Elmer according to standard protocols. PCR products were eluted from agarose gel using the Elu-quik DNA purification kit from Schleicher & Schuell. Automated DNA sequencing was performed by the Medical College of Virginia-Virginia Commonwealth University Nucleic Acids Core Facility.
General methods.
All DNA manipulations and transformation of E. coli were performed according to standard protocols (20). Plasmid transformation of S. glaucescens protoplasts was also carried out by following standard protocols (11). Analysis of fabH expression was carried out using sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis (SDS–15% PAGE). For protein purification, cells were lysed and purified by metal chelate chromatography according to the methodology supplied by Novagen.
Fatty acid profile analysis.
Fatty acid profiles of Streptomyces were analyzed by following standard protocols (29).
Malonyl-ACP decarboxylation.
Assay mixtures contained 50 μM malonyl-ACP, 1 mM β-mercaptoethanol, 100 μM substrate (butyryl-CoA or isobutyryl-CoA), 0.1 M sodium phosphate buffer (pH 7.0), and different amounts of wild-type or mutant KASIII in a final volume of 20 μl. The mixtures were incubated at 37°C for 1 h, and the reaction was stopped by placing the mixtures in an ice bath. The conversion of malonyl-ACP to acetyl-ACP was assayed by loading the mixture on a 15% polyacrylamide gel containing 0.5 M urea. Electrophoresis was performed at 25°C and 23 mA.
Perdeuterated acetate incorporation.
In feeding experiments perdeuterated acetate was added to Streptomyces growth media to a final concentration of 10 mM. Streptomyces was grown in the presence of perdeuterated acetate in YEME for 4 days at 30°C and then harvested for fatty acid analysis. The fatty acid quantities were estimated by integrating the area of each major fatty acid peak and expressed as percentages of the total fatty acid pool. The determination of the percent labeling of the palmitate pool from an intact D3-acetate was based on the relative intensities of molecular ions (m/z) at 273 (methyl palmitate with three deuterium atoms) and 270 (unlabeled methyl palmitate) after correction for the natural abundance of 13C and incorporation of deuterated malonate (28).
RESULTS
Expression of wild-type and KASIII mutants in S. glaucescens.
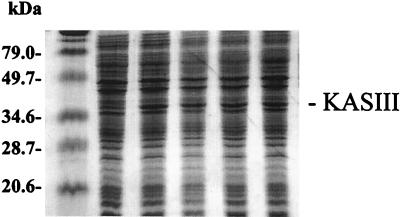
Analysis of the amino acid sequence of the S. glaucescens KASIII revealed only one cysteine residue (Cys122). This residue was shown by an alignment of the amino acid sequences of other KASs to be homologous to active-site Cys112 of the E. coli KASIII (5, 18). We considered that replacement of the Cys122 residue in the S. glaucescens KASIII with glutamine or other residues would reduce the KAS condensation activity of KASIII but not the malonyl decarboxylase activity. The acetyl-ACP generated by malonyl-ACP decarboxylation has been suggested previously to be an efficient primer for SCFA in organisms producing predominantly BCFAs (14, 21). Three mutant fabH genes capable of generating Cys122Ala, Cys122Ser, and Cys122Gln KASIII mutants were created by site-directed mutagenesis and cloned into Streptomyces expression vector pSE34 under the control of the PermE∗ promoter. The resulting plasmids and pSW7 (expressing the wild-type KASIII) were used to transform S. glaucescens. Analysis by SDS-PAGE of the resulting transformed strains revealed an enhanced protein band at the predicted molecular mass of 34 kDa compared to the S. glaucescens/pSE34 control (Fig. 2). A visual comparison did not reveal a distinctly distinguishable level of enhancement of this protein for each transformant, suggesting comparable levels of expression for the mutant and wild-type KASIIIs.
FIG. 2.
SDS-PAGE analysis of S. glaucescens cell extracts. Cells were grown in YEME supplemented with 5 μg of thiostrepton/ml at 30°C for 4 days. Lanes are numbered from left to right starting with the second lane from the left. Extracts were from S. glaucescens carrying pSE34 (lane 1), pSG14 (lane 2), pSS4 (lane 3), pSA11 (lane 4), and pSW7 (lane 5). The far left lane contains molecular mass standards.
Alteration of S. glaucescens fatty acid profiles by KASIII mutants.
The fatty acid compositions of the S. glaucescens transformants were analyzed by standard methodologies (Fig. 3.). The fatty acid profiles of S. glaucescens strains carrying the control pSE34 plasmid or pSW7 (wild-type KASIII expression) were essentially identical and exhibited no differences from profiles obtained with the wild-type S. glaucescens. It has previously been reported that high-level expression of the E. coli and Brassica napus KASIIIs in E. coli leads to significant shortening of the fatty acid chain lengths (24, 26). Several possible explanations for this observation have been advanced. None of these, however, can account for the observation that this phenomenon occurs only when certain KASIII enzymes are used and does not occur with expression of the S. glaucescens KASIII in either S. glaucescens (Fig. 3.) or E. coli (data not shown).
FIG. 3.
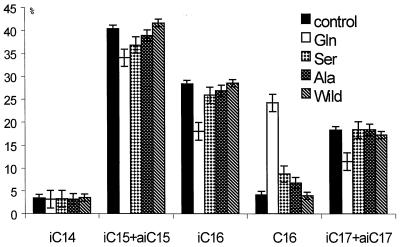
Relative abundances of the major fatty acids produced by S. glaucescens carrying plasmids pSE34 (control), pSG14 (Gln), pSS4 (Ser), pSA11 (Ala), and pSW7 (wild). pSG14, pSS4, pSA11, and pSW7 allow expression of S. glaucescens fabH122CQ, fabH122CS, fabH122CA, and fabH, respectively. Fatty acid abbreviations: iC14, isomyristate; iC15, isopentadecanoate; aiC15, anteisopentadecanoate; iC16, isopalmitate; C16, palmitate; iC17, isoheptadecanoate; aiC17, anteisoheptadecanoate.
Fatty acid analyses revealed that plasmid-based expression of the KASIII mutants in S. glaucescens led to various effects on the ratio of SCFAs and BCFAs. The pSA11 and pSS4 transformants, expressing the Cys122Ala and Cys122Ser mutants, respectively, generated slightly higher levels (increased by 75 to 100%) of their fatty acids as SCFA palmitate. The levels of palmitate in the pSG14 transformant (expressing the Cys122Gln mutant) were significantly higher (increased by more than 500%) (Fig. 3 and 4). Significant decreases in the major BCFAs of the pSA11 and pSS4 transformants were difficult to observe (large changes in the amounts of these fatty acids would not be expected if the levels of minor SCFA components were to increase less than twofold). Significant and reproducible decreases, however, were observed for the major BCFAs produced by the pSG14 transformant (Fig. 3 and 4). The observations with the pSG14 transformant are consistent with the notion that the Cys122Gln (KASIIIQ) mutant generates acetyl-ACP specifically for initiating SCFA biosynthesis and competes with the wild-type KASIII-catalyzed process for initiation of BCFA and SCFA biosynthesis. These competing pathways for initiation of fatty acid biosynthesis presumably also occur to a lesser degree in the strains carrying the Cys122Ser and Cys122Ala mutants.
FIG. 4.
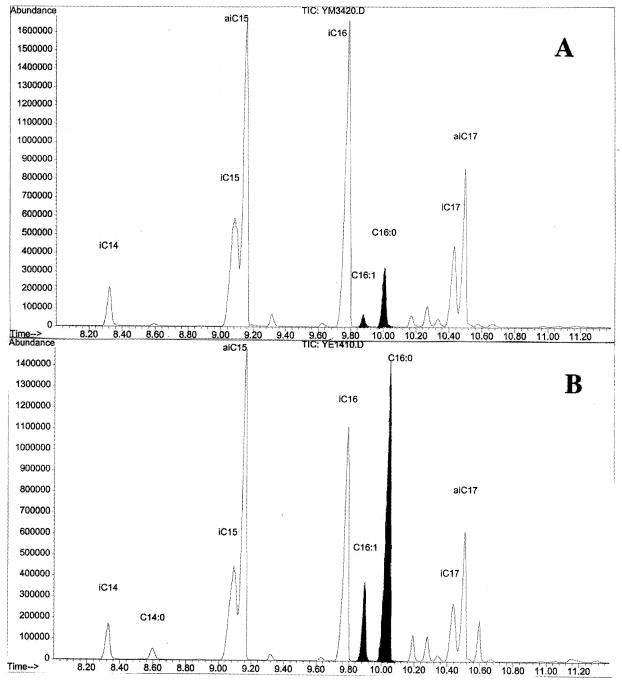
Fatty acid profiles of S. glaucescens carrying pSE34 (A) and pSG14 (B) (allows expression of S. glaucescens fabH122CQ). Fatty acid abbreviations: iC14, isomyristate; C14:0, myristate; iC15, isopentadecanoate; aiC15, anteisopentadecanoate; iC16, isopalmitate; C16:1, palmitoleate; C16:0, palmitate; iC17, isoheptadecanoate; aiC17, anteisoheptadecanoate. TIC, total ion chromatogram.
Incorporation experiments with perdeuterated acetate.
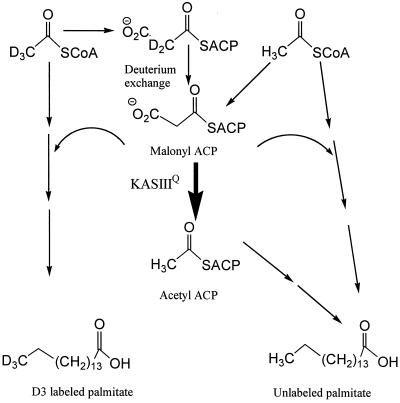
Evidence that the KASIII mutants catalyze fatty acid initiation via decarboxylation of malonyl-ACP was provided by growing S. glaucescens pSE34 and pSG14 transformants in the presence of perdeuterated acetate. Under these conditions perdeuterated acetyl-CoA is generated within the cell and competes with unlabeled acetyl-CoA for initiating biosynthesis of SCFAs such as palmitoyl-CoA. In the fatty acid analyses the hydrolysis and subsequent methylation of this acyl thioester generate methyl palmitate with a molecular ion of 270 (29). Labeled palmitate with a molecular ion of 273 elutes at a slightly earlier retention time in the gas chromatography-mass spectrometry analyses, and this is indicative of the intact use of perdeuterated acetyl-CoA as a starter unit (28). It is well established that significantly lower levels of deuterium are incorporated into palmitate via the malonyl-CoA extender units (formed by carboxylation of the perdeuterated acetyl-CoA) (28). This observation has been attributed to a combination of “washout” of the deuterium label (exchange of deuterium with hydrogen) from the malonyl-CoA and/or malonyl-ACP substrate (Fig. 5) and the corresponding β-ketoacyl-ACP product, as well as the loss of deuterium in the subsequent dehydration step (19). Thus any contributions to the labeled palmitate with a molecular ion of 273 by multiple incorporations of deuterated malonyl-ACP are low and can be readily corrected for (28).
FIG. 5.
Introduction of KASIIIQ into S. glaucescens creates a new pathway (boldface arrow) for initiation of SCFA biosynthesis. Acetyl-ACP is generated by the malonyl-ACP decarboxylase activity of KASIIIQ. Perdeuterated acetyl-CoA retains three deuteriums if used directly for palmitate biosynthesis but lost them if processed through a malonyl-ACP intermediate to acetyl-ACP.
In experiments with the pSE34 transformant 9% ± 1% of the palmitate pool was generated from a perdeuterated acetyl-CoA starter unit, compared with 3.6% ± 0.5% with the pSG14 transformant. Thus, while the amount of palmitate increases dramatically in the pSG14 transformant, the amount directly generated from perdeuterated acetyl-CoA decreases. This observation is consistent with the use of an alternative unlabeled starter unit such as acetyl-ACP in this process. Conversion of perdeuterated acetyl-CoA to acetyl-ACP via the actions of acetyl-CoA carboxylase and malonyl-CoA–ACP transacylase and the malonyl-ACP decarboxylase activity of the Cys122Gln KASIII mutant would result in loss of at least one deuterium. Analysis of the palmitate pool in the pSG14 transformant showed no evidence of significant labeling of the palmitate pool with a dideuteroacetyl starter unit, indicating that under the conditions of the experiment extensive deuterium washout must occur at the level of the malonyl thioester (Fig. 5).
Malonyl-ACP decarboxylase activity of mutant and wild-type KASIII.
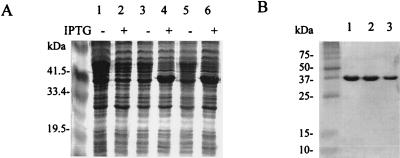
The malonyl-ACP decarboxylase activity of the wild-type S. glaucescens KASIII and mutants in vitro was studied. With E. coli as the host, the corresponding genes were expressed (Fig. 6A) and the resulting His-tagged proteins were purified by following standard protocols (Fig. 6B). The Cys122Ser mutant accumulated predominantly as an insoluble form in E. coli, preventing further study. This observation was somewhat surprising as SDS-PAGE analysis and fatty acid profiles of the pSS4 transformant did not indicate that there was a significant difference between the solubility of this mutant and those of the other mutants or wild-type KASIII in S. glaucescens.
FIG. 6.
Expression (A) and purification (B) of S. glaucescens KASIII wild type and mutants (B). Extracts prepared from E. coli BL21(DE3)/pLysS carried pET15b (A, lanes 1 and 2), pLH14 (A, lanes 3 and 4), and pEG4 (A, lanes 5 and 6) harvested 3 h after IPTG induction at 30°C. Control cultures (A, lanes 1, 3, and 5) were grown without IPTG. The wild type (B, lane 1) and Cys122Gln (B, lane 2), and Cys122Ala (B, lane 3) mutants were purified by Ni2+ chelate affinity chromatography as described in Materials and Methods. Samples were analyzed by using SDS-PAGE.
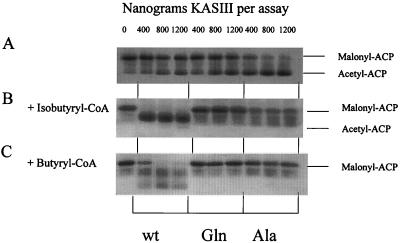
Malonyl-ACP decarboxylase assays were carried out at 37°C for 1 h using purified malonyl-ACP and different amounts of the enzymes. The conversion of malonyl-ACP to acetyl-ACP was analyzed on a conformationally sensitive gel (Fig. 7). Under these conditions the Cys122Ala mutant exhibited the highest in vitro decarboxylase activity. No significant differences between the wild type and the Cys122Gln mutant were observed (Fig. 7A).
FIG. 7.
Decarboxylation of malonyl-ACP by wild-type (wt) and Cys122Gln (Gln) and Cys122Ala (Ala) mutant KASIII. Assay mixtures contained malonyl-ACP and three different amounts (400 to 1,200 ng) of each KASIII (A). Assays were also conducted in the presence of isobutyryl-CoA (B) and isobutyryl-CoA (C) as described in Materials and Methods. The reaction mixtures were analyzed by using electrophoresis through a 15% polyacrylamide gel containing 0.5 M urea.
The assay was then repeated in the presence of isobutyryl-CoA and butyryl-CoA, (known acyl-CoA substrates for S. glaucescens KASIII [7]) (Fig. 7B and C). Under these conditions the decarboxylase activities of both mutants were extensively inhibited. In contrast, almost a complete loss of the malonyl-ACP substrate was observed with the wild-type enzyme (even at the lowest [400-ng] level). No significant levels of acetyl-ACP were generated; rather 3-ketoacyl-ACP and related degradation products were observed (it is well established that these compounds have poor stability under these assay conditions [9]). Thus in the presence of acyl-CoA substrates the wild-type enzyme preferentially catalyzes the coupled rather than the uncoupled decarboxylation process. Furthermore, under these assay conditions the efficiency of the malonyl-ACP decarboxylase activity in the coupled reaction of the wild-type KASIII is greater than the corresponding activity in the uncoupled reactions of either mutant, even in the absence of an acyl-CoA substrate (where the lowest [400-ng] level of protein only partially converted the malonyl-ACP substrate to acetyl-ACP).
DISCUSSION
KASIII initiates fatty acid biosynthesis in type II dissociable FASs by catalyzing a Claisen condensation using an acyl-CoA and a malonyl-ACP as substrates (5) (Fig. 1A). The first half of this double-displacement mechanism involves the binding of the acyl-CoA substrate and subsequent formation of an acyl-enzyme intermediate on a Cys residue with release of CoA (18). In the second half of the process malonyl-ACP binds to the acylated enzyme and a subsequent decarboxylative condensation releases the 3-ketoacyl-ACP product. It has been well established that chemical (iodoacetamide) (16, 22) or site-directed (substitution with glutamine) modifications of active-site cysteine in other KAS enzymes (which use acyl-ACP substrates) (2, 30) lead to a loss of the corresponding KAS activity. In many of these cases it has also been demonstrated that these modifications lead to an increase in the rate of the uncoupled malonyl thioester decarboxylase activity. We hypothesized that such a mutation in KASIII might have a similar effect and might lead to the generation of a mutant able to generate acetyl-ACP.
S. glaucescens was chosen as a host for these studies as it makes predominantly BCFAs (7). This strain like other streptomycetes makes a small amount of SCFAs, presumably because of the preference of KASIII for BCFA acyl-CoA precursors over acetyl-CoA (7). Studies of cell extracts of microorganisms that produce predominantly BCFAs have reported that acetyl-ACP functions more effectively than acetyl-CoA for initiating palmitate biosynthesis (14, 21). This difference presumably reflects an ability of the acetyl-ACP to be directly elongated using a different KAS, circumventing the substrate specificity of the KASIII-catalyzed initiation step. We reasoned that significant quantities of acetyl-ACP could be generated in S. glaucescens by expression of a Cys122Gln KASIII mutant (KASIIIQ) with malonyl-ACP decarboxylase activity and that this should therefore lead to a significant increase in the levels of SCFAs relative to BCFAs. As described above these levels increased more than fivefold in the S. glaucescens/pSG14 KASIIIQ expression strain.
Experiments using perdeuterated acetate demonstrated that a significantly smaller amount of the SCFA pool was generated directly from an acetyl-CoA starter unit in the pSG14 transformant than was generated in a control (a strain carrying pSE34). These observations support the notion that the increase in SCFA biosynthesis is the result of an increase in the acetyl-ACP pool. Significant increases in SCFA biosynthesis (with concomitant decreases in the levels of BCFA) have previously been observed for addition of nonlethal doses of type II FAS inhibitor thiolactomycin to cultures of S. glaucescens and Streptomyces collinus (7, 28). In this case an increase, rather than a decrease, in the intact utilization of a perdeuterated acetyl-CoA starter unit was observed (28). A different mechanism must, therefore, account for the alteration of fatty acid profiles of S. glaucescens grown in the presence of this type II FAS inhibitor.
Clear evidence that both type I and type II polyketide biosynthetic processes can be initiated using an uncoupled decarboxylative process with either malonyl- or methylmalonyl-ACP has been presented (1–3). It has been proposed that decarboxylation of malonyl-ACP by KASI and KASII might also under some circumstances play a role in initiation of fatty acid biosynthesis in E. coli (17). That such a process occurs to any significant degree in type II fatty acid biosynthetic processes has not been proven and would appear to be discounted by the observation that no viable fabH deletion mutants have been reported for E. coli. All attempts by us to generate a fabH mutants in S. glaucescens have failed, suggesting that the KASIII catalysis of fatty acid synthesis is an essential process (unpublished data).
The first clear example of a decarboxylative pathway for initiation of fatty acid biosynthesis was therefore observed in a strain carrying the Cys122Gln KASIII mutant. Interestingly this pathway does not occur with the increased levels of wild-type enzyme, despite the fact that both enzymes exhibit similar in vitro malonyl-ACP decarboxylase activities. In S. glaucescens/pSG14 the new decarboxylative pathway for initiation of SCFA biosynthesis must function alongside the normal KASII-catalyzed initiation of the SCFA and BCFA pathways (the host contains the wild-type fabH). It will be interesting to determine if an S. glaucescens mutant capable of generating only SCFAs can be created by replacement of the wild-type fabH with fabH122CQ.
Conversion of the active-site cysteine to glutamine in the KAS domain of the rat type I FAS has recently been reported to increase malonyl-ACP decarboxylase activity by more than 2 orders of magnitude (30). A substantial increase in malonyl-ACP decarboxlase activity with glutamine substitution of the active-site cysteine in KSα, a component of the actinorhodin type II PKS, has also recently been reported (2). It has long been speculated that KASs exist in two distinct conformations, one favoring the initial acylation step and the other favoring the binding and subsequent decarboxylation of malonyl-ACP (16, 30). It has been suggested that the structure of the Cys-to-Gln replacement mutant KAS of the rat FAS is homologous to the acyl enzyme intermediate and thus allows a conformation that favors the productive binding of the malonyl-ACP (30). Such an argument cannot be extended the S. glaucescens KASIII, where the uncoupled malonyl-ACP decarboxylase activity is unaffected by an analogous replacement and remains substantially lower than the corresponding activity in the coupled reaction of the wild-type enzyme. Decarboxylation activity of the S. glaucescens KASIII was, however, notably higher with the cysteine-to-alanine replacement. Interestingly a Cys112Ser mutant E. coli KASIII has also been reported to have an increased (400%) decarboxylase activity (5) compared to that of the wild-type enzyme. The homologous mutations of the KAS domain of the rat FAS, on the other hand, did not yield marked differences in the malonyl-ACP decarboxylase activity (30). It is evident from these studies that the type and magnitude of effects of mutations of the active-site cysteine on malonyl-ACP decarboxylase activity are very much dependent on the KAS used.
Inhibition of the malonyl-ACP decarboxylase activity of the two KASIII mutants by isobutyryl-CoA and butyryl-CoA was somewhat surprising in that the enzymes do not contain an active-site cysteine required for formation of an acyl enzyme intermediate. The mutations, however, might not completely disrupt the noncovalent enzyme substrate complex that precedes the formation of the acyl enzyme intermediate. Thus high concentrations of isobutyryl-CoA or butyryl-CoA could bind in this form, blocking malonyl-ACP binding. Within this context it is reasonable to suggest that a KASIII mutant replacing the cysteine with a longer glutamine residue would have a lower affinity for acyl-CoA compounds than mutants generated using alanine or serine. In this case the Cys122Gln mutant could act more effectively as a malonyl-ACP decarboxylase in vivo than the Cys122Ala mutant, despite having lower in vitro activity. A decreased inhibition by acyl-CoA compounds might similarly be a contributing factor for the selection of a glutamine residue, in place of cysteine, in the active site of the polyketide synthase KS domains which catalyze initiation by decarboxylative condensation. Unfortunately the use of a conformationally sensitive gel in an assay for malonyl-ACP decarboxylation prevented attempts to accurately determine if there was differential inhibition of various KASIII mutants with acyl-CoA compounds.
There is very little information available regarding the pools of substrates for either polyketide or fatty acid biosynthesis in streptomycetes. The experiments described in this study place some limits on the concentrations that must exist during fatty acid biosynthesis under the tested conditions. The wild-type KASIII does not appear to act as a decarboxylase in vivo but can do so in vitro. The levels of isobutyryl-CoA and methylbutyryl-CoA (precursors for the major fatty acid made in this organism) must be sufficient to allow this enzyme to function as a KAS rather than a decarboxylase. The Km of the S. glaucescens KASIII has previously been reported to be 0.4 μM (7). The observation that the KASIIIQ mutant functions as a decarboxylase in vivo must indicate that the levels of these substrates are not substantially higher than 100 μM. A preliminary analysis of acyl-CoA pools in streptomycetes has been found to be consistent with this prediction (G. Florova and K. A. Reynolds, unpublished results).
In conclusion, KASIII plays a critical role in controlling fatty acid biosynthesis in a type II FAS. The unique features of this pathway, and of this enzyme in particular, have attracted considerable interest as targets for both antibiotic development and the engineering of altered seed oil composition in transgenic plants (5, 25). In this paper we have clearly demonstrated that introduction of a KASIIIQ into a streptomycete can provide a new pathway for fatty acid initiation. This pathway, which proceeds by decarboxylation of a malonyl-ACP extender unit and which appears not to use the wild-type KASIII, leads to a significant alteration in the fatty acid profile of this organism.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (GM50542 and AI44772) and Pfizer Incorporated.
We thank Galina Florova and Xin He for help in preparation of malonyl-ACP.
REFERENCES
- 1.Bao W, Wendt-Pienkowski E, Hutchinson C R. Reconstitution of the iterative type II polyketide synthase for tetracenomycin F2 biosynthesis. Biochemistry. 1998;37:8132–8138. doi: 10.1021/bi980466i. [DOI] [PubMed] [Google Scholar]
- 2.Bisang C, Long P F, Cortes J, Westcott J, Crosby J, Matharu A-L, Simpson T J, Leadley P F. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature. 1999;401:502–505. doi: 10.1038/46829. [DOI] [PubMed] [Google Scholar]
- 3.Carreras C W, Khosla C. Purification and in vitro reconstitution of the essential protein components of an aromatic polyketide synthase. Biochemistry. 1998;37:2084–2088. doi: 10.1021/bi972919+. [DOI] [PubMed] [Google Scholar]
- 4.Choi K-H, Heath R J, Rock C O. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J Bacteriol. 2000;182:365–370. doi: 10.1128/jb.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies C, Heath R J, White S W, Rock C O. The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from Escherichia coli. Struct Fold Des. 2000;8:185–195. doi: 10.1016/s0969-2126(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 6.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Modular organization of the genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 7.Han L, Lobo S, Reynolds K A. Characterization of 3-ketoacyl acyl carrier protein synthase III from Streptomyces glaucescens: its role in the initiation of fatty acid biosynthesis. J Bacteriol. 1998;180:4481–4486. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He X, Mueller J P, Reynolds K A. Development of a scintillation proximity assay for beta-ketoacyl-acyl carrier protein synthase III. Anal Biochem. 2000;282:107–114. doi: 10.1006/abio.2000.4594. [DOI] [PubMed] [Google Scholar]
- 9.Heath R J, Rock C O. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271:1833–1836. doi: 10.1074/jbc.271.4.1833. [DOI] [PubMed] [Google Scholar]
- 10.Ho S H, Hunt H D, Horton R M, Pollen J K, Pease L R. Site-directed mutagenesis by overlapping extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 11.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Smith J M, Ward J M, Schrempf H S. Genetic manipulation of Streptomycetes, a laboratory manual. Norwich, United Kingdom: John Innes Institute; 1985. [Google Scholar]
- 12.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci USA. 1999;96:9509–9514. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakavas S J, Katz L, Stassi D. Identification and characterization of the niddamycin polyketide synthase genes from Streptomyces caelestis. J Bacteriol. 1997;179:7515–7522. doi: 10.1128/jb.179.23.7515-7522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz L. Manipulation of modular polyketide synthases. Chem Rev. 1997;97:2557–2575. doi: 10.1021/cr960025+. [DOI] [PubMed] [Google Scholar]
- 16.Kresze G B, Steber L, Oesterhelt D, Lynen F. Reaction of yeast fatty acid synthetase with iodoacetamide. 3. Malonyl-coenzyme A decarboxylase as product of the reaction of fatty acid synthetase with iodoacetamide. Eur J Biochem. 1977;79:191–199. doi: 10.1111/j.1432-1033.1977.tb11797.x. [DOI] [PubMed] [Google Scholar]
- 17.Magnusson K, Jackowski S, Rock C O, Cronan J E. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu X, Janson C A, Konstantinidis A K, Nwagwu S, Silverman C, Smith W W, Khandekar S, Lonsdale J, Abdel-Meguid S S. Crystal structure of beta-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis. J Biol Chem. 1999;274:36465–36471. doi: 10.1074/jbc.274.51.36465. [DOI] [PubMed] [Google Scholar]
- 19.Saito K, Kawaguchi A, Seyama Y, Yamakawa T, Okuda S. Steric course of deuterium incorporation from [2-2H]malonyl CoA into fatty acids by fatty acid synthases. J Biochem. 1981;90:1697–1704. doi: 10.1093/oxfordjournals.jbchem.a133646. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Suutari M, Laasko S. Changes in fatty acid branching and unsaturation in Streptomyces griseus and Brevibacterium fermentans as a response to growth temperature. Appl Environ Microbiol. 1992;58:2338–2430. doi: 10.1128/aem.58.7.2338-2340.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomoda H, Kawaguchi A, Omura S, Okuda S. Cerulenin resistance in a cerulenin-producing fungus. II. Characterization of fatty acid synthetase from Cephalosporium caerulens. J Biochem (Tokyo) 1984;95:1705–1712. doi: 10.1093/oxfordjournals.jbchem.a134784. [DOI] [PubMed] [Google Scholar]
- 23.Tsay J-R, Oh W, Larson T J, Jackowski S, Rock C O. Isolation and characterization of the b-keto-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem. 1992;267:6807–6814. [PubMed] [Google Scholar]
- 24.Tsay J T, Rock C O, Jackowski S. Overproduction of β-ketoacyl-acyl carrier protein synthase I imparts thiolactomycin resistance to Escherichia coli K-12. J Bacteriol. 1992;174:508–513. doi: 10.1128/jb.174.2.508-513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verwoert I I, Verbree E C, van der Linden K H, Nijkamp H J, Stuitje A R. Cloning, nucleotide sequence, and expression of the Escherichia coli fabD gene, encoding malonyl coenzyme A-acyl carrier protein transacylase. J Bacteriol. 1992;174:2851–2857. doi: 10.1128/jb.174.9.2851-2857.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verwoert I I G S, van der Linden K H, Walsh M C, Nijkamp H J J, Stuitje A R. Modification of Brassica napus seed oil by expression of the Escherichia coli fabH gene, encoding 3-ketoacyl-acyl carrier protein synthase III. Plant Mol Biol. 1995;27:875–886. doi: 10.1007/BF00037016. [DOI] [PubMed] [Google Scholar]
- 27.Wakil S J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 28.Wallace K K, Han L, Lobo S, McArthur H A I, Reynolds K A. In vivo and in vitro effect of thiolactomycin on fatty acid biosynthesis in Streptomyces collinus. J Bacteriol. 1997;179:3884–3891. doi: 10.1128/jb.179.12.3884-3891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace K K, Zhao B, McArthur H A I, Reynolds K A. In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol Lett. 1995;131:227–234. doi: 10.1111/j.1574-6968.1995.tb07781.x. [DOI] [PubMed] [Google Scholar]
- 30.Witkowski A, Joshi A K, Lindqvist Y, Smith S. Conversion of a beta-ketoacyl synthase to a malonyl decarboxylase by replacement of the active-site cysteine with glutamine. Biochemistry. 1999;38:11643–11650. doi: 10.1021/bi990993h. [DOI] [PubMed] [Google Scholar]
- 31.Xue Y, Zhao L, Liu H-W, Sherman D H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezulae: architecture of metabolic diversity. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]