Abstract
Tick salivary glands secrete a complex saliva into their hosts which modulates vertebrate hemostasis, immunity and tissue repair mechanisms. Transcriptomic studies revealed a large number of transcripts coding for structural and secreted protein products in a single tick species. These transcripts are organized in several large families according to their products. Not all transcripts are expressed at the same time, transcription profile switches at intervals, characterizing the phenomenon of “sialome switching”. In this work, using transcriptomic and proteomic analysis we explored the sialome of Rhipicephalus sanguineus (s.l.) adult female ticks feeding on a rabbit. The correlations between transcriptional and translational results in the different groups were evaluated, confirming the “sialome switching” and validating the idea that the expression switch may serve as a mechanism of escape from the host immunity. Recombination breakpoints were identified in lipocalin and metalloprotease families, indicating this mechanism could be a possible source of diversity in the tick sialome. Another remarkable observation was the identification of host-derived proteins as a component of tick salivary gland content. These results and disclosed sequences contribute to our understanding of tick feeding biology, to the development of novel anti-tick methods, and to the discovery of novel pharmacologically active products.
1. Introduction
Ticks are exclusive blood sucking mites belonging to three different families, the Argasidae, or soft ticks, which feeds for minutes to hours in their vertebrate hosts, the Ixodidae, or hard ticks, which feeds for several days and the Nuttalliellidae family comprising of a single extant species [1].
The salivary glands of ticks are important in several ways for their survival, by helping the tick to collect water from humid air (when the tick spreads a hygroscopic saliva over its palps, re-ingesting it later with the absorbed water), during blood feeding (by antagonizing host hemostasis and modulating host immunity and tissue repair reactions), and to maintain ion and water metabolism during blood feeding (when most of the ingested water contained in the blood meal is reinjected in the host as saliva), and to assist tick reproduction (when saliva is spread over the sperm collected by male ticks before insertion into the female vaginal pore, a function similar to that performed by invertebrate accessory glands or vertebrate prostate glands) [2–5].
The feeding process of hard ticks is classically divided in three phases, the attachment phase, usually lasting one day, the slow feeding phase lasting several days, and the fast feeding, or “big sip”, usually lasting one day [4]. The name “slow feeding”, however, is a misnomer, since most of the blood removed from the host occurs during this phase but the majority of the meal protein is excreted in the feces and the blood water and ions is reinjected into the host as saliva [6].
Recent transcriptomic studies of the salivary glands of hard ticks have revealed a surprisingly large number of transcripts coding for many different families of proteins, totaling over one thousand different salivary secreted polypeptides per tick species [5,7–27]. Their coding genes appear to be evolving at a fast pace of evolution, due to relaxed and/or positive selection [28,29]. Interestingly, these transcripts are not all expressed at once, but rather at different stages of feeding [8,22,26,30], or when feeding on different hosts [31]. This process of sialome switching has been interpreted as a mechanism of immune evasion [22,26,30]. Proteomic studies of the tick salivary glands at different stages of feeding and when ticks were exposed to different hosts have also been performed [32–36], indicating in some cases a disagreement between the proteomic and transcriptomic findings [35]. These pioneer studies, however, were done with few developmental stages of feeding, and in most cases had only one biological replicate.
Rhipicephalus sanguineus sensu lato (s.l.) is a cosmopolitan tick species parasitizing domestic animals such as dogs and cats, and, occasionally, humans [37,38]. R. sanguineus can act as a vector of Rickettsia rickettsii to humans [39] and Babesia spp. to dogs and cats [40,41]. A limited Sanger-based sialotranscriptome has been previously performed [21], using salivary glands from ticks feeding for 3–5 days, and 5 days, leading to the description of 1024 transcripts, 26% of which were considered as coding for salivary secreted proteins. Moreover, a transcriptome of larvae was described more recently focusing in genes involved in acaricide resistance [42]. Using Illumina technology, we herein analyze the sialotranscriptome of R. sanguineus of adult female ticks at six different stages of feeding, and with three biological replicates. Uniquely, instead of grouping the different feeding stages by days of feeding, we grouped the ticks by their weight, as it is a better indicator of the physiological status of the tick feeding process [43]. Using the coding sequences derived from the transcriptome assembly to form a database for proteomic studies, we pursued a proteome analysis of the salivary glands, which confirmed the sialome switching at the proteomic level, and with a high degree of agreement between transcriptome and proteome expression levels.
2. Material and methods
2.1. Ethics statement
Animals used in the experiments were housed at Faculdade de Veterinária, Universidade Federal do Rio Grande do Sul (UFRGS) facilities. This study was conducted according to the ethical and methodological norms prescribed by the International and National Directives and Norms by the Animal Experimentation Ethics Committee of UFRGS. Protocol (number 27559) was approved by the Comissão de Ética no Uso de Animais – CEUA – UFRGS.
2.1.1. Ticks, tick feeding and salivary gland dissection
R. sanguineus (s.l.) strain used in this study belongs to the tropical lineage [44] and were collected in Rio de Janeiro, Brazil [45], and kept at the UFRGS. Unfed ticks were maintained at 28 °C and 85% relative humidity before infestation on rabbits. Adults ticks used for salivary gland extraction were restricted to feed onto the outer part of the ear of four naïve female New Zealand rabbits with orthopedic stockinet’s glued. A total of 25 adult females and 10 males (35 ticks per ear, 70 ticks per animal) were placed into the tick containment apparatus and allowed to attach. To compose the groups of ticks by a blood feeding index, partially fed ticks were collected randomly from different hosts during feeding, selected by their engorgement size, sorted by weight, and collected into three independent replicates, including: group unfed (UF) (n = 5 females per replicate), group G1 (n = 5 females per replicate, average 1.8 mg, collected at day 2 of feeding), group G2 (n = 5 females per replicate, average 3.6 mg, collected at day 6 of feeding), group G3 (n = 4 females per replicate, average 7.0 mg, collected at day 6 of feeding), group G4 (n = 2 females per replicate, average 10.9 mg, collected at day 8 of feeding), group G5 (n = 3 females per replicate, average 24 mg, collected at days 8 and 11 of feeding), group G6 (n = 2 females per group, average 36 mg. collected at days 6, 10 and 13 of feeding). Supplemental fig. 1 displays a sample of the ticks collected for each group. Ticks from this experiment were used exclusively to dissect salivary glands for extraction of total RNA used for RNAseq analysis.
A second independent infestation was performed to dissect salivary glands to obtain protein for LC-MS/MS analysis. For this second infestation, two naïve female New Zealand rabbits were used. A total of 20 adult females and 10 males (30 ticks per ear, 60 ticks per animal) were placed into the tick containment apparatus and allowed to attach. Partially fed ticks were collected from host during feeding, selected by their engorgement size, sorted by weight, and divided: group UF (n = 10 females), group G1 (n = 13 females, average 1.8 mg), group G2 (n = 5 females, average 3.6 mg), group G3 (n = 5 females, average 7.0 mg), group G4 (n = 4 females, average 10.9 mg), group G5 (n = 5 females, average 24 mg), group G6 (n = 4 females, average 36 mg). The measurements of the scutum length and tick length were made using the ImageJ software [46].
After removal from the host, ticks were rinsed with ethanol 70% following a second rinsing with nuclease-free water. Ticks were dissected within two hours after removal from the host. Tick salivary glands (SGs) were dissected in a fresh ice-cold PBS, pH 7.4. After dissection, salivary glands were washed gently in a fresh ice-cold PBS. After washing, dissected SGs were stored immediately in RNAlater (Invitrogen, Carlsbad, CA, USA) prior to extracting total RNA and protein.
2.1.2. RNA and protein extractions
After removal from RNAlater, SGs were gently washed in nuclease- free phosphate-buffered saline pH 7.4 (Thermo Fisher Scientific), and immediately transferred to TRIzol® reagent (Thermo Fisher Scientific). Total RNA and protein were isolated according to manufacturer’s specifications. Protein pellet was resuspended into Tris-HCl 100 mM, urea 8 M, pH 8.0. Protein concentration was measured using BCA Protein Assay Reagent Kit (Thermo Scientific Pierce), following the manufacturer’s recommendations and samples were stored at −80 °C upon usage.
2.1.3. Library preparation and sequencing
Tissue samples were submitted to the North Carolina State Genomic Sciences Laboratory (Raleigh, NC, USA) for Illumina RNA library construction and sequencing, as detailed in [47], except that single ended libraries were sequenced. Briefly, RNA samples were analyzed with an Agilent 2100 Bioanalyzer with an RNA 6000 Nano Chip (Agilent Technologies, USA). mRNA purification used the NEBNExt Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, USA). Libraries were constructed using the NEBNext Ultra Directional RNA Library Prep Kit (NEB) and NEBNext Mulitplex Oligos for Illumina (NEB). The libraries were sequenced in an Illumina HiSeq 2500 DNA sequencer, utilizing 125bp single end sequencing flow cell with a HiSeq Reagent Kit v4 (Illumina, USA).
2.1.4. Bioinformatic analysis
Bioinformatic analyses were conducted following the methods described previously [47], with some modifications. Briefly, the fastq files were trimmed of low quality reads (< 20), removed from contaminating primer sequences and concatenated for single-ended assembly using the Abyss (using k parameters from 21 to 91 in 5 fold increments) [48] and Trinity [49] assemblers. The combined fasta files were further assembled using a iterative blast and CAP3 pipeline as previously described [23]. CDS were extracted based on the existence of a signal peptide in the longer open reading frame (ORF) and by similarities to other proteins found in the Refseq invertebrate database from the National Center for Biotechnology Information (NCBI), proteins from Acari deposited at NCBI’s Genbank and from SwissProt.
Reads for each library were mapped on the deducted CDS using the RSEM software [50]. Heat maps were made using the software gplots [51] and statistical tests used the package edgeR [52], both running under the R environment [53].
Protein alignments were done using ClustalX [54], and phylogenies were inferred using the Mega v.7 package [55]. The Maximum Likelihood method based on the best nucleotide substitution matrix available for the alignment was used to infer the evolutionary history, as discovered by the Mega package. Recombination breakpoints in aligned coding sequences were determined by the RDP4 software [56].
2.1.5. LC-MS/MS analysis and data analysis
Approximately 6 μg of protein from each sample was adjusted to a final volume of 30 μL with 50 mM HEPES, pH 8.5 in 8 M urea. The samples were reduced in 5 mM DTT for 1 h at room temperature followed by alkylation with 15 mM iodoacetamide for 20 min. The concentration of urea was reduced to 1.5 M by the addition of 50 mM HEPES, pH 8.5 and 0.5 μg of LysC protease was added and incubated for 15 h at 30 °C. The urea concentration was reduced to 0.8 M with 100 mM HEPES, pH 8.0, 1 μg of trypsin was added and incubated for 6 h at 30 °C. The pH was adjusted to 2.5 with trifluoroacetic acid (TFA) and the samples were desalted on an Oasis HLB micro-elution plate. The peptides were eluted with 0.1% TFA, 50% acetonitrile (AcCN) and the solvent was removed under vacuum at 50 °C. The residue was dissolved in 0.1% formic acid, 3% AcCN for injection.
Data were collected using an Orbitrap Fusion Lumos mass spectrometer equipped with an EASY-Spray Ion Source and an EASY-nLC 1200 liquid chromatography system (Thermo Fisher Scientific). The mobile phase solvent contains water and 0.1% formic acid. The peptides (5 μL) were loaded onto trap column (PepMap 100 C18, particle size 3 μm, length 2 cm, inner diameter 75 μm, Thermo Fisher Scientific), and separated on analytical column (PepMap 100 C18, particle size 2 μm, length 25 cm, inner diameter 75 μm, Thermo Fisher Scientific) with a linear gradient of 0–40% acetonitrile for 80 min, followed by 40–80% for 5 min, holding at 80% for 5 min, 80–0% for 5 min, and holding at 0% for 5 min. Throughout this 100-min data acquisition, the flow rate was set at 300 nL/min and the analytical column temperature was set at 50 °C. The data acquisition was done with the standard data-dependent acquisition strategy, where the survey MS1 scan was done every 2 s with Orbitrap mass analyzer at 120,000 resolution and the data-dependent MS2 scans were done with Linear Ion Trap mass analyzer for multiply charged precursor ions isolated with the 1.6 m/z window using Quadrupole and fragmented by CID at 35% collision energy. The dynamic exclusion period was set at 15 s, and the EASY-IC internal calibration was utilized for Orbitrap scans.
Tandem mass spectra were extracted from Thermo RAW files using RawExtract 1.9.9.2 [57] and searched with ProLuCID [58] against R. sanguineus database (71,643 entries) concatenated with Oryctolagus cuniculus from Uniprot [59] reference database (21,176 entries) and reverse sequences of all entries. The search space included all fully- tryptic and half-tryptic peptide candidates. Carbamidomethylation of cysteine was used as static modification. Data was searched with 50 ppm precursor ion tolerance and 0.4 Da fragment ion tolerance. The validity of the peptide spectrum matches (PSMs) generated by ProLuCID was assessed using Search Engine Processor (SEPro) module from PatternLab for Proteomics platform [60]. A cutoff score was established to accept a protein false discovery rate (FDR) of 1% based on the number of decoys. Results were post processed to only accept PSMs with < 10 ppm precursor mass error and proteins with a unique peptide. Normalized spectral abundance factors (NSAF) was used to represent relative abundance and secretion dynamics. Values were normalized calculating Z-score and values were used to generate heat maps using the heatmap2 function from the ggplot2 library in R.
2.1.6. Data availability
The transcriptome data was deposited to the National Institute for Biotechnology Information (NCBI) under Bioproject PRJNA606595 and Biosample accessions SAMN14115946, SAMN14115947, SAMN14115948, SAMN14115949, SAMN14115950, SAMN14115951, SAMN14115952, SAMN14115953, SAMN14115954, SAMN14115955, SAMN14115956, SAMN14115957, SAMN14115958, SAMN14115959, SAMN14115960, SAMN14115961, SAMN14115962, SAMN14115963, SAMN14115964 and SAMN14115965. The reads were deposited to the Short Reads Archive of the NCBI under accessions SRR11109985, SRR11109984, SRR11109973, SRR11109972, SRR11109971, SRR11109970, SRR11109969, SRR11109968, SRR11109967, SRR11109966, SRR11109983, SRR11109982, SRR11109981, SRR11109980, SRR11109979, SRR11109978, SRR11109977, SRR11109976, SRR11109975 and SRR11109974. This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GINV00000000. The version described in this paper is the first version, GINV01000000.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018964.
3. Results
3.1. Overall description of the transcriptomes
Following primer removal and trimming of low-quality bases, we obtained over 687 million reads from 20 libraries, including two biological replicates for the unfed group (UF), and three replicates for each of groups 1–6. A minimum of 17,854,945 reads was found for the third replicate of Group 4, and a maximum of 144,760,365 reads was found for replicate 2 of Group 6 (Supplemental table 1). As indicated in the methods section, the groups were organized according to the tick weight following attachment to a rabbit, not to days of feeding as it is usual.
Following assembly of the reads, 71,643 coding sequences (CDS) were extracted. The RSEM software was used to map the reads from each library to these transcripts, 28,921 of which had a TPM (transcripts per million) of 10 or larger in at least one library. Notice that a TPM value of 1000 indicates the transcript to be represented by 1000 in one million total transcripts, or one in one thousand transcripts, which is equal to 0.1%. A TPM value of 10 then indicates the transcript expression to be 0.001% of the totality of transcripts. These transcripts and their comparisons to several databases are available as a hyperlinked spreadsheet (Supplemental spreadsheet 1).
The total number of reads mapped to these CDS totalled 357,388,554 reads, or 51.98% of the 687,502,190 reads. The unmapping of nearly half of the reads could be due to reads mapping to the 5′ or 3’ UTR of the CDS, which were not extracted, and also due to many reads deriving from non-coding RNA’s. These results were similar to those obtained following analysis of the sialotranscriptome of Rhipicephalus zambeziensis [19].
Functional classification of the 28,921 transcripts with TPM values larger than 10 showed that the two major functional groups are represented by CDS of the unknown class and those representing CDS coding for proteins that are probably secreted (Table 1). These two classed accrued nearly 50% of all mapped reads, as follows: The secreted class accounted for 4039 CDS, or 14% of the total CDS, accruing over 80 million reads, or 22.5% of the total reads. The unknown class accounted for 20,743 CDS, or 71% of all 28,921 CDS, and these accrued over 99 million reads, or 28% of all mapped reads. Twenty-four additional classes are shown in Table 1, which also includes 238 CDS probably deriving from transposable elements, and 16 CDS that are of probable viral origin.
Table 1.
Functional classification of coding sequences (CDS) originating from the Rhipicephalus sanguineus sialotranscriptome. Classes were sorted according to their accreted number of reads, shown in background color from red (maximum value) to blue (minimum value). Yellow background represents average values.
| t1 |
Within the secreted class of transcripts, we found 42 families that have 4 or more members, including only those near full length in size. These 915 transcripts accrued ~60 million reads (Table 2). The top five families were identified as lipocalins, with 245 members and accruing 18.7% of the reads from this secreted group, Kunitz family of protease inhibitors, with 82 members and accruing 14.1% of the reads, glycine- rich protein family, with 67 members and accruing 38% of the reads, metalloprotease family, with 61 members and accruing 2.5% of the reads, and 8.9 kDa family, with 47 members and accruing 2.3% of the reads.
Table 2.
Families of secreted proteins within the sialotranscriptome of Rhipicephalus sanguineus. Protein families with 4 or more members are shown, ordered from the most abundant family and including the average number of reads accrued per each member, and the totality of reads accrued by the family.
| Class | Average Number of Reads per CDS | SE | N | Total number of reads | Percentage of this group |
|---|---|---|---|---|---|
| Lipocalin | 45,877 | 9530 | 245 | 11,239,766 | 18.71 |
| Kunitz | 103,602 | 45,692 | 82 | 8,495,355 | 14.14 |
| GRP | 341,156 | 106,131 | 67 | 22,857,468 | 38.04 |
| Metalloprotease | 24,959 | 3980 | 61 | 1522,472 | 2.53 |
| 8.9 kDa | 29,749 | 17,901 | 47 | 1,398,198 | 2.33 |
| BTSP | 33,885 | 9372 | 44 | 1,490,951 | 2.48 |
| AlaRich | 7985 | 1193 | 34 | 271,500 | 0.45 |
| DAP-36 | 35,187 | 16,422 | 23 | 809,295 | 1.35 |
| Evasin | 27,745 | 16,615 | 21 | 582,646 | 0.97 |
| M13_peptidase | 32,070 | 9163 | 18 | 577,258 | 0.96 |
| 28 kDa | 31,532 | 6607 | 17 | 536,052 | 0.89 |
| Defensin | 7207 | 3246 | 17 | 122,527 | 0.20 |
| Ixodegrin | 13,508 | 4518 | 16 | 216,131 | 0.36 |
| Mucin | 95,544 | 36,085 | 15 | 1,433,163 | 2.39 |
| OneOfEach | 27,022 | 11,343 | 15 | 405,329 | 0.67 |
| TIL | 25,574 | 9702 | 15 | 383,612 | 0.64 |
| Cystatin | 3603 | 1746 | 13 | 46,840 | 0.08 |
| Carboxypeptidase_inhibitor | 2713 | 981 | 13 | 35,271 | 0.06 |
| Antigen-5 | 17,443 | 3562 | 12 | 209,317 | 0.35 |
| Serine carboxypeptidase | 8541 | 3030 | 12 | 102,490 | 0.17 |
| Cytotoxin | 12,159 | 1916 | 10 | 121,595 | 0.20 |
| 23–24 kDa | 7152 | 1693 | 10 | 71,518 | 0.12 |
| Serpin | 14,320 | 3982 | 8 | 114,562 | 0.19 |
| Rhiapp specific protein | 639 | 220 | 8 | 5110 | 0.01 |
| Down syndrome cell adhesion molecule | 58,904 | 30,921 | 7 | 412,326 | 0.69 |
| Toll-like | 34,472 | 12,061 | 6 | 206,833 | 0.34 |
| Ficolin/Ixoderin | 32,515 | 12,359 | 6 | 195,091 | 0.32 |
| Transposon | 26,131 | 13,231 | 6 | 156,784 | 0.26 |
| Superoxide dismutase | 19,797 | 6965 | 6 | 118,780 | 0.20 |
| Microplusin | 11,645 | 5430 | 6 | 69,872 | 0.12 |
| Rapp-40-287 | 1522 | 462 | 6 | 9135 | 0.02 |
| SPARC/Kazal | 77,755 | 35,892 | 5 | 388,774 | 0.65 |
| Endonuclease | 38,671 | 24,806 | 5 | 193,354 | 0.32 |
| Mys-30-94 | 20,766 | 12,113 | 5 | 103,829 | 0.17 |
| Thyropin | 19,202 | 7985 | 5 | 96,009 | 0.16 |
| Mys-30-60 | 1593 | 377 | 5 | 7965 | 0.01 |
| 35–224 Madanin | 1,208,799 | 542,032 | 4 | 4,835,197 | 8.05 |
| Serine protease | 24,451 | 8522 | 4 | 97,804 | 0.16 |
| Insulin_growth_factor | 18,747 | 9464 | 4 | 74,988 | 0.12 |
| IPPase | 13,388 | 5220 | 4 | 53,552 | 0.09 |
| Serum amyloid A | 2560 | 166 | 4 | 10,242 | 0.02 |
| 12 kDa | 641 | 178 | 4 | 2563 | 0.00 |
| Total | 915 | 60,081,522 | 100 |
SE = standard error of the number of reads per coding sequence (CDS); N = number of sequences.
3.2. An insight into the structure of the families coding for secreted salivary proteins in R. sanguineus
3.2.1. Lipocalins
The lipocalin family in ticks have been associated with a kratagonist function towards biogenic amines [61,62] and eicosanoids [63,64], as well as having toxin properties in soft ticks [65]. This family has the highest number of transcripts identified in tick sialotranscriptomes [30,66,67]. Of the 245 transcripts attributed to the lipocalin family found in this sialotranscriptome, 55 are full length or near full-length members of the subfamily characterized by having the PFAM domain “pfam02098, His_binding, Tick histamine binding protein”. Phylogenetic analysis of these 55 coding sequences showed remarkable low bootstrap support for the branches containing most of the transcripts. Indeed, only 15 of the 55 sequences were within a clade with three or more members, totalling five clades (Supplemental fig. 2).
3.2.2. Metalloproteases
Transcripts coding for salivary zinc-dependent metalloproteases are abundantly found in tick sialotranscriptomes [30,66,67], and their function was associated with a fibrinolytic and anti-angiogenic activities [68–71]. The sialotranscriptome of R. sanguineus here described allowed for the identification of 46 full length or near full length coding for metalloproteases. The phylogenetic analysis produces a tree that is deeper than that observed for the lipocalins. Forty of the 46 sequences are organized within seven clades containing three or more sequences (Supplemental fig. 3).
3.2.3. Cystatins
Tick salivary cystatins were first discovered in Ixodes scapularis and found to affect immunity and inflammation by counteracting cysteine proteases found in leukocytes [72–78]. They are normally expressed in much lower levels than lipocalins or metalloproteases, and the family is not as numerous. The phylogram of 13 cystatin coding sequences indicated four divergent clades (Supplemental fig. 4).
3.3. An insight on the sialotranscriptome switch of R. sanguineus
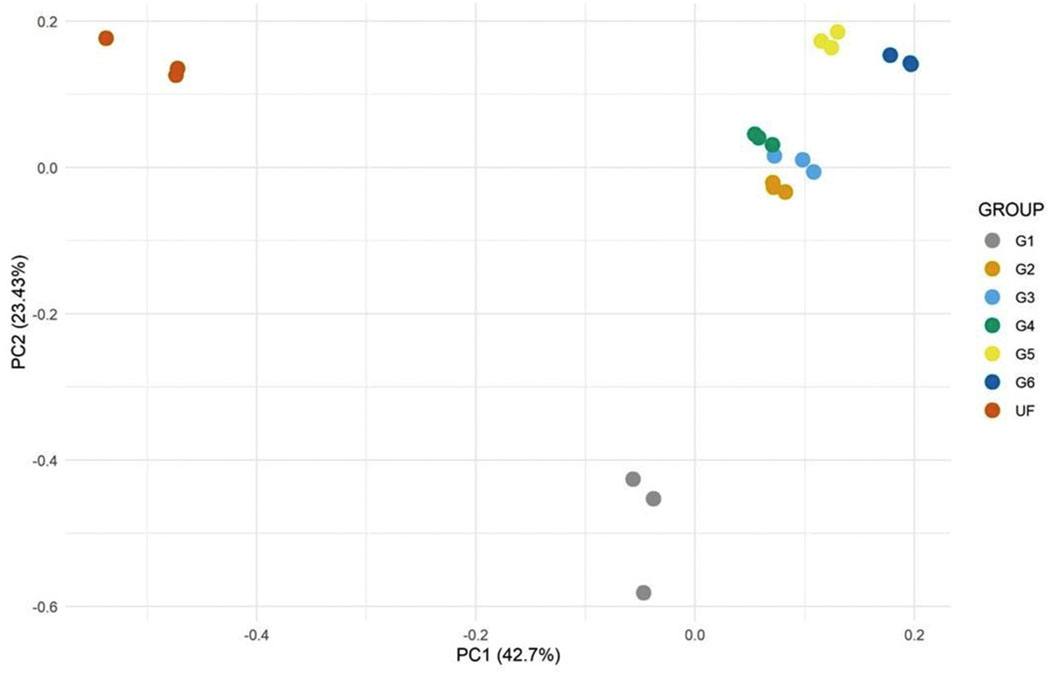
The edgeR package was used to determine the differential expressed CDS in paired comparisons (UF × SG1, SG1 × SG2, SG2 × SG3, SG4 × SG5 and SG5 × SG6). The multi-dimensional scaling plot shows good clustering of the biological replicates, indicating that the consolidation of samples by weight was a good choice, instead of days of feeding (Fig. 1). Out of 28,921 CDS having a TPM > 10 in at least one library, 17,590 were found differentially expressed (DE) (Fig. 2), including 1945 DE CDS found in paired comparisons at a FC larger than 128 (Table 3). When comparing the transcript class frequencies with FC > 4 with those with FC > 128 by a Χ2 test (adjusted for multiple testing with the Bonferroni correction), it is evident that the secreted class was represented in the FC > 128 at a higher frequency (28%) than the frequency found for the FC > 4 group (19%) (false discovery rate (FDR) = 2.07e−11). Other statistically significant changes occurred in the groups: protein export machinery, energy metabolism, protein modification machinery, proteasome machinery, transporters/storage, carbohydrate metabolism and lipid metabolism, all significantly decreasing their frequency in the FC > 128 group when compared to the FC > 4 group (Tables 3 and 4).
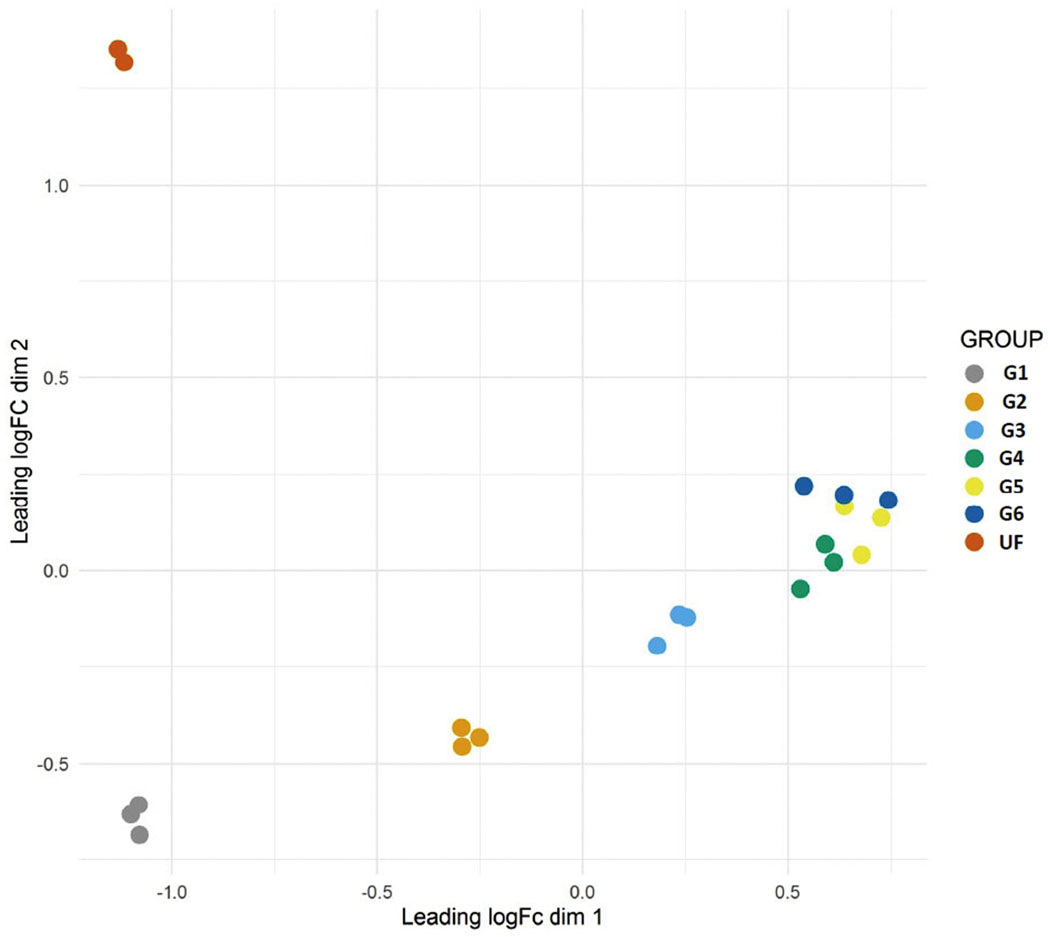
Fig. 1.
Principal component analysis of the transcriptome of Rhipicephalus sanguineus when the different libraries (determined by Groups) were contrasted using the edgeR program. Group unfed female (UF), partially fed ticks groups G1 (1.8 mg), G2 (3.6 mg), G3 (7.0 mg), G4 (10.9 mg), G5 (24 mg) and G6 (36 mg).
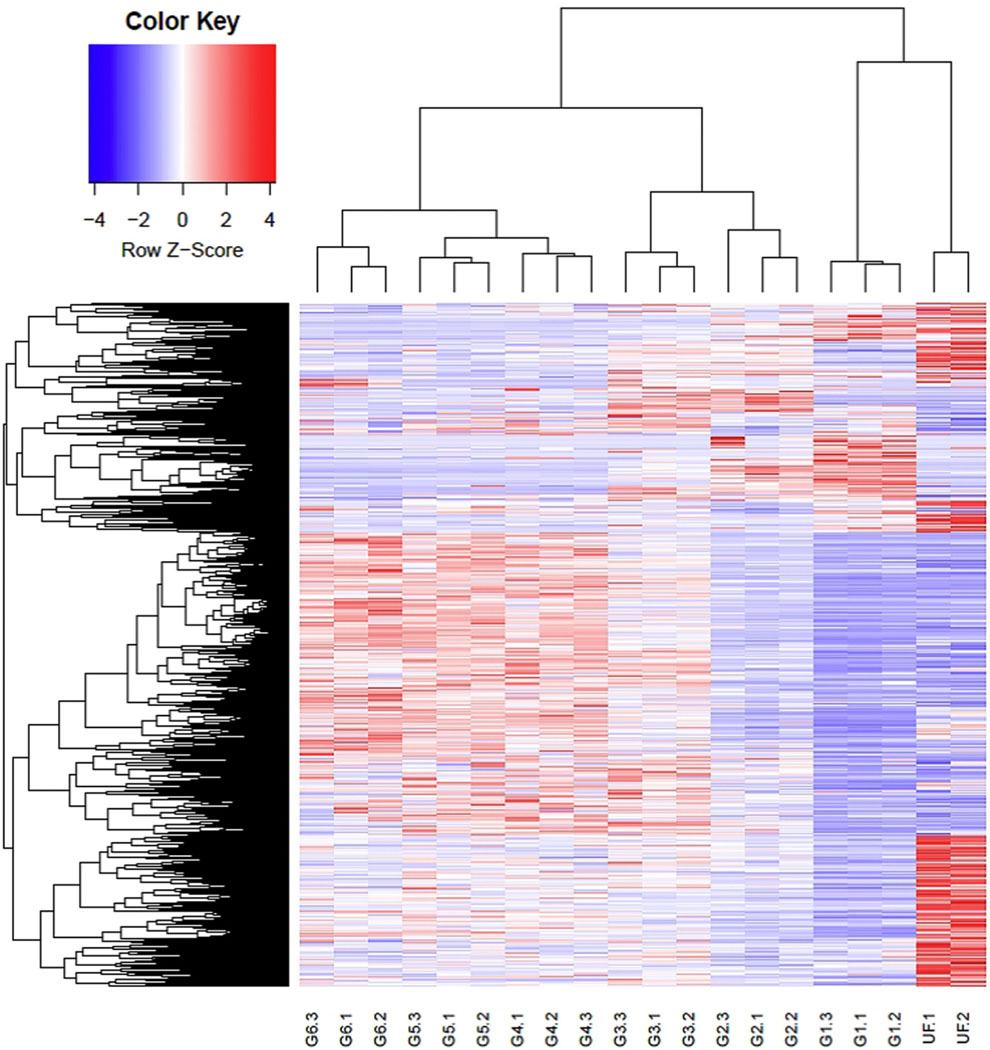
Fig. 2.
Heat plot of differentially expressed transcripts from the sialotranscriptome of Rhipicephalus sanguineus obtained from unfed ticks (UF) or at 6 partially fed ticks with different weights (G1 - G6). Two biological replicates are shown for the unfed (UF) group, and three for each of the six feeding groups.
Table 3.
Functional classification of the Rhipicephalus sanguineus transcripts for all coding sequences and for the differentially expressed CDS, including all those that are significantly differentially expressed, and within these, those achieving more than 4× fold change (FC), more than 16 × FC, etc. up to 128 × FC.
| Class | Number of transcripts | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| All | All Significant | 4× FC | 8× FC | 16× FC | 32× FC | 64× FC | 128× FC | |
| Unknown | 20,743 | 11,580 | 4734 | 2922 | 2146 | 1759 | 1482 | 1247 |
| Secreted | 4039 | 2519 | 1371 | 1108 | 940 | 794 | 680 | 554 |
| Unknown, conserved | 439 | 361 | 100 | 43 | 23 | 15 | 13 | 11 |
| Signal transduction | 394 | 326 | 86 | 42 | 27 | 20 | 14 | 8 |
| Transcription machinery | 421 | 350 | 71 | 24 | 11 | 7 | 6 | 6 |
| Protein synthesis machinery | 381 | 340 | 66 | 20 | 5 | 1 | 1 | 1 |
| Extracellular matrix/cell adhesion | 192 | 170 | 105 | 87 | 67 | 50 | 38 | 33 |
| Transposable element | 238 | 172 | 79 | 61 | 44 | 37 | 26 | 19 |
| Protein export machinery | 287 | 250 | 68 | 19 | 7 | 3 | 2 | 2 |
| Metabolism, energy | 264 | 227 | 58 | 17 | 11 | 5 | 3 | 3 |
| Protein modification machinery | 199 | 170 | 68 | 39 | 26 | 14 | 5 | 3 |
| Proteasome machinery | 188 | 150 | 42 | 11 | 4 | 3 | 3 | 3 |
| Transporters/storage | 142 | 129 | 44 | 26 | 14 | 13 | 10 | 8 |
| Immunity | 95 | 84 | 50 | 38 | 32 | 29 | 23 | 20 |
| Metabolism, carbohydrate | 124 | 107 | 47 | 22 | 13 | 6 | 3 | 3 |
| Metabolism, lipid | 129 | 114 | 48 | 19 | 8 | 2 | 2 | 2 |
| Oxidant metabolism/detoxification | 97 | 86 | 42 | 23 | 21 | 16 | 13 | 8 |
| Nuclear regulation | 130 | 105 | 24 | 8 | 3 | 3 | 2 | 2 |
| Cytoskeletal | 130 | 102 | 21 | 8 | 4 | 4 | 3 | 3 |
| Metabolism, amino acid | 86 | 77 | 22 | 11 | 9 | 6 | 5 | 2 |
| Metabolism, nucleotide | 68 | 57 | 20 | 12 | 8 | 7 | 6 | 4 |
| Transcription factor | 70 | 54 | 7 | 2 | 2 | 2 | 2 | 2 |
| Metabolism, intermediate | 27 | 25 | 9 | 2 | 2 | 1 | 0 | 0 |
| Viral | 16 | 16 | 10 | 8 | 5 | 3 | 1 | 1 |
| Nuclear export | 15 | 14 | 3 | 0 | 0 | 0 | 0 | 0 |
| Storage | 7 | 5 | 4 | 4 | 3 | 3 | 1 | 0 |
| Total | 28,921 | 17,590 | 7199 | 4576 | 3435 | 2803 | 2344 | 1945 |
Table 4.
Functional classification of the sialotranscriptome of Rhipicephalus sanguineus represented as percentage of transcripts according to their ascribed class. The columns represent results for All transcripts, for all differentially expressed transcripts predicted by edgeR, and those having a fold change (FC) higher than 4×, 8×,..128× . The last column shows the false discovery rate (FDR) resulting from a X2 test contrasting the expressions 4× and 128× for each functional class. The color gradients shows background colors from red (maximum value) to blue (minimum value). Yellow background represents average values.
| t2 |
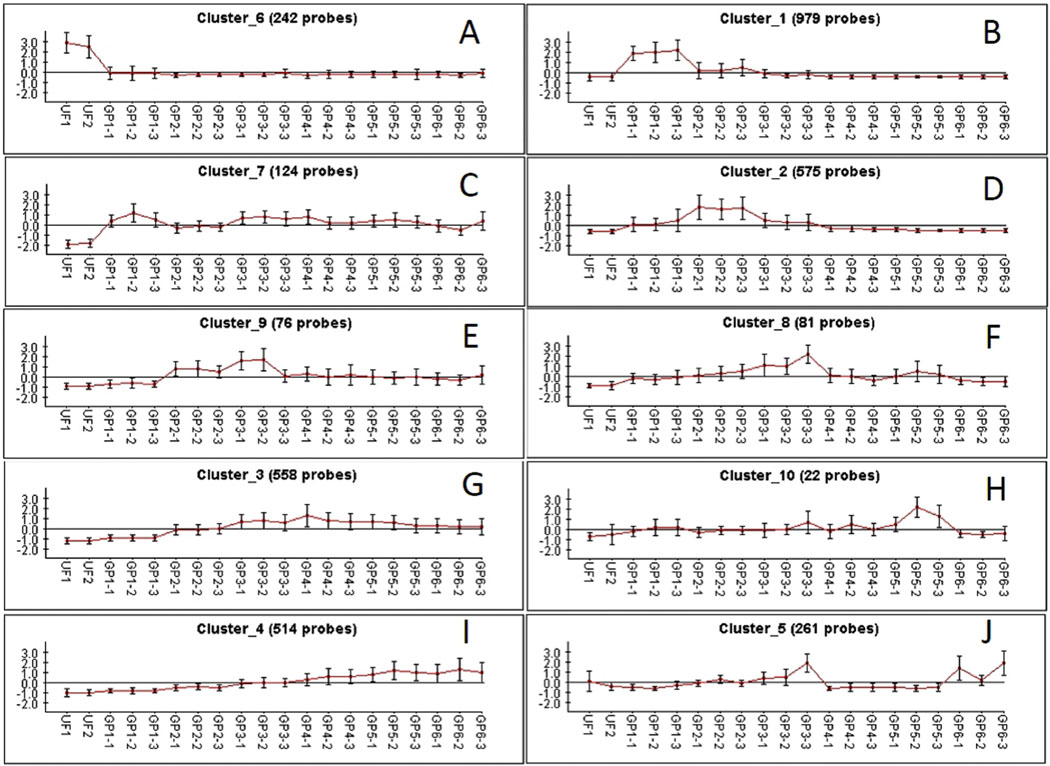
To verify whether the DE transcripts were organized in clusters associated with the different library groups, the normalized TPM values of 3435 transcripts having a significant FC > 16 were submitted to the CLICK algorithm of the expander program, which identified 10 clusters of transcripts (Fig. 3). Clusters that were unique to the UF, SG1, SG2, SG3 and SG5 are seen in Fig. 3 A, B, D, F, H. Other clusters contained transcripts of 2 or more groups. The results of the paired contrasts and Expander clusterization (Tables 3 and 4) indicated the occurrence of sialome switch within R. sanguineus, with remarkable values of differential expression, exceeding in many cases over 100-fold.
Fig. 3.
Clusters of 16 x differentially expressed transcripts from the sialotranscriptome of Rhipicephalus sanguineus according to their library groups, as determined by the click algorithm of the program Expander.
3.3.1. An insight into the sialome switch of the lipocalins of R. sanguineus
The assembled transcriptome of R. sanguineus identified 245 transcripts coding for lipocalins, 216 of which (88%) were found significantly DE in at least one paired comparison. To visualise the expression patterns of these DE transcripts, we made graphs of their normalized average TPM values found within each of the 7 groups of ticks. Supplemental fig. 5 displays the graphed results for 53 lipocalins that peak their expressions within tick group 1, and Supplemental fig. 6 displays the results for 30 lipocalin coding transcripts that peak within tick group 2. Notice that while some lipocalin transcripts could be found expressed in two or more contiguous groups, some were narrowly expressed in a single group. This narrow distribution was mostly seen in the lipocalins peaking within group 1, which shows the longer distance from its neighbours in the MDS plot (Fig. 1). Finally, Supplemental fig. 7 displays 77 lipocalins with DE FC > 16 that were found in expander clusters 3–9.
3.3.2. An insight into the sialome switch of the Kunitz-coding transcripts of R. sanguineus
The assembled transcriptome of R. sanguineus identified 82 transcripts coding for proteins containing Kunitz domains, 74 of which (90%) were found significantly DE in at least one paired comparison. Of these 74, 37 are at least DE with a FC > 16. Supplemental figs. 8 and 9 display the plots of the normalized average TPM values for these transcripts according to their tick groups.
3.3.3. An insight into the sialome switch of the cystatin-coding transcripts of R. sanguineus
There were 13 transcripts coding for cystatins in the assembled sialotranscriptome, all of which were DE in at least one paired comparison. The graph of the DE transcripts coding for cystatins is shown in Supplemental fig. 10.
3.3.4. Transcripts possibly involved with sialome switching in R. sanguineus
The mechanism of sialome switching in ticks remains a mystery. How are the genes of the same family turned on and off? It has been suggested that either epigenetic mechanisms or more classical signal-transduction/transcription factors could be involved in controlling the sialome switch [26]. Table 5 displays the transcripts found in the sialotranscriptome of R. sanguineus that are associated with epigenetic regulation (DNA methylation and histone modification machinery) [79]. Table 6 displays transcripts associated with transcription factors that might be associated with the regulation of gene expression in tick salivary glands.
Table 5.
Transcripts possibly associated with epigenetic gene expression within the salivary glands of Rhipicephalus sanguineus.
| Transcript name | Annotation |
|---|---|
| Rs-116658_FR3_1-2041 | Chromatin remodeling complex WSTF-ISWI small subunit |
| Rs-182989_FR3_48-542 | DNA methyltransferase 1-associated protein-1 |
| Rs-119673_FR3_78-274 | heterochromatin-associated protein hp1 |
| Rs-138721_FR3_6-538 | Histone deacetylase complex catalytic component RPD3 |
| Rs-89535_FR2_92-653 | histone-lysine N-methyltransferase PRDM9-like |
| Rs-88148_FR3_105-361 | Methyl-CpG binding transcription regulators |
| Rs-133231_FR2_1-702 | NAD-dependent histone deacetylases and class I sirtuins (SIR2 family) |
| 93106_FR2_23-447 | Phosphatidylserine-specific receptor PtdSerR contains JmjC domain |
| 91046_FR1_1-320 | Sirtuin 4 |
| Rs-76172_FR2_23-420 | Sirtuin 5 |
Table 6.
Transcription factor-coding transcripts found in the salivary glands of Rhipicephalus sanguineus.
| Transcript name | Annotation |
|---|---|
| 150871_FR1_1-632 | Transcription factor CA150 |
| 167428_FR2_41-174 | Transcription factor PBX and related HOX domain proteins |
| 88577_FR3_1-153 | Phosphatidylserine-specific receptor PtdSerR, contains JmjC domain |
| 92854_FR2_29-739 | Heat shock transcription factor |
| 96041_FR3_1-152 | RNA polymerase II transcription elongation factor DSIF/SUPT5H/SPT5 |
| 96060_FR3_66-187 | Transcription factor e(y)2 |
| Rs-100916_FR2_1-328 | Transcriptional activator FOSB/c-Fos and related bZIP transcription factors |
| Rs-100931_FR3_1-341 | Transcription factor GT-2 and related proteins |
| Rs-103965_FR3_6-947 | Transcription factor NFAT, subunit NF90 |
| Rs-105917_FR3_339-722 | Transcription factor LIM3, contains LIM and HOX domains |
| Rs-107136_FR1_40-299 | RNA polymerase II transcription factor complex subunit |
| Rs-112269_FR2_1-381 | Basic region leucine zipper transcription factor |
| Rs-112270_FR3_14-407 | Basic region leucine zipper transcription factor |
| Rs-113329_FR1_61-366 | Transcription factor XBP-1 |
| Rs-115972_FR3_1-274 | CREB/ATF family transcription factor |
| Rs-116088_FR2_41-465 | Transcription factor PBX and related HOX domain proteins |
| Rs-117324_FR3_27-552 | CREB/ATF family transcription factor |
| Rs-117326_FR3_14-540 | CREB/ATF family transcription factor |
| Rs-119605_FR3_37-278 | bHLHZip transcription factor BIGMAX |
| Rs-119650_FR1_31-170 | Transcription factor, subunit of SRB subcomplex of RNA polymerase II |
| Rs-120403_FR1_6-193 | bZIP transcription factor MafK |
| Rs-121027_FR1_55-1220 | Transcription factor TMF, TATA element modulatory factor |
| Rs-121251_FR1_1-575 | Apoptosis antagonizing transcription factor/protein transport protein |
| Rs-121258_FR2_1-464 | Apoptosis antagonizing transcription factor/protein transport protein |
| Rs-126695_FR3_68-392 | Transcription factor of the Forkhead/HNF3 family |
| Rs-127921_FR1_18-193 | Transcription factor MBF1 |
| Rs-128460_FR3_69-821 | Regulatory protein MLP and related LIM proteins |
| Rs-131367_FR2_52-256 | Basic region leucine zipper transcription factor |
| Rs-131369_FR1_30-270 | Basic region leucine zipper transcription factor |
| Rs-133834_FR3_313-885 | Hypoxia-inducible factor 1/Neuronal PAS domain protein NPAS1 |
| Rs-139212_FR1_68-294 | Transcription factor containing NAC and TS-N domains |
| Rs-145690_FR2_39-687 | Alternative splicing factor ASF/SF2 (RRM superfamily) |
| Rs-147431_FR1_17-245 | Basic region leucine zipper transcription factor |
| Rs-151159_FR3_33-393 | cAMP response element binding protein and related transcription factors |
| Rs-152328_FR1_1-439 | HMG-box transcription factor |
| Rs-158588_FR1_1-268 | Nucleosome-binding factor SPN, POB3 subunit |
| Rs-160619_FR1_304-853 | Splicing factor 3b, subunit 1 |
| Rs-167325_FR3_105-531 | Activating transcription factor 4 |
| Rs-169240_FR3_154-301 | Transcription factor NERF and related proteins, contain ETS domain |
| Rs-169255_FR3_67-287 | Transcription factor NERF and related proteins, contain ETS domain |
| Rs-169445_FR3_1-403 | Basic region leucine zipper transcription factor |
| Rs-170419_FR1_127-306 | RNA polymerase II general transcription factor BTF3 and related proteins |
| Rs-175974_FR3_68-493 | C2H2-type Zn-finger protein |
| Rs-183043_FR3_1-407 | Transcription factor NFAT, subunit NF45 |
| Rs-68622_FR2_34-231 | Transcription factor IIB |
| Rs-76207_FR4_79-197 | Transcription factor e(y)2 |
| Rs-76208_FR1_121-238 | Transcription factor e(y)2 |
| Rs-81156_FR3_88-263 | Nucleosome-binding factor SPN, POB3 subunit |
| Rs-81157_FR2_51-290 | Nucleosome-binding factor SPN, POB3 subunit |
| Rs-85500_FR3_97-942 | WD40 repeat protein |
3.3.5. Proteome studies reveal sialome switching in R. sanguineus
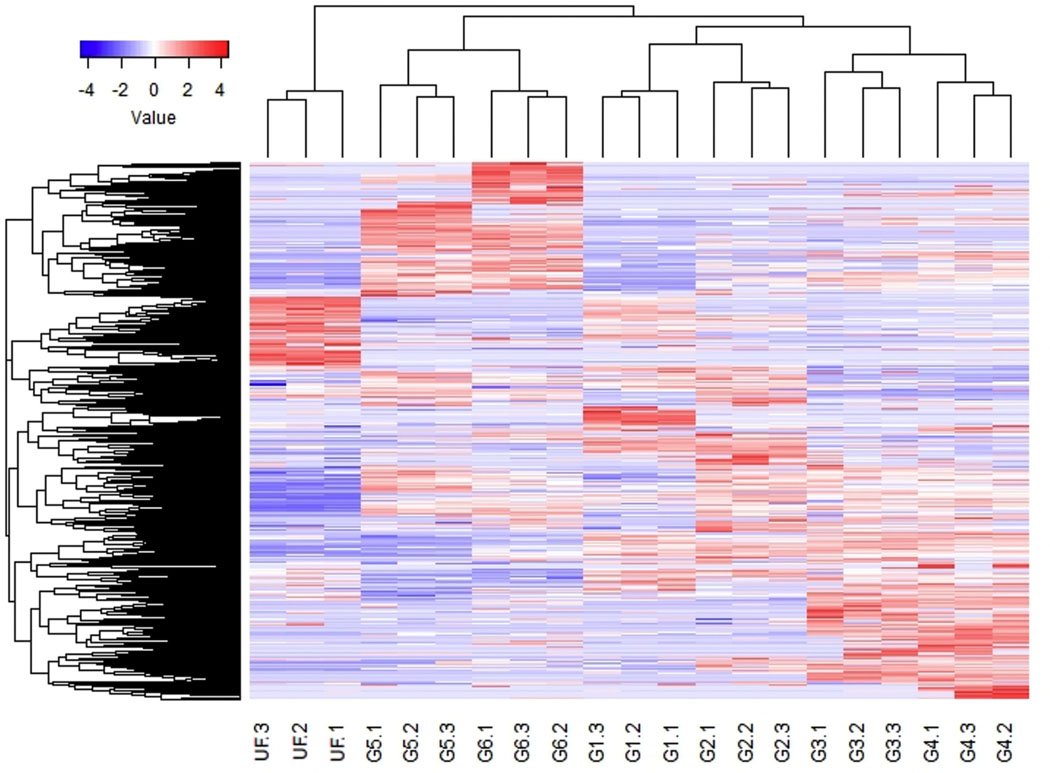
In parallel with the transcriptome studies, proteome analysis of the salivary glands of adult female R. sanguineus was performed, using groups of ticks achieving the same range of weight gain after attachment to a rabbit. Following a shotgun proteomic approach, the generated raw data was used to query the “de novo” transcriptome assembly described above using the PatternLab software [60] which allows normalization by the normalized spectral abundance factor (NSAF) approach, which takes into account a protein’s length during the normalization process [80]. A total of 2125 transcripts were identified as coding for the peptides found by the proteomic study (Supplemental spreadsheet 2), 1745 of which matched transcript translations with a TPM value of 10 or more. To gain insight into broad relationships of secretion dynamics of tick proteins with the tick feeding processes, Z-score statistics normalized NSAF values were visualized on heat maps (Fig. 4). Indeed, the PCA plot (Fig. 5) showed remarkable clustering of the replicates, revealing the sialome switching at protein level as well.
Fig. 4.
Heat map of the normalized spectral abundance factor (NSAF) values for 2125 peptides identified by mass spectrometry in seven triplicate groups of Rhipicephalus sanguineus ticks (unfed ticks (UF) or 6 partially fed ticks with different weights (G1 - G6)). For more details, see text.
Fig. 5.
Principal component analysis plot of the normalized spectral abundance factor (NSAF) values of 2125 polypeptides identified by mass spectrometry in seven triplicate groups (unfed ticks (UF) or 6 different weights (G1 - G6)) of Rhipicephalus sanguineus ticks. For more details, see text.
To get insights on the relation between transcriptomic and proteomic data, and using solely the transcripts with a TPM value of 10 or more, we normalized the transcriptome TPM values as well as the proteome NSAF values within each tick group, taking as a value of 100 the largest TPM or NSAF value for each measured sample in all tick groups. There was a positive correlation between the maximum TPM value for a transcript and the maximum NSAF value for the same transcript, with a correlation coefficient of 0.549 (P < 1e-6), and a coefficient of determination (R2) equal to 0.301. A correlation analysis between the TPM and NSAF values of the 1745 transcripts identified 221 that were positively correlated when their TPM values where compared to their NSAF values for each tick group (Supplemental fig. 11 having a p value smaller than 0.1). Notice that many of these correlations refer to an expression that is found in a single group. Thirty per cent of these transcripts coded for secreted proteins. Conversely, there were 17 transcripts where the correlation was negative with a p value smaller than 0.1 (Supplemental fig. 12). Of these 17 transcripts, 13 had a maximum TPM on the unfed group, with no or little NSAF values, which increased in values within group 1, indicating that the transcript was present in unfed ticks but with little protein expression, which took over after the tick started feeding. All these negatively correlated transcripts belong to the housekeeping class.
Of the 1745 transcript products that were identified by MS/MS and correlated with TPM value, 1507 showed no statistically significant correlation between the TPM and NSAF results as a function of the 7 tick groups. Inspection of the graphs plotting the normalized TPM and NSAF results (Supplemental spreadsheet 2, column TU) indicated that despite the non-significant statistical correlation result, the curves were relatively similar. Indeed, if we compare the peaks of the TPM and NSAF results, 49% of the 1507 results are concordant or found in the neighbor tick group (Supplemental spreadsheet 2 and Supplemental fig. 13). A X2 test contrasting the observed peak differences with an expected uniform peak distance indicated a highly significant departure of the random expectation (P < .00001).
3.3.6. Host-derived proteins are present into R. sanguineus salivary glands
Presence of host-derived proteins was already described in several species as a component of tick saliva [81–83]. This observation raises questions regarding of whether host-derived proteins could be originated by regurgitation and/or mouthpart contamination. Here we investigated for the presence of host proteins in the tick salivary gland homogenate, not saliva, excluding the possibility of regurgitation or mouthpart contamination. Host-derived proteins were identified as a component of the tick salivary gland content (Supplemental spreadsheet 2, worksheet “Host”). Out of the 72 matches of rabbit proteins found by MS/MS, we excluded eight that were at least 90% identical to R. sanguineus proteins (tubulins, actin beta, elongin A, tyrosine 3- monooxygenase and proteasome 26S subunit) and another 17 that matched skin proteins (keratins, desmoglein and plakoglobin), which are common contaminants in MS/MS experiments [84]. The NSAF levels for the remaining 47 host proteins, which includes serum albumin, immunoglobulin G chains, hemoglobin, hemopexin, lactoferrin, annexin, fibrinogen, antithrombin, among others are shown in Supplemental fig. 14. These results suggest that the presence of host proteins in tick saliva may be a real and common recycling system present in ticks, and not a result of contamination during saliva collection.
3.3.7. An insight on the diversity of salivary proteins of R. sanguineus
On a recent review (Ribeiro and Mans, submitted) it was suggested that intra-gene recombination events could be driving the diversification of the tick sialome, as evidenced in the abundant protein families of lipocalins and metalloproteases which had indications of several breakpoints as determined by the RDP4 pipeline [56]. The less abundant and less expressed cystatin protein family, however, showed no signs of recombination. Here, using the coding sequences for lipocalins (Supplemental fig. 2), metalloproteases (Supplemental fig. 3) and cystatins (Supplemental fig. 4) we found four recombination events for the lipocalins, five for the metalloproteases and zero for the cystatin family (Supplemental file 3).
4. Discussion
In this work we explored the sialotranscriptome and sialoproteome of adult female R. sanguineus tick, aiming at (1) disclosing its salivary repertoire of transcripts and polypeptides and (2) shedding light into the rate and mechanisms associated with the tick sialome switch. Accordingly, from a total of 71,643 transcripts, 28,921 had a TPM of 10 or larger in at least one library (Supplemental spreadsheet 1). The translated peptides from these transcripts served as a target database for proteomic studies, leading to the identification of a total of 2125 proteins. From these, 1745 polypeptides derived from transcripts having a minimum TPM value of 10 in any library. Of these, 221 had their group expression values for TPM and NSAF significantly correlated, indicating a synchronicity, or better, a “syngroupicity” of transcript and peptide expression. On the other hand, there were 17 transcripts that were deemed negatively correlated, the majority of which had a peak transcription in the unfed group, and a peak translation in the first tick group, indicating the transcript was “waiting” for the feeding to start to be translated. The non-correlated transcripts were found to have a non-random departure of their peak values: The NSAF peaks were significantly associated with the corresponding vicinal TPM peaks.
Previous tick sialotranscriptomes done at different days of feeding indicated vast changes in transcript composition as a function of feeding time, characterizing the sialome switching phenomenon [22,85]. This variability was also observed within proteomic studies of tick saliva. The salivary composition is time-dependent, and it changes in quantity and quality during the feeding process [86,87]. Moreover, the composition of saliva is differentially expressed when ticks are exposed to different host species [31,32].
Transcriptomes done with salivary glands of single ticks, artificially fed or fed on a rabbit, demonstrated a large variance on transcript expression and suggested that the switches occurred in a time frame well below 12 h [26]. Aiming at reducing this variance, we planned our libraries to be built from ticks having similar weights instead of similar times of feeding, as the physiological status of the tick may be better defined by its weight gain rather than the time it has been since commencement of feeding, as previously indicated by the “critical weight” of ticks that determine their host detachment and salivary gland degeneration [43]. Indeed, the PCA plots for the transcripts and polypeptides (Figs. 1 and 5) showed remarkable clustering of the replicates, even though individual groups contained ticks feeding at several days’ difference from each other but having similar weights. The distance between these clusters indicates that the larger difference occurred between the unfed and fed groups, and, within the fed groups, between group 1 vs group 2, followed by groups 2 vs 3 and 3 vs 4, 5 and 6. The last three groups formed distinguished clusters, but were located very near each other in the PCA plots. The statistically significant differentially expressed transcripts as well as the clusters identified by the Expander program followed this pattern. Notably, there were many transcripts and peptides that were found in a single group, most often at groups 1 and 2. These results indicate that several sialome switches must have occurred between groups 1 and 2, 2 and 3 and 3 and 4, raising the possibility that there are hundreds of novel transcripts yet to be discovered. A future and improved experimental design would be to have the transcriptome and proteome done on single gland pairs, similar to performed by Perner et al. [26], or with an specific type of acini, since each type has a specific function and composition [88,89], from ticks weighting from 1.8 mg to 40 mg at 10% weight increments, a minimum of 35 ticks or libraries, not an impossible task.
It has been proposed that the sialome switching mechanisms in ticks could be associated with classical transcription factor regulation and/or epigenetic regulation [26]. Tables 5 and 6 presents transcripts associated with these processes. RNAi experiments targeting these transcripts may shed light into the mechanism of sialome switching.
In addition to tick proteins, many host proteins have been described as components of tick saliva in different tick species [81–83,90–92], suggesting the presence of a recycling system of host proteins during tick feeding [83]. Immunoglobulins, serum albumin, enzymes, serine protease inhibitors, and other host blood proteins were found [33,34]. The pattern of appearance of host proteins in the salivary glands of R. sanguineus is intriguing (Supplemental spreadsheet 2 and Supplemental fig. 14). First, it implies a mechanism of midgut transportation of the host proteins to hemolymph, and from the hemolymph to the salivary glands. Digestive cells of the cattle tick, Rhipicephalus microplus, were shown to transport hemoglobin and albumin by distinct mechanisms [93], indicating the possibility of direct transport of host blood proteins to the hemocoel. Lacking still is the knowledge of how these host proteins are acquired by the salivary glands, and how their voyage is accomplished through the cell. Are they inserted into the endosomal compartment and later secreted together with salivary gland-synthesized proteins? If so, can the host proteins be glycosylated in this process? Can the anti-tick immunoglobulins bind with the newly synthesized tick proteins? Will this binding trigger a misfold reaction [94]? Another intriguing observation is the presence of host haptoglobin, hemopexin and lactoferrin in the tick’s saliva. These proteins may act as scavengers of hemoglobin degradation products that are the substrate of heme-oxygenase, an enzyme associated with tissue repair [95] which was found activated at skin sites that were fed by sand flies [96]. Finally, while the majority of the 47 host proteins were found in all tick groups, some of them had distinct peaks that could reflect a selective control of protein transport through the midgut or salivary gland uptake.
Finally, the finding of intra-genic recombination breakpoints on the abundantly expressed lipocalin and metalloprotease families raises the possibility that non-homologous recombination events may be a mechanism increasing the diversity of salivary transcripts in ticks. Whether these events are meiotic or somatic remain to be elucidated. If the events are somatic, then the recombinant transcripts would only exist transiently in the salivary glands and attempts to map their genomic location would fail. In meiotic recombination the recombinants should be mappable to the genome. However, the diversity or heterozygosity of these recombinants may be so large as to be very difficult to map all recombinant products. For example, if a gene has one recombinant breakpoint, we could imagine one chromosome representing A-B and another one a-b, thus four types Ab, aB, ab and AB would be produced, thus a single heterozygotic tick has only the maximum ability to recover 50% of the possible forms. With two recombination breakpoints, nine recombinant forms would exist, and with three breakpoints we would have 16 possible forms, etc. If we multiply these considerations by all genes having intragenic recombination breakpoints, several thousand different genomes would emerge. Accordingly, mapping of these recombinant coding sequences to a single genome would be futile. Indeed, a calculation of the successful genome mapping of transcripts coding for lipocalins and ribosomal proteins from Ixodes scapularis is consistent with this scenario: the program BLAT [97] was used to map the transcripts to the available genome then the exons were summed up to find the percent coverage for the transcripts. While we found an average of 51% genomic coverage for 721 transcripts coding for ribosomal proteins, we found only 31% genomic coverage for lipocalins (Ribeiro, in preparation). Notice that the published genome is a draft that was expected to cover 57% of the tick genome [98]. Thus, the mapping of 51% of the ribosomal proteins is an expected result. This indicates that the lipocalins are underrepresented in the published genome in relation to the more conserved ribosomal proteins. Working with single pair of glands it would be possible to use its mRNA to build a transcriptome library and save its DNA for attempting salivary genomic mapping by PCR methods. Carcass DNA from the same tick would serve as a non-salivary gland genomic control. This experimental design hopefully will allow to distinguish between somatic and meiotic recombination as driving the diversity of transcripts in tick salivary glands.
Supplementary Material
Significance:
Ticks are a burden by themselves to humans and animals, and vectors of viral, bacterial, protozoal and helminthic diseases. Their saliva has anti-clotting, anti-platelet, vasodilatory and immunomodulatory activities that allows successful feeding and pathogen transmission. Previous transcriptomic studies indicate ticks to have over one thousand transcripts coding for secreted salivary proteins. These transcripts code for proteins of diverse families, but not all are transcribed simultaneously, but rather transiently, in a succession. Here we explored the salivary transcriptome and proteome of the brown dog tick, Rhipicephalus sanguineus. A protein database of over 20 thousand sequences was “de novo” assembled from over 600 million nucleotide reads, from where over two thousand polypeptides were identified by mass spectrometry. The proteomic data was shown to vary in time with the transcription profiles, validating the idea that the expression switch may serve as a mechanism of escape from the host immunity. Analysis of the transcripts coding for lipocalin and metalloproteases indicate their genes to contain signals of breakpoint recombination suggesting a new mechanism responsible for the large diversity in tick salivary proteins. These results and the disclosed sequences contribute to our understanding of the success ticks enjoy as ectoparasites, to the development of novel anti-tick methods, and to the discovery of novel pharmacologically active products.
Acknowledgements
JMCR was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (Vector-Borne Diseases: Biology of Vector Host Relationship, Z01 AI000810-20). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This study was financed in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES): # Procad 88881.068421/2014-01; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq): 405763/2018-2, 302360/2018-2 and 441092/2016-0.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jprot.2020.103899.
References
- [1].Guglielmone AA, Robbins RG, Apanaskevich DA, Petney TN, Estrada-Pena A, Horak IG, Shao RF, Barker SC, The Argasidae, Ixodidae and Nuttalliellidae (Acari: Ixodida) of the world: a list of valid species names, Zootaxa 2528 (2010) 1–28. [Google Scholar]
- [2].Sauer JR, Acarine salivary glands - physiological relationships, J Med Ent 14 (1977) 1–9. [DOI] [PubMed] [Google Scholar]
- [3].Binnington KC, Kemp DH, Role of tick salivary glands in feeding and disease transmission, Adv. Parasitol. 18 (1980) 316–340. [DOI] [PubMed] [Google Scholar]
- [4].Kemp DH, Stone BF, Binnington KC, Tick attachment and feeding: Role of mouthparts, feeding apparatus, salivary gland secretions and the host response, in: Obenchain FD, Galun R. (Eds.), Physiology of ticks, Pergamon Press, Ltd, Oxford, 1982, pp. 119–168. [Google Scholar]
- [5].Tan AW, Francischetti IM, Slovak M, Kini RM, Ribeiro JM, Sexual differences in the sialomes of the zebra tick, Rhipicephalus pulchellus, J. Proteome 117 (2015) 120–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jellison WL, Kohls GM, Tick-host anemia: a secondary anemia induced by Dermacentor andersoni stiles, J. Parasitol. 24 (1938) 143–154. [Google Scholar]
- [7].Francischetti IM, My Pham V, Mans BJ, Andersen JF, Mather TN, Lane RS, Ribeiro JM, The transcriptome of the salivary glands of the female western black-legged tick Ixodes pacificus (Acari: Ixodidae), Insect Biochem. Mol. Biol. 35 (10) (2005) 1142–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ribeiro JM, Alarcon-Chaidez F, Francischetti IM, Mans BJ, Mather TN, Valenzuela JG, Wikel SK, An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks, Insect Biochem. Mol. Biol. 36 (2) (2006) 111–129. [DOI] [PubMed] [Google Scholar]
- [9].Ribeiro JM, Anderson JM, Manoukis NC, Meng Z, Francischetti IM, A further insight into the sialome of the tropical bont tick, Amblyomma variegatum, BMC Genom. 12 (2011) 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Francischetti IM, Anderson JM, Manoukis N, Pham VM, Ribeiro JM, An insight into the sialotranscriptome and proteome of the coarse bontlegged tick, Hyalomma marginatum rufipes, J. Proteome 74 (12) (2011) 2892–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ribeiro JMC, Anderson JM, Manoukis NC, Meng ZJ, Francischetti IMB, A further insight into the sialome of the tropical bont tick, Amblyomma variegatum, BMC Genomics 12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schwarz A, von Reumont BM, Erhart J, Chagas AC, Ribeiro JM, Kotsyfakis M, De novo Ixodes ricinus salivary gland transcriptome analysis using two next-generation sequencing methodologies, FASEB J. 27 (12) (2013) 4745–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Garcia GR, Gardinassi LG, Ribeiro JM, Anatriello E, Ferreira BR, Moreira HN, Mafra C, Martins MM, Szabo MP, de Miranda-Santos IK, et al. , The sialotranscriptome of Amblyomma triste, Amblyomma parvum and Amblyomma cajennense ticks, uncovered by 454-based RNA-seq, Parasit. Vectors 7 (2014) 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kotsyfakis M, Schwarz A, Erhart J, Ribeiro JM, Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host, Sci. Rep. 5 (2015) 9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ribeiro JM, Slovak M, Francischetti IM, An insight into the sialome of Hyalomma excavatum, Ticks and tick-borne diseases, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Alarcon-Chaidez FJ, Sun JX, Wikel SK, Transcriptome analysis of the salivary glands of Dermacentor andersoni Stiles (Acari : Ixodidae), Insect Biochem. Mol. Biol. 37 (1) (2007) 48–71. [DOI] [PubMed] [Google Scholar]
- [17].de Castro MH, de Klerk D, Pienaar R, Latif AA, Rees DJG, Mans BJ, De novo assembly and annotation of the salivary gland transcriptome of Rhipicephalus appendiculatus male and female ticks during blood feeding, Ticks Tick-borne Dis. 7 (4) (2016) 536–548. [DOI] [PubMed] [Google Scholar]
- [18].Esteves E, Maruyama SR, Kawahara R, Fujita A, Martins LA, Righi AA, Costa FB, Palmisano G, Labruna MB, Sa-Nunes A, et al. , Analysis of the salivary gland transcriptome of unfed and partially fed Amblyomma sculptum ticks and descriptive proteome of the saliva, Front. Cell. Infect. Microbiol. 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].de Castro MH, de Klerk D, Pienaar R, Rees DJG, Mans BJ, Sialotranscriptomics of Rhipicephalus zambeziensis reveals intricate expression profiles of secretory proteins and suggests tight temporal transcriptional regulation during blood-feeding, Parasite Vector 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aljamali MN, Hern L, Kupfer D, Downard S, So S, Roe BA, Sauer JR, Essenberg RC, Transcriptome analysis of the salivary glands of the female tick Amblyomma americanum (Acari: Ixodidae), Insect Mol. Biol. 18 (2) (2009) 129–154. [DOI] [PubMed] [Google Scholar]
- [21].Anatriello E, Ribeiro JM, de Miranda-Santos IK, Brandao LG, Anderson JM, Valenzuela JG, Maruyama SR, Silva JS, Ferreira BR, An insight into the sialotranscriptome of the brown dog tick, Rhipicephalus sanguineus, BMC Genom. 11 (2010) 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Karim S, Ribeiro JM, An insight into the sialome of the Lone Star tick, Amblyomma americanum, with a glimpse on its time dependent gene expression, PLoS One 10 (7) (2015) e0131292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Karim S, Singh P, Ribeiro JM, A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum, PLoS ONE 6 (12) (2011) e28525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mercado-Curiel RF, Palmer GH, Guerrero FD, Brayton KA, Temporal characterisation of the organ-specific Rhipicephalus microplus transcriptional response to Anaplasma marginale infection, Int. J. Parasitol. 41 (8) (2011) 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Narasimhan S, Deponte K, Marcantonio N, Liang X, Royce TE, Nelson KF, Booth CJ, Koski B, Anderson JF, Kantor F, et al. , Immunity against Ixodes scapularis salivary proteins expressed within 24 hours of attachment thwarts tick feeding and impairs Borrelia transmission, PLoS One 2 (5) (2007) e451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Perner J, Kropackova S, Kopacek P, Ribeiro JMC, Sialome diversity of ticks revealed by RNAseq of single tick salivary glands, PLoS Negl. Trop. Dis. (2018) 12(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rodriguez-Valle M, Moolhuijzen P, Barrero RA, Ong CT, Busch G, Karbanowicz T, Booth M, Clark R, Koehbach J, Ijaz H, et al. , Transcriptome and toxin family analysis of the paralysis tick, Ixodes holocyclusee, Int. J. Parasitol. 48 (1) (2018) 71–82. [DOI] [PubMed] [Google Scholar]
- [28].Mans BJ, Featherston J, de Castro MH, Pienaar R, Gene duplication and protein evolution in tick-host interactions, Front. Cell. Infect. Microbiol. 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mans BJ, Louw AI, Neitz AW, Evolution of hematophagy in ticks: common origins for blood coagulation and platelet aggregation inhibitors from soft ticks of the genus Ornithodoros, Mol. Biol. Evol. 19 (10) (2002) 1695–1705. [DOI] [PubMed] [Google Scholar]
- [30].Nuttall PA, Wonders of tick saliva, Ticks and tick-borne diseases 10 (2) (2019) 470–481. [DOI] [PubMed] [Google Scholar]
- [31].Narasimhan S, Booth CJ, DePonte K, Wu MJ, Liang XP, Mohanty S, Kantor F, Fikrig E, Host-specific expression of Ixodes scapularis salivary genes, Ticks Tick- borne Dis. 10 (2) (2019) 386–397. [DOI] [PubMed] [Google Scholar]
- [32].Tirloni L, Kim TK, Pinto AFM, Yates JR, Vaz ID, Mulenga A, Tick-host range adaptation: changes in protein profiles in unfed adult Ixodes scapularis and Amblyomma americanum saliva stimulated to feed on different hosts, Front. Cell. Infect. Microbiol. 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tirloni L, Islam MS, Kim TK, Diedrich JK, Yates JR, Pinto AFM, Mulenga A, You MJ, Vaz ID, Saliva from nymph and adult females of Haemaphysalis longicornis: a proteomic study, Parasite Vector 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tirloni L, Reck J, Terra RM, Martins JR, Mulenga A, Sherman NE, Fox JW, Yates JR 3rd, Termignoni C, Pinto AF, et al. , Proteomic analysis of cattle tick Rhipicephalus (Boophilus) microplus saliva: a comparison between partially and fully engorged females, PLoS One 9 (4) (2014) e94831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schwarz A, Tenzer S, Hackenberg M, Erhart J, Gerhold-Ay A, Mazur J, Kuharev J, Ribeiro JM, Kotsyfakis M, A systems level analysis reveals transcriptomic and proteomic complexity in Ixodes ricinus midgut and salivary glands during early attachment and feeding, Mol. Cell. Proteomics 13 (10) (2014) 2725–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bensaoud C, Aounallah H, Sciani JM, Faria F, Chudzinski-Tavassi AM, Bouattour A, Mghirbi Y, Proteomic informed by transcriptomic for salivary glands components of the camel tick Hyalomma dromedarii, BMC Genomics (2019) 20(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dantas-Torres F, The brown dog tick, Rhipicephalus sanguineus (Latreille, 1806) (Acari: Ixodidae): from taxonomy to control, Vet. Parasitol. 152 (3–4) (2008) 173–185. [DOI] [PubMed] [Google Scholar]
- [38].Beugnet F, Marie JL, Emerging arthropod-borne diseases of companion animals in Europe, Vet. Parasitol. 163 (4) (2009) 298–305. [DOI] [PubMed] [Google Scholar]
- [39].Demma LJ, Traeger M, Blau D, Gordon R, Johnson B, Dickson J, Ethelbah R, Piontkowski S, Levy C, Nicholson WL, et al. , Serologic evidence for exposure to Rickettsia rickettsii in eastern Arizona and recent emergence of Rocky Mountain spotted fever in this region, Vector Borne Zoonotic Dis. (Larchmont NY) 6 (4) (2006) 423–429. [DOI] [PubMed] [Google Scholar]
- [40].Dantas-Torres F, Figueredo LA, Canine babesiosis: a Brazilian perspective, Vet. Parasitol. 141 (3–4) (2006) 197–203. [DOI] [PubMed] [Google Scholar]
- [41].Solano-Gallego L, Baneth G, Babesiosis in dogs and cats-expanding parasitological and clinical spectra, Vet. Parasitol. 181 (1) (2011) 48–60. [DOI] [PubMed] [Google Scholar]
- [42].De Marco L, Epis S, Comandatore F, Porretta D, Cafarchia C, Mastrantonio V, Dantas-Torres F, Otranto D, Urbanelli S, Bandi C, et al. , Transcriptome of larvae representing the Rhipicephalus sanguineus complex, Mol. Cell. Probes 31 (2017) 85–90. [DOI] [PubMed] [Google Scholar]
- [43].Kaufman WR, Correlation between Hemolymph Ecdysteroid Titer, salivary-gland degeneration and ovarian development in the Ixodid tick, Amblyomma-Hebraeum Koch, J. Insect Physiol. 37 (2) (1991) 95–99. [Google Scholar]
- [44].Moraes J, Marcili A, Nieri-Bastos FA, Richtzenhain LJ, Labruna MB, Genetic analysis of ticks belonging to the Rhipicephalus sanguineus group in Latin America, Acta Trop. 117 (1) (2011) 51–55. [DOI] [PubMed] [Google Scholar]
- [45].Cunha NC, Fonseca AH, Rezende J, Rozental T, Favacho ARM, Barreira JD, Massard CL, Lemos ERS, First identification of natural infection of Rickettsia rickettsii in the Rhipicephalus sanguineus tick, in the State of Rio de Janeiro, Pesqui Vet Brasil 29 (2) (2009) 105–108. [Google Scholar]
- [46].Schneider CA, Rasband WS, Eliceiri KW, NIH image to ImageJ: 25 years of image analysis, Nat. Methods 9 (7) (2012) 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Scarpassa VM, Debat HJ, Alencar RB, Saraiva JF, Calvo E, Arca B, Ribeiro JMC, An insight into the sialotranscriptome and virome of Amazonian anophelines, BMC Genomics 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE, et al. , De novo transcriptome assembly with ABySS, Bioinformatics (Oxford, England) 25 (1) (2009) 2872–2877. [DOI] [PubMed] [Google Scholar]
- [49].Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome, Nat. Biotechnol. 29 (7) (2011) 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinform. 12 (2011) 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, et al. , gplots: Various R programming tools for plotting data, R Package Version 2 (4) (2009). [Google Scholar]
- [52].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics (Oxford, England) 26 (1) (2010) 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Team RC, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2013. [Google Scholar]
- [54].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. , Clustal W and Clustal X version 2.0, Bioinformatics (Oxford, England) 23 (21) (2007) 2947–2948. [DOI] [PubMed] [Google Scholar]
- [55].Kumar S, Stecher G, Tamura K, MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol. 33 (7) (2016) 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Martin DP, Murrell B, Golden M, Khoosal A, Muhire B: RDP4: Detection and analysis of recombination patterns in virus genomes, Virus Evol 1 (1) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR, MS1, MS2, and SQT - three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications, Rapid Commun Mass Sp 18 (18) (2004) 2162–2168. [DOI] [PubMed] [Google Scholar]
- [58].Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, et al. , ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity, J. Proteome 129 (2015) 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bateman A, Martin MJ, Orchard S, Magrane M, Alpi E, Bely B, Bingley M, Britto R, Bursteinas B, Busiello G, et al. , UniProt: a worldwide hub of protein knowledge, Nucleic Acids Res. 47 (D1) (2019) D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Carvalho PC, Lima DB, Leprevost FV, Santos MDM, Fischer JSG, Aquino PF, Moresco JJ, Yates JR, Barbosa VC, Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0, Nat. Protoc. 11 (1) (2016) 102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Paesen GC, Adams PL, Harlos K, Nuttall PA, Stuart DI, Tick histamine-binding proteins: isolation, cloning, and three- dimensional structure, Mol. Cell 3 (5) (1999) 661–671. [DOI] [PubMed] [Google Scholar]
- [62].Paesen GC, Adams PL, Nuttall PA, Stuart DL, Tick histamine-binding proteins: lipocalins with a second binding cavity, Biochim. Biophys. Acta 1482 (1–2) (2000) 92–101. [DOI] [PubMed] [Google Scholar]
- [63].Mans BJ, Ribeiro JM, Function, mechanism and evolution of the moubatin-clade of soft tick lipocalins, Insect Biochem. Mol. Biol. 38 (9) (2008) 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mans BJ, Ribeiro JM, A novel clade of cysteinyl leukotriene scavengers in soft ticks, Insect Biochem. Mol. Biol. 38 (9) (2008) 862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mans BJ, Louw AI, Neitz AW, The major tick salivary gland proteins and toxins from the soft tick, Ornithodoros savignyi, are part of the tick Lipocalin family: implications for the origins of tick toxicoses, Mol. Biol. Evol. 20 (7) (2003) 1158–1167. [DOI] [PubMed] [Google Scholar]
- [66].Chmelar J, Kotal J, Karim S, Kopacek P, Francischetti IM, Pedra JH, Kotsyfakis M, Sialomes and Mialomes: A Systems-Biology View of Tick Tissues and Tick-Host Interactions, Trends Parasitol 32 (3) (2016) 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM, The role of saliva in tick feeding, Front. Biosci. 14 (2009) 2051–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ali A, Khan S, Ali I, Karim S, Vaz ID, Termignoni C, Probing the functional role of tick metalloproteases, Physiol. Entomol. 40 (3) (2015) 177–188. [Google Scholar]
- [69].Ali A, Parizi LF, Guizzo MG, Tirloni L, Seixas A, Vaz ID, Termignoni C, Immunoprotective potential of a Rhipicephalus (Boophilus) microplus metalloprotease, Vet. Parasitol. 207 (1–2) (2015) 107–114. [DOI] [PubMed] [Google Scholar]
- [70].Francischetti IM, Mather TN, Ribeiro JM, Tick saliva is a potent inhibitor of endothelial cell proliferation and angiogenesis, Thromb. Haemost. 94 (1) (2005) 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Francischetti IM, Mather TN, Ribeiro JM, Cloning of a salivary gland metalloprotease and characterization of gelatinase and fibrin(ogen)lytic activities in the saliva of the Lyme disease tick vector Ixodes scapularis, Biochem. Biophys. Res. Commun. 305 (4) (2003) 869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kotal J, Stergiou N, Busa M, Chlastakova A, Berankova Z, Rezacova P, Langhansova H, Schwarz A, Calvo E, Kopecky J, et al. , The structure and function of Iristatin, a novel immunosuppressive tick salivary cystatin, Cell. Mol. Life Sci. 76 (10) (2019) 2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lieskovska J, Palenikova J, Langhansova H, Campos Chagas A, Calvo E, Kotsyfakis M, Kopecky J, Tick sialostatins L and L2 differentially influence dendritic cell responses to Borrelia spirochetes, Parasit Vectors 8 (2015) 275, 10.1186/s13071-13015-10887-13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Klein M, Bruhl TJ, Staudt V, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Bohn T, Ulges A, Stergiou N, et al. , Tick Salivary Sialostatin L Represses the Initiation of Immune Responses by Targeting IRF4-Dependent Transcription in Murine Mast Cells, J. Immunol 195 (2) (2015) 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lieskovska J, Palenikova J, Sirmarova J, Elsterova J, Kotsyfakis M, Campos Chagas A, Calvo E, Ruzek D, Kopecky J, Tick salivary cystatin sialostatin L2 suppresses IFN responses in mouse dendritic cells, Parasite Immunol. 37 (2) (2015) 70–78. [DOI] [PubMed] [Google Scholar]
- [76].Horka H, Staudt V, Klein M, Taube C, Reuter S, Dehzad N, Andersen JF, Kopecky J, Schild H, Kotsyfakis M, et al. , The tick salivary protein sialostatin L inhibits the Th9-derived production of the asthma-promoting cytokine IL-9 and is effective in the prevention of experimental asthma, J. Immunol. 188 (6) (2012) 2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kotsyfakis M, Horka H, Salat J, Andersen JF, The crystal structures of two salivary cystatins from the tick Ixodes scapularis and the effect of these inhibitors on the establishment of Borrelia burgdorferi infection in a murine model, Mol. Microbiol. 77 (2) (2010) 456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sa-Nunes A, Bafica A, Antonelli LR, Choi EY, Francischetti IM, Andersen JF, Shi GP, Chavakis T, Ribeiro JM, Kotsyfakis M, The immunomodulatory action of sialostatin L on dendritic cells reveals its potential to interfere with autoimmunity, J. Immunol. 182 (12) (2009) 7422–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jaenisch R, Bird A, Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals, Nat. Genet. 33 (3) (2003) 245–254. [DOI] [PubMed] [Google Scholar]
- [80].Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP, Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae, J. Proteome Res. 5 (9) (2006) 2339–2347. [DOI] [PubMed] [Google Scholar]
- [81].Diaz-Martin V, Manzano-Roman R, Valero L, Oleaga A, Encinas-Grandes A, Perez-Sanchez R, An insight into the proteome of the saliva of the argasid tick Ornithodoros moubata reveals important differences in saliva protein composition between the sexes, J. Proteome 80 (2013) 216–235. [DOI] [PubMed] [Google Scholar]
- [82].Madden RD, Sauer JR, Dillwith JW, A proteomics approach to characterizing tick salivary secretions, Exp. Appl. Acarol. 32 (1–2) (2004) 77–87. [PubMed] [Google Scholar]
- [83].Oliveira CJ, Anatriello E, de Miranda-Santos IK, Francischetti IM, Sa-Nunes A, Ferreira BR, Ribeiro JM, Proteome of Rhipicephalus sanguineus tick saliva induced by the secretagogues pilocarpine and dopamine, Ticks Tick-borne Dis. 4 (6) (2013) 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hodge K, Ten Have S, Hutton L, Lamond AI, Cleaning up the masses: exclusion lists to reduce contamination with HPLC-MS/MS, J. Proteome 88 (2013) 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Mather TN, Ribeiro JMC, Exploring the sialome of the tick, Ixodes scapularis, J. Exp. Biol. 205 (2002) 2843–2864. [DOI] [PubMed] [Google Scholar]
- [86].Kim TK, Tirloni L, Pinto AFM, Diedrich JK, Moresco JJ, Yates JR, Vaz ID, Mulenga A, Time-resolved proteomic profile of Amblyomma americanum tick saliva during feeding, PLoS Negl. Trop. Dis. (2020) 14(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kim TK, Tirloni L, Pinto AFM, Moresco J, Yates JR, Vaz ID, Mulenga A, Ixodes scapularis Tick Saliva Proteins Sequentially Secreted Every 24 h during Blood Feeding, PLoS Negl. Trop. Dis. (2016) 10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bowman AS, Sauer JR, Tick salivary glands: function, physiology and future, Parasitology 129 (Suppl) (2004) S67–S81. [DOI] [PubMed] [Google Scholar]
- [89].Simo L, Kazimirova M, Richardson J, Bonnet SI, The essential role of tick salivary glands and saliva in tick feeding and pathogen transmission, Front. Cell. Infect. Microbiol. 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Francischetti IMB, Anderson JM, Manoukis N, Pham VM, Ribeiro JMC, An insight into the sialotranscriptome and proteome of the coarse bontlegged tick, Hyalomma marginatum rufipes, J. Proteome 74 (12) (2011) 2892–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wang H, Nuttall PA, Excretion of host immunoglobulin in tick saliva and detection of IgG-binding proteins in tick haemolymph and salivary glands, Parasitology 109 (Pt 4) (1994) 525–530. [DOI] [PubMed] [Google Scholar]
- [92].Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Mather TN, Ribeiro JM, Exploring the sialome of the tick Ixodes scapularis, J. Exp. Biol. 205 (Pt 18) (2002) 2843–2864. [DOI] [PubMed] [Google Scholar]
- [93].Lara FA, Lins U, Bechara GH, Oliveira PL, Tracing heme in a living cell: hemoglobin degradation and heme traffic in digest cells of the cattle tick Boophilus microplus, J. Exp. Biol. 208 (16) (2005) 3093–3101. [DOI] [PubMed] [Google Scholar]
- [94].Ramirez MU, Hernandez SR, Soto-Pantoja DR, Cook KL, Endoplasmic Reticulum Stress Pathway, the Unfolded Protein Response, Modulates Immune Function in the Tumor Microenvironment to Impact Tumor Progression and Therapeutic Response, Int. J. Mol. Sci. (2020) 21(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lee GR, Shaefi S, Otterbein LE, HO-1 and CD39: it takes two to protect the realm, Front. Immunol. 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Luz NF, DeSouza-Vieira T, De Castro W, Vivarini AC, Pereira L, Franca RR, Silveira-Mattos PS, Costa DL, Teixeira C, Meneses C, et al. , Lutzomyia Iongipalpis saliva induces Heme Oxygenase-1 expression at bite sites, Front. Immunol. 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kent WJ, BLAT–the BLAST-like alignment tool, Genome Res. 12 (4) (2002) 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, de la Fuente J, Ribeiro JM, Megy K, et al. , Genomic insights into the Ixodes scapularis tick vector of Lyme disease, Nat. Commun. 7 (2016) 10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data was deposited to the National Institute for Biotechnology Information (NCBI) under Bioproject PRJNA606595 and Biosample accessions SAMN14115946, SAMN14115947, SAMN14115948, SAMN14115949, SAMN14115950, SAMN14115951, SAMN14115952, SAMN14115953, SAMN14115954, SAMN14115955, SAMN14115956, SAMN14115957, SAMN14115958, SAMN14115959, SAMN14115960, SAMN14115961, SAMN14115962, SAMN14115963, SAMN14115964 and SAMN14115965. The reads were deposited to the Short Reads Archive of the NCBI under accessions SRR11109985, SRR11109984, SRR11109973, SRR11109972, SRR11109971, SRR11109970, SRR11109969, SRR11109968, SRR11109967, SRR11109966, SRR11109983, SRR11109982, SRR11109981, SRR11109980, SRR11109979, SRR11109978, SRR11109977, SRR11109976, SRR11109975 and SRR11109974. This Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GINV00000000. The version described in this paper is the first version, GINV01000000.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018964.