Although today the majority of scientific data including microscopy and imaging data are available in digital format, a real benefit from easy sharing and re-using digital data according to the FAIR principles1 only exists if data are understandable and unambiguously interpretable. Collecting and maintaining the relevant metadata is key to ensuring that data are reliable, reusable and can be found and accessed by the scientific community. Imaging data are usually extremely rich data files as they report on various parameters in a multidimensional space and are acquired with complex microscopy instruments. The metadata or data models are very diverse due to the wide range of e.g. modalities, scales, experimental setups and file formats. Therefore, the appropriate use of suitable standardized metadata and data models is a challenge2,3. Accordingly, flexible tools for capturing a complete set of metadata are in great demand by researchers applying microscopy techniques. Moreover, it is important for imaging core facilities to be able to provide different standards with one tool and still be flexible enough for dynamic developments. Many tools fail to strike this balance and therefore oscillate between using rigid data models and free text input without semantic context. Similarly, the integration and referencing of existing metadata is often lacking.
Our tool, MDEmic (MetaData Editor for microscopy), provides an easy and comfortable way for editing metadata of microscopic imaging data and at the same time offers high flexibility in terms of adjustment of metadata sets and their data models. As the standardization process regarding the metadata of microscopic experiments is in full swing, MDEmic offers high flexibility to follow this process. This is achieved through the dynamic configurability of both the queried or integrated metadata and predefined values. The underlying data model can be extended sequentially by these specified objects. The additional integration of ontology databases increases the interpretability of the data in general and ensures the discipline-specific integration of imaging data. MDEmic reads the technical metadata stored in the image file using Bio-Formats3, a software library for reading proprietary microscopy image (meta)data and presents this metadata in the form of the OME (Open Microscopy Environment) Data Model4,5,6. Visualizing this model as input forms allows the researcher to adjust or correct the technical metadata. In addition, based on the default in a configuration file associated with MDEmic, input forms for further metadata can be generated dynamically, integrating, or extending the OME Data Model for technical metadata. The specification of metadata can include: i) type and category of metadata, ii) fixed terms as selectable input values loading from subclasses by specifying ontology class identifiers, iii) definition of relation to other metadata categories. For all metadata, different sets of predefinitions can be integrated via the configuration file and selected by the user according to the respective scientific application or image technology. For example, fully described components of the existing microscope can be added or replaced by the user in the metadata collection. MDEmic is part of the standard installation package of the image database OMERO7 and is integrated in the OMERO.importer as OMERO.mde. The OMERO.importer can be used independent from a local OMERO instance by referring to a public OMERO instance like the Image Data Repository, IDR8,9. All metadata descriptions created in OMERO.mde can be saved and reloaded by the user for later reuse and adaptation or can be exported to different textual formats. This functionality allows the output to be easily integrated with other needs. For example, this increases interoperability with other research data management tools to support integration with other data types or preparation of image data for publications or upload to public repositories10 like the IDR (supplementary figure 5,6).
In the following we describe a use case scenario where image data from samples treated with various membrane dyes are made available in a „Membrane Dye Database“ hosted in the institutional OMERO instance and shared between the members of a collaborative research center (CRC) with a focus in membrane research. Here, we utilized the OMERO.mde for customized metadata annotation (supplementary figure 2–4). To this purpose we have defined a metadata object „Membrane Dye” in OMERO.mde. This object is available with different sub-objects describing the membrane dyes in more detail like „Effects On Sample” and „Internalization“ of the dye. This object is provided together with the technical description and essential OME Data Model objects as input forms. All input forms are summarized in the setup „Membrane Dye Database”. This adjustment for the specific object „Membrane Dye” can be done by editing the configuration file of OMERO.mde e.g., by a data steward of the CRC or a scientist of the imaging core facility.
The use case described shows the direct benefit of a metadata annotation tool like MDEmic (in particular here: OMERO.mde) for researchers, as it illustrates a clearly defined purpose. It will help to increase the overall awareness for the importance of metadata annotation and consistent research data management in general. Tools like MDEmic, which significantly improves the interoperability of bioimaging data with other data types and within discipline specific data management environments, have a high impact on the acceptance of FAIR research data management.
Supplementary Material
Figure 1. OMERO.importer with integrated MDEmic as OMERO.mde.
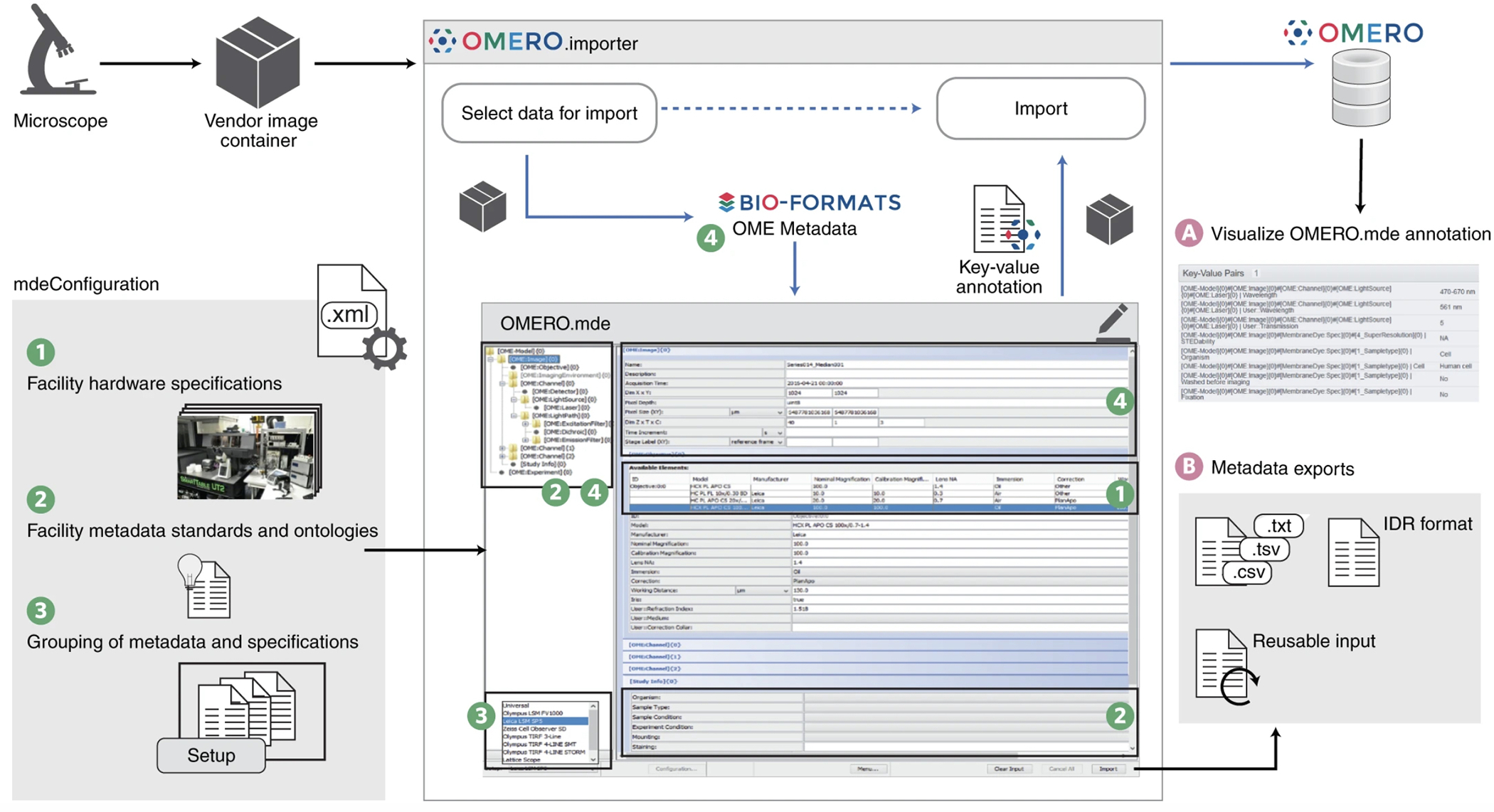
In the OMERO.importer the MDEmic is integrated as an intermediate step for the selection of data for the import and the import itself. Metadata can now be added, which is then transferred to the repository together with the image data (A), or the annotations can be exported in different formats in this step (B). The MDEmic can be customised via a configuration file and loads the specifications from this file dynamically when the OMERO.importer is started (1,2,3). All technical metadata of the images marked in the previous step of data selection are read out by Bio-Format (4) and provided as values in the MDEmic respectively.
Acknowledgements
We would like to thank the OME team for constant support and helpful discussion, in particular Josh Moore, Dominik Lindner and Jean-Marie Burel. We received funding by: the Deutsche Forschungsgemeinschaft: grants SFB 1208 (Ref: 267205415 INF/Z02 (S.W.-P.) and SFB 944 (Ref: 180879236 INF) (S.K.); Wellcome Trust grant (Ref: 212962/Z/18/Z) and the BBSRC (Ref: BB/R015384/1) (F.W.); NIH grant (Ref: 2U01CA200059-06) and grant (Ref: 2019–198155 (5022) by Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation, as part of their Imaging Scientist Program) (C.S.D.C).
Footnotes
Competing interests
The authors declare no competing interests.
Code availability
All code is accessible at Github (https://github.com/ome/omero-insight). The fully open-source code link is listed in Supplementary Table 1.
Additional information
Supplementary information is available for this paper.
Data availability
All data available under links listed in Supplementary Table 1.
References:
- 1.Wilkinson MD et al. Scientific Data 3.1, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarkans U et al. Nat. Methods (2021). doi: 10.1038/s41592-021-01166-8. [DOI] [Google Scholar]
- 3.Linkert M et al. J. Cell Biol. 189.5, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg IG et al. Genome Biol. 6.5, 1–13 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S et al. Methods 96, 27 (2016). doi: 10.1016/j.ymeth.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swedlow J et al. Science 300.5616, 100–102 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allan C et al. Nat. Methods 9.3, 245–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellenberg J et al. Nat. Methods 15, 849–854 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams E et al. Nat. Methods 14.8, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swedlow JR et al. Nat. Methods (2021). doi: 10.1038/s41592-021-01113-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data available under links listed in Supplementary Table 1.


