Abstract
The current inhabitants of the Canary Islands have a unique genetic makeup in the European diversity landscape due to the existence of African footprints from recent admixture events, especially of North African components (> 20%). The underrepresentation of non-Europeans in genetic studies and the sizable North African ancestry, which is nearly absent from all existing catalogs of worldwide genetic diversity, justify the need to develop CIRdb, a population-specific reference catalog of natural genetic variation in the Canary Islanders. Based on array genotyping of the selected unrelated donors and comparisons against available datasets from European, sub-Saharan, and North African populations, we illustrate the intermediate genetic differentiation of Canary Islanders between Europeans and North Africans and the existence of within-population differences that are likely driven by genetic isolation. Here we describe the overall design and the methods that are being implemented to further develop CIRdb. This resource will help to strengthen the implementation of Precision Medicine in this population by contributing to increase the diversity in genetic studies. Among others, this will translate into improved ability to fine map disease genes and simplify the identification of causal variants and estimate the prevalence of unattended Mendelian diseases.
Subject terms: Genotype, Population genetics
Introduction
The Canary Islands are a Spanish archipelago of seven main islands located in the Atlantic Ocean, a hundred kilometers off the Northwest African coast, with Cape Juby (Morocco) being the closest mainland point. Before the XV century, when the archipelago was fully incorporated into the European world, it was inhabited by aborigines1 with their most likely origin in the Berber population from North Africa2,3. This and subsequent historic events, including the European colonization of diverse origins and the slave trade from western sub-Saharan African populations4, have shaped the genetic makeup of Canary Islanders, as has been established by the historical burial remains and the diverse ancient DNA studies5–7. Early genetic studies have supported that the current Canary Islanders could be modeled as descendants of a recent three-way admixture event. Note, however, that these studies just considered the major continental populations and did not assess the substructure components in the parental populations. Sexual asymmetry in the admixture event has been invoked to explain the observed unbalanced proportions of indigenous parental lineages from the non-recombining portion of the Y chromosome (NRY) and the mitochondrial DNA (mtDNA) in current inhabitants8. This has been explained by a steady increase of European male lineages soon after the conquest, whereas the indigenous founder mtDNA lineages have remained at roughly constant frequencies until the present days9,10. Overall, while there is a large interindividual variability in the ancestry proportions in the current inhabitants, they have been estimated to an average of 75–83% European (EUR), 17–23% North African (NAF), and 3% or less sub-Saharan Africa (SSA)11,12. The most recent analyses based on genome-wide single nucleotide polymorphism (SNP) array data evidenced that these numbers can be as high as up to 29.9% NAF and 9.2% SSA ancestries in some individuals13. Most importantly, they have also evidenced broad genomic regions of Canary Islanders that tend to concentrate African alleles and that are enriched in genes involved in diverse complex diseases, which is highly suggestive of characteristic footprints of local adaptations.
The success of Next Generation DNA Sequencing (NGS) represents a milestone in how genetic variation is discovered and analyzed nowadays. It has opened new horizons to improve disease diagnosis, prognosis, and treatment, and constitutes a central element of the Precision Medicine paradigm14. Nevertheless, the genetic knowledge of human traits and diseases remain to be almost entirely based on results from studies in European populations15,16. This results in an underrepresentation of ethnic diversity and a conspicuous lack of accuracy in the understanding of the genetic architecture and the biology of human traits, thus challenging the translation of this knowledge into generalizable clinical applications17–19. While there are differences in allele frequencies across populations20, the rarer the variant, the more likely for it to be more locally circumscribed to populations21. The estimations indicate that while the proportion of rare variants (minor allele frequency [MAF] < 0.5%) shared among populations from the same continent is 70–80%, the proportion of shared rare variants drops down to 10–30% among populations from different continents, and are, therefore, poorly represented in the reference catalogs of genetic variation22,23. Most importantly, deleteriousness is known to accumulate on the lower end of the allele frequency spectrum24. The application of NGS, both through whole-exome sequencing (WES) and whole-genome sequencing (WGS), has drastically increased the diagnostic yield in patients of European ancestry, such as for autosomal dominant retinitis pigmentosa25, severe intellectual disability26, and Mendelian conditions in a broad sense27,28. Because of that, the substantial benefits of incorporating participants from diverse ancestries and recently admixed populations to improve the discovery of disease genes have been evidenced15. This highlights the urgent need for building local population reference catalogs of genetic diversity to efficiently facilitate the identification of disease genes29. Developing these population catalogs of genetic variation is such of importance that multiple countries have made huge efforts to develop their own based on the study of a representative control strata of the populations while preserving genetic diversity specificities, many of which have been recently integrated in the Genome Aggregation Database (gnomAD)30. Iran31, Japan32, Korea33, Finland34, Spain35, the United Kingdom36, or the Netherlands37 are some of the countries which have seen the necessity to develop their own catalogs of genetic variation. The availability of population-specific catalogs of variation allows to identify genetic peculiarities of the population38 as is key for identifying disease-causing variants in both rare diseases and complex human traits32,36. This has been recently exemplified by the striking detection of recessive deficiencies in two genes of the type I interferon pathway, critically involved in life-threatening viral diseases including COVID-19, at relatively high frequency (> 1%) in Polynesia and Inuits, while these deficient are extremely rare or absent from other regions of the world39,40. For the particular case of the Canary Islands, one of the early examples of its benefit has been recently demonstrated to support the underdiagnosis of Wilson disease, a rare difficult-to-diagnose disease41. The benefits have been also shown for complex traits, which is more evident for the case of isolated populations, as the cases of the Northern Greek populations of the Pomak villages and the Mylopotamos villages in Crete42,43, or the population of Cilento from Southern Italy44, highlighting for example the increase in allele frequency of variants involved in haematological traits, among others. Besides these applications, there are substantial gains of incorporating information from the population of interest to reference panels during variant imputation, as it improves the power to identify disease variants and enables fine-mapping in genome-wide association studies of complex traits18,42,43,45. Thus, developing a population-specific catalog of genetic variation is an essential step to optimally develop generalizable clinical applications.
The historical conquest and admixture events, jointly with the isolation and inbreeding, as well as the likely local adaptation processes, have shaped the current genetic background of the Canary Islands population, constituting the population with the largest proportion of North African ancestry among Southwestern Europeans12,13. Despite the increase in awareness of the necessity of including more diversity in the genetic studies46,47, the currently available catalogs of genetic variation have a strong bias towards the representation of northern, western, and central European populations. In particular, it has been shown that the African genomic ancestry has important biomedical implications for European populations12 and it has been associated with risk in cardiovascular, renal, and respiratory diseases, as well as in diabetes, among others48–51. Strikingly, for some of them, the estimates of prevalence and/or their complications are higher in the Canary Islands than in other mainland regions of Spain. This is the case of asthma and allergic diseases in children52,53, and of diabetes, obesity, and hypertension in all age groups53. Besides, not only the morbidity but also the mortality due to diabetes is increased, being three-fold higher among Canary Islands compared to the rest of the Spanish populations54. Despite the interest considering that the Canary Islanders exhibit the largest proportion of NAF ancestry known to date in the European diversity landscape13, there is a lack of data from North African populations on the public catalogs representing human genetic diversity55, not even being covered by the African Genome Variation Project46.
Here we aimed to establish the foundations to develop a reference genetic catalog of the Canary Islands population (termed CIRdb). Providing an unbiased catalog of natural genetic variation of this population, preserving unique genetic African ancestry footprints, is a first necessary resource for optimal development of Precision Medicine in this population56.
Materials and methods
Study samples and genotyping
The study was approved by the Research Ethics Committee of the Hospital Universitario Nuestra Señora de Candelaria and performed according to The Code of Ethics of the World Medical Association (Declaration of Helsinki).
The samples were obtained from the cohort study ‘CDC of the Canary Islands’57, which constitutes the most extensive general population cohort for epidemiological studies of the Canary Islands archipelago. Briefly, this cohort involves health survey data and samples from nearly 7000 randomly selected donors providing informed consent through personal interviews, aged between 18 and 75 years from the seven main islands and without gender bias. Despite it is well-known to properly represent the Islands’ population58, the CDC cohort lacks deep genetic assessment despite the recognized necessity53. A subset of 416 individuals from the cohort was previously assessed to characterize inbreeding, selection, and the mosaic nature of the Canarian genomes13. We nested the current study in that cohort, particularly focusing on a subset of 1024 donors (483 males and 541 females), fulfilling that they self-reported absence of cardiovascular, metabolic, immunologic, or cancer diseases, and that the four grandparents were born on the same island. The latter criterion was relaxed to accommodate donors from Fuerteventura, where the selection imposed self-reporting three grandparents born on the island. The samples were pseudonymized for the purposes of this study.
DNA was extracted from peripheral blood using the Blood genomicPrep Mini Spin Kit (Cytiva, Marlborough, MA) following the manufacturer’s recommendations. We relied on the Axiom® Genome-Wide Human CEU 1 Array (Affymetrix, Santa Clara, CA) to obtain genotypes from 587,352 variants with the support of the National Genotyping Center (CeGen), Universidad de Santiago de Compostela Node. The AffyPipe v2.10.0 open-source pipeline59 was used to process the image files and to run the first genotyping quality controls (QCs) based on a Dish-QC (i.e. an own statistic from the tool which allows to evaluate the signal of non-polymorphic positions) with values higher than 0.82, and samples with call rate (CR) above 0.93. Additionally, after running AffyPipe, more SNP QCs were implemented to select accurate variants by means of SNPolisher package in R 3.2.2 environment60 and according to next parameters which were extracted from ‘ps.performance.txt’ file: Fisher’s Linear Discriminant (FLD) > 4.375, Heterozygous Strength Offset (HetSO) ≥ − 0.5, SNP CR ≥ 95, and variants with assigned rsID. Next, the R environment and PLINK v1.0761 were used for additional standard QC steps, including the identification of variants in non-autosomal chromosomes, and variants with large deviations from Hardy–Weinberg equilibrium (p < 1.0 × 10−6), large missingness rate (CR < 0.95), or MAF < 0.01, and the identification of samples with gender discordances (self-declared vs. genetically inferred), outlier heterozygosity rate, and family relationships with other study donors (PIHAT > 0.2). For the purpose of this particular study, we only considered the donors declaring four grandparents born on the same island (three grandparents in the context of Fuerteventura). After variant and sample QCs, the dataset was ready for ulterior population analysis.
Reference datasets and analyses
The genetic background of the Canary Islanders was evaluated under two different scenarios: one focusing only the Canary Islanders, and another comparing the Canary Islanders against reference populations. Each scenario involved a different number of samples and variants (based on the use of different filters) (Table 1).
Table 1.
Summary of the population analyses with indications of the number of samples and SNPs involved.
| Analysis | Samples | Filters involved in SNP selection | #SNPs used | |
|---|---|---|---|---|
| Canary Islanders | Reference population | |||
| PCA | 863 | 522 | LD SNPs (r2 > 0.5) and regionsa | 101,271 |
| 863 | – | LD SNPs (r2 > 0.15) and regionsa | 116,959 | |
| 617 | – | LD SNPs (r2 > 0.15) and regionsa | 116,959 | |
| ADMIXTURE | 690 | 522* | LD SNPs (r2 > 0.5) and regionsa | 101,271 |
| ELAI | 690 | 522 | Not pruned | 114,929 |
PCA, Principal Component Analysis; LD, linkage disequilibrium. a Regions of long-range linkage disequilibrium were defined elsewhere13. *As a sanity check, some analysis included other European populations from the south of the continent (i.e., Toscani in Italy [TSI, N = 106] and Iberian Populations in Spain [IBS, N = 106] from 1KGP).
Reference population datasets
To place the genetic variation of Canary Islanders in context, we accessed reference data from The 1000 Genomes Project (1KGP) Phase 362 for EUR and SSA populations. Europeans included data from Finnish in Finland (FIN) (N = 99), British in England and Scotland (GBR) (N = 91), and Utah Residents with Northern and Western European ancestry (CEU) (N = 99). Note, however, that for specific analysis (i.e., for ADMIXTURE), we also included other European populations from the south of the continent (i.e., Toscani in Italy [TSI, N = 106] and Iberian Populations in Spain [IBS, N = 106] from 1KGP). Given that using alternative African populations from 1KGP or smaller subsets of individuals from the parental populations provide equivalent admixture results in Canary Islanders13, we used only the Yoruba population in Ibadan (Nigeria) (YRI) (N = 108) as representatives of the SSA populations. NAF populations were represented by the 125-individual dataset that is publicly available and genotyped using Genome-Wide Human SNP Array 6.0 (Affymetrix) for 732,532 variants63. The intersection of the reference datasets (N = 522) with those of Canary Islanders (N = 863) was implemented using ‘–bmerge’ command on PLINK v1.964 to merge into one file including only the variants that were shared among populations. This process left us with data from 1385 individuals and 114,929 variants as the final filtered dataset for the downstream analyses involving both Canary Islanders and the reference population datasets.
Principal component analysis
Principal Component Analysis (PCA) among Canary Islanders and reference population datasets (N = 1385) was computed with PLINK based on a dataset of 101,271 variants, which excluded variants in high linkage disequilibrium (LD) and those that were located in regions of long-range LD as performed elsewhere13. For the comparisons within the Canary Islands populations, a pairwise r2 threshold of 0.15 was used to maintain 116,959 variants for the analyses in the 863 individuals collected from the archipelago.
Ancestry inferences
Two approaches were conducted to assess the genetic ancestry partitions of the subjects under study: 1) a direct global ancestry estimation using ADMIXTURE v1.3.065; and 2) a global ancestry estimation from local ancestry inference for admixed individuals by means of ELAI v1.0166. We have shown that, compared to other local ancestry estimators, ELAI offers the least biased estimates for this population13,67. Note that ancestry estimations inferred for the reference populations (i.e., Europeans and North Africans) should be considered with caution due to the existence of genetic drift effects that have not been properly modelled13. Nevertheless, this does not affect the ancestry inferences obtained for the Canary Islanders.
ADMIXTURE implements a maximum likelihood estimation to calculate the individual ancestries averaged across the genome. For this approach, 2 to 7 ancestral populations (K) and 10-times cross-validation were tested to estimate the best fitting K. In order to avoid spurious clusters in the ADMIXTURE results due to the existence of inbreeding, which we have evidenced in the populations from smaller islands13, we further pruned the Canary Islands dataset to be considered for this particular analysis. For that, we calculated the runs of homozygosity (ROHs) with PLINK following a sliding window approach68. We allowed that a minimum window density of 50 kb/SNP was asserted, as well as one heterozygous variant and up to five missing calls per window. For tracts with a minimum length of 500 kb, 50 was the minimum number of homozygous SNPs to consider a tract as a ROH, and 100 kb was the maximum gap allowed between two consecutive SNPs to include them in the same ROH. The hit rate of all scanning windows containing a variant must be at least 0.05 to comprise a certain ROH. Finally, the samples with ROH lengths above the 80th percentile (1.08 Mb) were excluded from the analysis. This left us with 690 Canary Islanders for this particular analysis, providing a final dataset of 1212 samples (including 552 individuals from the reference populations) and the 101,271 pruned variants which have been previously used for the PCA. Additionally, some European populations from the south of the continent (TSI and IBS from 1KGP) were included into the ADMIXTURE analysis as a sanity check.
For local ancestry estimation, ELAI uses a two-layer hidden Markov model to assess the local ancestry in the individuals. The same subset of 690 Canary Islanders was evaluated to match the results from both ancestry inferences approaches. Based on our previous observations, we assumed a three-way admixture model of EUR, NAF, and SSA to calculate the structure of local haplotypes given that both approaches (i.e., ADMIXTURE and ELAI) provided similar estimates13. Moreover, 14 generations since the last admixture event was assumed based on our previous findings13. We excluded SNPs with MAF < 0.01 or with missing position information in any of the reference datasets. Subsequently, the global ancestry estimation was calculated considering the average of each ancestry per individual per chromosome and summarizing all the information into a unique value per individual per ancestry. Therefore, global inferences based on ELAI algorithm were implemented using 1212 individuals (690 Canary Islanders and 522 individuals from reference datasets) and 114,929 variants (given that independence of SNPs is not a requirement for this approach).
Results
Samples included in the CIRdb catalog
Samples from a total of 1024 donors were selected and utilized for SNP array genotyping. After QC filters based on the obtained genotypes (Fig. 1), we identified 863 unrelated individuals (406 males, 457 females) and 514,561 variants for further assessments. We also identified samples where, albeit all grandparents were born in the Canaries, they were not from the same island. The data from these samples was excluded from the analyses described in this study, although they will be considered in further development of the CIRdb catalog. The distribution of samples per island that will be considered in the analyses is as follows: 105 from El Hierro, 93 from La Palma, 141 from La Gomera, 156 from Tenerife, 210 from Gran Canaria, 47 from Fuerteventura, and 111 from Lanzarote.
Figure 1.
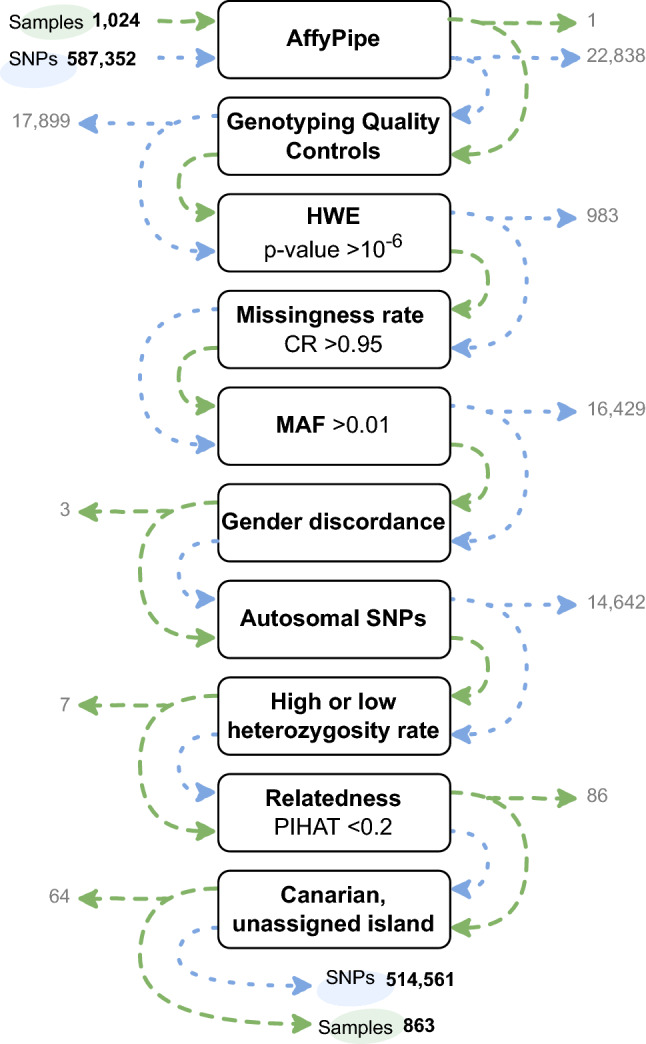
Schematic representation of the variants (blue) and samples (green) that were filtered out based on quality control steps. SNPs, single nucleotide polymorphisms; HWE, Hardy–Weinberg Equilibrium; MAF, Minor Allele Frequency; PIHAT, proportion of identity-by-descent. Created with draw.io v16.2.7 (https://github.com/jgraph/drawio).
Population characteristics based on SNP arrays included in CIRdb catalog
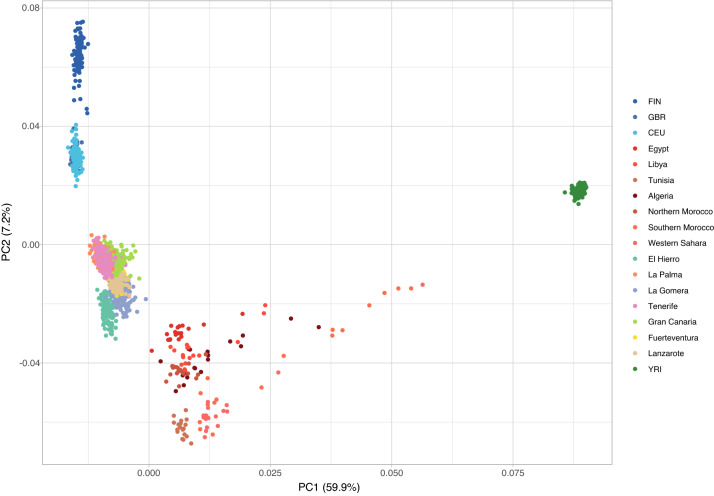
In the PCA, including Canary Islanders and the reference populations (101,271 variants, 1385 samples), the first two principal components (PCs) encompassed 67.1% of the variation and revealed a distinctive separation in four main clusters (Fig. 2). Similar to what has been described recently by us13, the EUR and SSA individuals dominate the PC1 axis of differentiation, forming compact and well-separated clusters, also revealing a scattering pattern of clustering of NAF individuals from diverse populations. PC2 portrays the differentiation between NAF and EUR. In this axis of differentiation, the samples from the different Canary Islands clustered tightly between each other, but separately from the EUR, NAF, and SSA populations. In the cluster, within-Archipelago affinities are somehow evident, with some island populations plotting closer to NAF (El Hierro, La Gomera, Fuerteventura, and Lanzarote) while others situated closer to EUR (La Palma, Gran Canaria, and Tenerife).
Figure 2.
Representation of the first two principal components comprising 67.1% of genetic variation in Canary Islanders and reference populations from Europe (GBR, FIN, and CEU), North Africa (Algeria, Egypt, Libya, Northern Morocco, Southern Morocco, Western Sahara, and Tunisia), and Sub-Saharan Africa (YRI). A total of 101,271 variants and 1385 samples were used. Created with R v3.2.2 (https://www.r-project.org/).
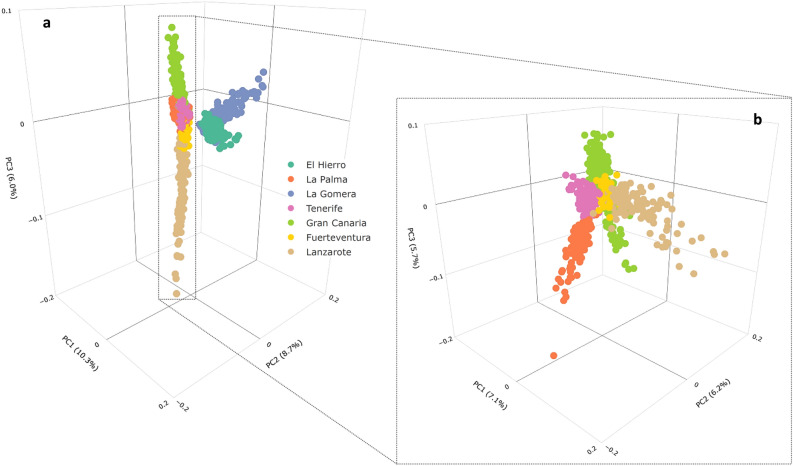
We also assessed the 863 unrelated Canary Islands donors (Fig. 3A) (116,959 variants, 863 samples) by PCA, where the first two PCs clearly distinguished the donors from El Hierro and La Gomera, the two smallest islands, from the rest of the islands. The rest of the island populations followed a continuum in the PC3 axis of differentiation, with a tendency to locate the samples from Gran Canaria, La Palma, and Tenerife on one side, and those from Fuerteventura and Lanzarote on the other. This differentiation is more evident when excluding the samples from El Hierro and La Gomera from the PCA analysis (Fig. 3B).
Figure 3.
Representation of the first three principal components from PCA of Canary Islanders (a total of 116,959 variants were used). a) Including all unrelated Canary Islands samples (N = 863), where the first three PCs explain 25.0% of variability. b) Excluding the samples from El Hierro and La Gomera (N = 617), where the first three PCs explain 19.0% of variability. Created with R v3.2.2 (https://www.r-project.org/).
Represented genetic ancestries in the CIRdb catalog
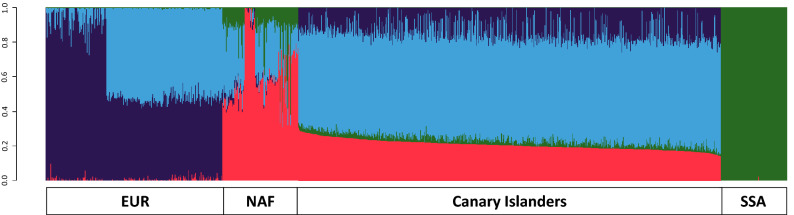
ADMIXTURE ancestries for the dataset indicated that the best fitting model was obtained for K = 4 (Fig. 4). This is in agreement with previous results11,12 assessing the best fitting based on badMIXTURE residuals which ensured that K = 4 provided robust ancestry proportions13. Note, however, that the two EUR ancestries were aggregated into one for the rest of assessments to avoid relying on unstable subcontinental ancestry estimates given the small number of variants (Table 2). By aligning the identified clusters with the most abundant components identified in the references, they supported that the largest contribution to the genetic background of Canary Islanders is, on average, EUR ancestry (76.4%; composed of two ancestries aligned with the European northwest–southeast axis of differentiation69), followed by NAF (20.8%), and SSA (2.8%) (Table 2). Alternative analysis including also European populations from the south of the continent (TSI and IBS from 1KGP) to have information from the main European axis of differentiation barely changed the overall results (see Supplementary Fig. S1, Supplementary Table S1, and Supplementary Table S2 online). ELAI ancestries provided a similar scenario of admixture composed mainly by EUR (71.4%), followed by NAF (26.7%), and SSA (1.9%) (Table 2). However, we observed a much wider interindividual variation in the admixture proportions in Canary Islanders in this study with more samples from the geography (Fig. 4), so that the NAF and SSA ancestry assignations in Canary Islanders could be as high as 38.2% and 9.5%, respectively.
Figure 4.
ADMIXTURE estimates for the best fitting model (K = 4) for the Canary Islanders and the reference populations. EUR, Europeans; NAF, North Africans; SSA, sub-Saharan Africans. A total of 1212 samples and 101,271 variants were used. Colors represent ancestry components aligned with different populations (dark blue, Northwestern Europe; light blue, Southeastern Europe; pink, North Africa; green, sub-Saharan Africa). Created with R v3.2.2 (https://www.r-project.org/).
Table 2.
Percentage of genomic ancestry proportions obtained by ADMIXTURE (K = 4, samples = 1212, variants = 101,271) and ELAI (14 generations, samples = 1212, variants = 114,929) in the Canary Islanders.
| Ancestry (%) | ADMIXTURE | ELAI | ||||
|---|---|---|---|---|---|---|
| Min | Average | Max | Min | Average | Max | |
| EUR | 66.5 | 76.4 ± 5.7* | 84.9 | 59.4 | 71.4 ± 4.9 | 84.5 |
| NAF | 14.3 | 20.8 ± 3.0 | 30.6 | 15.0 | 26.7 ± 4.6 | 38.2 |
| SSA | 0.0 | 2.8 ± 1.6 | 9.5 | 0.0 | 1.9 ± 1.3 | 8.3 |
EUR, European; NAF, North African; SSA, sub-Saharan African. For Average columns, numbers refer to average ± standard deviation (in percentage). *European ancestry represents the sum of percentages from both Northwestern and Southeastern components.
When island populations were considered individually, the largest average NAF ancestries were obtained for El Hierro, La Gomera, Fuerteventura, and Lanzarote for both admixture estimators. La Gomera was also the island population with the largest SSA proportion on average (Table 3).
Table 3.
Mean ancestry proportions obtained with ADMIXTURE (K = 4, samples = 1212, variants = 101,271) and ELAI (14 generations, samples = 1212, variants = 114,929) per island population.
| Canary Islands | ADMIXTURE | ELAI | ||||
|---|---|---|---|---|---|---|
| EUR* | NAF | SSA | EUR | NAF | SSA | |
| El Hierro | 77.7 ± 2.9 | 20.0 ± 2.1 | 2.3 ± 0.7 | 68.3 ± 2.6 | 31.0 ± 2.6 | 0.7 ± 0.4 |
| La Palma | 79.7 ± 2.6 | 18.8 ± 1.9 | 1.5 ± 0.8 | 76.5 ± 2.8 | 22.4 ± 2.7 | 1.1 ± 0.6 |
| La Gomera | 73.7 ± 2.9 | 21.6 ± 2.3 | 4.8 ± 1.3 | 65.8 ± 2.4 | 31.0 ± 2.6 | 3.2 ± 1.2 |
| Tenerife | 78.7 ± 2.4 | 19.7 ± 2.0 | 1.6 ± 0.9 | 75.3 ± 2.8 | 23.6 ± 2.6 | 1.1 ± 0.6 |
| Gran Canaria | 77.4 ± 2.5 | 19.3 ± 2.1 | 3.3 ± 1.5 | 73.7 ± 3.1 | 23.6 ± 2.5 | 2.7 ± 1.3 |
| Fuerteventura | 72.6 ± 2.8 | 24.6 ± 2.1 | 2.9 ± 1.1 | 67.2 ± 3.1 | 31.1 ± 2.8 | 1.7 ± 0.8 |
| Lanzarote | 72.3 ± 2.5 | 24.7 ± 2.4 | 3.1 ± 1.0 | 67.1 ± 2.8 | 30.9 ± 2.6 | 2.0 ± 0.7 |
EUR, European; NAF, North African; SSA, sub-Saharan African. All numbers refer to average ± standard deviation (in percentage). *European ancestry represents the sum of percentages from both Northwestern and Southeastern components.
CIRdb: the first step for cataloging the natural genetic variation of the Canary Islands
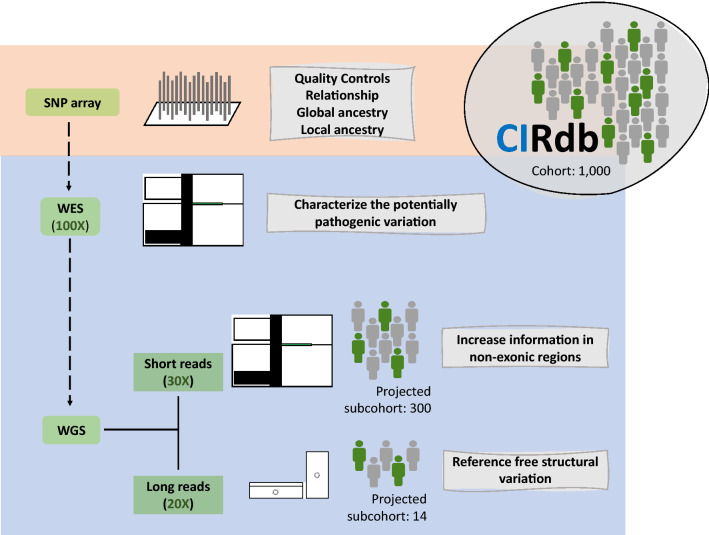
Considering SNP array data analyses as the starting point, the design for the reference genetic catalog of the Canary Islands population (CIRdb) is presented here for the first time. This catalog will be based on all the unrelated individuals identified in this study (irrespective of whether they declared that the four grandparents were born in the same island) and has been envisaged as a combination of data from three different technologies where each one provides its advantages for the genetic characterization of Canary Islanders. The conceptual design of CIRdb is shown in Fig. 5 and will involve the use of SNP array data (this study), as well as whole-exome, and whole-genome sequencing studies. In this regard, in-house bioinformatic pipelines for detecting single nucleotide variants, small insertions and deletions, and structural variants in whole-exome and whole-genome data are being developed and benchmarked against Genome In a Bottle standard materials70. Laboratory intercomparisons and updates are deposited in a publicly available repository (https://github.com/genomicsITER/benchmarking).
Figure 5.
Overall schematic representation of the technologies and sample estimates projected for developing the catalog of natural genetic variation in the Canary Islanders. The sections corresponding to the data presented in this study (orange) and the work in progress (blue) are shown. CIRdb, Canary Islands Reference database; WES, whole-exome sequencing; WGS, whole-genome sequencing.
Discussion
This study provides the largest genomic study of current Canary Islanders conducted to date, revealing unique population features and particular ancestry patterns based on SNP array data. In line with our previous studies with fewer samples13, we identify genetic peculiarities that differentiate the current Canary Islands populations from mainland populations8–12. Besides, with a larger sample size, we now evidence a clear pattern of genetic differentiation among islands not observed previously, where donors from El Hierro, La Gomera, Fuerteventura, and Lanzarote exhibited the largest average ancestry that can be assigned to NAF. We also evidenced the existence of more extreme individual NAF ancestries in the population (i.e., 38.2%) compared to previous estimates. An important isolation pattern of the populations from El Hierro and La Gomera, and the existence of further substructuring in the Canaries was also evident in this study. Taken together, this study evidenced the unique admixed makeup of current Canary Islanders and thus, establish the grounds for developing a catalog of genetic variation for this population that will be useful for the transition to Precision Medicine in the region.
There have been significant advances in Precision Medicine. From Archibald Garrod71 and the precursors of Precision Medicine, through one of the first examples of prevention, detection, and treatment of diseases tailored to individual profiles based on pharmacokinetics (i.e., warfarin)72, nowadays this paradigm encompasses diverse areas such as epigenetics, environmental exposures, imaging and radiology, and genetics and genomics, among others73,74. In this context, Genomic Medicine has emerged as a key discipline that has demonstrated important benefits in oncology75, pharmacology76, and rare and undiagnosed diseases77, to name a few, and including the possibility to improve the turnaround time, and in reducing the costs and the uncertainty of the diagnostic odyssey of the patients and their relatives78,79. In this context, many countries have seen the benefits of the pioneering implementation of genomic medicine within their healthcare system80–82. The first successful use of whole-exome sequencing to identify a disease-causing genetic mutation was reported about ten years ago, by Worthey in 201183. A 15-month-old child was diagnosed with presumptive Crohn’s disease and treated accordingly without improvements in symptoms. After several years of diagnostic odyssey, a WES analysis identified a novel, hemizygous missense mutation in the X-linked inhibitor of apoptosis. This landmark study was followed by others based on the same concept and techniques but considering more patients and controls, sometimes without a clear clinical diagnosis in place before the analysis78,84–88. For whole-genome sequencing, several fruitful studies have also been carried out28,89–91. Nowadays, the routine implementation of NGS in clinical settings has drastically improved the average diagnostic yield from 10 to 36% (WES) or 41% (WGS), and the rate of clinical utility from 6 to 17% (WES) or 27% (WGS). Based on these benefits, many countries have extended these studies to comprise global, population-scale analyses including population controls, so that the natural genetic variation of the population could be also deeply characterized. In some cases, population classification is not entirely accurate and a more fine-scale analysis, based on ancestry, is needed92.
Following on this idea, here we present the study sample for the establishment of a reference genetic catalog for the current Canary Islanders. As the greatest fraction of rare genetic variation, which accumulates the most clinically relevant genetic variation, would remain understudied unless NGS technologies are in place, with CIRdb we envisage the use of a combination of several technologies to efficiently develop the population-specific catalog. As a starting point, we have assessed all samples to be included with the Axiom® Genome-Wide Human CEU 1 array as a first stage to efficiently characterize the global and local ancestry components, local substructure and inbreeding patterns, but also for the detection of samples that could be difficult to sequence or that had unknown family relationships with others in the cohort, allowing us to prioritize the samples for more expensive ulterior approaches. Considering the next steps, CIRdb plans to run WES in all the prioritized samples to efficiently examine the fraction of the genome that includes ~ 85% of all described disease-causing variants93. Using WES at population scale, it has been possible to detect an enrichment of risk variants for Panic Disorder in the Faroese population94, specific genetic loci associated with longevity in Bulgarian centenarians95, or study the metabolic impact of candidate effector genes in Southwestern American Indian population96, to name a few. WGS theoretically targets the entire DNA sequence of donors, offering the optimal solution for unbiased genetic studies although at higher costs per sample. Because of that, the use of WGS is projected in CIRdb as a complementary approach that will be used in subsets of the samples to improve the catalog and allowing to improve the imputation of genetic variation97 in the biomedical studies conducted in the Canary Islanders, as has been evidenced in Estonian and Native Hawaiian populations18,38. CIRdb aims to leverage two technologies for WGS, namely short-read sequencing (SRS) (Illumina, San Diego, CA, USA) and long-read sequencing (LRS) (Oxford Nanopore Technologies, Oxford, UK). The former will allow us to enrich the catalog with genetic information beyond the exome regions with high accuracy while containing the project costs. The latter will specifically enable the analysis of other types of genetic variation (e.g. structural variants, SVs)98,99, particularly beneficial for medically-relevant genes100 and assess the benefits of de novo assembly of genomes to assist in improving the population101,102. Studies in patients with Bardet-Biedl syndrome103 or Carney complex104 have shown the benefits of using LRS which would still be unsolved otherwise using SRS technologies.
Despite the forthcoming studies to build CIRdb will deepen in the genetic characterization of this population, we recognize some major issues of the study. Firstly, the number of evaluated SNPs (up to 114,929 in total in comparative studies) and a focus on autosomal variation limited our ability to assess the existent subcontinental influences63,105 in the ancestry analyses. This is the main reason for us to focus on the three continental ancestry components following our previous observations13. Forthcoming studies incorporating a much higher number of variants and the analysis of maternal (mtDNA) or paternal (NRY) lineages will be optimal to assess the subcontinental components of the admixture. Secondly, although relying on SNP arrays benefits from standardized pipelines and highly reproducible and reliable genotyping data, one of the most pronounced drawbacks of the study is the focus on one type of genetic variation (i.e., SNPs) and on alleles in the higher end of frequency spectrum. Information from structural and rare variation will provide new clues for disentangling the recent evolutionary history of this population and identify novel genetic links with disease. Filling these gaps will be the aim of leveraging different sequencing technologies for the establishment of the CIRdb catalog.
In summary, here we deepen into the genetic characterization of current Canary Islanders and establish the grounds for developing CIRdb to put forward a catalog of genetic variation for this population. CIRdb will be developed with complementary technologies and the tools and resources are currently under active development to create a precise public and available database for researchers and healthcare professionals.
Supplementary Information
Acknowledgements
We would like to thank the support from our colleagues from the Teide-HPC Supercomputing facility (http://teidehpc.iter.es/en), which was funded by INP-2011-0063-PCT-430000-ACT (INNPLANTA program) from the Spanish Ministry of Economy and Competitiveness.
Author contributions
Conceptualization: C.F.; Methodology: C.F., I.M.-R., D.J. and A.D.-d.U.; Investigation: M.C.R.-P., A.C.-d.-L., B.G.-G., I.M.-R., A.C. and A.Í.-C.; Formal Analysis: A.D.-d.U. and I.M.-R.; Data Curation: A.D.-d.U., I.M.-R., and L.A.R.-R.; Writing—Original Draft: A.D.-d.U. and C.F.; Writing—Review & Editing: all authors; Supervision: C.F.; Funding Acquisition: C.F.
Funding
This research was funded by Ministerio de Ciencia e Innovación (RTC-2017–6471-1; AEI/FEDER, UE ) and the Instituto de Salud Carlos III (CD19/00231), which were co-financed by the European Regional Development Funds ‘A way of making Europe’ from the European Union; Fundación CajaCanarias and Fundación Bancaria “La Caixa” (2018PATRI20); Cabildo Insular de Tenerife (CGIEU0000219140); and by the agreement OA17/008 with Instituto Tecnológico y de Energías Renovables (ITER) to strengthen scientific and technological education, training, research, development and innovation in Genomics, Personalized Medicine and Biotechnology. A.D.-d.U. was supported by a fellowship from the Spanish Ministry of Education and Vocational Training (grant number FPU16/01435).
Data availability
The data generated as part of this study has been deposited in the European Genome-Phenome Archive (EGA, https://ega-archive.org/studies/EGAS00001006050).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-20442-x.
References
- 1.Crosby, A. W. Imperialismo ecológico. La expansión biológica de Europa, 900–1900 (ed. Barcelona: Crítica) (1988).
- 2.Hooton, E. A. The Ancient inhabitants of the Canary Islands. (ed. Peabody Museum of Harvard University. Kraus Reprint Co. New York) (1970 [1925]).
- 3.Arauna LR, et al. Recent historical migrations have shaped the gene pool of Arabs and Berbers in North Africa. Mol. Biol. Evol. 2017;34:318–329. doi: 10.1093/molbev/msw218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lobo-Cabrera ML. esclavitud en Fuerteventura en los Siglos XVI y XVII. V Jorn. de estudios sobre Fuertevent. y Lanzarote. 1993;1:13–40. [Google Scholar]
- 5.Maca-Meyer N, et al. Mitochondrial DNA diversity in 17th-18th century remains from Tenerife (Canary Islands) Am. J. Phys. Anthropol. 2005;127:418–426. doi: 10.1002/ajpa.20148. [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez-Varela R, et al. Genomic analyses of pre-European conquest human remains from the Canary Islands reveal close affinity to modern North Africans. Curr. Biol. 2017;27:3396–3402. doi: 10.1016/j.cub.2017.09.059. [DOI] [PubMed] [Google Scholar]
- 7.Fregel R, et al. Mitogenomes illuminate the origin and migration patterns of the indigenous people of the Canary Islands. PLoS ONE. 2019;14(3):e0209125. doi: 10.1371/journal.pone.0209125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flores C, et al. The origin of the Canary Island aborigines and their contribution to the modern population: A molecular genetics perspective. Curr. Anthropol. 2001;42:749–755. [Google Scholar]
- 9.Flores C, et al. A predominant European ancestry of paternal lineages from Canary Islanders. Ann. Hum. Genet. 2003;67:138–152. doi: 10.1046/j.1469-1809.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- 10.Fregel R, et al. Demographic history of Canary Islands male gene-pool: Replacement of native lineages by European. BMC Evol. Biol. 2009;9(1):181. doi: 10.1186/1471-2148-9-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pino-Yanes M, et al. North African influences and potential bias in case-control association studies in the Spanish population. PLoS ONE. 2011;6(3):e18389. doi: 10.1371/journal.pone.0018389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botigué LR, et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Nat. Acad. Sci. U. S. A. 2013;110:11791–11796. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillen-Guio B, et al. Genomic analyses of human European diversity at the southwestern edge: Isolation, African influence and disease associations in the Canary Islands. Mol. Biol. Evol. 2018;35:3010–3026. doi: 10.1093/molbev/msy190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morash M, Mitchell H, Beltran H, Elemento O, Pathak J. The role of next-generation sequencing in precision medicine: A review of outcomes in oncology. J. Pers. Med. 2018;8(3):30. doi: 10.3390/jpm8030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wojcik GL, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills MC, Rahal C. The GWAS diversity monitor tracks diversity by disease in real time. Nat. Genet. 2020;52:242–243. doi: 10.1038/s41588-020-0580-y. [DOI] [PubMed] [Google Scholar]
- 17.Eisfeldt J, Mårtensson G, Ameur A, Nilsson D, Lindstrand A. Discovery of novel sequences in 1,000 Swedish genomes. Mol. Biol. Evol. 2020;37:18–30. doi: 10.1093/molbev/msz176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin M, et al. Population-specific reference panels are crucial for genetic analyses: An example of the CREBRF locus in Native Hawaiians. Hum. Mol. Genet. 2020;29:2275–2284. doi: 10.1093/hmg/ddaa083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbujani G, Magagni A, Minch E, Cavalli-Sforza LL. An apportionment of human DNA diversity. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4516–4519. doi: 10.1073/pnas.94.9.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gravel S, et al. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am. J. Hum. Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Merida I, et al. Toward the mutational landscape of autosomal dominant retinitis pigmentosa: A comprehensive analysis of 258 Spanish families. Invest. Ophthalmol. Vis. Sci. 2018;59:2345–2354. doi: 10.1167/iovs.18-23854. [DOI] [PubMed] [Google Scholar]
- 26.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 27.Lee H, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor JC, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat. Genet. 2015;47:717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Y, et al. Comprehensive genetic testing of Chinese SNHL patients and variants interpretation using ACMG guidelines and ethnically matched normal controls. Eur. J. Hum. Genet. 2020;28:231–243. doi: 10.1038/s41431-019-0510-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fattahi Z, et al. Iranome: A catalogue of genomic variations in the Iranian population. Hum. Mutat. 2019;40:1968–1984. doi: 10.1002/humu.23880. [DOI] [PubMed] [Google Scholar]
- 32.Nagasaki M, et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat. Commun. 2015;6(1):8018. doi: 10.1038/ncomms9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, et al. KoVariome: Korean national standard reference variome database of whole genomes with comprehensive SNV, indel, CNV, and SV analyses. Sci. Rep. 2018;8(1):5677. doi: 10.1038/s41598-018-23837-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chheda H, et al. Whole-genome view of the consequences of a population bottleneck using 2926 genome sequences from Finland and United Kingdom. Eur. J. Hum. Genet. 2017;25:477–484. doi: 10.1038/ejhg.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dopazo J, et al. 267 Spanish exomes reveal population-specific differences in disease-related genetic variation. Mol. Biol. Evol. 2016;33:1205–1218. doi: 10.1093/molbev/msw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature. 526, 82–90 (2015). [DOI] [PMC free article] [PubMed]
- 37.The Genome of the Netherlands Consortium Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 38.Mitt M, et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet. 2017;25:869–876. doi: 10.1038/ejhg.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastard P, et al. A loss-of-function IFNAR1 allele in Polynesia underlies severe viral diseases in homozygotes. J. Exp. Med. 2022;219(6):e20220028. doi: 10.1084/jem.20220028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duncan CJA, et al. Life-threatening viral disease in a novel form of autosomal recessive IFNAR2 deficiency in the Arctic. J. Exp. Med. 2022;219(6):20212427. doi: 10.1084/jem.20212427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorente-Arencibia P, et al. Wilson disease prevalence: Discrepancy Between clinical records, registries and mutation carrier frequency. J. Pediatr. Gastroenterol. Nutr. 2022;74:192–199. doi: 10.1097/MPG.0000000000003322. [DOI] [PubMed] [Google Scholar]
- 42.Panoutsopoulou K, et al. Genetic characterization of Greek population isolates reveals strong genetic drift at missense and trait-associated variants. Nat. Commun. 2014;5:5345. doi: 10.1038/ncomms6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Southam L, et al. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nat. Commun. 2017;8:15606. doi: 10.1038/ncomms15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nutile T, et al. Whole-exome sequencing in the isolated populations of cilento from South Italy. Sci. Rep. 2019;9(1):4059. doi: 10.1038/s41598-019-41022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu K, et al. Meta-imputation: An efficient method to combine genotype data after imputation with multiple reference panels. Am. J. Hum. Genet. 2022 doi: 10.1016/j.ajhg.2022.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gurdasani D, et al. The African genome variation project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malaria Genomic Epidemiology Network. Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa, Asia and Oceania. Nat. Commun. 10 1 5732; 10.1038/s41467-019-13480-z (2019). [DOI] [PMC free article] [PubMed]
- 48.Freedman BI. End-stage renal failure in African Americans: Insights in kidney disease susceptibility. Nephrol. Dial. Transplant. 2002;17:198–200. doi: 10.1093/ndt/17.2.198. [DOI] [PubMed] [Google Scholar]
- 49.Kumar R, et al. Genetic ancestry in lung-function predictions. N. Engl. J. Med. 2010;363:321–330. doi: 10.1056/NEJMoa0907897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flores C, et al. African ancestry is associated with asthma risk in African Americans. PLoS ONE. 2012;7(1):e26807. doi: 10.1371/journal.pone.0026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Go AS, et al. Heart disease and stroke statistics–2014 update: A report from the American heart association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sánchez-Lerma B, et al. High prevalence of asthma and allergic diseases in children aged 6 to [corrected] 7 years from the Canary Islands. [corrected] J. Investig. Allergol. Clin. Immunol. 2009;19:383–390. [PubMed] [Google Scholar]
- 53.Marcelino-Rodríguez I, et al. On the problem of type 2 diabetes-related mortality in the Canary Islands, Spain. The DARIOS study. Diabetes Res. Clin. Pract. 2016;111:74–82. doi: 10.1016/j.diabres.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 54.Lorenzo V, et al. Disproportionately high incidence of diabetes-related end-stage renal disease in the Canary Islands. An analysis based on estimated population at risk. Nephrol. Dial. Transplant. 2010;25:2283–2288. doi: 10.1093/ndt/gfp761. [DOI] [PubMed] [Google Scholar]
- 55.Serra-Vidal G, et al. Heterogeneity in palaeolithic population continuity and Neolithic expansion in North Africa. Curr. Biol. 2019;29:3953–3959. doi: 10.1016/j.cub.2019.09.050. [DOI] [PubMed] [Google Scholar]
- 56.Martin AR, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cabrera de León A, et al. Presentación de la cohorte “CDC de Canarias”. Objetivos, diseño y resultados preliminares. Rev. Esp. Salud Publica. 2008;82:519–534. doi: 10.1590/s1135-57272008000500007. [DOI] [PubMed] [Google Scholar]
- 58.Cabrera de León A, et al. Leptin and altitude in the cardiovascular diseases. Obes. Res. 2004;12:1492–1498. doi: 10.1038/oby.2004.186. [DOI] [PubMed] [Google Scholar]
- 59.Nicolazzi EL, Iamartino D, Williams JL. AffyPipe: An open-source pipeline for Affymetrix Axiom genotyping workflow. Bioinformatics. 2014;30:3118–3119. doi: 10.1093/bioinformatics/btu486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R Core Team. R: A language and environment for statistical computing. The R Project for Statistical Computing. Available online at https://www.r-project.org/ (2020).
- 61.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 526, 68–74 (2015). [DOI] [PMC free article] [PubMed]
- 63.Henn BM, et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guan Y. Detecting structure of haplotypes and local ancestry. Genetics. 2014;196:625–642. doi: 10.1534/genetics.113.160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guillen-Guio, B. et al. Admixture mapping of asthma in southwestern Europeans with North African ancestry influences. Am. J. Physiol. Lung Cell. Mol. Physiol. 318 5 L965–L975 (2020). [DOI] [PubMed]
- 68.Kirin M, et al. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE. 2010;5(11):e13996. doi: 10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seldin MF, et al. European population substructure: Clustering of northern and southern populations. PLoS Genet. 2006;2(9):e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olson ND, et al. precisionFDA truth challenge V2: Calling variants from short and long reads in difficult-to-map regions. Cell Genomics. 2022;2(5):100129. doi: 10.1016/j.xgen.2022.100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garrod A. The incidence of alkaptonuria: A study in chemical individuality. Lancet. 1902;160:1616–1620. [Google Scholar]
- 72.Lee MTM, Klein TE. Pharmacogenetics of warfarin: Challenges and opportunities. J. Hum. Genet. 2013;58:334–338. doi: 10.1038/jhg.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patel CJ, et al. Whole genome sequencing in support of wellness and health maintenance. Genome Med. 2013;5(6):58. doi: 10.1186/gm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carlsten C, et al. Genes, the environment and personalized medicine: We need to harness both environmental and genetic data to maximize personal and population health. EMBO Rep. 2014;15:736–739. doi: 10.15252/embr.201438480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong M, et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat. Med. 2020;26:1742–1753. doi: 10.1038/s41591-020-1072-4. [DOI] [PubMed] [Google Scholar]
- 76.van der Lee M, et al. Toward predicting CYP2D6-mediated variable drug response from CYP2D6 gene sequencing data. Sci. Trans. Med. 2021;13(603):eabf3637. doi: 10.1126/scitranslmed.abf3637. [DOI] [PubMed] [Google Scholar]
- 77.East KM, et al. A state-based approach to genomics for rare disease and population screening. Genet. Med. 2021;23:777–781. doi: 10.1038/s41436-020-01034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valencia, C. A. et al. Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: A pediatric center’s experience. Front. Pediatr. 3, 67; 10.3389/fped.2015.00067 (2015). [DOI] [PMC free article] [PubMed]
- 79.Hu X, et al. Proband-only medical exome sequencing as a cost-effective first-tier genetic diagnostic test for patients without prior molecular tests and clinical diagnosis in a developing country: The China experience. Genet. Med. 2018;20:1045–1053. doi: 10.1038/gim.2017.195. [DOI] [PubMed] [Google Scholar]
- 80.Stark Z, et al. Australian genomics: A federated model for integrating genomics into healthcare. Am. J. Hum. Genet. 2019;105:7–14. doi: 10.1016/j.ajhg.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sperber NR, et al. Strategies to integrate genomic medicine into clinical care: Evidence from the IGNITE Network. J. Pers. Med. 2021;11(7):647. doi: 10.3390/jpm11070647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vidgen ME, et al. Queensland Genomics: An adaptive approach for integrating genomics into a public healthcare system. NPJ Genom. Med. 2021;6(1):71. doi: 10.1038/s41525-021-00234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Worthey EA, et al. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet. Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 84.Chen Y-Z, et al. Gain-of-function ADCY5 mutations in familial dyskinesia with facial myokymia. Ann. Neurol. 2014;75:542–549. doi: 10.1002/ana.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farwell KD, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: Results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 87.Wright CF, et al. Genetic diagnosis of developmental disorders in the DDD study: A scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trujillano D, et al. Clinical exome sequencing: Results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 2017;25:176–182. doi: 10.1038/ejhg.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stavropoulos DJ, et al. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. NPJ Genom. Med. 2016;1(1):15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farnaes L, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med. 2018;3(1):10. doi: 10.1038/s41525-018-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lionel AC, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belbin GM, et al. Toward a fine-scale population health monitoring system. Cell. 2021;184:2068–2083. doi: 10.1016/j.cell.2021.03.034. [DOI] [PubMed] [Google Scholar]
- 93.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gregersen, N. O. et al. Whole-exome sequencing implicates DGKH as a risk gene for panic disorder in the Faroese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171 8 1013 1022 (2016). [DOI] [PubMed]
- 95.Serbezov D, et al. Novel genes and variants associated with longevity in Bulgarian centenarians revealed by whole exome sequencing DNA pools: A pilot study. J. Transl. Genet. Genom. 2020;4(4):446. [Google Scholar]
- 96.Kim HI, et al. Characterization of exome variants and their metabolic impact in 6,716 American Indians from Southwest US. Am. J. Hum. Genet. 2020;107:251–264. doi: 10.1016/j.ajhg.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quick C, et al. Sequencing and imputation in GWAS: Cost-effective strategies to increase power and genomic coverage across diverse populations. Genet. Epidemiol. 2020;44:537–549. doi: 10.1002/gepi.22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mantere, T., Kersten, S. & Hoischen, A Long-read sequencing emerging in medical genetics. Front. Genet. 10, 426; 10.3389/fgene.2019.00426 (2019). [DOI] [PMC free article] [PubMed]
- 99.Pauper M, et al. Long-read trio sequencing of individuals with unsolved intellectual disability. Eur. J. Hum. Genet. 2021;29:637–648. doi: 10.1038/s41431-020-00770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wagner J, et al. Curated variation benchmarks for challenging medically relevant autosomal genes. Nat. Biotechnol. 2022;40:672–680. doi: 10.1038/s41587-021-01158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim H-S, et al. Chromosome-scale assembly comparison of the Korean Reference Genome KOREF from PromethION and PacBio with Hi-C mapping information. Gigascience. 2019;8(12):giz125. doi: 10.1093/gigascience/giz125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nagasaki M, et al. Construction of JRG (Japanese reference genome) with single-molecule real-time sequencing. Hum. Genome Var. 2019;6(1):27. doi: 10.1038/s41439-019-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reiner J, et al. Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet-Biedl Syndrome 9 (BBS9) deletion. NPJ Genom. Med. 2018;3(1):3. doi: 10.1038/s41525-017-0042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Merker JD, et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med. 2018;20:159–163. doi: 10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choudhury A, et al. High-depth African genomes inform human migration and health. Nature. 2020;586:741–748. doi: 10.1038/s41586-020-2859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated as part of this study has been deposited in the European Genome-Phenome Archive (EGA, https://ega-archive.org/studies/EGAS00001006050).