Background:
Cardiac involvement is an important determinant of mortality among sarcoidosis patients. Although granulomatous inflammation is a hallmark finding in cardiac sarcoidosis, the precise immune cell populations that comprise the granuloma remain unresolved. Furthermore, it is unclear how the cellular and transcriptomic landscape of cardiac sarcoidosis differs from other inflammatory heart diseases.
Methods:
We leveraged spatial transcriptomics (GeoMx digital spatial profiler) and single-nucleus RNA sequencing to elucidate the cellular and transcriptional landscape of cardiac sarcoidosis. Using GeoMX digital spatial profiler technology, we compared the transcriptomal profile of CD68+ rich immune cell infiltrates in human cardiac sarcoidosis, giant cell myocarditis, and lymphocytic myocarditis. We performed single-nucleus RNA sequencing of human cardiac sarcoidosis to identify immune cell types and examined their transcriptomic landscape and regulation. Using multichannel immunofluorescence staining, we validated immune cell populations identified by single-nucleus RNA sequencing, determined their spatial relationship, and devised an immunostaining approach to distinguish cardiac sarcoidosis from other inflammatory heart diseases.
Results:
Despite overlapping histological features, spatial transcriptomics identified transcriptional signatures and associated pathways that robustly differentiated cardiac sarcoidosis from giant cell myocarditis and lymphocytic myocarditis. Single-nucleus RNA sequencing revealed the presence of diverse populations of myeloid cells in cardiac sarcoidosis with distinct molecular features. We identified GPNMB (transmembrane glycoprotein NMB) as a novel marker of multinucleated giant cells and predicted that the MITF (microphthalmia-associated transcription factor) family of transcription factors regulated this cell type. We also detected additional macrophage populations in cardiac sarcoidosis including HLA-DR (human leukocyte antigen-DR)+ macrophages, SYTL3 (synaptotagmin-like protein 3)+ macrophages and CD163+ resident macrophages. HLA-DR+ macrophages were found immediately adjacent to GPMMB+ giant cells, a distinct feature compared with other inflammatory cardiac diseases. SYTL3+ macrophages were located scattered throughout the granuloma and CD163+ macrophages, CD1c+ dendritic cells, nonclassical monocytes, and T cells were located at the periphery and outside of the granuloma. Finally, we demonstrate mTOR (mammalian target of rapamycin) pathway activation is associated with proliferation and is selectively found in HLA-DR+ and SYLT3+ macrophages.
Conclusions:
In this study, we identified diverse populations of immune cells with distinct molecular signatures that comprise the sarcoid granuloma. These findings provide new insights into the pathology of cardiac sarcoidosis and highlight opportunities to improve diagnostic testing.
Keywords: giant cells, granuloma, inflammation, macrophages, myocarditis, sarcoidosis
Novelty and Significance.
What Is Known?
Cardiac sarcoidosis (CS) is the heart involvement of sarcoidosis, a multisystemic autoinflammatory disease, and is difficult to diagnose and can lead to mortality.
Granulomatous inflammation, a histologic hallmark for CS, is primarily comprised of macrophages.
What New Information Does This Article Contribute?
CS granulomas are comprised of a complex compilation of macrophage populations with distinct transcriptional profiles, spatial distribution, and biological functions.
A distinct organization of GPNMB (transmembrane glycoprotein NMB)+ multinucleated giant cells cuffed by HLA-DR (human leukocyte antigen-DR isotype)+ epithelioid macrophages can differentiate CS from other forms of inflammatory cardiac disease.
Diagnosis of CS presents clinical challenges caused by phenotypic and histological overlaps with other inflammatory heart conditions. In this current study, we uncovered remarkably complicated and heterogeneous macrophage populations within CS and implicated that MITF (microphthalmia-associated transcription factor) and mTOR (mammalian target of rapamycin) pathway might service as key molecular pathway for CS. This work could be translated to clinical practice, developing novel immunohistochemistry staining strategies for routine diagnostic pathology of CS. Our findings deepened the understanding of the specification and formation of giant cells and granuloma inflammation in CS, and identified macrophage marker candidates to develop new tracers for noninvasive molecular imaging.
In This Issue, see p 651
Meet the First Author, see p 652
Editorial, see p 670
Sarcoidosis is a multisystemic autoinflammatory disease characterized by granulomatous inflammation on histology.1 Although lung and lymph node are the most common sites of involvement, cardiac sarcoidosis (CS) has recently gained increasing attention, as it portends an unfavorable prognosis accounting for nearly 85% of deaths in sarcoidosis patients.2,3 The common clinical manifestations of CS include congestive heart failure, arrhythmias and sudden cardiac death. It is imperative to diagnose and treat CS in a timely manner to avoid these adverse outcomes.
Sarcoidosis is remarkably heterogeneous with respect to anatomic site of involvement, rate of disease progression, and response to immune modulating therapies, making accurate diagnosis and disease monitoring clinically challenging. Recent data suggest that CS is under diagnosed and the true incidence and prevalence might be higher than generally perceived. Although CS is identified in 20% to 30% of living sarcoidosis patients, the true prevalence may be as high as 70% as reported in autopsy studies with sudden cardiac death as the first manifestation.4,5 This discrepancy stems from the limitations of current diagnostic modalities: metabolic imaging and tissue sampling. Endomyocardial biopsy has poor sensitivity due to the patchy distribution of granulomatous inflammation. The pathognomonic granulomatous inflammation with multinucleated giant cells can be found in only up to 20% of the biopsies.6,7 Imaging modalities (positron emission tomography/computed tomography or cardiac magnetic resonance imaging) for sarcoidosis diagnosis suffer from suboptimal specificity as increased metabolic activity on imaging may be seen in various infectious and inflammatory conditions.8–10
Various causes can exhibit overlapping clinical and pathological features with CS. Clinically, the patient may manifest as heart failure, and with overlapping features of other type of cardiomyopathies. Ischemic cardiomyopathy (ICM), a sequela of obstructive coronary artery disease, accounts for a significant portion of heart failure cases.11 Non-ICM (NICM) is a heterogeneous group comprised of various cause, such as valvular disease, underlying genetic mutation, medication-induced, or idiopathic. Moreover, other types of myocarditis, such as giant cell myocarditis (GCM) and lymphocytic myocarditis, share similar histological features of admixed lymphocytic and histiocytic inflammatory infiltration in routine pathology examination of the heart biopsy specimens.12 It is clinically challenging to distinguish these various disease entities based on clinical presentation, imaging studies, and pathology examination. A better understanding of the cellular composition of CS may lead to new and improved diagnostic modalities including molecular pathology and imaging strategies.
Mechanisms and pathogenesis of CS remain unknown, partially due to the lack of reliable experimental models that recapitulate the molecular and morphological features of sarcoid granulomas.13 Current models utilize infectious agents to trigger granulomatous inflammation. However, it is unclear whether infectious-related granulomatous inflammation exhibit the same cellular composition and molecular features as sarcoidosis. Moreover, granulomatous inflammation may differ based on anatomical site. A deeper understanding of the CS microenvironment is critical to advance our understanding of disease pathogenesis and improve diagnostic clinical practice.
Here, we utilized spatial transcriptomics to explore the molecular features of inflammatory lesions in different types of myocarditis and the potential of transcriptomic analysis in our understanding of these complex entities. We then applied single-nucleus RNA sequencing to characterize the cellular composition and transcriptional landscape of human CS. We identified diverse and geographically organized populations of immune cells that comprise regions of granulomatous inflammation in CS. These studies identify a novel marker of multinucleated giant cells and yield a strategy to distinguish CS from other inflammatory cardiac pathologies. Collectively, our findings provide new insights into CS pathogenesis and highlight opportunities to improve diagnosis.
Methods
Data Availability
All the codes used in the manuscript are deposited in GitHub (pma22wustl/transcriptional-and-immune-landscape-of-cardiac-sarcoidosis [github.com]). The single-nuclei raw expression matrices and raw sequence files that support the findings of this study are available on the Gene Expression Omnibus (GSE183852 [donor; dilated cardiomyopathy (DCM)]; GSE 205734 [ICM; Sarcoidosis]). Alignment was performed to the publicly available transcriptome GRCh38 (Genome Reference Consortium Human Build 38)-1.2.0. The nanostring dataset is available in Supplemental Material. Please see the Major Resources Table in the Supplemental Material.
Study Approval and Human Samples
This study was approved by the Institutional Review Board of Washington University in St. Louis, Stanford University, and Duke University. The 4 frozen CS samples for single-nucleus RNA sequencing (snRNA-seq) were from Duke University biobank. The frozen ICM, NICM, and donor heart samples for snRNA-seq were from Washington University in St. Louis TCBR biobank. The samples used for GeoMx Digital Spatial transcriptomic analysis (sarcoidosis, lymphocytic myocarditis, and GCM) and immunofluorescence stains (sarcoidosis, lymphocytic myocarditis and GCM, ICM [ischemic cardiomyopathy] NICM [nonischemic cardiomyopathy]) were from Washington University in St. Louis archival pathology formalin-fixed, paraffin-embedded samples. Additional sarcoidosis samples for immunofluorescence stains were from Stanford University archival pathology formalin-fixed, paraffin-embedded samples. All the cases were identified by searching biobank/archival pathology database for specific diagnosis. Hematoxylin and eosin stained slides were reviewed by cardiovascular pathologists (C.Y.L. and C.G.) to confirm the diagnosis. The clinical and demographic data of cases used for snRNA-seq (single-nucleus RNA sequencing) is summarized in Table S1.
GeoMx Digital Spatial Profiler
Tissues sections from archival formalin-fixed paraffin-embedded specimens were arrayed on independent slides. Digital spatial profiler (DSP) was performed using nCounter barcoding technology (NanoString Technologies, Seattle, WA). Briefly, formalin-fixed, paraffin-embedded sections were deparaffinized, subjected to antigen retrieval procedures, then incubated overnight with pan-macrophage primary antibody CD68 containing a photocleavable indexing oligonucleotide and fluorescent-labeled visualization antibody. The stained slides were scanned using the GeoMx DSP instrument (NanoString Technologies, Seattle, WA) to produce a digital fluorescent image of the tissues. Next, individual regions of interest covering the most abundant CD68+ inflammation were identified. Oligonucleotides from regions of interests with positively stained CD68+ cells were released upon exposure to ultraviolet light in a sequential manner, collected by microcapillary aspiration, dispensed into a microtiter plate, and hybridized to optical barcodes for subsequent quantitation in the nCounter system (NanoString Technologies). Digital counts from barcodes corresponding to protein probes were first normalized to internal spike-in controls and then to the area of their compartment. Compartments with <10 nuclei or with an area of illumination <100 μm2 were excluded. The count matrices after quality control were analyzed in R v4.0.1 for further differential expression analysis and data visualization.
Single-Nucleus Suspensions Preparation and Sorting
Frozen heart tissues were minced with a razor blade, then homogenized gently in a 5 mL Dounce homogenizer containing 1 to 2 mL of chilled lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40 [nonidet P-40] in nuclease-free water). Lysate was gently filtered through a 40μm filter after a 15 minutes incubation on ice. Nuclei were resuspended in 1 mL Nuclei Wash Buffer (1× PBS with 2% bovine serum albumin and 0.2 U/μL RNase inhibitor) after centrifuging at 500× g for 5 minutes at 4 °C, and filtered through a 20 μm pluristrainer into a fresh 15 mL conical tube. Nuclei were resuspended in 300 μL Nuclei Wash Buffer after a second wash with Nuclei Wash Buffer and transferred to 5 mL FACS (fluorescence-activated cell sorting) tube for flow sorting. 1 μL DRAQ5 (deep red anthraquinone 5) (5 mM solution; Thermo Catalog no. 62251) was added and gently mixed before sorting. DRAQ5+ nuclei were sorted into Nuclei Wash Buffer on BD FACS Melody (BD Biosciences, San Jose, CA) using a 100 µM nozzle. Recovered nuclei after centrifuging at 500×g for 5 minutes at 4 °C were gently resuspended in Nuclei Wash Buffer to a target concentration of 1000 nuclei/μL after counting with a Hemocytometer.
snRNA-seq and Analysis
The Chromium Single-Cell 5’ Reagent V1.1 Kit from 10X Genomics was used to capture the transcriptome of individual nuclei. For each of the samples, 10 000 nuclei were loaded into a single well of a Chip G kit for GEM generation. Reverse transcription, barcoding, cDNA amplification, and purification were performed according to the Chromium 10x V1.1 protocol as the library preparation step. Each sample was sequenced on a NovaSeq 6000 S4 platform with a sequencing depth of ≈20 000–3 000 reads per nucleus. Sequencing reads were aligned to the whole genome pre-mRNA reference generated from the GRCh38 transcriptome using the Cell Ranger V3 software (10X Genomics) according to the 10X Genomics instructions. The count matrices were pre-processed and analyzed using R package Seurat (v3.2.3). Nuclei with <1000 or >20 000 unique molecular identifier counts, as well as nuclei with >2.5 mitochondrial read percentage, were excluded from all subsequent analysis. For each sample, counts were transformed and normalized using SCTransform with default thresholds, and integration of the samples was performed on the filtered and normalized objects. Principal component analysis was performed on the integrated object using RunPCA (principal component analysis). The number of significant principal components (PCs) (ndims [number of dimensions]=30) was determined based on the ElbowPlot and DimHeatmap generated in Seurat. Nonlinear dimensional reduction and visualization were performed using RunUMAP (uniform manifold approximation and projection). Cells showing coexpression of multiple cell type–specific genes were identified as doublets and removed from any downstream analysis. Differentially expressed genes were identified using the FindAllMarkers function on the normalized RNA assay as recommended by Seurat, specifically returning only upregulated genes with a Log2FC cutoff of 0.25 and a P value of 0.05.
Pathway Enrichment Analysis
A list of output genes by Seurat different expression analysis with Log2FC>1 and adjusted P<0.05 were used to identify enrichment in CS, myeloid clusters. Gene ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene set enrichment analysis were performed using the ClusterProfiler package (version 2.4.3).
Trajectory Analysis
Trajectory analysis was performed using the Python package Palantir. The count matrix and meta data sheet from the integrated object were exported as the input. Principal components and the diffusion components were calculated using the imported matrix as an estimate of the low dimensional phenotypic manifold of the data. Then, the Palantir algorithm was run with a specified start cell state (the presumed progenitor cell in the dataset). The algorithm then returned with the calculated terminal cells, entropy values, pseudotime values, and the probability of ending up in each of the terminal states for all cells.
Transcription Factor Analysis
Single-Cell Regulatory Network Inference and Clustering is a robust clustering method for the identification of stable cell states from scRNA-seq data based on the underlying gene regulatory networks. It was performed as described14 (Single-Cell Regulatory Network Inference and Clustering 0.1.5, GENIE3 0.99.3, and AUCell 0.99.5) using the 20-thousand motifs database for RcisTarget (RcisTarget.hg19.motifDatabases.20k). The input data was the size-factor normalized expression matrix in myeloid cells based on Seurat analysis filtered for Log2FC>0.5 and adjusted P<0.05. From those genes that passed the default filtering (rowSums > 5×0.03×760 and detected in at least 1% of the cells), only the protein coding genes were kept in the coexpression modules from GENIE3 (Gene Network Inference with Ensemble of Trees) and analyzed for motif enrichment with RcisTarget. AUCell score was used to visualize which cells are enriched for the gene set. TFs expression levels were visualized as colored projections onto uniform manifold approximation and projection (UMAP). The primary results were further loaded as a matrix in R v4.0.1 for generation of heatmap and regulons network.
Cell to Cell Communication
Cell to cell communication analysis was performed using the R package CellCall, to infer intercellular and intracellular crosstalk between certain cell types based on scRNA-seq data.15 It deciphers cell to cell communication and related internal gene regulatory networks by integrating paired ligand-receptor and TF (transcription factor) activity. Basically, normalized expression matrix of myeloid cells and natural killer (NK) and T cells was inputted into a biological and statistical model. The cell-cell communication score of a ligand-receptor pair is calculated by integrating intercellular signaling (expression of ligand and receptor) and intracellular signaling (activity score of downstream TFs).
Reference Mapping and Annotating Query Datasets
We mapped snRNA-seq data from human donor, ICM and NICM hearts (queries) onto our CS dataset (reference) using the Azimuth workflow.16,17 A subset of donor/NICM sample has been reported previously by our lab.18 Briefly, we first generated an integrated query dataset (workflow described in more detail in snRNA-seq and analysis). After integration, we identified a set of anchors, or pairwise correspondences between cells predicted to be in a similar biological state, between queries and reference datasets. We then computed mapping cell prediction scores of query cells to decipher the robustness of label transfer and mapping. Cell prediction scores reflect the confidence associated with each assigned annotation. Cells with high-confidence annotations (prediction scores >0.75) reflect predictions that are supported by multiple consistent anchors.
Immunofluorescence Staining
Paraffin-embedded sections were dewaxed in xylene, rehydrated, endogenous peroxide activity quenched in 10% methanol and 3% hydrogen peroxide, processed for antigen retrieval by boiling in citrate buffer pH 6.0 for 15 minutes, then blocked in 10% bovine serum albumin containing 0.05% Tween-20, and stained with the following primary antibodies overnight at 4 °C: CD68 (KP1, 1:2000; Bio-Rad), Ki67 (ab15580, 1:1000; Abcam), HLA-DR (human leukocyte antigen-DR isotype,L243,1:750;Abcam) CD163 (MRQ-26, 1:1000; Cell marque), CD1C (OTI2F4, 1:500; Abcam), SYTL3 (synaptotagmin-like protein 3; HPA030586, 1:500; Sigma-Aldrich), Phospho-S6 (2211S, 1:1000; Cell Signaling Technology), FCGR3A (low-affinity immunoglobulin gamma Fc region receptor III-A; SP189, 1:200; Abcam), GPNMB (transmembrane glycoprotein NMB; AF2550, 1:500; R&D Systems), CD4 (1: 400; Creative diagnostics), CD8 (1:500; Abcam). The primary antibody was detected using Opal Polymer HRP (horseradish peroxidase conjugated anti-mouse +anti-rabbit) Ms+Rb (PerkinElmer Opal Multicolor IHC system) or biotinylated anti-goat secondary antibody (BP-9500-50, Vector Labs) in conjunction with streptavidin horseradish peroxidase (ABC Elite, Vector Labs). The PerkinElmer Opal Multicolor IHC system was utilized to visualize antibody staining per manufacturer protocol. Immunofluorescence was visualized with Zeiss confocal microscopy and Zeiss Axio Scan Z1. The area with granulomas (clusters of macrophages with multinucleated giant cells) were identified by the morphology and these areas were selected for quantification and representative images were presented in the figures.
Cell Culture and Transfection
THP1 (human leukemia monocytic cell line) cells (American Type Culture Collection) were grown in RPMI (Roswell Park Memorial Institute)-1640 (+L-glutamine), 10% FBS, 1% HEPES buffer, and 1% penicillin/streptomycin supplement (all from Invitrogen). Transfections were performed with Lipofectamine 3000 Transfection Kit with 5 μg of plasmid DNA, 7.5 μL of Lipofectamine 3000, 10 μL of P3000, and 250 μL of Opti-MEM (Invitrogen) per well of 4×105cells in 12-wells plate. pEGFP (enhanced GFP)-N1-MITF (microphthalmia-associated transcription factor)-M plasmid (no. 38131) were purchased from Addgene. Plasmid were amplified and extracted with Qiagen Plasmid Maxi Kit.
Real-Time Quantitative Polymerase Chain Reaction
RNA (1 µg) from treated THP1 cells was reverse-transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primers from IDT were used for specific SYBR Green real-time polymerase chain reaction. Expression was normalized to ACTB, GAPDH, RPL32. Alterations in the expression levels were calculated in relation to mock treated cells. The sequences of primers used in the study are listed below:
GPNMB F: CAAGAAGAGGCGGGATACTTT R: AGTGGGATTCAGTTAGCTTGG
tumor protein P63 regulated 1 (TPRG1) F: ACATCCTGTAGCTGTGGTTTAG R: TGCAGACCAAAGGTGAGTTTA
SNTB1 F: TGACGGGAAATACACAGCATAG R: GCCTAGGAACTGGGATGATTAAG
GAPDH F: CCATGTTCGTCATGGGTGTGAACCA R: GCCAGTAGAGGCAGGGATGATGTTG
ACTB F: AGGCCAACCGCGAGAAG R: ACAGCCTGGATAGCAACGTACA
RPL32 F: GCCCAAGATCGTCAAAAAGAGA R: TCCGCCAGTTACGCTTAATTT
Statistical Analyses
The statistical analysis used for each experiment was specified in the figure legend. All methods are included as below. Normal distribution of data was tested by the Kolmogorov-Smirnov normality test as well as the Shapiro-Wilk test and D’Agostino-Pearson normality test. For the analysis to in vitro transfected THP1 cells, the assumption of normality is assumed by the central limit theorem. Significance of the quantification results was tested by one-way ANOVA with Dunn post hoc test, and Kruskal-Wallis with Dunn post hoc test, Mann-Whitney test or Student t test using Prism 9.0 (GraphPad Software, San Diego, CA). P values of the NanoString data were calculated by false discovery rate (FDR) corrected empirical Bayes moderated T statistics using R package Limma, hypergeometric distribution using R package ClusterProfiler or permutation test using R package ClusterProfiler. P values of the snRNA-seq data were calculated using R package Seurat, hypergeometric distribution using R package ClusterProfiler or permutation test using R package ClusterProfiler. P<0.05 was considered as significant.
Results
Distinct Transcriptomic Signatures of Myeloid Cell Aggregates in CS
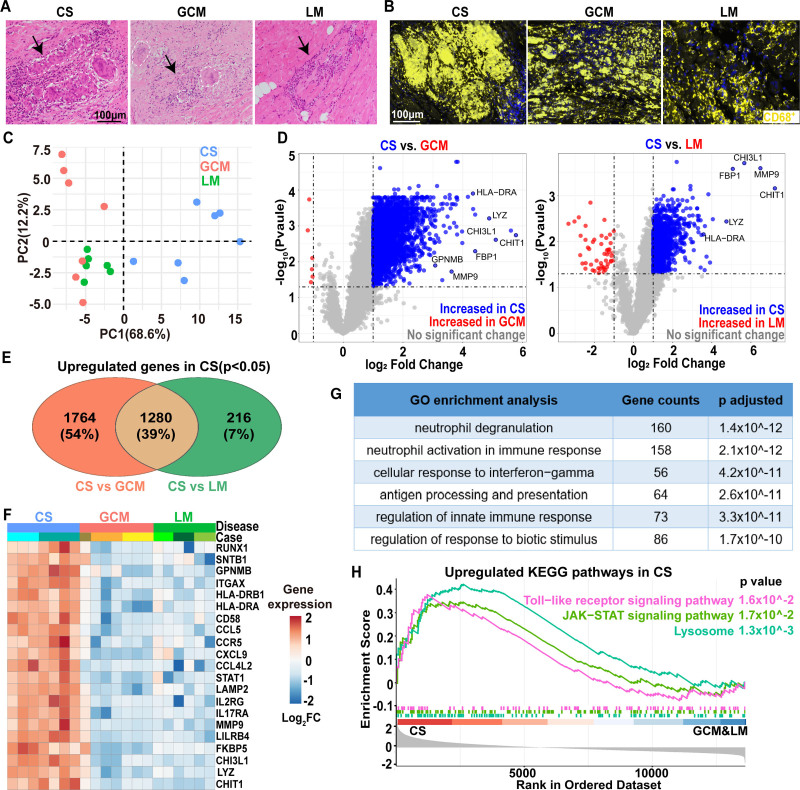
CS, GCM, and lymphocytic myocarditis share overlapping morphological features including abundant populations of myeloid cells. Granulomatous inflammation composed of epithelioid histiocytes and scattered multinucleated giant cells are present in CS and GCM, while a mixed lymphohistiocytic inflammation in an interstitial pattern is commonly found in lymphocytic myocarditis (Figure 1A). To understand the transcriptional differences of myeloid cells in these disease entities, we investigated the transcriptomic landscape of CD68+ cell aggregates in each using digital spatial profiles (DSP) technology (NanoString; Figure 1B and Figure S1). For CS and GCM cases, areas with granulomatous inflammation were selected. For lymphocyte myocarditis, areas with dense lymphohistiocytic infiltrates identified by both histology and immunofluorescence were selected. Principal component analysis revealed that CD68+ aggregates in CS exhibited a distinct transcriptional profile compared to GCM and lymphocytic myocarditis (Figure 1C). Differentially expressed gene analysis revealed 3044 and 1496 genes upregulated in CS compared to GCM and lymphocytic myocarditis, respectively (Figure 1D), with 1280 shared upregulated genes (Figure 1E). Genes selectively upregulated in CS were associated with monocyte/macrophage differentiation (CHIT-1, CHI3L-1, RUNX1 and GPNMB), antigen presentation (HLA-DRA and HLA-DRB1), ECM-degradation (MMP9), lysosomes (LAMP2), and inflammation (LYZ, CCR5, CCL5, CXCL9, FKBP5, and STAT1). These transcriptional changes were observed across multiple subjects and regions of interest (Figure 1F). Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis using the 1280 shared differentially expressed genes showed enrichment for inflammatory, lysosome, JAK-STAT (Janus kinases-signal transducer and activator of transcription), and Toll-like receptor signaling pathways (Figure 1G and 1H). In summary, these data showed that myeloid cells in CS exhibit a distinct transcriptomic profile compared with those in GCM and lymphocytic myocarditis.
Figure 1.
Distinct transcriptomic signatures of myeloid cell aggregates in cardiac sarcoidosis. A, Representative histology (by hematoxylin and eosin staining) of cardiac sarcoidosis (CS), giant cell myocarditis (GCM), and lymphocytic myocarditis (LM). Black arrows: areas with inflammation; dashed lines: multinucleated giant cells. Scale bar: 100 μm. B, Representative images of immunofluorescence staining for CS, GCM, and LM specimens in the region of interests (ROIs) for NanoString analysis. CD68: yellow; DAPI (4′,6-diamidino-2-phenylindole): blue. Scale bar: 100 μm. C, Principal Component Analysis of transcriptome of CD68+ cells in CS, GCM, and LM. Each dot represents a ROI. CS: 7 ROIs from 2 cases; GCM: 7 ROIs from 3 cases; LM: 6 ROIs from 3 case. D, Volcano plots showing differentially expressed genes (DEGs) between CS vs GCM (left) and CS vs LM (right). Gray: not significantly differentially expressed; blue: upregulated in CS (log2 fold change>1, false discovery rate [FDR] P<0.05); red: upregulated in GCM or LM (log2 fold change<−1, FDR P<0.05). P values were obtained by FDR corrected empirical Bayes moderated T statistics in R package Limma. E, Venn diagram showing overlap of DEGs that are upregulated in CS using DEGs from D. F, Heatmap of representative gene expression (population averages) for CS, GCM, and LM. G, Gene ontology (GO) pathway analysis based on 1280 shared genes in E showing top three pathways upregulated in CS compared with GCM and LM. P values were calculated by hypergeometric distribution using R package ClusterProfiler. H, Gene set enrichment analysis plot showing selected significant enrichment pathways for sarcoidosis compared to GCM and LM. P values were obtained by permutation test using R package ClusterProfiler. CHIT1 indicates chitinase 1; CHI3L-1, chitinase-3-like protein 1; FBP, Fructose-1,6-bisphosphatase; FC, Fold Change; GPNMB, transmembrane glycoprotein NMB; ITAGX, Integrin Subunit Alpha X; JAK-STAT,Janus kinases-signal transducer and activator of transcription; KEGG, Kyoto Encyclopedia of Genes and Genomes; LYZ, lysosomal enzyme; MMP, Matrix metallopeptidase; PC, principle component; RUNX1, runt-related transcription factor 1; and SNTB1, beta-1-syntrophin.
Cellular and Transcriptomic Landscape of Inflammation in CS
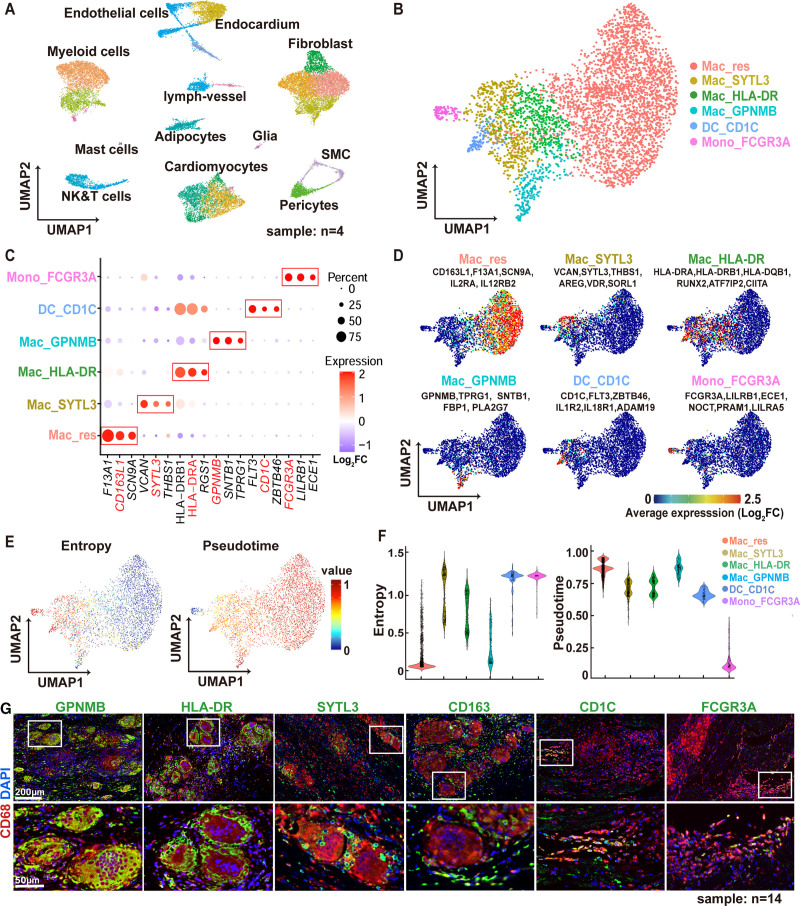
To dissect the composition of CD68+ cells in CS, we performed snRNA-seq from 4 CS samples. After excluding low-quality reads and doublets (Figure S2A), unsupervised clustering revealed 12 major cell types (Figure 2A and Figure S2B). Cell identities were annotated using cell type–specific markers (Figure S2C). The predominant inflammatory cells type identified are myeloid cells and NK-T cells. Further subclustering of the NK-T cells showed 3 cell types: NK cells, CD4+ T cells, and CD8+ T cells (Figure S3A and S3B). Immunostaining of human CS tissue showed that the CD4+ and CD8+ T cells are mostly resided at the periphery of granulomas (Figure S3C). Further subclustering of myeloid cells (Figure 2B and 2D and Figure S4A and S4B) demonstrated 6 distinct myeloid subpopulations. In addition to nonclassical monocytes (Mono_FCGR3A) and dendritic cells (DC_CD1C), 4 groups of macrophages (defined by the expression of canonical macrophage genes, such as CD68, MRC1 (Mannose receptor C-type 1), and C1QA (complement C1q A chain); Figure S4C) were identified. Cardiac resident macrophages (Mac_res) expressed canonical markers of tissue-resident macrophages, such as F13A1 (Coagulation Factor XIII A Chain), SCN9A (Sodium Voltage-Gated Channel Alpha Subunit 9) and CD163L1 (CD163 Molecule Like 1).19,20 The other 3 macrophage subpopulations, Mac_GPNMB, Mac_HLA-DR, and Mac_SYTL3, expressed distinct signatures as shown in the dot map and UMAP (Figure 2C and 2D). The other representative marker genes of each cluster were shown in Table S2. Based on Palantir pseudotime trajectory analysis,21 these 6 myeloid subclusters demonstrated different entropy and cell differentiation potential (Figure 2E and 2F), with Mac_res and Mac_GPNMB clusters showing the lowest entropy and highest pseudotime values suggesting that they represent the most differentiated cell states.
Figure 2.
Cellular and transcriptomic landscape of cardiac sarcoidosis. A, The uniform manifold approximation and projection (UMAP) projection of 23 772 cells from 4 cardiac sarcoidosis patients, showing 12 major cell types. B, The UMAP projection of 3970 myeloid cells, showing 6 myeloid subclusters. C, Dot plot of expression levels of the signature markers for each myeloid subcluster. Red boxes highlight the prominent patterns defining each myeloid cell subcluster. Color scale denotes log2 fold change (log2FC) of relative gene expression. D, Mean expression feature plots of selected marker genes in each myeloid cell subcluster. Color scale denotes average gene expression. E, UMAP projection of differentiation potential/entropy (left) and palantir pseudotime (right) for myeloid cells. F, Violin plots showing the entropy (left) and pseudotime (right) for the 6 myeloid cell subclusters. G, Immunofluorescence staining for CD68 (red) and signature myeloid subcluster markers (GPNMB [transmembrane glycoprotein NMB], human leukocyte antigen–DR isotype [HLA-DR (human leukocyte antigen-DR isotype,L243,1:750;Abcam)], SYTL3 [synaptotagmin-like protein 3], CD163, CD1C, CD16a, green) in archival cardiac sarcoidosis (CS) human heart samples. Blue: DAPI (4′,6-diamidino-2-phenylindole). Representative images from 14 independent samples. White boxes indicate zoomed-in areas displayed on the bottom row. Scale bars: 200 μm (top row) and 50 µm (bottom row). ADAM19 indicates ADAM Metallopeptidase Domain 19; ATF7IP2, Activating Transcription Factor 7 Interacting Protein 2; AREG, Amphiregulin; CD163L1,CD163 Molecule Like 1; CIITA, Class II Major Histocompatibility Complex Transactivator; ECE1, Endothelin Converting Enzyme 1; FBP1, Fructose-Bisphosphatase 1; FC, Fold Change; FCGR3A, low-affinity immunoglobulin gamma Fc region receptor III-A; F13A1,Coagulation Factor XIII A Chain; FLT3,Fms Related Receptor Tyrosine Kinase 3; GPNMB, Glycoprotein Nmb; HLA-DR, human leukocyte antigen–DR isotype; HLA-DRB1, Major Histocompatibility Complex, Class II, DR Beta 1; HLA-DQB1, Major Histocompatibility Complex, Class II, DQ Beta 1; IL1R, Interleukin 1 Receptor; IL2RA, Interleukin 2 Receptor Subunit Alpha; IL12RB2, Interleukin 12 Receptor Subunit Beta 2; IL18R1, Interleukin 18 Receptor Type 1; LILRA5,Leukocyte Immunoglobulin Like ADAM19 indicates ADAM Metallopeptidase Domain 19; ATF7IP2, Activating Transcription Factor 7 Interacting Protein 2; AREG, Amphiregulin; CD163L1,CD163 Molecule Like 1; CIITA, Class II Major Histocompatibility Complex Transactivator; ECE1, Endothelin Converting Enzyme 1; FBP1, Fructose-Bisphosphatase 1; FC, Fold Change; FCGR3A, low-affinity immunoglobulin gamma Fc region receptor III-A; F13A1,Coagulation Factor XIII A Chain; FLT3,Fms Related Receptor Tyrosine Kinase 3; GPNMB, Glycoprotein Nmb; HLA-DR, human leukocyte antigen–DR isotype; HLA-DRB1, Major Histocompatibility Complex, Class II, DR Beta 1; HLA-DQB1, Major Histocompatibility Complex, Class II, DQ Beta 1; IL1R, Interleukin 1 Receptor; IL2RA, Interleukin 2 Receptor Subunit Alpha; IL12RB2, Interleukin 12 Receptor Subunit Beta 2; IL18R1, Interleukin 18 Receptor Type 1; LILRA5,Leukocyte Immunoglobulin Like Receptor A5 LILRB1, Leukocyte Immunoglobulin Like Receptor B1; mac, macrophage; Mac_res, resident macrophages; NOCT, Nocturnin; PLA2G7,Phospholipase A2 Group VII; PRAM1, PML-RARA Regulated Adaptor Molecule 1; RUNX2, RUNX Family Transcription Factor 2; SCN9A, Sodium Voltage-Gated Channel Alpha Subunit 9; SMC, smooth muscle cells; SNTB1, Syntrophin Beta 1; SORL1, Sortilin Related Receptor 1; SYTL3,Synaptotagmin Like 3; THBS1,Thrombospondin 1; TPRG1, Tumor Protein P63 Regulated 1; VCAN,Versican; VDR, Vitamin D Receptor; and ZBTB46, Zinc Finger And BTB Domain Containing 46.
Although snRNA-seq is a powerful tool to reveal the diversity of myeloid cells in CS, it lacks the ability to resolve the spatial relationship of these cells. To this end, we performed multichannel immunofluorescence staining to visualize the location and spatial relationships of the 6 identified myeloid subclusters using their most discriminative markers (Mac_res: CD163; Mac_HLA-DR: HLA-DR; Mac_SYTL3: SYTL3, Mac_GPNMB: GPNMB; DC_CD1C: CD1c; mono_FCGR3A: CD16a), along with the pan-myeloid marker CD68 (Figure 2G, n=14 patients). This analysis uncovered a specific spatial arrangement of myeloid cell types in CS. GPNMB staining was present in multinucleated giant cells, indicating that the Mac_GPNMB subcluster represented multinucleated giant cells. HLA-DR staining identified a rim of epithelioid macrophages immediately adjacent to the multinucleated giant cells. SYTL3 and CD163 immunostaining highlighted epithelioid macrophages dispersed throughout the granuloma between multinucleated giant cells. CD1c and CD16a showed that dendritic cells and nonclassical monocytes were located at the periphery of the granuloma (Figure 2G). This pattern was consistently observed across all samples. Together, these data highlight the complex heterogeneity and spatial distribution of myeloid cell populations in CS. Moreover, we have identified GPNMB as a molecular marker for multinucleated giant cells in CS.
Mammalian Target of Rapamycin Pathway Activation and Macrophage Proliferation in CS
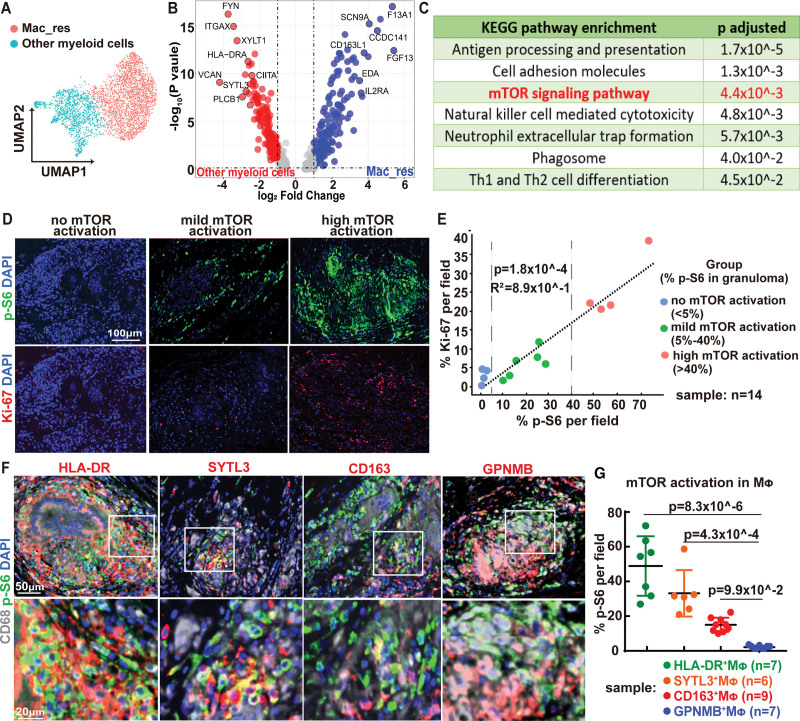
To further understand the role of myeloid cells in CS, we compared the transcriptional profiles of Mac_HLA-DR, Mac_SYTL3, Mac_GPNMB, DC_CD1C, and mono_FCGR3A to Mac_res (Figure 3A). We reasoned that cardiac resident macrophages were unlikely to contribute to disease pathology. We identified 621 upregulated differentially expressed genes that were associated with several immunemodulating pathways, including mTOR (mammalian target of rapamycin) signaling (Figure 3B and 3C). The mTOR pathway has been implicated in the pathogenesis of pulmonary sarcoidosis,22 although its role in CS remains unclear. p-S6 (phospho-S6 ribosomal protein) is a marker of mTOR pathway activation. Immunostaining revealed variable levels of p-S6 expression in the granulomas of 14 CS samples (Figure 3D through 3E). A subset of cases showed robust p-S6 staining (n=4, >40% of cells staining p-S6), whereas others showed substantially less (n=6, 5%–40% of cells staining p-S6) or no p-S6 staining (n=4, <5% of cells staining p-S6). The degree of p-S6 staining in the granuloma correlated with cell proliferation, as assessed by Ki67 staining (r2=0.89, Figure 3E). We then examined p-S6 staining in the different macrophages subclusters defined by snRNA-seq (Figure 3F). Mac_HLA-DR and Mac_SYTL3 showed high levels of p-S6 staining, while multinucleated giant cells (Mac_GPNMB) showed minimal p-S6 staining (Figure 3G). Cardiac resident macrophages showed low levels of p-S6 staining. Taken together, these results suggest that the cardiac sarcoid granuloma is comprised of diverse myeloid cell types arranged in a stereotypic pattern. Moreover, our findings raise the possibility that mTOR activation may regulate expansion of specific macrophage subsets including Mac_HLA-DR and Mac_SYTL3 subclusters.
Figure 3.
mTOR (mammalian target of rapamycin) pathway activation and macrophage proliferation in cardiac sarcoidosis. A, Uniform manifold approximation and projection (UMAP) projection of resident macrophages (Mac_res) and other myeloid cells. B, Volcano plot showing the differentially expressed genes (DEGs) between Mac_res and other myeloid cells. P values were obtained by Wilcoxon Rank Sum test using R package Seurat (v4). Gray dot: no significant change; Red dot: upregulated in other myeloid cells (log2 fold change>1, FDR P<0.05); Blue dot: upregulated in resident macrophages (log2 fold change<−1, false discovery rate [FDR] P<0.05). C, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis based on DEGs showing top upregulated pathways in other myeloid cells compared to resident macrophages. P values calculated by hypergeometric distribution using R package ClusterProfiler. D, Immunofluorescence staining of p-S6 (phospho-S6 ribosomal protein; green) and Ki67 (red) of cardiac sarcoidosis tissue. Representative images from no-, mild-, and high-mTOR activation cases. Blue: DAPI (4′,6-diamidino-2-phenylindole). Scale bar: 100 μm. E, Quantification of p-S6 and Ki67 immunoreactivity in cardiac sarcoidosis (CS) granulomas with strong correlation (R2=0.886) and significance (P=1.8×10-4). p-S6 and Ki67 staining was scored in 14 cardiac sarcoidosis samples. The relationship was investigated using Pearson correlation coefficient test. 14 CS samples were divided into 3 groups according to the percent of p-S6 positive cells in the granuloma (no mTOR activation: %p-S6 positive cells in granuloma <5%; mild mTOR activation: %p-S6–positive cells in granuloma between 5% to 40%; high-mTOR activation: %p-S6 positive cells in granuloma >40%). F, Immunofluorescence staining for CD68 (white), p-S6 (green), and macrophage subcluster markers (HLA-DR [Human Leukocyte Antigen – DR isotype], SYTL3 [synaptotagmin-like protein 3], CD163, GPNMB [transmembrane glycoprotein NMB], red) of granulomas in cardiac sarcoidosis. DAPI: blue. White boxes indicate the zoomed-in areas displayed on the bottom row. Scale bar represents 50 μm (top row) and 20 µm (bottom row). G, Dot plots showing the percentage of p-S6 positive cells in macrophage subclusters (Mac_res [CD163+MΦ, n=9]; Mac_HLA-DR [HLA-DR+MΦ, n=7]; Mac_SYTL3 [SYTL3+MΦ, n=6]; and Mac_GPNMB [GPNMB+MΦ, n=6]) in cardiac sarcoidosis granulomas with mean±SD shown in lines. Statistics was performed using one-way ANOVA with Dunn post hoc test. CCDC141 indicates coiled-coil domain containing 141; CD163L1, CD163 molecule like 1; CIITA, class II major histocompatibility complex transactivator; EDA, ectodysplasin A; FGF13, fibroblast growth factor 13; FYN, Src family tyrosine kinase; F13A1, coagulation factor XIII A chain; HLA-DR, human leukocyte antigen – DR isotype; IL2RA, interleukin 2 receptor subunit alpha; ITGAX, integrin subunit alpha X; PLCB1, phospholipase C beta 1; SCN9A, sodium voltage-gated channel alpha subunit 9; SYTL3, synaptotagmin-like protein 3; Th, T helper; VCAN, versican; and XYLT1, xylosyltransferase 1.
Multinucleated Giant Cells Display a Specific Transcriptional Regulatory Network
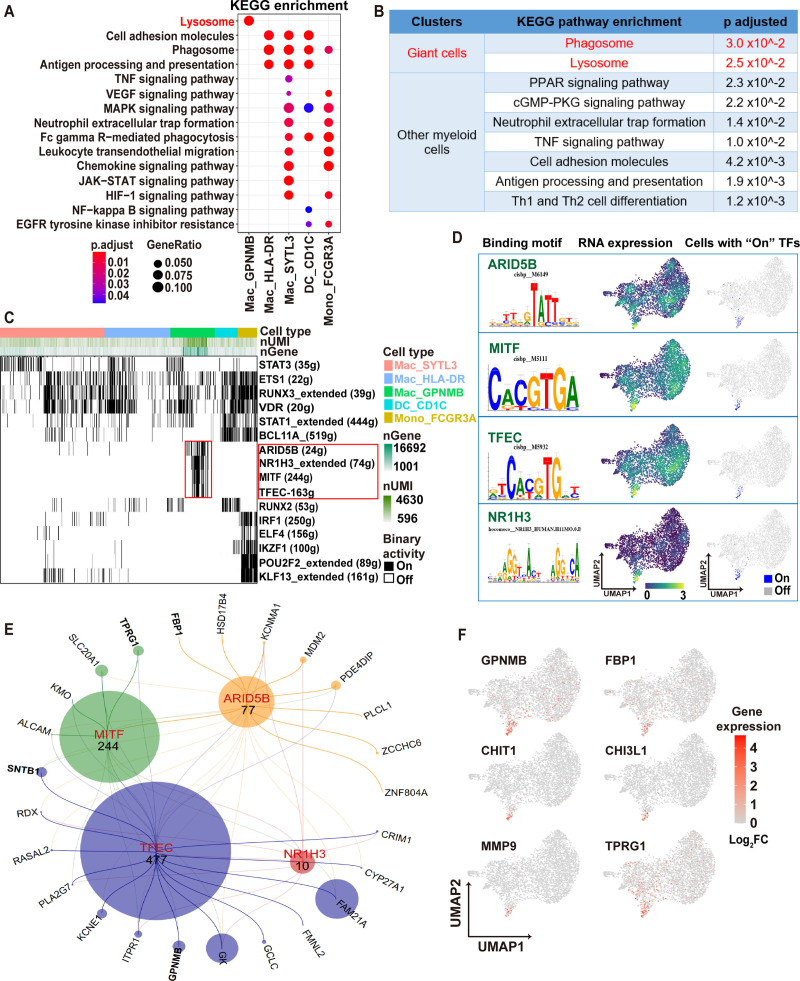
To understand putative functions of the Mac_HLA-DR, Mac_SYTL3, Mac_GPNMB, DC_CD1C, and mono_FCGR3A to Mac_res subclusters, we performed Kyoto Encyclopedia of Genes and Genomes enrichment analysis. The Mac_GPNMB cluster (multinucleated giant cells) showed exclusively enrichment for lysosomal and PPAR pathways (Figure 4A and 4B). The other subclusters showed somewhat overlapping signatures including cell adhesion, phagosome, and antigen presentation (Figure 4A). To further investigate these myeloid populations, we performed gene regulatory network analysis using the Single-Cell Regulatory Network Inference and Clustering pipeline, which identifies modules of transcription factors co-expressed with their target genes, referred to as regulons.14 On average, 10 gene regulons were identified per cell, with a subset of cells possessing significantly more regulons, up to more than 3 times the average level (Figure S5). Several regulons and their transcription factors were enriched in Mac_GPNMB (multinucleated giant cells), including MITF (Microphthalmia-associated transcription factor), TFEC (Transcription factor EC), ARID5B (AT-rich interactive domain-containing protein 5B), and NR1H3 (Liver X receptor alpha) (Figure 4C and 4D). The RNA transcripts of these TFs were present in the Mac_GPNMB subcluster (Figure 4D). TF regulatory network analysis predicted several downstream genes regulated by these TFs in the Mac_GPNMB subcluster, including GPNMB, SNTB1(beta-1-syntrophin), TPRG1 (tumor protein P63 regulated 1), FBP1 (fructose-1,6-bisphosphatase 1) (Figure 4E). Furthermore, several genes significantly upregulated in CD68+ myeloid cells of CS compared with GCM or lymphocytic myocarditis in DSP analysis, such as GPNMB, FBP1, CHIT-1 (chitinase 1), CHI3L1 (chitinase-3-like protein 1) (Figure 1F), were specifically expressed in Mac_GPNMB (Figure 4F). To validate the role of MITF transcription in Mac_GPNMB specification, we transfected THP1 cells with MITF. Flow cytometry and polymerase chain reaction (PCR) confirmed MITF expression and transfection efficiency (Figure S6A and S6B). Consistent with the snRNA-seq data, MITF expression led to upregulation of predicted downstream genes, including of GPNMB and TPRG1 (Figure S6C).
Figure 4.
Multinucleated giant cells display a specific transcriptional regulatory network. A, Dot plot of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis based on cell type–specific marker genes showing top pathways upregulated in myeloid subclusters other than resident macrophages. P values were calculated by hypergeometric distribution using R package ClusterProfiler. Detailed P values were provided in Table S4. B, KEGG pathway enrichment showing significant enrichment pathways for giant cells (red font) compared to other nonresident myeloid cells (black font). P values calculated by hypergeometric distribution using R package ClusterProfiler. C, Binary heatmap generated from the binary activities of the TF (transcription factor) regulons for myeloid cells, with black blocks representing cells that are on (with activated TF regulon). The identity of cell type was assigned by established cluster information in single-nucleus RNA sequencing (snRNA-seq) data. Red boxes highlight the specific and activated TF regulons in giant cells. Top bars show the cell type information, the number of genes and unique molecular identifiers. D, Four specific regulons in giant cells are highlighted: ARID5B (AT-rich interactive domain-containing protein 5B), MITF (microphthalmia-associated transcription factor), TFEC (transcription factor EC), and NR1H3 (liver X receptor alpha). For each TF identified, the specific motif along with the promotor binding motif (left), the RNA expression values of that TF (middle), and the cells passed the binary threshold set in the AUC histogram (right) are shown. E, A network generated with iRegulon using ARID5B, MITF, TFEC, and NR1H3 target genes identified by Single-Cell Regulatory Network Inference and Clustering (SCENIC) as an input. The nodes, representing the TF regulons sized by the number of their motifs. The edges, representing the connections between each of the 5 TFs, and their target genes and shown as a line colored based on the TFs. Input target genes shown in the network are identified as marker genes for giant cells in snRNA-seq analysis. F, Z score normalized mean expression of 6 genes identified in digital spatial profiler analysis (Figure 1). Color scale denotes log2FC of gene expression. ALCAM, activated leukocyte cell adhesion molecule; ARID5B, AT-Rich interaction domain 5B; BCL11A, BAF chromatin remodeling Complex Subunit BCL11A; CHIT1, chitinase 1; CHI3L-1 indicates chitinase-3-like protein 1; CRIM1, cysteine rich transmembrane BMP regulator 1; CYP27A1, cytochrome P450 family 27 subfamily A member 1; ELF4, E74 like ETS transcription factor 4; EGFR, epidermal growth factor receptor; ETS1,ETS proto-oncogene 1; FAM21A, WASH complex subunit 2A; FBP, fructose-bisphosphatase; FBP1, fructose-bisphosphatase 1; FC, fold change; FCGR3A, low-affinity immunoglobulin gamma Fc region receptor III-A; GCLC, glutamate-cysteine ligase catalytic subunit; GK, glycerol kinase; GPNMB, glycoprotein Nmb; HIF, hypoxia Inducible factor; HSD17B4, hydroxysteroid 17-beta dehydrogenase 4; IKZF1, IKAROS family zinc finger 1; IRF1, interferon regulatory factor 1; ITPR1, Inositol 1,4,5-trisphosphate receptor type 1; JAK-STAT, janus kinase-signal transducer and activator of transcription; KCNE1, potassium voltage-gated channel subfamily E regulatory subunit 1; KCNMA1, potassium calcium-activated channel subfamily M alpha 1; KLF13, KLF transcription factor 13; KMO, kynurenine 3-monooxygenase; Mac, macrophage; MAPK, mitogen-activated protein kinase; MDM2, MDM2 proto-oncogene; MITF, melanocyte inducing transcription factor; MMP, matrix metalloproteinase; NF, nuclear factor; NMB; HLA-DR, human leukocyte antigen–DR isotype; NR1H3, nuclear receptor subfamily 1 group H member 3; PDE4DIP, phosphodiesterase 4D interacting protein; PKG; PKG protein kinase G; PLA2G7, phospholipase A2 group VII; PLCL1, phospholipase C like 1; POU2F2, POU class 2 homeobox 2; PPAR, peroxisome proliferator activated receptor; RASAL2, RAS protein activator like 2; RDX, radixin; RUNX2, RUNX family transcription factor 2; RUNX3, RUNX family transcription factor 3; SLC20A1, solute carrier family 20 member 1 SNTB1, syntrophin beta 1; STAT1, signal transducer and activator of transcription 1; STAT3, signal transducer and activator of transcription 3; SYTL3, synaptotagmin-like protein 3. TFEC, transcription factor EC; Th, T helper; TNF, tumor necrosis factor; TPRG, tumor protein P63 regulated 1; TPRG1,tumor protein P63 regulated 1; UMAP, uniform manifold approximation and projection; VDR, vitamin D receptor; VEGF, vascular endothelial growth factor A; ZCCHC6, terminal uridylyl transferase 7; and ZNF804A, zinc finger protein 804A.
We next investigated possible cell-cell communication between giant cells (Mac_GPNMB), monocytes (Mono_FCGR3A), other macrophages (Mac_res, Mac_HLA-DR, Mac_SYTL3), dendritic cells (DC_CD1C), T cells (T_CD4, T_CD8), and NK cells (NK_GZMB [granzyme B]) using Cell Call (Figure S7A). This analysis predicted cell-cell interactions between multinucleated giant cells and other myeloid cells. Several pathways were enriched in these predicted intercellular communication events including cellular senescence and MAPK (mitogen-activated protein kinase) signaling (Figure S7B). Fewer predicted cell-cell interactions were evident with T cells or NK cells. This finding is concordant with the spatial orientation of macrophages, T cells, and NK cells in relation to the granuloma, whereby macrophages are located in close proximity within and immediately surrounding the granuloma and T cells are found at the periphery of the granuloma (Figure S3).
The origin of multinucleated giant cells remains unclear in the literature. Pseudotime analysis suggested that multinucleated giant cells in CS (Mac_GPNMB) are in a relatively differentiated state as they display low entropy values. Further analysis of entropy and pseudotime of nonresident macrophages in CS (Figure S8) showed that Mac_SYTL3 may represent an intermediate state of macrophage differentiation, and could give rise to multinucleated giant cells (Mac_GPNMB) and more matured epithelioid histiocytes (Mac_HLA-DR) that immediately surrounding the giant cells (Figure 2G).
Mac_GPNMB Subcluster Is Specifically Identified in CS
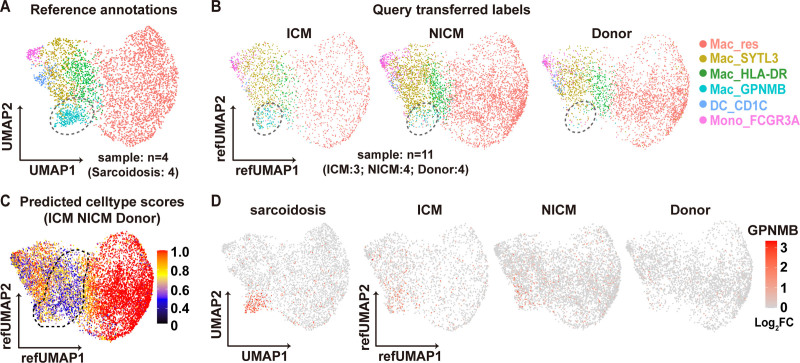
The snRNA-seq data from human CS demonstrated distinct populations of macrophages and their transcriptomic signatures (Figure 2). To demonstrate that the Mac_GPNMB subcluster (multinucleated giant cell population) is specifically present or enriched in CS, we compared the snRNA-seq data from nondiseased human hearts (donor heart), human ICM, and human NICM. We integrated human donor, ICM and NICM single-nucleus RNA sequencing data (Figure S9A) and mapped these query datasets onto the CS reference (Figure S9B through S9D). We then projected query (ICM, NICM, and donor) myeloid cells onto the reference (CS) myeloid UMAP (Figure S9E, Figure 5A and 5B). We computed cell type prediction scores, which predict how closely the query cells resembles the reference cells (Figure S10). Resident macrophages (Mac_res) had the highest prediction scores across datasets. In contrast, the cell prediction scores for Mac_GPNMB, Mac_HLA-DR, and Mac_SYTL3 were quite low indicating that these subpopulations cannot be identified within the query datasets with high confidence (Figure 5C, Figure S10B). We further examined the expression of feature genes used to identify each CS myeloid subcluster within the query datasets (Figure S11). Although we observed strong correspondence between Mac_res cells across datasets, transcriptional signatures of Mac_GPNMB, Mac_HLA-DR, and Mac_SYTL3 were weakly expressed in ICM, NICM, and donor myeloid cells. Furthermore, GPNMB expression was weakly expressed in ICM, NICM, and donor myeloid cells compared with CS and was not concentrated in the Mac_GPNMB cluster (Figure 5D). These data support the conclusion that the Mac_GPNMB cluster is specific to CS. GPNMB and HLA-DR immunostaining distinguish CS from other forms of myocarditis.
Figure 5.
Reference mapping of myeloid subpopulations in query datasets. A and B, Integration and reference mapping of query data from donor (n=4), nonischemic cardiomyopathy (NICM; n=4) and ICM (n=3) hearts (B) onto the cardiac sarcoidosis (n=4) myeloid reference uniform manifold approximation and projection (UMAP) (A). The circle area indicates the Mac_GPNMB (transmembrane glycoprotein NMB) cluster. C, Feature plot of prediction scores for various myeloid subpopulation in the query datasets (NICM, donor and ICM). The circled area indicates the Mac_human leukocyte antigen–DR isotype (HLA-DR) and Mac_GPNMB clusters. D, Mean expression feature plot of GPNMB in cardiac sarcoidosis, ICM, NICM, and donor myeloid clusters. Color scale denotes log2FC of gene expression. FC indicates fold change; FCGR3A, low affinity immunoglobulin gamma Fc region receptor III-A; Mac, macrophage; and SYTL3, synaptotagmin-like protein 3.
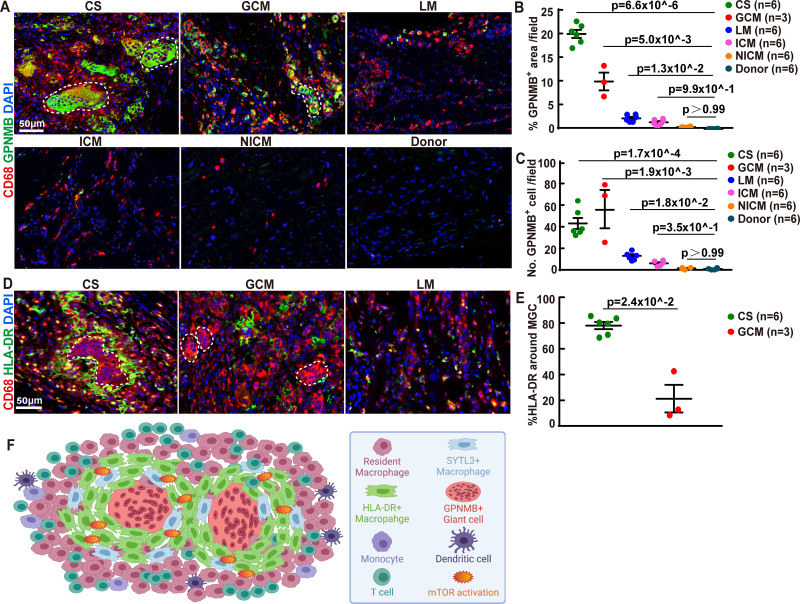
We identified a specific staining pattern for CS granulomas: GPNMB+ multinucleated giant cells cuffed by HLA-DR positive epithelioid histiocytes (Figure 2G). Next, we explored the clinical utility of these features in differentiating various cardiac diseases. GPNMB immunostaining was specific to CS and GCM with giant cells exhibiting diffuse cytoplasmic positivity (Figure 6A and 6B). GPNMB staining was only evident in a small proportion of macrophages in lymphocytic myocarditis and ICM, and was undetectable in NICM or donor hearts (Figure 6A through 6C). HLA-DR immunostaining further distinguished CS from GCM. In CS, a large number of HLA-DR+ macrophages were found to closely surround multinucleated giant cells. This pattern of HLA-DR signal was distinct from what was observed in GCM and lymphocytic myocarditis, in which HLA-DR+ macrophages were scattered throughout areas of inflammation (Figure 6D and 6E). These data highlight a distinct organization of GPNMB+ and HLA-DR+ macrophages in CS (Figure 6F), which may provide diagnostic utility in differentiating CS from other forms of myocarditis including GCM.
Figure 6.
GPNMB (transmembrane glycoprotein NMB) and HLA-DR (human leukocyte antigen-DR isotype) immunostaining distinguishes cardiac sarcoidosis from other forms of myocarditis. A, Immunofluorescence staining of CD68 (red) and GPNMB (green) in heart tissues from patients with cardiac sarcoidosis (CS), giant cell myocarditis (GCM), lymphocytic myocarditis (LM), ischemic cardiomyopathy (ICM), nonischemic cardiomyopathy (NICM), and normal subjects. Blue: DAPI. Representative images from 6 independent samples. Scale bar represents 50 µm. B, Dot plots showing the percentage of GPNMB+ area in heart tissue from patients with CS (n=6), GCM (n=3), LM (n=6), ICM (n=6), NICM (n=6), and normal subject (n=6) with Mean±SD shown in lines. Statistics was performed using Kruskal-Wallis with Dunn post hoc test. C, Dot plots showing GPNMB+ cell numbers per 20X field in heart tissue from patients with CS (n=6), GCM (n=3), LM (n=6), ICM (n=6), NICM (n=6), and normal subject (n=6) with Mean±SD shown in lines. Statistics was performed using Kruskal-Wallis with Dunn post hoc test. D, Immunofluorescence staining of CD68 (red) and HLA-DR (green) in cardiac tissue from patients with CS, GCM, and LM. Blue: DAPI (4′,6-diamidino-2-phenylindole). Representative images from 6 independent samples. Scale bar: 50 µm. White dotted line: multinucleated giant cells. E, Dot plots showing cell numbers of HLA-DR+ cell around the giant cell in cardiac tissue from patients with CS (n=6) and GCM (n=3) with mean±SD shown in lines. Significance was tested using Mann-Whitney test, two-tailed. F, Pictorial representation of the cellular diversity and spatial pattern of the typical granuloma in cardiac sarcoidosis. mTOR indicates mammalian target of rapamycin; and SYTL3, synaptotagmin-like protein 3.
Discussion
Various mechanisms and causes pertaining to how granulomatous inflammation is formed in sarcoidosis have been proposed.23 Granulomatous inflammation is a histologic hallmark for CS, which is primarily comprised of macrophages. Macrophages are extremely plastic cells that have the capacity to acquire a broad array of functional phenotypes involved in inflammation, host defense, inflammatory resolution, and repair.24,25 In general, inflammatory macrophage subsets are largely derived from circulating monocytes and are associated with disease pathogenesis, whereas tissue-resident macrophages self-renew locally and promote tissue repair.26,27 Using a combination of techniques, we demonstrate that cardiac sarcoid granulomas are comprised of a complex compilation of macrophage populations with distinct transcriptional profiles, spatial distribution, and predicted functions. Intriguingly, we found important and clinically relevant distinctions between granulomatous inflammation in CS and GCM.
Multinucleated giant cells are a classic finding in CS. However, their origin and function remain unclear. It was previously proposed that multinucleated giant cells formed through membrane fusion and cytoplasmic expansion of monocytes/macrophages within the granuloma.28 Subsequent in vitro studies suggested that, rather than cell fusion, modified cell divisions, and mitotic defects might contribute to multinucleated giant cell formation through Toll-like receptor 2 signaling.29 An alternative possibility is that multinucleated giant cells represent a distinct and specialized macrophage population. Consistent with this possibility, snRNA-seq revealed that multinucleated giant cells display a transcriptional signature that is strikingly distinct from other macrophage populations within the granuloma. GPNMB, TPRG1, and SNTB1 were among the genes specifically expressed in multinucleated giant cells. Pseudotime trajectory and gene regulatory network analyses provided informative predictions regarding multinucleated giant cell origins and differentiation. Multinucleated giant cells were predicted to represent a differentiated cell state, possibly through an intermediate status SYLTL3+ macrophages, and supported by a gene regulatory network driven by MITF and TFEC transcription factors. TFEC is a direct transcriptional target of MITF, physically interacts with MITF, and may coordinately function in a feed-forward transcriptional circuit. MITF plays a crucial role in driving lysosomal biogenesis.30,31 Lysosomal pathway enrichment was evident in multinucleated giant cells (Figure 4A and 4B). MITF regulates GPNMB expression in macrophages by directly binding to and transactivating the GPNMB promoter.32 Downstream immunostaining assays confirmed that GPNMB was a robust marker for multinucleated giant cells. These observations suggest a potential route leading to the differentiation of multinucleated giant cells, which will require evaluation in suitable model systems. Several other immune cell populations were found in cardiac sarcoid lesions. SYLT3+ macrophages and HLA-DRhi macrophages were identified within the granuloma surrounding multinucleated giant cells. CD3+ T cells, CD16a+ monocytes, CD1c+ dendritic cells, and CD163+ resident cardiac macrophages were located at the periphery of the granuloma. Each of these populations were transcriptionally distinct. Their exact roles and contribution to the pathogenesis of CS is a topic of future investigation.
GPMNB is a highly glycosylated type I transmembrane protein localized to the cell surface and in endosome and lysosomes. The function of GPNMB in immune response and inflammation remains unresolved with reports of both inflammatory and anti-inflammatory effects. In the previous studies, GPNMB is found to be highly expressed in osteoclasts, and controls the differentiation and activities of osteoclasts by modulating cell fusion and nuclei number in osteoclasts.33,34 Although the biological functions of osteoclast, mainly resorbing mineralized bone,35 is distinct from the multinucleated giant cells identified in sarcoidosis, they share striking morphological similarities. Moreover, cathepsin K and TRAP (tartrate-resistant acid phosphatase), two protein markers that has been used to identify osteoclasts, are also positive in multinucleated giant cells found in multiple disease, including sarcoidosis and GCM.36 These data, although without direct mechanistic evidence, suggests that multinucleated giant cells found in cardiac disease might share similar developmental origin or molecular regulation.
GPNMB immunostaining specifically identified giant cells in CS and GCM. Prior studies have suggested possible overlap between these two diseases including similar pathological characteristics.37,38 Intriguingly, we found that HLA-DR staining provided a robust approach to distinguish CS from GCM. In CS, GPMNB+ multinucleated giant cells were encased by a dense collection of HLA-DRhi macrophages. Although GPMNB+ multinucleated giant cells and HLA-DRhi macrophages were present in GCM, these cells types were scattered throughout the inflammatory lesion and not found in close proximity. This observation may prove useful in improving diagnosis of CS and distinguishing it from other inflammatory cardiomyopathies. The diagnostic utility of GPMNB and HLA-DR immunostaining will require independent validation.
A previous study reported increased mTOR signaling in a subset of pulmonary sarcoidosis patients. Furthermore, macrophage-specific activation of mTORC1 in mice can induce sarcoidosis-like granulomatous inflammation in the lung and liver.22 mTOR is a crucial metabolic checkpoint and autophagy suppressor,39 responsible in part for the rapid response of monocytes and macrophages.40 It was hypothesized that mTOR may inhibit autophagy, decrease antigen clearance, and promote macrophage proliferation and granuloma formation.41 Consistent with these findings, we observed a strong association between mTOR activation and cell proliferation within the granuloma. Interestingly, mTOR activation was restricted to SYLT3+ macrophages and HLA-DRhi macrophages with lower activity in CD163+ macrophages. Although mTOR inhibition is not an established therapeutic option for sarcoidosis patients, these data suggest a potential clinical role. This concept is supported by clinical case reports showing possible efficacy.42–44 Further studies are needed to correlate pathologic evidence of mTOR activation with disease progression and response to mTOR inhibitors.
Diagnosis of CS presents challenges caused by phenotypic overlaps and dependence on often elusive histopathological features. In this current study, we have uncovered remarkably complicated and heterogeneous myeloid cell populations within CS, exhibiting distinct transcriptomic and spatial characteristics comparing with other types of inflammatory heart diseases. With further validation studies, these findings could be translated to clinical practice, developing novel immunohistochemistry staining strategies for routine anatomic pathology practice and to increase the diagnostic sensitivity and accuracy for inflammatory cardiac disease. Moreover, identified macrophage subset markers could be considered as candidates to develop new tracers for noninvasive molecular imaging that could more precisely identify CS. In addition to these diagnostic implications, our findings provide new mechanistic insights. We found that a subset of myeloid cells within the cardiac sarcoid granuloma (HLA-DR+ and SYTL3+ macrophages) display marked mTOR pathway activation. This finding raises the possibility that mTOR activation in specific macrophage subsets is sufficient to drive CS pathogenesis. Such a hypothesis could be tested in suitable experimental models and may provide information regarding a novel treatment approach. Furthermore, mTOR activity or the abundance of HLA-DR+ and SYTL3+ macrophages may provide information regarding granuloma activity and serve as a potential biomarker to distinguish active from quiescent sarcoidosis. Finally, we identified MITF as a putative regulator of multinucleated giant cell gene expression deepening our understanding of the specification and formation of giant cells.
Conclusions
This study is not without limitations related to sample size and need for external validation. Predictions regarding the functions, origin, and differentiation of each macrophage population will require investigation in appropriate model systems. In conclusion, by dissecting the cellular and transcriptional landscape of CS, we elucidated the presence of diverse and distinct populations of monocytes, macrophages and dendritic cells within areas of granulomatous inflammation. Multinucleated giant cells in CS express GPNMB and are surrounded by epithelioid histiocytes composed of HLA-DR+ and SYTL3+ macrophages that display robust mTOR activation. Diagnostically, we found that GPMNB and HLA-DR staining might provide an approach to distinguish CS from other inflammatory cardiomyopathies.
Article Information
Sources of Funding
C.-Y. Lin and K.J. Lavine are supported by the Washington University in St. Louis Rheumatic Diseases Research Resource-Based Center grant (National Institutes of Health [NIH] P30AR073752). K.J. Lavine is supported by the NIH (R01 HL138466, R01 HL139714, R01 HL151078, R01 HL161185), Leducq Foundation Network (no. 20CVD02), Burroughs Welcome Fund (1014782), and Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CH-II-2015-462, CH-II-2017-628, PM-LI-2019-829) and Foundation of Barnes-Jewish Hospital (8038-88). J. Liu was supported by the China Scholarship Council. P. Ma is supported by American Heart Association Postdoctoral Fellowship (916955).
Disclosures
None.
Supplemental Materials
Figures S1–S12
Table S1–S5 (Table S3 as a separate excel file)
Major Resources Table
Supplementary Material
Nonstandard Abbreviations and Acronyms
- CCL5
- chemokine(C-C motif) ligand 5
- CCR5
- C-C chemokine receptor type 5
- CS
- cardiac sarcoidosis
- CXCL9
- chemokine (C-X-C motif) ligand 9
- DSP
- digital spatial profiler
- FCGR3A
- low-affinity immunoglobulin gamma Fc region receptor III-A
- GCM
- giant cell myocarditis
- GPNMB
- transmembrane glycoprotein NMB
- HLA-DR
- human leukocyte antigen–DR isotype
- ICM
- ischemic cardiomyopathy
- MAPK
- mitogen-activated protein kinase
- MITF
- microphthalmia-associated transcription factor
- mTOR
- mammalian target of rapamycin
- NICM
- nonischemic cardiomyopathy
- NK-cell
- natural killer cell
- p-S6
- phospho-S6 ribosomal protein
- SYTL3
- synaptotagmin-like protein 3
- TF
- transcription factor
- TFEC
- transcription factor EC
- TPRG1
- tumor protein P63 regulated 1
- TRAP
- tartrate-resistant acid phosphatase
J. Liu and P. Ma contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.121.320449.
For Sources of Funding and Disclosures, see page 668.
References
- 1.Iannuzzi MC, Rybicki BA, Teirstein AS. Sarcoidosis. N Engl J Med. 2007;357:2153–2165. doi: 10.1056/NEJMra071714 [DOI] [PubMed] [Google Scholar]
- 2.Yatsynovich Y, Dittoe N, Petrov M, Maroz N. Cardiac sarcoidosis: a review of contemporary challenges in diagnosis and treatment. Am J Med Sci. 2018;355:113–125. doi: 10.1016/j.amjms.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 3.Ardehali H, Howard DL, Hariri A, Qasim A, Hare JM, Baughman KL, Kasper EK. A positive endomyocardial biopsy result for sarcoid is associated with poor prognosis in patients with initially unexplained cardiomyopathy. Am Heart J. 2005;150:459–463. doi: 10.1016/j.ahj.2004.10.006 [DOI] [PubMed] [Google Scholar]
- 4.Matsui Y, Iwai K, Tachibana T, Fruie T, Shigematsu N, Izumi T, Homma AH, Mikami R, Hongo O, Hiraga Y., et al. Clinicopathological study of fatal myocardial sarcoidosis. Ann N Y Acad Sci. 1976;278:455–469. doi: 10.1111/j.1749-6632.1976.tb47058.x [DOI] [PubMed] [Google Scholar]
- 5.Perry A, Vuitch F. Causes of death in patients with sarcoidosis. A morphologic study of 38 autopsies with clinicopathologic correlations. Arch Pathol Lab Med. 1995;119:167–172. [PubMed] [Google Scholar]
- 6.Uemura A, Morimoto S, Hiramitsu S, Kato Y, Ito T, Hishida H. Histologic diagnostic rate of cardiac sarcoidosis: evaluation of endomyocardial biopsies. Am Heart J. 1999;138:299–302. doi: 10.1016/s0002-8703(99)70115-8 [DOI] [PubMed] [Google Scholar]
- 7.Cooper LT, Baughman KL, Feldman AM, Frustaci A, Jessup M, Kuhl U, Levine GN, Narula J, Starling RC, Towbin J., et al. The role of endomyocardial biopsy in the management of cardiovascular disease: a scientific statement from the American Heart Association, the American College of Cardiology, and the European Society of Cardiology. Circulation. 2007;116:2216–2233. doi: 10.1161/CIRCULATIONAHA.107.186093 [DOI] [PubMed] [Google Scholar]
- 8.Ohira H, Tsujino I, Yoshinaga K. (1)(8)F-Fluoro-2-deoxyglucose positron emission tomography in cardiac sarcoidosis. Eur J Nucl Med Mol Imaging. 2011;38:1773–1783. doi: 10.1007/s00259-011-1832-y [DOI] [PubMed] [Google Scholar]
- 9.Kim SJ, Pak K, Kim K. Diagnostic performance of F-18 FDG PET for detection of cardiac sarcoidosis; A systematic review and meta-analysis. J Nucl Cardiol. 2020;27:2103–2115. doi: 10.1007/s12350-018-01582-y [DOI] [PubMed] [Google Scholar]
- 10.Osborne MT, Hulten EA, Murthy VL, Skali H, Taqueti VR, Dorbala S, DiCarli MF, Blankstein R. Patient preparation for cardiac fluorine-18 fluorodeoxyglucose positron emission tomography imaging of inflammation. J Nucl Cardiol. 2017;24:86–99. doi: 10.1007/s12350-016-0502-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gheorghiade M, Bonow RO. Chronic heart failure in the United States: a manifestation of coronary artery disease. Circulation. 1998;97:282–289. doi: 10.1161/01.cir.97.3.282 [DOI] [PubMed] [Google Scholar]
- 12.Baughman KL. Diagnosis of myocarditis: death of Dallas criteria. Circulation. 2006;113:593–595. doi: 10.1161/CIRCULATIONAHA.105.589663 [DOI] [PubMed] [Google Scholar]
- 13.Jeny F, Grutters JC. Experimental models of sarcoidosis: where are we now?. Curr Opin Pulm Med. 2020;26:554–561. doi: 10.1097/MCP.0000000000000708 [DOI] [PubMed] [Google Scholar]
- 14.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Liu T, Hu X, Wang M, Wang J, Zou B, Tan P, Cui T, Dou Y, Ning L., et al. CellCall: integrating paired ligand-receptor and transcription factor activities for cell-cell communication. Nucleic Acids Res. 2021;49:8520–8534. doi: 10.1093/nar/gkab638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart T Butler A Hoffman P Hafemeister C Papalexi E Mauck WM, 3rd, Hao Y Stoeckius M Smibert P and Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YS, Chiu WT, Chang FP, Chen YL. Decline of chlorinated hydrocarbon insecticides residues in the tea-garden soils of Taiwan. Proc Natl Sci Counc Repub China B. 1988;12:9–13. [PubMed] [Google Scholar]
- 18.Koenig AL, Shchukina I, Amrute JM, Andhey PS, Zaitsev K, Lai L, Bajpai G, Bredemeyer AL, Smith G, Jones C., et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat Cardiovasc Res. 2022;1:263–280. doi: 10.1038/s44161-022-00028-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moeller JB, Nielsen MJ, Reichhardt MP, Schlosser A, Sorensen GL, Nielsen O, Tornoe I, Gronlund J, Nielsen ME, Jorgensen JS., et al. CD163-L1 is an endocytic macrophage protein strongly regulated by mediators in the inflammatory response. J Immunol. 2012;188:2399–2409. doi: 10.4049/jimmunol.110315 [DOI] [PubMed] [Google Scholar]
- 20.Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L., et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol. 2019;20:29–39. doi: 10.1038/s41590-018-0272-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, Pe’er D. Characterization of cell fate probabilities in single-cell data with Palantir. Nat Biotechnol. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linke M, Pham HT, Katholnig K, Schnoller T, Miller A, Demel F, Schutz B, Rosner M, Kovacic B, Sukhbaatar N., et al. Chronic signaling via the metabolic checkpoint kinase mTORC1 induces macrophage granuloma formation and marks sarcoidosis progression. Nat Immunol. 2017;18:293–302. doi: 10.1038/ni.3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakthivel P, Bruder D. Mechanism of granuloma formation in sarcoidosis. Curr Opin Hematol. 2017;24:59–65. doi: 10.1097/MOH.0000000000000301 [DOI] [PubMed] [Google Scholar]
- 24.Gosselin D, Glass CK. Epigenomics of macrophages. Immunol Rev. 2014;262:96–112. doi: 10.1111/imr.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavin Y, Mortha A, Rahman A, Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol. 2015;15:731–744. doi: 10.1038/nri3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D., et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19:514–522. doi: 10.1016/j.tcb.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 29.Herrtwich L, Nanda I, Evangelou K, Nikolova T, Horn V, Sagar Erny D, Stefanowski J, Rogell L, Klein C, et al. DNA Damage signaling instructs polyploid macrophage fate in granulomas. Cell. 2016;167:1264–1280 e18. doi: 10.1016/j.cell.2016.09.054 [DOI] [PubMed] [Google Scholar]
- 30.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS., et al. A gene network regulating lysosomal biogenesis and function. Science. (New York, NY). 2009;325:473–477. doi: 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- 31.Rehli M, Lichanska A, Cassady AI, Ostrowski MC, Hume DA. TFEC is a macrophage-restricted member of the microphthalmia-TFE subfamily of basic helix-loop-helix leucine zipper transcription factors. J Immunol. 1999;162:1559–1565. [PubMed] [Google Scholar]
- 32.Gabriel TL, Tol MJ, Ottenhof R, van Roomen C, Aten J, Claessen N, Hooibrink B, de Weijer B, Serlie MJ, Argmann C., et al. Lysosomal stress in obese adipose tissue macrophages contributes to MITF-dependent Gpnmb induction. Diabetes. 2014;63:3310–3323. doi: 10.2337/db13-1720 [DOI] [PubMed] [Google Scholar]
- 33.Sheng MH, Wergedal JE, Mohan S, Lau KH. Osteoactivin is a novel osteoclastic protein and plays a key role in osteoclast differentiation and activity. FEBS Lett. 2008;582:1451–1458. doi: 10.1016/j.febslet.2008.03.030 [DOI] [PubMed] [Google Scholar]
- 34.Abdelmagid SM, Sondag GR, Moussa FM, Belcher JY, Yu B, Stinnett H, Novak K, Mbimba T, Khol M, Hankenson KD., et al. Mutation in Osteoactivin Promotes Receptor Activator of NFkappaB Ligand (RANKL)-mediated Osteoclast Differentiation and Survival but Inhibits Osteoclast Function. J Biol Chem. 2015;290:20128–20146. doi: 10.1074/jbc.M114.624270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung G, Helm RM, Carraway CA, Carraway KL. Mechanism of concanavalin A-induced anchorage of the major cell surface glycoproteins to the submembrane cytoskeleton in 13762 ascites mammary adenocarcinoma cells. J Cell Biol. 1984;98:179–187. doi: 10.1083/jcb.98.1.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JK, Rosen A, Saffitz JE, Asimaki A, Litovsky SH, Mackey-Bojack SM, Halushka MK. Expression of cathepsin K and tartrate-resistant acid phosphatase is not confined to osteoclasts but is a general feature of multinucleated giant cells: systematic analysis. Rheumatology (Oxford). 2013;52:1529–1533. doi: 10.1093/rheumatology/ket184 [DOI] [PubMed] [Google Scholar]
- 37.Ekström K, Räisänen-Sokolowski A, Lehtonen J, Nordenswan HK, Mäyränpää MI, Kupari M. Idiopathic giant cell myocarditis or cardiac sarcoidosis? a retrospective audit of a nationwide case series. ESC heart failure. 2020;7:1362–1370. doi: 10.1002/ehf2.12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okura Y, Dec GW, Hare JM, Kodama M, Berry GJ, Tazelaar HD, Bailey KR, Cooper LT. A clinical and histopathologic comparison of cardiac sarcoidosis and idiopathic giant cell myocarditis. J Am Coll Cardiol. 2003;41:322–329. doi: 10.1016/s0735-1097(02)02715-8 [DOI] [PubMed] [Google Scholar]
- 39.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weichhart T, Hengstschläger M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol. 2015;15:599–614. doi: 10.1038/nri3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacheco Y, Lim CX, Weichhart T, Valeyre D, Bentaher A, Calender A. Sarcoidosis and the mTOR, Rac1, and autophagy Triad. Trends Immunol. 2020;41:286–299. doi: 10.1016/j.it.2020.01.007 [DOI] [PubMed] [Google Scholar]
- 42.Manzia TM, Bellini MI, Corona L, Toti L, Fratoni S, Cillis A, Orlando G, Tisone G. Successful treatment of systemic de novo sarcoidosis with cyclosporine discontinuation and provision of rapamune after liver transplantation. Transplant Int. 2011;24:e69–e70. doi: 10.1111/j.1432-2277.2011.01256.x. [DOI] [PubMed] [Google Scholar]
- 43.Damsky W, Thakral D, Emeagwali N, Galan A, King B. Tofacitinib treatment and molecular analysis of cutaneous sarcoidosis. N Engl J Med. 2018;379:2540–2546. doi: 10.1056/NEJMoa1805958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan JL, Fong HK, Birati EY, Han Y. Cardiac Sarcoidosis. Am J Cardiol. 2019;123:513–522. doi: 10.1016/j.amjcard.2018.10.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the codes used in the manuscript are deposited in GitHub (pma22wustl/transcriptional-and-immune-landscape-of-cardiac-sarcoidosis [github.com]). The single-nuclei raw expression matrices and raw sequence files that support the findings of this study are available on the Gene Expression Omnibus (GSE183852 [donor; dilated cardiomyopathy (DCM)]; GSE 205734 [ICM; Sarcoidosis]). Alignment was performed to the publicly available transcriptome GRCh38 (Genome Reference Consortium Human Build 38)-1.2.0. The nanostring dataset is available in Supplemental Material. Please see the Major Resources Table in the Supplemental Material.