Abstract
Background
While cardiac-specific troponin (cTn) allows for rapid diagnosis of acute type 1 myocardial infarction (T1MI), its performance to differentiate acute myocardial injury (AI) or type 2 myocardial infarction (T2MI) is limited. The objective was to combine biomarkers to improve discrimination of different myocardial infarction (MI) aetiologies.
Methods
We determined levels of cardiac troponin T and I (cTnT, cTnI), cardiac myosin-binding protein C (cMyBP-C), NT-proBNP and ten miRNAs, known to be associated with cardiac pathology in a total of n = 495 serial plasma samples at three time points (on admission, after 1 h and 3 h) from 57 NSTEMI (non-ST-elevation myocardial infarction), 18 AI, and 31 STEMI patients, as defined by fourth universal definition of MI (UDMI4) and 59 control individuals. We then applied linear mixed effects model to compare the kinetics of all molecules in these MI sub-types.
Results
Established (cTnT, cTnI) and novel (cMyBP-C) cardiac necrosis markers failed in differentiating T1MI vs T2MI at early time points. All cardiac necrosis markers were higher in T1MI than in T2MI at 3 h after admission. Muscle-enriched miRNAs (miR-1 and miR-133a) were correlated with cardiac necrosis protein markers and did not offer better discrimination. Established cardiac strain marker NT-proBNP differentiated AI and T1MI at all time points but failed to discriminate T2MI from T1MI. However, the combination of NT-proBNP and cTnT along with age returned an overall AUC of 0.76 [95 % CI 0.67–0.84] for differentiating T1MI, T2MI and AI.
Conclusions
Rather than using single biomarkers of myocardial necrosis, a combination of clinical biomarkers for cardiac necrosis (troponin) and cardiac strain (NT-proBNP) might aid in differentiating T1MI, T2MI and AI.
Keywords: Myocardial infarction type 2, Biomarkers, Troponin, microRNA, Cardiac myosin-binding protein C, Acute injury, NT-proBNP
Graphical abstract
Schematic of study design and summary of key findings.
Abbreviations
- AI
acute myocardial injury
- AUC
area under the curve
- cMyBP-C
cardiac myosin binding protein-C
- cTn
cardiac troponin
- cTnT
cardiac troponin T
- cTnI
cardiac troponin I
- MI
myocardial infarction
- miRNA/miR
microRNA
- NSTEMI
non-ST-elevation myocardial infarction
- PEA
proximity extension assay
- ROC
receiver operating characteristic
- STEMI
ST-elevation myocardial infarction
- T1MI
myocardial infarction type 1
- T2MI
myocardial infarction type 2
- UDMI4
fourth universal definition of myocardial infarction
1. Introduction
The diagnosis of myocardial infarction (MI) relies on the quantification of cardiac troponins (cTn). High-sensitive cTn assays offer rapid rule-in and rule-out for MI [1], [2]. The recent fourth Universal Definition of Myocardial Infarction (UDMI4) re-defined the categories of MI [3] by emphasising the differences of NSTEMI (non-ST-elevation myocardial infarction) type 1 (T1MI) and NSTEMI type 2 (T2MI) and including a novel subgroup of acute myocardial injury (AI). While plaque disruption is the main cause of T1MI, T2MI is defined by a general imbalance of oxygen demand and supply. T2MI comprises numerous different pathomechanisms and lacks a reliable biomarker for diagnosis. T2MI has a similar, if not worse, long-term outcome as T1MI [4], [5]. Thus, there is a need to identify novel biomarker candidates, particularly with UDMI4 redefining T2MI to exclude patients with abnormal cTn values without acute ischaemia and grouping them into AI.
Our previous proteomics analysis led to the discovery that cardiac myosin-binding protein C (cMyBP-C) can serve as novel cardiac necrosis marker [6]. This prompted further studies in defined clinical cohorts to evaluate if cMyBP-C can differentiate ST-elevation myocardial infarction (STEMI) in addition to cTn [7]. We have previously compared the diagnostic accuracy of non-coding RNAs, such as cardiac and muscle microRNAs (miRNAs) to cMyBP-C and clinical troponin measurements for STEMI (ST-elevation myocardial infarction) and T1MI [8]. In the Biomarkers in Acute Cardiac Care (BACC) study, cardiac or skeletal muscle miRNAs, lacked sensitivity for small infarcts but cMyBP-C eluted earlier than cTns [8]. Kaier et al. [2] have shown cMyBP-C 0/1 h-algorithm to be superior to ESC high-sensitivity (hs) TnT/I 0/1 h-algorithm for effective rule-in/rule-out of NSTEMI patients.
Nestelberger et al. have recently compared established cardiac biomarkers as well as cMyBP-C in their potential to differentiate T1MI and T2MI [9]. The authors concluded that current single biomarkers only provide moderate additional value in the discrimination of T1MI and T2MI. However, patients with AI were excluded from their analysis. AI represents a clinically important subgroup, including predominantly patients with heart failure and heart failure-like phenotypes. More recently, Neumann et al. compared 29 biomarkers measured at baseline to discriminate MI and myocardial injury [10]. The authors propose a panel of 4 biomarkers to discriminate T1MI and T2MI and a combination of 6 biomarkers to distinguish NSTEMI from myocardial injury. However, AI was not segregated from chronic injury and biomarker combinations were not explored to differentiate all the 3 subtypes i.e., T1MI, T2MI and AI. Additionally, the protein biomarker combinations were measured only at baseline [10] despite biomarker kinetics being important in the clinical evaluation of myocardial damage [11].
The objective of this study was to compare the discriminative ability of biomarker trajectories for MI subtypes including AI. Measurements of established and novel protein biomarkers alongside miRNA-based biomarker candidates were performed at three serial time points: on admission, after 1 h and after 3 h. We then applied linear mixed effects model to compare patient subgroups and biomarker kinetics. Finally, we harnessed machine learning methods to combine independent biomarkers and identify the best serial biomarker combination in discriminating T1MI, T2MI and AI.
2. Methods
2.1. Patient and sample selection
For the study, we assessed human plasma samples from a chest pain cohort comprising of patients presenting with suspected MI (BACC study, see below). This sub-cohort was derived from the BACC cohort as a selection of patients comprising 1) STEMI, 2) T1MI, 3) T2MI, 4) AI and 5) non-cardiac chest pain. This sub-cohort was weighed to select those MI patients with initially low and over the course of the first 3 h steeply rising hs-TnT levels. The distribution of the available samples is as follows: 1) n = 31 STEMI patients with serial sampling across 3 time points totalling 93 samples; 2) n = 26 T1MI patients with sequential specimens across 3 time points adding up to 78 samples; 3) n = 31 T2MI patients with serial sampling across 3 time points making 93 samples; 4) n = 18 AI patients with serial specimens across 3 time points totalling 54 samples; 5) n = 59 non-cardiac chest pain patients with repeat sampling across 3 time points adding up to177 samples.
2.2. The Biomarkers in Acute Cardiac Care (BACC) study
The BACC study is an ongoing, prospective cohort study including patients presenting to the emergency department of the University Hospital Hamburg with suspected MI [12]. The inclusion criteria were suspected acute MI, age above 18 and the ability to provide written informed consent. All patients were triaged according to local standard of care: a standard ECG was collected at admission. The self-reported onset of pain was obtained from a study-specific questionnaire or medical records and then categorised to time intervals as follows: 0–1 h, 1–3 h, 3–6 h, 6–12 h, 12–24 h, 24–72 h. The final diagnosis was adjudicated based on the UDMI4 by two cardiologists independently, considering the hs-TnT results (Elecsys, Roche Diagnostics) and all available clinical and imaging results, ECG and routine laboratory testing. In cases of disagreement, a third cardiologist reviewed the case. A subset of patients was selected for this study. The BACC study was registered at www.clinicaltrials.gov (NCT02355457), complied with the Declaration of Helsinki and was approved by the local Ethics Committee.
2.3. Protein measurements
A panel of four cardiac specific protein biomarkers (hs-TnI, hs-TnT, cMyBP-C and NT-proBNP) were selected for serial quantification based on previous studies in the BACC cohort [8], [10]. hs-TnI, hs-TnT and cMyBP-C were quantified on assays proprietary to the manufacturer. cMyBP-C was measured by Merck Millipore using the Erenna platform with a lower limit of detection (LLoD) of 0.4 ng/l and a lower limit of quantification (LLoQ) of 1.2 ng/l. The 99th percentile cut-off point was previously determined at 87 ng/l [13]. Troponin was routinely measured using the local standard of care hs-TnT (Elecsys® troponin T high-sensitive, Roche Diagnostics, Basel, Switzerland, lower limit of detection 5.0 ng/l, values are reported up until the limit of blank 3.0 ng/l) at admission and after 3 h. A study specific additional blood draw was ascertained 1 h after admission for hs-TnT and for hs-TnI (Troponin I hs STAT Abbott Architect, lower limit of detection 1.9 ng/l) at all time points. The 99th percentile is 14 ng/l (hs-TnT) and 27 ng/l (hs-TnI) respectively [14], [15]. We used proximity extension assays (PEA, Olink®, Olink Proteomics, Uppsala, Sweden) to measure NT-proBNP in serial samples. In the PEA, oligonucleotide-labelled monoclonal or polyclonal antibodies (PEA probes) are used to bind target proteins in a pair-wise manner using only 1 μl of sample. The PEA was used to address the high levels of 44 % missingness in baseline clinical NT-proBNP measurements. NT-proBNP is not routinely measured in patients with suspected acute MI. Additionally, serial measurements (1 h and 3 h) of NT-proBNP were not available. Importantly, results from the PEA showed a positive and significant correlation to clinical NT-proBNP (Pearson r = 0.64, P value < 0.001; Spearman rho = 0.82, P value < 0.001, Supplemental Fig. 1).
2.4. RNA extraction
Total RNA was extracted using the miRNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations, with some modifications. In brief, 100 μl of plasma was combined with 694.75 μl of Qiazol lysis reagent, 4 μl of diluted synthetic Caenorhabditis elegans miR-39 (cel-miR-39-3p) spike-in and 1.25 μl carrier RNA from bacteriophage MS2 (Roche). Following brief incubation at room temperature, 140 μl of chloroform was added and the solution was mixed vigorously. Samples were then centrifuged at 13,500 ×g for 15 min at 4 °C. 280 μl of the upper (aqueous) phase were transferred to a new tube and mixed with 1.5 volumes (420 μl) of 100 % ethanol and applied to columns and washed according to the manufacturer's protocol. Total RNA was eluted in 35 μl of nuclease-free H2O by centrifugation at 8500 ×g for 1 min at 4 °C.
2.5. RNA extraction quality control and heparinase treatment of RNA
To assess the reliability and efficiency of the RNA extractions, cel-miR-39-3p spike-in raw Cq values were assessed for a standard deviation <0.5 and a coefficient of variation <2 % in both groups, confirming good RNA processing and consistency of the RNA isolation. Prior to reverse transcription, the extracted RNA was treated with heparinase 1 from Flavobacterium (Sigma) as previously reported [8]. In brief, 5 μl of each sample were combined with 1.25 μl heparinase, 0.25 μl of RNase inhibitor (Ribo Lock 40 U/μl, Thermofisher) and 3.5 μl of heparinase buffer (pH 7.5) and thoroughly mixed, then incubated at 25 °C for 3 h. The samples were then immediately used for reverse transcription. For comparison, a buffer-only group was treated with heparinase buffer devoid of heparinase, which was incubated under the same conditions as the heparinase-treated samples. The untreated group received neither heparinase nor buffer, nor was it left for incubation, but instead was used for further reverse transcriptase together with the treated samples.
2.6. Reverse transcription and real-time PCR assays
3 μl of RNA from plasma/serum RNA was used as input in each reverse transcription (RT) reaction. RT reactions were set up according to the manufacturer's recommendations. Briefly, miRNAs were reverse-transcribed using the miRCURY LNA RT kit (Exiqon), combining 3 μl RNA with 5× reaction buffer, 1 μl enzyme mix, 0.5 μl UniSp6 synthetic spike-in and 3.5 μl nuclease-free water. The RT-PCR reaction was set as follows: reverse transcription, 42 °C for 60 min; inactivation, 95 °C for 5 min using a Veriti Thermal Cycler (Applied Biosystems). miRCURY SYBR Green qPCR in combination with miRCURY LNA miRNA PCR Assays (both Exiqon) were used to assess relative expression levels of miRNAs. cDNA was diluted 1:30 according to the manufacturer's recommendations, then 3 μl of the diluted cDNA were combined with 5 μl miRCURY SYBR Green Mastermix, 0.05 μl ROX reference dye, 1 μl PCR Primer Mix and 0.95 μl of RNase-free water to a 10 μl reaction volume. Reactions were loaded using a Bravo Automated Liquid Handling Platform (Agilent). qPCR was performed on a ViiA7 Real-Time PCR System (Applied Biosystems) at 95 °C for 2 min followed 40 cycles of 95 °C for 10 s and 56 °C for 1 min for miRCURY SYBR Green.
2.7. RNA quantification
In the analyses of raw Cq data, any value measurements beyond 35 cycles were considered undetectable and marked as missing value. The threshold was set so that we exclude Cq values which follow uniform distribution and can thus be considered random noise. To make these calculations we used the one-way Kolmogorov-Smirnov test. For all four muscle/cardiac-enriched miRNAs of interest the p-value of the test for the Cq values >35 was smaller than 0.05 and bigger than 0.05 for the Cq values smaller than 35. The delta-delta Cq method was used for relative quantification, using cel-miR-39-3p as a normalization control. Quantification results were calibrated against the median of three identical replicates consisting of equal volumes from all MI samples. Relative quantification was performed with Microsoft Excel, version 15.32 for MacOS. For the four cardiac-specific miRNAs we conducted correlation analysis between the RQ values obtained from each one of the normalization strategies and in all cases, we found strong correlation (R2 > 0.5) indicating that the selection of the normalization method has no relevant influence on the overall RQ results.
2.8. Statistical analyses
The serial release kinetics of proteins and miRNAs were analysed using linear mixed effect model with individuals as the random effect to factor within subject variance. Linear mixed effect model was selected to allow heteroscedasticity and imbalanced repeats across various timepoints i.e., baseline (0 h), 1 h and at 3 h. Release kinetics model for proteins and miRNAs were performed without data scaling. Given the insignificant association between demographics and biomarkers (proteins and miRNAs), release kinetics were not adjusted for age and sex (Supplemental Fig. 2). R package lme4 was used to implement linear mixed effect model. Post-hoc pairwise comparisons were performed using R package ‘emmeans’ with “Tukey” adjustment for comparing a family of 9 estimates (3 groups, 3 timepoints). Satterthwaite degrees-of-freedom method was used. Correlation between continuous variables was performed using Spearman correlation adjusted for individual effects. Correlation between continuous and binary variables was performed using point-biserial correlation adjusted for individual effects. P values were adjusted for multiple-testing using Benjamini-Hochberg FDR correction. Correlation plot with dendrogram was generated using R package heatmaply [16] with Ward's minimum variance method (‘ward.D2’) as the hierarchical clustering method. Baseline characteristics significance test was undertaken using Mann-Whitney U test for continuous variables and Fisher exact test for binary variables. Box plots to show raw data distribution of biomarkers were generated using R package ggboxplot. Statistical analysis and associated figures were generated with R programming environment (version 4.0.2).
2.9. Machine learning
Biomarkers with statistically significant release kinetics (P value < 0.05) across the three time points in at least one pairwise comparison were assessed for their power to discriminate T1MI, T2MI and AI. This resulted in selection of hs-TnT and NT-proBNP. Hence, the predictive performance of hs-TnT, NT-proBNP and their combination were evaluated in T1MI, T2MI and AI. Boruta stability selection [17] was used as a wrapper to multi-class random forest for feature selection in individual as well as combination biomarkers design along with demographics i.e., age and sex. For internal validation, leave-one-out cross validation was performed such that samples from the same individual are grouped into the same fold and thus avoid leakage into model performance metrics. Majority voting was applied to aggregate sample level classification into patient level final grouping. Performance metrics were computed using one-vs-all comparison with weightage to account for data imbalance. The implementation of multi-class random forest was done using Scikit-learn 0.23.2 python package.
3. Results
3.1. Clinical characteristics
We analysed a total of n = 495 serial plasma samples across three time points i.e., on admission = baseline, 1 h after admission and 3 h after admission, respectively. This included n = 318 serial measurements from patients with STEMI (n = 31 patients), T1MI (n = 26 patients), T2MI (n = 31 patients), AI (n = 18 patients) and n = 177 sequential control samples from patients with non-cardiac chest pain (n = 59 patients). Based on UDMI4, non-ischaemic cardiac conditions such as acute heart failure and Takotsubo cardiomyopathy are classified as AI (Supplemental Fig. 3). Baseline characteristics are shown in Table 1. Patients with T2 and AI were older than patients with T1 (p = 0.022). AI was more common in females (p = 0.015). Grace score for 6-month mortality was higher in T2 and in AI (p = 0.003) compared to T1 (Table 1).
Table 1.
Baseline characteristics: STEMI vs. NSTEMI Type 1 (T1MI) vs. NSTEMI Type 2 (T2MI) vs. acute injury (AI).
| BACC Clinical characteristics |
All MI patients (N = 106) | Control (N = 59) | STEMI (N = 31) | T1MI (N = 26) | T2MI (N = 31) | AI (N = 18) | P value (T1MI vs T2MI) | P value (T1MI vs AI) |
|---|---|---|---|---|---|---|---|---|
| Demographics | ||||||||
| Sex (% female) | 34 % | 71 % | 16 % | 23 % | 45 % | 61 % | 0.101 | 0.015 |
| Age (in years) | 66 (54, 75) |
59 (50, 69) |
65 (49.5, 68.5) |
61 (52.3, 70.3) |
73 (58.5, 78) |
74 (64, 80.3) |
0.019 | 0.022 |
| BMI (kg/m2) | 27.5 (24.6, 29.9) |
26 (23.6, 27.7) |
26.9 (24.8, 30.4) |
28.3 (25.2, 29.2) |
27.9 (25.7, 29.8) |
24.8 (21.5, 28.0) |
0.88 | 0.037 |
| GRACE score (6-month mortality) | 105.5 (79.3, 133) |
81 (67.5, 105) |
79 (60, 104) |
93 (64.5, 118) |
134 (103, 149.5) |
122 (107.3, 145.5) |
0.0003 | 0.003 |
| Hypertension (%) | 70 % | 47 % | 71 % | 62 % | 81 % | 61 % | 0.144 | 1.00 |
| Hyperlipoproteinemia (%) | 30 % | 27 % | 32 % | 42 % | 29 % | 11 % | 0.405 | 0.043 |
| History of smoking (%) | 55 % | 39 % | 52 % | 81 % | 42 % | 44 % | 0.003 | 0.023 |
| Comorbidities and baseline clinical parameters | ||||||||
| History of AMI (%) | 17 % | 10 % | 16 % | 31 % | 13 % | 6 % | 0.117 | 0.060 |
| Congestive heart failure (%) | 11 % | 2 % | 3 % | 8 % | 19 % | 17 % | 0.269 | 0.386 |
| Stroke (%) | 3 % | 5 % | 0 % | 0 % | 6 % | 6 % | 0.495 | 0.409 |
| Atrial fibrillation (%) | 18 % | 7 % | 10 % | 4 % | 42 % | 11 % | 0.001 | 0.558 |
| Heart rate (/min) | 82.5 (72.8, 101.3) |
74 (64, 85) |
78 (71, 87) |
79 (68.3, 88.5) |
97 (79, 130.5) |
95 (80, 101) |
0.004 | 0.050 |
| Systolic blood pressure (mm Hg) | 150 (126, 164) |
144 (136, 155) |
150 (141, 170) |
151 (137, 161) |
140 (119, 158) |
150 (125, 157) |
0.147 | 0.970 |
| Baseline medication | ||||||||
| Anti-platelet drugs (%) | 39 % | 29 % | 33 % | 54 % | 42 % | 22 % | 0.431 | 0.061 |
| Anti-hypertensive drugs (%) | 56 % | 36 % | 43 % | 50 % | 81 % | 44 % | 0.023 | 0.767 |
| ACE-I/ARB (%) | 46 % | 27 % | 37 % | 42 % | 61 % | 39 % | 0.189 | 1.00 |
| Beta blocker (%) | 39 % | 22 % | 27 % | 46 % | 61 % | 11 % | 0.294 | 0.021 |
| Diuretics (%) | 19 % | 12 % | 10 % | 12 % | 32 % | 22 % | 0.111 | 0.419 |
| Calcium channel blocker (%) | 13 % | 14 % | 10 % | 12 % | 16 % | 17 % | 0.715 | 0.676 |
| Antidiabetics (%) | 11 % | 7 % | 17 % | 12 % | 10 % | 6 % | 1.000 | 0.634 |
| If coronary angiography: PCI (%) | 67 % | 0 % | 100 % | 92 % | 0 % | 8 % | <0.001 | <0.001 |
| Clinical cardiac biomarkers | ||||||||
| CK 0 h (U/l) | 154 (113,222) |
104 (72, 145) |
193 (128, 345) |
152.5 (104, 196) |
142.5 (106, 178) |
178 (129, 205) |
0.628 | 0.397 |
| CK 1 h (U/l) | 175 (122, 292) |
98 (69, 136) |
262 (165, 571) |
194 (147, 211) |
134 (90, 182) |
167.5 (135, 194) |
0.047 | 0.483 |
| CK 3 h (U/l) | 222 (137, 410) |
91 (67, 128.5) |
510 (239, 1182) |
261 (178, 325) |
142 (103, 205) |
165 (126, 248) |
0.003 | 0.075 |
| hs-TnT 0 h (ng/l) | 40 (16, 168) |
5 (4, 7) |
32 (16, 232) |
32 (12, 89) |
23 (13, 57) |
195 (133, 377) |
0.608 | 0.0001 |
| hs-TnT 1 h (ng/l) | 92 (42, 299) |
5 (3.5, 7) |
184 (95, 437) |
78.5 (43, 198) |
36 (26, 68) |
287.5 (165, 440) |
0.005 | 0.022 |
| hs-TnT 3 h (ng/l) | 178 (91, 517) |
5 (4, 6) |
519 (283, 1838) |
213 (104, 546) |
78 (42, 113) |
307 (152, 502) |
<0.001 | 0.685 |
| hs-TnI 0 h (ng/l) | 68 (18, 690) |
3 (2, 5.5) |
108 (22, 949) |
50 (18, 263) |
22 (10, 73) |
1149 (302, 2160) |
0.267 | 0.0002 |
| hs-TnI 1 h (ng/l) | 266 (65, 1021) |
3 (2, 4.3) |
476 (206, 2459) |
383 (112, 867) |
53 (33,211) |
1045 (155, 2306) |
0.0008 | 0.184 |
| hs-TnI 3 h (ng/l) | 1072 (286, 4288) |
3 (2, 5) |
4775 (1423, 26,461) |
2198 (482, 4746) |
241 (52, 525) |
1902 (845, 3358) |
<0.001 | 0.952 |
Continuous variables are presented as median (25th and 75th percentile). P value computed for NSTEMI type1 (T1MI) vs NSTEMI type2 (T2MI) and NSTEMI type1 (T1MI) vs acute injury (AI) using Mann-Whitney test for continuous variables and Fisher exact test for binary variables. Abbreviations: MI: Myocardial infarction; T1MI: NSTEMI type1; T2MI: NSTEMI type2; AI: Acute injury; GRACE: Global Registry of Acute Coronary Events; CK: creatine kinase; hs-TnT: high-sensitive troponin T; hs-TnI: high-sensitive troponin I. Baseline characteristic comparisons with P-value < 0.05 are highlighted in bold.
3.2. Protein release kinetics
Baseline measurements of cardiac troponins I and T and cMyBP-C were not different between T1MI and T2MI (Fig. 1). All cardiac necrosis markers showed significant differences between T1MI and T2MI at 3 h after admission (Fig. 1). The release kinetics of NT-proBNP also did not show discriminative ability between T1MI and T2MI (Fig. 1). cTnT differentiated AI from T1MI, albeit only on admission (Fig. 1). However, NT-proBNP levels remained higher in AI than T1MI at all time points (Fig. 1). hs-TnT showed discriminative power for AI versus T2MI at all time points (Fig. 1). Pairwise statistical comparison of hs-TnT and NT-proBNP at each time point for MI subtypes and control patients with non-cardiac chest pain is presented in Supplemental Fig. 4. Box plots include hs-TnT and NT-proBNP values in STEMI patients for reference.
Fig. 1.
Protein kinetics in MI subtypes using linear mixed effects model. Y-axis shows fitted value using linear mixed effects regression (lmer). X-axis shows sampling time in hours. Interaction style plots were generated using R package ‘emmip’ with dots in the plot indicating the estimated marginal mean i.e., mean adjusted for individual random effect for within subject variance and lines show the 95 % confidence interval. n numbers indicate the serial measurements quantified across the 3 time points for each of the MI sub-types. Satterthwaite degrees-of-freedom method was used. Contrasts were generated using R package ‘emmeans’. P values were adjusted using Tukey's method for comparing a family of 9 estimates. */†P value < 0.05, **<0.01 and ***/###<0.001. MI: Myocardial infarction; AI: Acute injury; T1MI: NSTEMI type1; T2MI: NSTEMI type2; NPX: Normalized Protein eXpression, Olink's® arbitrary unit in Log2 scale. hsTnT: high-sensitive cardiac troponin T; hsTnI: high-sensitive cardiac troponin I; cMyBP-C: cardiac myosin-binding protein C; NT-proBNP: N-terminal pro-brain natriuretic peptide.
3.3. Kinetics of microRNAs
miRNAs with cardiac and muscle origin as well as plasma miRNAs previously associated with cardiovascular diseases (CVD) (Supplemental Table 1) were quantified in the 495 samples from all three time points. The selection of miRNAs was based on 1) known tissue origin (cardiac/muscle vs circulating cells or other organs) and 2) detectability in the circulation. Compared to STEMI, cardiac-specific (miR-208 and miR-499) and muscle-enriched miRNAs (miR-1 and miR-133a) were less well detectable in NSTEMI. Detectability for cardiac-specific miR-208 and miR-499 was <50 %, even at 3 h. Correlation analysis revealed clustering of muscle-enriched miRNAs with cardiac necrosis markers in MI (Fig. 2), consistent with our previous study in STEMI [8]. However, release kinetics of miR-1 and miR-133a did not offer discrimination between T1MI, T2MI and AI (Fig. 3). Other abundant plasma miRNAs clustered together (Fig. 2) but their release kinetics also failed to discriminate T1MI, T2MI and AI (Fig. 3).
Fig. 2.
Correlation of protein and miRNA biomarkers. The pairwise Spearman correlation was calculated between proteins and miRNAs adjusted for individual effects (repeated measure). Hierarchical clustering analysis and heatmap matrix illustrates positive and negative co-expression and clusters. Red and blue colours indicate a positive and negative correlation, respectively (P value < 0.05). White indicates no significant correlation (P value > 0.05). P values were adjusted using the Benjamini-Hochberg FDR correction.
Fig. 3.
Circulating miRNA kinetics using linear mixed effects model. Y-axis shows fitted value using linear mixed effects regression (lmer). X-axis shows sampling time in hours. Interaction style plots were generated using R package ‘emmip’ with dots in the plot indicating the estimated marginal mean i.e., mean adjusted for individual random effect for within subject variance and lines show the 95 % confidence interval. n numbers indicate the serial measurements quantified across the 3 time points for each of the MI sub-types. Satterthwaite degrees-of-freedom method was used. Contrasts were generated using R package ‘emmeans’. P value shows adjusted value using Tukey method for comparing a family of 9 estimates. MI: Myocardial infarction; AI: Acute injury; T1MI: NSTEMI type1; T2MI: NSTEMI type2.
3.4. Combining NT-proBNP with cardiac necrosis markers aids in discriminating T1MI, T2MI and AI
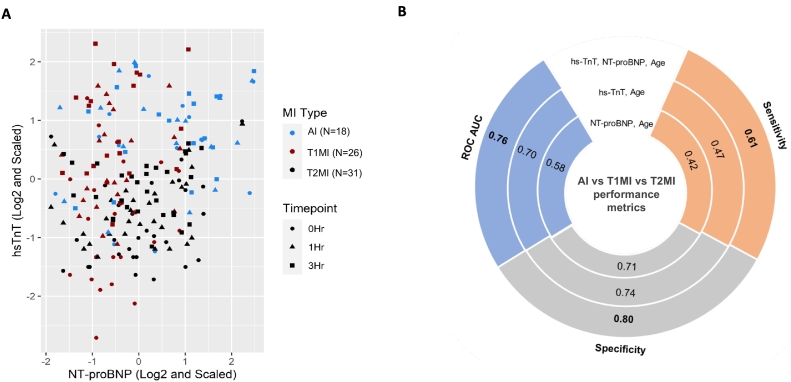
Given the distinct release kinetics of cardiac necrosis markers and NT-proBNP, we analysed their biomarker potential in a combinatorial approach. The scatterplot of hs-TnT and NT-proBNP displays how a combination of hs-TnT and NT-proBNP separates MI patients into T1MI (high hs-TnT, low NT-proBNP), T2MI (low hs-TnT, high NT-proBNP) and AI (high hs-TnT and high NT-proBNP) (Fig. 4A). This discrimination is also apparent in the individual kinetics of hs-TnT and NT-proBNP (Fig. 1). Hence, the integrated signature of hs-TnT, NT-proBNP and age was compared to standalone signatures of ‘hs-TnT, age’ and ‘NT-proBNP, age’ using one-vs-all multi-class random forest model in a leave-one-out cross validation with bootstrapping (Fig. 4B and Supplementary Fig. 5). One-vs-all comparison was undertaken for T1MI, T2MI and AI, thus presenting not only the overall performance of the model but also the comparative performances for T1MI vs (T2MI + AI), T2MI vs (T1MI + AI) and AI vs (T1MI + T2MI). Sex as a feature was not supported by Boruta feature selection. Hence, only age was retained in the prediction signature with individual and combination biomarkers. The AUC was 0.70 [95 % CI: 0.60–0.78] with ‘hs-TnT, age’ and 0.58 [95 % CI: 0.47–0.69] with ‘NT-proBNP, age’ (Fig. 4B and Supplementary Fig. 5B). Further, the low sensitivity (high false negatives) of 0.47 with ‘hs-TnT, age’ and 0.42 with ‘NT-proBNP, age’ makes these signatures unsuitable in discriminating T1MI, T2MI and AI (Fig. 4B and Supplementary Fig. 5A). The combined signature of hs-TnT, NT-proBNP and age returned an overall AUC of 0.76 [95 % CI: 0.67–0.84] for discriminating T1MI (AUC: 0.69), T2MI (AUC: 0.78) and AI (AUC: 0.81) with a more balanced overall sensitivity of 0.61, outperforming measurements of either hs-TnT or NT-proBNP alone (Fig. 4B, Supplementary Figs. 5 and 6).
Fig. 4.
Discriminative value of hs-TnT and NT-proBNP to differentiate T1MI, T2MI and AI. A. Scatterplot for hs-TnT and NT-proBNP illustrates the ability of combined biomarkers in clustering T1MI (high hs-TnT, low NT-proBNP), T2MI (low hs-TnT, high NT-proBNP) and AI (high hs-TnT and high NT-proBNP). Legends shown are in combination of shape (triangle, square and circle) for timepoints and colour (red, blue, black) for MI sub-types. For example, a blue square denotes AI at 3 h while a red square indicates T2MI at 3 h. B. Sensitivity, specificity and ROC AUC comparing predictive power of biomarkers in discriminating T1MI, T2MI and AI. ROC AUC of 0.58 with ‘NT-proBNP, age’ was inferior compared to ROC AUC of 0.76 with the combined signature of ‘hs-TnT, NT-proBNP, age’. The low sensitivity (high false negatives) of 0.47 with ‘hs-TnT, age’ and 0.42 with ‘NT-proBNP, age’ makes these signatures unsuitable in discriminating T1MI, T2MI and AI. The combined signature of hs-TnT, NT-proBNP and age returned an overall AUC of 0.76 for discriminating T1MI, T2MI and AI with a more balanced overall sensitivity of 0.61, outperforming measurements of either hs-TnT or NT-proBNP alone. Abbreviations - ROC AUC: Receiver operating characteristic area under the curve; AI: Acute injury; T1MI: NSTEMI type1; T2MI: NSTEMI type2.
4. Discussion
Our study suggests that a combination of NT-proBNP and cTnT could aid in differentiating T1MI, T2MI and AI. Established (cTnT, cTnI) and novel (cMyBP-C) cardiac necrosis markers failed in differentiating T1MI vs T2MI at early time points. The combination of cardiac necrosis markers with NT-proBNP, however, helped to identify patients with T2MI and to differentiate MI subtypes with heart failure-like pathologies, which are common among patients with AI but infrequent in patients with T1MI.
4.1. Cardiac necrosis markers in MI subtypes
Cardiac troponins are the gold standard biomarkers in the evaluation of acute MI [3], [18]. While mortality and morbidity in T2MI is similar or even higher than in T1MI [19], [20], there are currently no biomarkers to differentiate between T1MI and T2MI. This is in agreement with a recent study by Eggers et al. [21] who evaluated the prognostic potential of cTn in T2MI. A similar conclusion was reached by Nestelberger et al. [9] supporting the observation in our study that cTns do not differentiate between T1MI and T2MI. The study by Nestelberger et al. [9], however, did not assess AI. AI patients have a worse outcome than T1MI patients as reflected by their elevated GRACE score that is comparable to T2MI (Table 1). Therefore, we assessed not only T2MI patients but also AI patients to account for this important MI subtype. None of the cardiac necrosis markers was able to differentiate all three MI subtypes at any given time point. While cMyBP-C shows an earlier release kinetics upon myocardial injury compared with cTn [6], [7], cMyBP-C was also unable to discriminate T1MI and T2MI at early time points. Both, cTn and cMyBP-C are sarcomeric proteins of cardiomyocytes and are predominantly released upon cardiomyocyte death. All necrosis markers including cMyBP-C showed significant differences between T1MI and T2MI only 3 h after admission. This information extends the study by Nestelberger et al. [9] who assessed biomarkers up to 2 h after admission and reported no significant differences for cardiac biomarkers between T1MI and T2MI. Our dataset comprises biomarker measurements up to 3 h after admission. Only at 3 h, cardiac necrosis markers had discriminative power for T1MI versus T2MI. Thus, biomarker combinations may be required to differentiate T1MI, T2MI and AI upon a patient's presentation at the emergency department.
4.2. miRNA biomarkers in MI subtypes
The performance of miRNAs has not been evaluated in T2MI and AI to date. This study extends the biomarker measurements beyond proteins to also include miRNAs. Our group has previously studied miRNAs as biomarker candidates for prediction of primary [22] and secondary [23] CVD events. Many of those miRNAs, however, are not released from cardiomyocytes [8], but from platelets and other circulating cells. A systemic response is the most likely explanation for post-MI changes of these miRNAs [24], [25]. In the present study, neither cardiac- and/or muscle miRNAs, nor other selected miRNAs returned discriminative power in distinguishing T1MI versus T2MI or AI. For cardiac miRNAs, their assessment is hampered by less cardiac release due to the smaller infarct size in NSTEMI compared to STEMI patients. Similarly, muscle miRNA showed lower detectability. Other plasma miRNAs are well-detectable, but their tissue/cell origin may not reflect the underlying patho-mechanisms of most T2MI cases.
4.3. NT-proBNP in MI subtypes
NT-proBNP is actively secreted by cardiomyocytes upon increased wall stress and the current gold standard biomarker in the diagnosis and prognosis of heart failure. While NT-proBNP is a strong prognostic marker post-MI [26], [27], NT-proBNP is not routinely measured during ACS. Interestingly, NT-proBNP has been reported to facilitate the diagnosis of low-troponin ACS when used in combination with cTnT [28]. Another report suggested that NT-proBNP identified a ‘previously unrecognised group of patients’ among patients presenting to the emergency department for cardiac symptoms [29]. These early reports highlight that NT-proBNP may have diagnostic value in subtypes of MI, although the differential diagnosis of T2MI was not yet established at the time. Nestelberger et al. also reported that baseline NT-proBNP was among the best performing biomarkers although only ranged in moderate levels of predictive power [9]. While the study includes more patients, their analysis was focussed on the performance of single biomarkers, predominantly on admission. Also, AI, comprising patients with heart failure-like diagnoses, was not assessed. Neumann et al. [10] reported baseline NT-proBNP in combination with cardiac troponin and other non-cardiac specific protein biomarkers showed most discriminatory potential between T1MI and T2MI or NSTEMI and myocardial injury. While the study by Neumann et al. [10] includes more patients, the analysis was once again focused on the performance of biomarkers at baseline i.e., on admission, not addressing the biomarker kinetics in the clinical evaluation of myocardial damage [11]. Our dataset comprises biomarker serial measurements up to 3 h after admission, thus providing contrasts of their release kinetics.
4.4. Biomarker combinations for differentiation of MI subtypes
Recent studies indicate modest ability of individual routine biomarkers to differentiate T2MI [30], while biomarker combinations have only been tested at baseline, but not in serial measurements [10]. In our BACC sub cohort, serial measurements of all biomarkers were performed at all time points providing an opportunity for a direct comparison of their release kinetics. Despite the limited value of cardiac necrosis markers in the early differentiation of T2MI, a combination with NT-proBNP depicted different patterns with high hs-TnT and low NT-proBNP in T1MI, but low hs-TnT and high NT-proBNP in T2MI and high hs-TnT and high NT-proBNP in AI. These results highlight potential for a combinatory use of hs-TnT and NT-proBNP in the diagnosis of all three MI subtypes i.e., T1MI, T2MI and AI. cTnT and NT-proBNP are established clinical cardiac necrosis and strain biomarkers, respectively. cTnT but not NT-proBNP is routinely measured in STEMI/NSTEMI patients. A combination of measurements could be readily implemented in clinical settings if the combination were to aid in the differentiation of MI subtypes. An early diagnosis of T2MI and AI would have important clinical implications, i.e., 1) guidance on the necessity of invasive diagnostic procedures and 2) focus on specific treatment options for T2MI and/or AI.
4.5. Strengths and limitations
Our study is exploratory, searching for novel approaches to discriminate T1MI, T2MI and AI, and awaits further validation in independent cohorts. Although the NT-proBNP PEA correlated significantly with clinical NT-proBNP measurements, further studies are required to evaluate cTn and NT-proBNP thresholds to differentiate NSTEMI from AI. The finding that the cardiac strain marker NT-proBNP and cardiac necrosis marker cTnT combined with age may provide better discrimination between NSTEMI and AI than either of the biomarkers on its own offers an interesting observation with potential clinical relevance. Biomarker performance was compared for all 3 MI sub-types i.e., T1MI, T2MI and AI which has not been evaluated thus far. The causes of T2MI and AI are multifactorial and comprise systemic responses to circulatory insufficiency [3]. T2MI is defined as a mismatch of cardiac oxygen demand and supply in the absence of acute coronary plaque rupture. Thus, other biomarkers beyond the ones tested in the present study may better capture the distinct patho-mechanisms leading to cardiac ischemia in T2MI distinguishing it from AI and T1MI.
5. Conclusion
A combination of cTnT with NT-proBNP was able to better distinguish T1MI, T2MI and AI compared to single biomarker measurements in the BACC cohort. The diagnosis of T2MI and its former sub-category AI may benefit from the combination of biomarker trajectories during close clinical observation in the acute setting of suspected MI. Whether this combination is sufficient to improve early differentiation of T2MI in acute clinical settings will require further validation in independent cohorts.
Declaration of competing interest
M. Mayr is named inventor on a licensed patent held by King's College London for the detection of cMyBP-C as a biomarker of myocardial injury (EP2430453B1, US8546089). M. Mayr filed and licensed patent applications on miRNAs as biomarkers (EP15193448.6, EP2776580 B1, DE112013006129T5, GB2524692A, EP2576826 B, JP2013-513740). JT Neumann received honoraria from Siemens and Abbott Diagnostics. PMH reports fees from Beckman Coulter. D. Westermann reports personal fees from Bayer, Boehringer-Ingelheim, Berlin Chemie, Astra Zeneca, Biotronik and Novartis. S. Blankenberg received honoraria from Abbott Diagnostics, Siemens, Thermo Fisher, and Roche Diagnostics and is a consultant for Thermo Fisher.
Acknowledgments
Acknowledgements
We acknowledge Mr. Christian Cassel for technical assistance. We thank Mrs. Alina Goßling and Dr. Francisco M. Ojeda for data handling of the BACC study.
Funding
M. Mayr is a British Heart Foundation (BHF) Chair Holder (CH/16/3/32406) with BHF programme grant support (RG/16/14/32397, RG/F/21/110053). M. Mayr was awarded a BHF Special Project grant to participate in the ERA-CVD Transnational Grant “MacroERA: Noncoding RNAs in cardiac macrophages and their role in heart failure” and is part of the Marie Skłodowska-Curie Innovative Training Network TRAIN-HEART (http://train-heart.eu) as well as the PlaqOmic network funded by the Foundation Leducq. K. Theofilatos and M. Mayr were awarded a BHF Project Grant named “Harnessing machine learning for a multi-omics approach to cardiovascular disease” (PG/20/10387). This work was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS (National Health Service) Foundation Trust and King's College London in partnership with King's College Hospital. CS is the recipient of a research fellowship by the Deutsche Forschungsgemeinschaft (DFG) (SCHU2983/1 and SCHU 2983/2). BS is funded by BHF PhD studentship FS/19/58/34895. TZ is funded by the German Centre for Cardiovascular Research (DZHK) (81Z0710102). The BACC study was supported by an unrestricted grant by Abbott Diagnostics. It was further funded in part by the German Centre of Cardiovascular Research (DZHK e.V.). NS and JTN were supported by grants from the German Heart Foundation/German Foundation of Heart Research. JTN was supported by a fellowship by the Deutsche Forschungsgemeinschaft (NE 2165/1-1)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmccpl.2022.100014.
Contributor Information
Christian Schulte, Email: c.schulte@uke.de.
Manuel Mayr, Email: manuel.mayr@kcl.ac.uk.
Appendix A. Supplementary data
Supplementary material
References
- 1.Twerenbold R., Neumann J.T., Sörensen N.A., Ojeda F., Karakas M., Boeddinghaus J., Nestelberger T., Badertscher P., Rubini Giménez M., Puelacher C., Wildi K., Kozhuharov N., Breitenbuecher D., Biskup E., du Fay J., de Lavallaz D., Flores D., Wussler Ò., Miró F.J.Martín, Sánchez B., Morawiec J., Parenica N., Geigy D.I., Keller T., Zeller T., Reichlin S., Blankenberg D., Westermann C.Mueller. Prospective validation of the 0/1-h algorithm for early diagnosis of myocardial infarction. J Am Coll Cardiol. 2018;72:620–632. doi: 10.1016/j.jacc.2018.05.040. [DOI] [PubMed] [Google Scholar]
- 2.Kaier T.E., Twerenbold R., Lopez-Ayala P., Nestelberger T., Boeddinghaus J., Alaour B., Huber I.-M., Zhi Y., Koechlin L., Wussler D., Wildi K., Shrestha S., Strebel I., Miro O., Martín-Sánchez J.F., Christ M., Kawecki D., Keller D.I., Gimenez M.Rubini, Marber M., Mueller C., Freese M., Ratmann P.D., Prepoudis A., Gualandro D.M., Geigy N., Reichlin T., Rentsch K., Maier M., Troester V., Gehrke J., Coscia T., Glarner N., Schoepfer H., Buechi M., Walter J., Sanchez A.Y., Puelacher C., du Fay de Lavallaz J., Sanzione A., Schäfer I., Hillinger P., López B., Adrada E.R., Muzyk P., Morawiec B., Parenica J., Ganovská E., Lohrmann J., Buser A., von Eckardstein A., Bingisser R., Nickel C. A 0/1h-algorithm using cardiac myosin-binding protein C for early diagnosis of myocardial infarction. Eur Heart J Acute Cardiovasc Care. 2022 doi: 10.1093/ehjacc/zuac007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thygesen K., Alpert J.S., Jaffe A.S., Chaitman B.R., Bax J.J., Morrow D.A., White H.D. Fourth universal definition of myocardial infarction (2018) Glob Heart. 2018;13:305–338. doi: 10.1016/j.gheart.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Chapman A.R., Shah A.S.V., Lee K.K., Anand A., Francis O., Adamson P., McAllister D.A., Strachan F.E., Newby D.E., Mills N.L. Long-term outcomes in patients with type 2 myocardial infarction and myocardial injury. Circulation. 2018;137:1236–1245. doi: 10.1161/CIRCULATIONAHA.117.031806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartikainen T.S., Sörensen N.A., Haller P.M., Goßling A., Lehmacher J., Zeller T., Blankenberg S., Westermann D., Neumann J.T. Clinical application of the 4th universal definition of myocardial infarction. Eur Heart J. 2020;41:2209–2216. doi: 10.1093/eurheartj/ehaa035. [DOI] [PubMed] [Google Scholar]
- 6.Jacquet S., Yin X., Sicard P., Clark J., Kanaganayagam G.S., Mayr M., Marber M.S. Identification of cardiac myosin-binding protein C as a candidate biomarker of myocardial infarction by proteomics analysis. Mol Cell Proteomics. 2009;8:2687–2699. doi: 10.1074/mcp.M900176-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaier T.E., Twerenbold R., Puelacher C., Marjot J., Imambaccus N., Boeddinghaus J., Nestelberger T., Badertscher P., Sabti Z., Giménez M.R., Wildi K., Hillinger P., Grimm K., Loeffel S., Shrestha S., Widmer D.F., Cupa J., Kozhuharov N., Miró Ò., Martín-Sánchez F.J., Morawiec B., Rentsch K., Lohrmann J., Kloos W., Osswald S., Reichlin T., Weber E., Marber M., Mueller C. Direct comparison of cardiac myosin-binding protein C with cardiac troponins for the early diagnosis of acute myocardial infarction. Circulation. 2017;136:1495–1508. doi: 10.1161/CIRCULATIONAHA.117.028084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulte C., Barwari T., Joshi A., Theofilatos K., Zampetaki A., Barallobre-Barreiro J., Singh B., Sörensen N.A., Neumann J.T., Zeller T., Westermann D., Blankenberg S., Marber M., Liebetrau C., Mayr M. Comparative analysis of circulating noncoding RNAs versus protein biomarkers in the detection of myocardial injury. Circ Res. 2019;125:328–340. doi: 10.1161/CIRCRESAHA.119.314937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nestelberger T., Boeddinghaus J., Lopez-Ayala P., Kaier T.E., Marber M., Gysin V., Koechlin L., Sanchez A.Y., Giménez M.R., Wussler D., Walter J.E., Strebel I., Zimmermann T., Glarner N., Miró Ò., Martin-Sanchez F.J., Zehnder T., Twerenbold R., Keller D.I., Mueller C., Schoepfer H., Hillinger P., Ratmann P.D., Gualandro D.M., Coscia T., Troester V., Gehrke J., Widmer V., Prepoudis A., Rentsch K., Badertscher P., Wildi K., Puelacher C., Potlukova E., Freese M., Michou E., von Eckardstein A., Kawecki D., Morawiec B., Muzyk P., Bürgler F., Geigy N., Reichlin T., Shrestha S., López B., Mañé Cruz H., Fuenzalida Inostroza C.I., Rodgriguez Adrada E., García Briñón M.A., Parenica J., Buser A. Cardiovascular biomarkers in the early discrimination of type 2 myocardial infarction. JAMA Cardiol 6. 2021:771. doi: 10.1001/jamacardio.2021.0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann J.T., Weimann J., Sörensen N.A., Hartikainen T.S., Haller P.M., Lehmacher J., Brocks C., Tenhaeff S., Karakas M., Renné T., Blankenberg S., Zeller T., Westermann D. A biomarker model to distinguish types of myocardial infarction and injury. J Am Coll Cardiol. 2021;78:781–790. doi: 10.1016/j.jacc.2021.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Wereski R., Kimenai D.M., Taggart C., Doudesis D., Lee K.K., Lowry M.T.H., Bularga A., Lowe D.J., Fujisawa T., Apple F.S., Collinson P.O., Anand A., Chapman A.R., Mills N.L. Cardiac troponin thresholds and kinetics to differentiate myocardial injury and myocardial infarction. Circulation. 2021;144:528–538. doi: 10.1161/CIRCULATIONAHA.121.054302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann J.T., Sörensen N.A., Schwemer T., Ojeda F., Bourry R., Sciacca V., Schaefer S., Waldeyer C., Sinning C., Renné T., Than M., Parsonage W., Wildi K., Makarova N., Schnabel R.B., Landmesser U., Mueller C., Cullen L., Greenslade J., Zeller T., Blankenberg S., Karakas M., Westermann D. Diagnosis of myocardial infarction using a high-sensitivity troponin I 1-hour algorithm. JAMA Cardiol. 2016;1:397. doi: 10.1001/jamacardio.2016.0695. [DOI] [PubMed] [Google Scholar]
- 13.Marjot J., Liebetrau C., Goodson R.J., Kaier T., Weber E., Heseltine P., Marber M.S. The development and application of a high-sensitivity immunoassay for cardiac myosin–binding protein C. Transl Res. 2016;170:17–25. doi: 10.1016/j.trsl.2015.11.008. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westermann D., Neumann J.T., Sörensen N.A., Blankenberg S. High-sensitivity assays for troponin in patients with cardiac disease. Nat Rev Cardiol. 2017;14:472–483. doi: 10.1038/nrcardio.2017.48. [DOI] [PubMed] [Google Scholar]
- 15.Zeller T., Ojeda F., Brunner F.J., Peitsmeyer P., Münzel T., Binder H., Pfeiffer N., Michal M., Wild P.S., Blankenberg S., Lackner K.J. High-sensitivity cardiac troponin I in the general population – defining reference populations for the determination of the 99th percentile in the gutenberg health study. Clin Chem Lab Med. 2015;53 doi: 10.1515/cclm-2014-0619. [DOI] [PubMed] [Google Scholar]
- 16.Galili T., O’Callaghan A., Sidi J., Sievert C. Heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2018;34:1600–1602. doi: 10.1093/bioinformatics/btx657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kursa M.B., Rudnicki W.R. Feature selection with the boruta package. J Stat Softw. 2010;36 doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 18.Roffi M., Patrono C., Collet J.-P., Mueller C., Valgimigli M., Andreotti F., Bax J.J., Borger M.A., Brotons C., Chew D.P., Gencer B., Hasenfuss G., Kjeldsen K., Lancellotti P., Landmesser U., Mehilli J., Mukherjee D., Storey R.F., Windecker S. ESC guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation. Eur Heart J. 2015;37(2016):267–315. doi: 10.1093/eurheartj/ehv320. [DOI] [PubMed] [Google Scholar]
- 19.Lippi G., Sanchis-Gomar F., Cervellin G. Cardiac troponins and mortality in type 1 and 2 myocardial infarction. Clin Chem Lab Med. 2017;55 doi: 10.1515/cclm-2016-0324. [DOI] [PubMed] [Google Scholar]
- 20.Arora S., Strassle P.D., Qamar A., Wheeler E.N., Levine A.L., Misenheimer J.A., Cavender M.A., Stouffer G.A., Kaul P. Impact of type 2 myocardial infarction (MI) on hospital-level MI outcomes: implications for quality and public reporting. J Am Heart Assoc. 2018;7 doi: 10.1161/JAHA.118.008661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggers K.M., Baron T., Gard A., Lindahl B. Clinical and prognostic implications of high-sensitivity cardiac troponin T concentrations in type 2 non-ST elevation myocardial infarction. Int J Cardiol Heart Vasc. 2022;39 doi: 10.1016/j.ijcha.2022.100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zampetaki A., Willeit P., Tilling L., Drozdov I., Prokopi M., Renard J.-M., Mayr A., Weger S., Schett G., Shah A., Boulanger C.M., Willeit J., Chowienczyk P.J., Kiechl S., Mayr M. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–299. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 23.Schulte C., Molz S., Appelbaum S., Karakas M., Ojeda F., Lau D.M., Hartmann T., Lackner K.J., Westermann D., Schnabel R.B., Blankenberg S., Zeller T. miRNA-197 and miRNA-223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLOS ONE. 2015;10 doi: 10.1371/journal.pone.0145930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long G., Wang F., Duan Q., Chen F., Yang S., Gong W., Wang Y., Chen C., Wang D.W. Human circulating MicroRNA-1 and MicroRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci. 2012;8:811–818. doi: 10.7150/ijbs.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devaux Y., Mueller M., Haaf P., Goretti E., Twerenbold R., Zangrando J., Vausort M., Reichlin T., Wildi K., Moehring B., Wagner D.R., Mueller C. Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J Intern Med. 2015;277:260–271. doi: 10.1111/joim.12183. [DOI] [PubMed] [Google Scholar]
- 26.Omland T., de Lemos J.A., Morrow D.A., Antman E.M., Cannon C.P., Hall C., Braunwald E. Prognostic value of N-terminal pro-atrial and pro-brain natriuretic peptide in patients with acute coronary syndromes. Am J Cardiol. 2002;89:463–465. doi: 10.1016/S0002-9149(01)02271-8. [DOI] [PubMed] [Google Scholar]
- 27.Omland T., Persson A., Ng L., O’Brien R., Karlsson T., Herlitz J., Hartford M., Caidahl K. N-terminal pro-B-type natriuretic peptide and long-term mortality in acute coronary syndromes. Circulation. 2002;106:2913–2918. doi: 10.1161/01.cir.0000041661.63285.ae. [DOI] [PubMed] [Google Scholar]
- 28.Truong Q.A., Bayley J., Hoffmann U., Bamberg F., Schlett C.L., Nagurney J.T., Koenig W., Januzzi J.L. Multi-marker strategy of natriuretic peptide with either conventional or high-sensitivity troponin-T for acute coronary syndrome diagnosis in emergency department patients with chest pain: from the “Rule out myocardial infarction using computer assisted tomography” (ROMICAT) trial. Am Heart J. 2012;163:972–979. doi: 10.1016/j.ahj.2012.03.010. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell D.J., Munir V., Hennessy O.F., Dent A.W. Plasma amino-terminal pro-brain natriuretic peptide levels in subjects presenting to the emergency department with suspected acute coronary syndrome: possible role in selecting patients for follow up? Intern Med J. 2001;31:211–219. doi: 10.1046/j.1445-5994.2001.00042.x. [DOI] [PubMed] [Google Scholar]
- 30.Nestelberger T., Boeddinghaus J., Giménez M.R., Lopez-Ayala P., Ratmann P.D., Badertscher P., Wildi K., Wussler D., Koechlin L., Arslani K., Zimmermann T., Freese M., Rinderknecht T., Miró Ò., Martin-Sanchez F.J., Kawecki D., Geigy N., Keller D., Twerenbold R., Müller C. Direct comparison of high-sensitivity cardiac troponin T and I in the early differentiation of type 1 vs. Type 2 myocardial infarction. Eur Heart J Acute Cardiovasc Care. 2022;11:62–74. doi: 10.1093/ehjacc/zuab039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material