Abstract
Ki67 has potential clinical importance in breast cancer but has yet to see broad acceptance due to inter-laboratory variability. Here we tested an open source and calibrated automated digital image analysis (DIA) platform to: (i) investigate the comparability of Ki67 measurement across corresponding core biopsy and resection specimen cases, and (ii) assess section to section differences in Ki67 scoring. Two sets of 60 previously stained slides containing 30 core-cut biopsy and 30 corresponding resection specimens from 30 estrogen receptor-positive breast cancer patients were sent to 17 participating labs for automated assessment of average Ki67 expression. The blocks were centrally cut and immunohistochemically (IHC) stained for Ki67 (MIB-1 antibody). The QuPath platform was used to evaluate tumoral Ki67 expression. Calibration of the DIA method was performed as in published studies. A guideline for building an automated Ki67 scoring algorithm was sent to participating labs. Very high correlation and no systematic error (p = 0.08) was found between consecutive Ki67 IHC sections. Ki67 scores were higher for core biopsy slides compared to paired whole sections from resections (p ≤ 0.001; median difference: 5.31%). The systematic discrepancy between core biopsy and corresponding whole sections was likely due to pre-analytical factors (tissue handling, fixation). Therefore, Ki67 IHC should be tested on core biopsy samples to best reflect the biological status of the tumor.
Subject terms: Pathology, Prognostic markers, Breast cancer
Introduction
It has been long acknowledged that the immunohistochemical (IHC) detection of Ki67 positive tumor cells provides important clinical information in breast cancer1. More recently, Ki67 gained clinical utility in the T1-2, N0-1, estrogen receptor-positive (ER) and HER2-negative patient group by allowing to identify those patients that are unlikely to benefit from adjuvant chemotherapy2. However, Ki67 has not been consistently adopted for clinical care due to unacceptable reproducibility across laboratories3–5.
Therefore, the International Ki67 in Breast Cancer Working Group (IKWG) originally published consensus recommendations in 2011 for best practices in the application of Ki67 IHC in breast cancer6. According to this consensus, parameters that predominantly influence Ki67 IHC results can be grouped into pre-analytical (type of biopsy, tissue handling), analytical (IHC protocol), interpretation and scoring, and data analysis steps6. As the scoring method was the largest contributor to test variability7, the IKWG has undertaken serious efforts to standardize the Ki67 scoring method of pathologists8,9. Although in multi-institutional studies, standardized Ki67 scoring methods reached pre-defined thresholds for adequate reproducibility9,10, this was only after completing calibration training and by using tedious counting methods. In this context, recently updated guidelines by the IKWG now recommend Ki67 IHC for clinical adoption in specific situations, including the identification of very low (<5) or very high proliferation (>30) indices, that render more expensive gene expression tests unnecessary2.
An important additional issue that can cause variability in Ki67 measurements is the type of specimen (core biopsy vs excision) and its effect on Ki67 scoring in a multi-center setting2. Indeed, the IKWG recommended use of core biopsies (CB), based on apparent superior results for Ki67 when visual evaluation was compared to that of whole sections (WS).
In this multi-observer and multi-institutional study, we aimed to investigate the comparability of Ki67 measurements across corresponding core biopsy and resection specimens from the same breast cancer cases, when evaluated using a calibrated, automated reading system. Furthermore, we assessed between-(consecutive) section differences in Ki67 scoring as no difference between sections will facilitate the selection of the tumor-block to perform the IHC staining on.
Materials and methods
Patients
Thirty cases of ER-positive breast cancer used in phase 3 of IKWG initiatives collecting 15 cases from the UK and 15 cases from Japan designed to cover a range of Ki67 scores9 were employed in this study. No outcome data were collected for this cohort. Patient selection was irrespective of patients’ age at diagnosis, grade, tumor size or lymph node status. The clinicopathological characteristics of these 30 cases can be found in our previous publications9,10.
Tissue preparation and immunohistochemistry (IHC)
Tissues from UK patients, both core biopsies and surgical resections were collected according to ASCO/CAP guidelines, while patients’ tissues from Japan were collected following ISO (International Organization for Standardization) 15189 approved by the Japan Accreditation Board. Preparation of the Ki67 slides of the first cohort has been previously described9. Briefly, the corresponding core-cut biopsy and surgical resection blocks were centrally cut and stained with Ki67, resulting in 60 Ki67 slides from 30 cases. The IHC was performed using monoclonal antibody MIB-1 at dilution 1:50 (DAKO UK, Cambridgeshire, UK) using an automated staining system (Ventana Medical Systems, Tucson, AZ, USA) according to the consensus criteria established by the International Ki67 Working Group6. Sections from the same block were stained in a single immunohistochemistry run, except for four cases where the staining was performed in two different runs. This approach effectively controls for any technical variation in staining.
Sample distribution
Twenty volunteer pathologists from 15 countries, most of whom participated in the previous Phase 3A study, were invited to participate. Four adjacent sections from each of the 60 blocks were centrally stained as follows: the first section with haematoxylin and eosin (H&E), the second with p63 (a myoepithelial marker, to assist the distinction of DCIS from invasive breast cancer) and the third to fourth with Ki67 (designated as slide sets 1–2).
The Aperio ScanScope XT platform was used at 20× magnification to digitize the slides (pixel size: 0.4987 µm × 0.4987 µm), which were uploaded to a server and distributed as digital images. Seventeen pathologists successfully completed the study (Fig. 1).
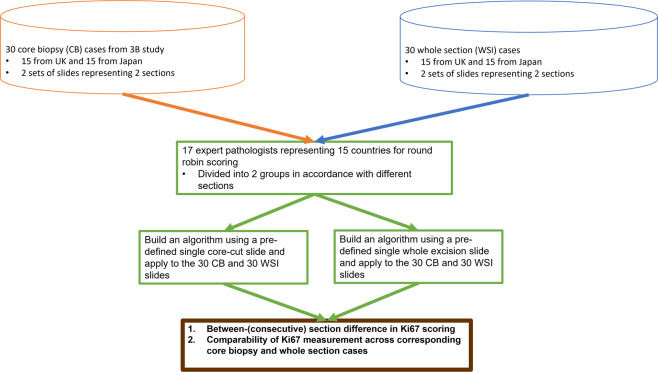
Fig. 1. Study design.
Thirty patients of ER-positive breast cancer were enrolled comprising 15 cases from UK and 15 cases from Japan. Corresponding core-cut biopsy and surgical resection blocks were centrally cut two adjacent sections per case and stained with Ki67. Seventeen pathologists from 15 countries were given 60 slides (30 Core cut biopsy slides and 30 surgical resection specimen slides) of Ki67 to score.
Digital image analysis (DIA)
The QuPath open-source software platform was used to build automated Ki67 scoring algorithms for breast cancer11. A detailed guideline for setting up and building an automated Ki67 scoring algorithm was sent to the participating labs. All the participating labs were requested to build their own Ki67 scoring algorithm following the instructions and apply them on these 60 slides. The complete step by step instructions are available in Supplementary File 1. The reason why we asked each lab to build their own algorithm instead of using the same pre-trained and locked down Ki67 algorithm was to mimic clinical practice. As of the date of the study, no generalizable Ki67 scoring algorithm was available that provides whole slide scoring. Thus, theoretically, all the labs would need to adjust/ optimize any such DIA approach to their lab characteristics (different fixation, different antibodies and IHC protocols etc.) necessitating a lab-specific DIA approach. Calibration of the DIA method/guideline was performed in our previous studies demonstrating very good reproducibility among users12,13. Briefly, after the whole invasive cancer area on a digitized slide was annotated, hematoxylin and DAB stain estimates for each case were refined using the “estimate stain vectors” command. We used watershed cell detection14 to segment the cells in the image with the following settings: Detection image: Optical density sum; requested pixel size: 0.5 µm; background radius: 8 µm; median filter radius: 0 µm; sigma: 1.5 µm; minimum cell area: 10 µm2; maximum cell area: 400 µm2; threshold: 0.1; maximum background intensity: 2. In order to classify detected cells into tumor cells, immune cells, stromal cells, necrosis and others (false detections, background) (Supplementary File 1), we used random trees as a supervised machine-learning method. The features used in the classification are described in Supplementary Table 1. After setting the optimal color deconvolution and cell segmentation, two independent classifiers were trained on a randomly selected, pre-defined core biopsy (CB classifier) and a resection specimen slide (WSI classifier). Both CB and WSI classifiers were run on both CB slides and resection specimen slides in order to adjust for potentially different characteristics of the two specimen types (Fig. 2).
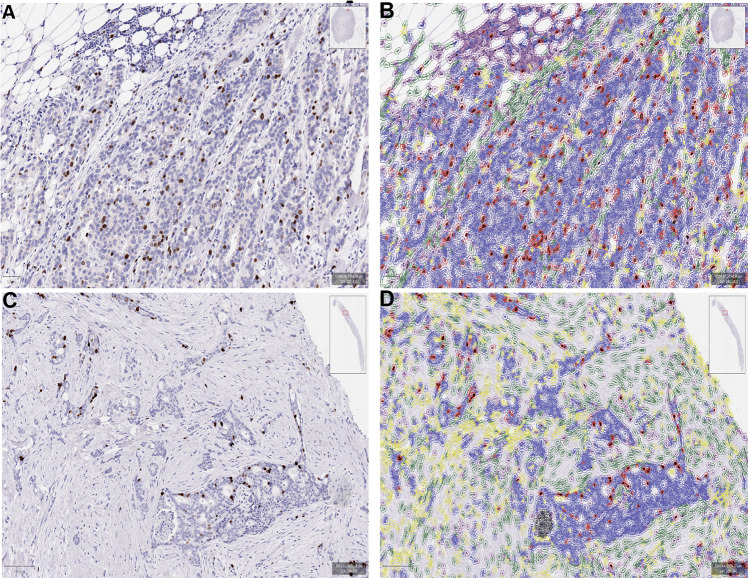
Fig. 2. Digital Image Analysis.
Representative pictures of digital image analysis (DIA) masks on a resection specimen (A, B) and a core biopsy case (C, D). Blue corresponds to Ki67 negative tumor cells, red indicates Ki67 positive tumor cells, green indicates stromal cells and purple marks immune cells. Black corresponds to necrosis and yellow marks other detections (false cell detections, noise).
Statistical analysis
For statistical analysis, SPSS 22 software (IBM, Armonk, USA) software was used. Degree of agreement was evaluated by Bland–Altman plot and linear regression. To assess differences between specimen type the Wilcoxon signed-rank test was applied, since the data were not normally distributed. Data were visualized using boxplot, spaghetti plot, and dot-plot.
Results
Between-(consecutive) section difference in Ki67 scoring
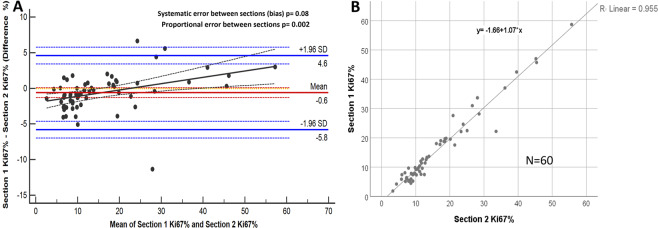
Very high correlation and no systematic error (bias: −0.6%; p = 0.08) was found between the two consecutive (serial) sections regarding Ki67 scores. If the Ki67 score is higher for a given case, the difference between the sections tends to be also greater (proportional error p = 0.002, Fig. 3.), however this difference (0.6% mean difference) does not reach clinical relevance.
Fig. 3. Between-(consecutive) section difference in Ki67 scoring.
Bland–Altman plot comparing Ki67 scores between consecutive sections (A). Orange dashed line corresponds to the expected mean zero difference between Ki67 scores of the two sections. Red line represents the observed mean difference between Ki67 scores of the two sections, namely the observed bias (red dashed lines are the CI of the observed mean difference). Blue lines illustrate the range of agreement (lower and upper limit of agreement) based on 95% of differences (blue dashed lines are the CI of the limits of agreement). Black line is the fitted regression line to detect potential proportional error (black dashed lines are the CI of the regression line). B represents the scatter plot with fitted regression between the Ki67 scores of the two consecutive sections.
Specimen type (CB vs resection specimen) difference in Ki67 scoring
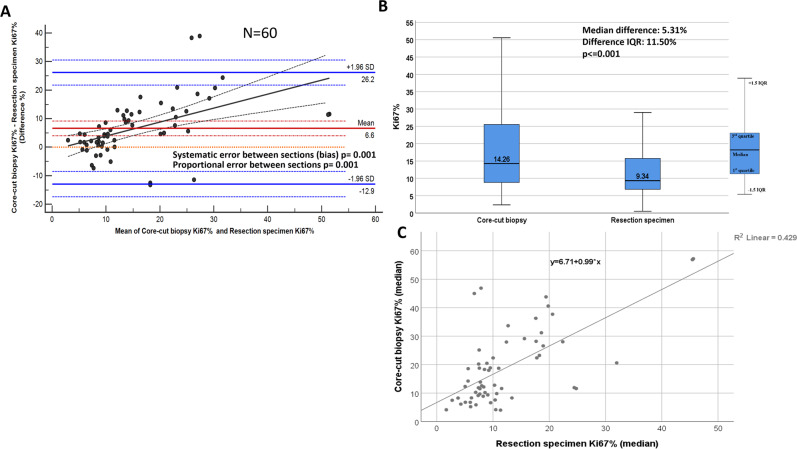
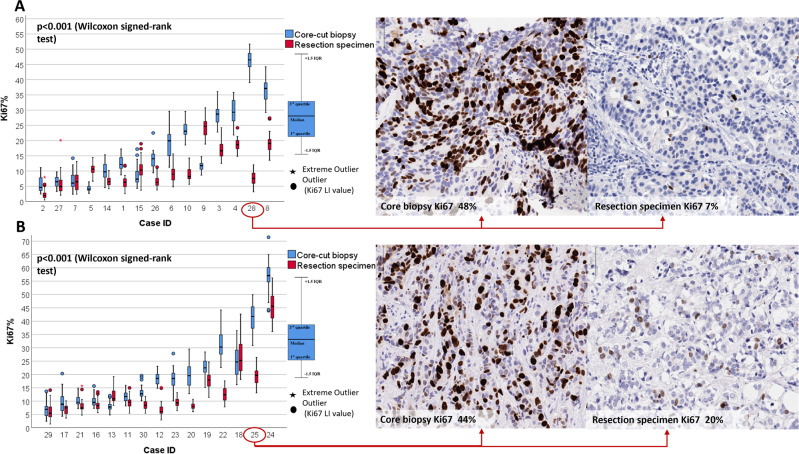
A low correlation was found between core biopsy and whole section excision images (Fig. 4). Ki67 scores were higher when determined on core biopsy slides compared to paired whole sections (p ≤ 0.001; median difference: 5.31%; IQR: 11.50%) from subsequent surgical excisions of the same tumor. Systematic error occurred between specimens from the same patient as core biopsy Ki67 scores were greater, with a clinically relevant mean difference of 6.6% (bias p = 0.001). The limits of agreement also have to be considered wide from a clinical perspective (between −13.7 and 27). Furthermore, Ki67 scores on CB were even higher compared to WS on cases with higher Ki67 scores (proportional error p = 0.001). Moreover, the variability of differences in Ki67 scores between CB and WS showed an increasing trend, proportional to the magnitude of Ki67 score (Fig. 4). The same results were found irrespective of the origin of the specimens (CB vs WS p < 0.001 for both UK and Japan cases Fig. 5).
Fig. 4. Between-specimen (CB vs resection specimen) difference in Ki67 scoring.
Bland–Altman plot comparing Ki67 scores between specimens (A). Orange dashed line corresponds the expected mean zero difference between Ki67 scores of the two sections. Red line represents the observed mean difference between Ki67 scores of the two sections, namely the observed bias (red dashed lines are the CI of the observed mean difference). Blue lines illustrate the range of agreement (lower and upper limit of agreement) based on 95% of differences (blue dashed lines are the CI of the limits of agreement). Black line is the fitted regression line to detect potential proportional error (black dashed lines are the CI of the regression line). B shows the distributions of Ki67 scores of the two specimens. The bottom/top of the boxes represent the first (Q1)/third (Q3) quartiles, the bold line inside the box represents the median and the two bars outside the box represent the lowest/highest datum still within 1.5× the interquartile range (Q3–Q1). C represents the scatter plot with fitted regression between the Ki67 scores of the two specimens.
Fig. 5. Between-specimen (CB vs resection specimen) difference in Ki67 scoring by case and by origin of the cases.
A represents cases collected in the United Kingdom with representative Ki67 IHC images of corresponding CB and resection specimens. B represents cases collected in Japan with representative Ki67 IHC images of corresponding CB and resection specimens. The bottom/top of the boxes represent the first (Q1)/third (Q3) quartiles, the bold line inside the box represents the median and the two bars outside the box represent the lowest/highest datum still within 1.5× the interquartile range (Q3–Q1). Outliers are represented with circles, extreme outliers with asterisk.
Discussion
In this study, we observed that clinically relevant and systematic discrepancies occurred in Ki67 scores between core biopsy and corresponding surgical specimens when evaluated with an automated reading system. Overall, Ki67 scores were higher on CB compared to WS samples. Furthermore, this discrepancy was even more pronounced in tumors that expressed higher levels of Ki67 in general.
Ki67 is one of the most promising yet controversial biomarkers in breast cancer with limited adoption into clinical practice due to its high inter- and intra-laboratory variability3,15. However, Ki67 is widely used in many countries, there is wide variability in its use (to distinguish luminal A-like vs B-like tumors; to determine whether to decide for gene-expression profiling or not; as an adjunct to mitotic counts, etc.), with still no uniformity between clinicians on how to use this biomarker, let alone which cut-off to use. Although the IKWG set up a guideline in 2011 to improve pre-analytical and analytical performance, inter-laboratory protocols still demonstrated low reproducibility related to different sampling, fixation, antigen retrieval, staining and scoring methods6,7. As the latter was the largest single contributor to assay variability, the IKWG has undertaken multi-institution efforts that have standardized visual scoring of Ki67 in a manner which requires on-line calibration tools and careful scoring of several hundred cells, which may or may not be ideal for pathologists in daily practice with time-constraints8,9. This result suggests that digital solutions may still be required to address this issue.
The rise of digital image analysis (DIA) platforms has improved capacity and automation in biomarker evaluation16,17. DIA platforms are able to assess nuclear IHC biomarkers such as Ki67, and numerous studies have been conducted to compare human visual scoring with DIA platforms12,18–28. Although the latest guideline of IKWG recommends Ki67 for clinical practice in specific situations, the type of specimen as a potential pre-analytical factor contributing to Ki67 variability was not specifically investigated in a multi-operator/multi-center setting. In this study we aimed to address these biospecimen questions including assessment by specimen type and between serial sections.
One explanation for our finding would be the presence of tumor heterogeneity, and the broader field of review in a whole section from resection specimen. However, one would expect that this cause of discrepancy would result in random discordance, not the consistent finding that Ki67 scores on core biopsies are higher than that of on resection specimens. Rather, we conclude that lower Ki67 in resection specimens is more easily explained by pre-analytical factors. For example, since longer times to fixation occur with resection specimens compared to CB, persistent cell division will occur even in an unfixed, hypoxic environment. Further, epitope degradation also occurs with prolonged time to fixation29–31.
In addition, one can expect that hot spot scoring might lead to less discrepancy between CB and WS because it considers only the hottest area of Ki67 positivity (highest percentiles of Ki67 distribution) on both specimen types, while global assessment evaluates the total Ki67 distribution which can be variable10. However, there remains a fundamental issue of exact hot spot definition and where pathologists set its boundaries. Moreover, the International Ki67 Working Group has recommended global scoring over hot spot as it did show a consistent trend towards increased reproducibility in both core biopsy9 and excision10 specimens.
Additional support for the conclusion that the difference in Ki67 between CB and WS is provided by the observation of clinically relevant differences between specimens in cases from different institutions used in this study, independently scored multiple times by 17 pathologists. Although many studies focused on assessing the level of agreement between CB and resection samples in Ki67 scoring; consensus was not possible due to lack of standardization32.
Our results are consistent with previous results showing poor/moderate concordance (κ = 0.195–0.814) occurring between CB and resection specimen in Ki67 scoring1,33–46. However, some studies showed higher Ki67 scores on resection samples35,36,38. This discrepancy among studies may be due to lack of standardization in methodology leading to different scoring methods, which we have previously demonstrated to be highly variable2. Moreover, inter-institutional discrepancies could also be the result of different antibodies and protocols used to detect Ki67, different tissue handling/fixation protocols and at some point tumor Ki67 heterogeneity since Ki67 is heterogeneous in tumors6. Thus, our findings provide further support to the latest IKWG recommendations and provide a consensus that Ki67 should be ideally tested on CB samples because it minimizes many fixation problems as Ki67 IHC is more sensitive than ER or HER2 to variabilities in fixation2. Since pre-analytical factors are critical in diagnostic pathology, the IKWG recommends that breast cancer samples for Ki67 testing should be processed in line with ASCO/CAP guidelines2.
There are a number of limitations in this study. This study only focused on analytical and preanalytical questions, therefore we cannot demonstrate the clinical validation of the calibrated tool. There are many other studies that address the prognostic or predictive value of this test, and that goal was beyond the scope of this effort. For the same reason, further clinical studies are needed to demonstrate how does this consistent difference in Ki67 between corresponding core-cuts and resection specimen impact on prognostic value or its clinical implication on the assessment of neoadjuvant endocrine therapy benefit. Furthermore, the low correlation suggests a critical difference between a core biopsy score and a whole section excision score, which can undermine the use of data on outcome, derived predominantly from resection samples, to identify patients at high risk using a score derived from a core biopsy. Therefore, this study suggests caution in this approach given that even without intervening therapy a clinically relevant change in Ki67 may occur. Further, the Ki67 assessments were based on biospecimens from only 2 central sites. While the participating pathologists within the IKWG represented 15 countries, specimens were centrally acquired and stained. Whereas other investigators have compared specimens from multiple different sites5,7,47 we limited the number of sites to remove the variables associated with the technical aspect of the stain. Finally, while the core cut biopsy and resection are from the same case, only a single core was assessed. Thus, we could be missing heterogeneity seen in larger resection specimens. The effect of heterogeneity could be decreased by taking multiple core cuts when clinical situation allows. However, since examination of a single core cut represents the clinical standard of care in several countries, we did not pursue multiple cores.
In conclusion, while we find no significant difference in digitally-assessed Ki67 index between serial sections, we do find a systematic discrepancy between core biopsy and corresponding whole sections – core biopsy samples yield higher scores (likely due to pre-analytical factors including more standard and prompt tissue handling, fixation, etc.). Therefore, this work suggests that Ki67 IHC tested on core biopsy samples should be preferred to excision specimens in clinical decision-making, because doing so will preclude many pre-analytical factors.
Supplementary information
Acknowledgements
We are grateful to the Breast International Group and North American Breast Cancer Group (BIG-NABCG) collaboration, including the leadership of Nancy Davidson, Thomas Buchholz, Martine Piccart, and Larry Norton.
Author contributions
BA, DLR, SCYL, TON, DFH and MD performed study concept and design; BA, DLR, SCYL performed development of methodology. All authors except DLR, TON, DFH, MD provided analysis, data acquisition and data curation. BA, SCYL, KMK provided statistical analysis. BA, DLR, SCYL, TON, DFH, MD, KMK provided interpretation of data. BA, DLR, SCYL, TON, DFH, MD wrote the first draft of the paper. All authors provided review, revision of the manuscript and approved the final paper.
Funding
BA is supported by The Swedish Society for Medical Research (Svenska Sällskapet för Medicinsk Forskning) Postdoctoral grant, Swedish Breast Cancer Association (Bröstcancerförbundet) Research grant 2021, The Fulbright Program and The Rosztoczy Foundation Scholarship Program. MD, DFH, RS (BCRF grant N° 17–194), (many others) and DLR are supported by the Breast Cancer Research Foundation. This work was supported by a generous grant from the Breast Cancer Research Foundation (DFH). Additional funding for the UK laboratories was received from Breakthrough Breast Cancer and the National Institute for Health Research Biomedical Research Centre at the Royal Marsden Hospital. Funding for the Ontario Institute for Cancer Research is provided by the Government of Ontario. JH is the Lilian McCullough Chair in Breast Cancer Surgery Research and the CBCF Prairies/NWT Chapter. Open access funding provided by Karolinska Institute.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
In the last 12 months, DLR has served as a consultant for advisor to Astra Zeneca, Agendia, Amgen, Cell Signaling Technology, Cepheid, Danaher, Konica-Minolta, Merck, PAIGE.AI, Regeneron, and Sanofi. TON reports a proprietary interest in PAM50/Prosigna and consultant work with Veracyte. JH was former member of the advisory board at Visiopharm A/S. JH has obtained speaker’s honoraria or advisory board remunerations from Roche, Novartis, AstraZeneca, Eli Lilly, Pfizer and MSD. JH is co-founder and shareholder of Stratipath AB. JH has received institutional research grants from Cepheid and Novartis. DFH reports no research or personal financial support related to this study. DFH does report research support unrelated to this study provided to his institution during conduct from Menarini/Silicon BioSystems, Astra Zeneca, Eli Lilly Company, Merrimack Pharmaceuticals, Inc. (Parexel Intl Corp), Veridex and Janssen Diagnostics (Johnson & Johnson), Pfizer, and Puma Biotechnology, Inc. (subcontract Wash Univ St. Louis to Univ Mich). DFH also reports that his institution holds a patent regarding circulating tumor cell characterization for which DFH is the named investigator that was licensed to Menarini Silicon Biosystems and from which both received annual royalties, ending in January 2021. DFH reports personal income related to consulting or advisory board activities from BioVeca, Cellworks, Cepheid, EPIC Sciences, Freenome, Guardant, L-Nutra, Oncocyte, Macrogenics, Predictus BioSciences, Salutogenic Innovations, Turnstone Biologics, and Tempus. DFH reports personally held stock options from InBiomotion. BvdV reports speaker’s honoraria or consultation/advisory board remunerations provided to his institution from Visiopharm, Philips, Merck/MSD and Diaceutics. GV reports receipt of grants/research supports from Roche/Genentech, Ventana Medical Systems, Dako/Agilent Technologies, and receipt of honoraria or consultation fees from Ventana, Dako/Agilent, Roche, MSD Oncology, AstraZeneca, Daiichi Sankyo, Pfizer, Eli Lilly. ZK has served in a paid advisory role to Eli Lilly. Unrelated to this study, SR is currently employed by Stratipath AB. RS reports non-financial support from Merck and Bristol Myers Squibb; research support from Merck, Puma Biotechnology, and Roche; and advisory board fees for Bristol Myers Squibb; and personal fees from Roche for an advisory board related to a trial-research project. RS has no COI related to this project. SF served as expert pathology consultant for Axdev Global Corp inc and as an expert advisory panel for Genomic Health in 2017. RML is co-founder and shareholder of MUSE Microscopy, Inc. and Histolix, Inc. RML served as consultant for Cell IDx, Inc., BriteSeed, Inc., ImmunoPhotonics, Inc., Pathology Watch, Inc. and Verily, Inc. FPL reports board meetings and conference support from Astrazeneca, Lilly, Novartis, Pfizer and Roche. JB served as consultant for Insight Genetics, Inc., BioNTech AG, Biotheranostics, Inc., Pfizer, Rna Diagnostics Inc., oncoXchange/MedcomXchange Communications Inc, Herbert Smith French Solicitors, OncoCyte Corporation. JB served as member of the scientific advisory board for MedcomXchange Communications Inc. JB reports honoraria from NanoString Technologies, Inc., Oncology Education, Biotheranostics, Inc., MedcomXchange Communications Inc. JB reports travel and accommodation expenses support from Biotheranostics, Inc., NanoString Technologies, Inc., Breast Cancer Society of Canada. JB received research funding from Thermo Fisher Scientific, Genoptix, Agendia, NanoString Technologies, Inc., Stratifyer GmbH, Biotheranostics, Inc. The remaining authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the British Columbia Cancer Agency’s Clinical Research Ethics Board (H10-03420).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Balazs Acs, Email: balazs.acs@ki.se.
David L. Rimm, Email: david.rimm@yale.edu
on behalf of the International Ki67 in Breast Cancer Working Group of the Breast International Group and North American Breast Cancer Group (BIG-NABCG):
Mitch Dowsett, Daniel F. Hayes, Lisa M. McShane, Kelley M. Kidwell, Torsten Nielsen, Samuel Leung, Balazs Acs, Indu Arun, Renaldas Augulis, Sunil S. Badve, Yalai Bai, Anita L. Bane, John M. S. Bartlett, Jane Bayani, Gilbert Bigras, Annika Blank, Signe Borgquist, Henk Buikema, Angela Chan, Martin C. Chang, Carsten Denkert, Robin L. Dietz, Andrew Dodson, Anna Ehinger, Matthew Ellis, Susan Fineberg, Margaret Flowers, Cornelia M. Focke, Chad Galderisi, Dongxia Gao, Abhi Gholap, Allen M. Gown, Carolina Gutierrez, Douglas J. Hartman, Johan Hartman, Judith C. Hugh, Anagha Jadhav, Elizabeth N. Kornaga, Zuzana Kos, Hans Kreipe, Anne-Vibeke Lænkholm, Arvydas Laurinavicius, Richard Levenson, Mauro Mastropasqua, Takuya Moriya, Sharon Nofech-Mozes, C. Kent Osborne, Hongchao Pan, Liron Pantanowitz, Ernesta Paola Neri, Frédérique M. Penault-Llorca, Mei-Yin Polley, Tammy Piper, Mary Anne Quintayo, Tilman T. Rau, David L. Rimm, Stefan Reinhard, Stephanie Robertson, Jason Ruan, Takashi Sakatani, Roberto Salgado, Lois Shepherd, Ian Smith, Joseph Sparano, Melanie Spears, Malini Srinivasan, Jane Starczynski, Tomoharu Sugie, Austin Todd, Bert van der Vegt, Giuseppe Viale, Shakeel Virk, Yihong Wang, Hua Yang, Lila A. Zabaglo, Zhiwei Zhang, and Inti Zlobec
Supplementary information
The online version contains supplementary material available at 10.1038/s41379-022-01104-9.
References
- 1.Smith I, Robertson J, Kilburn L, Wilcox M, Evans A, Holcombe C, et al. Long-term outcome and prognostic value of Ki67 after perioperative endocrine therapy in postmenopausal women with hormone-sensitive early breast cancer (POETIC): an open-label multicentre parallel-group randomised phase 3 trial. Lancet Oncol 21, 1443–1454 (2020) [DOI] [PMC free article] [PubMed]
- 2.Nielsen TO, Leung SCY, Rimm DL, Dodson A, Acs B, Badve S, et al. Assessment of Ki67 in breast cancer: updated recommendations from the International Ki67 in Breast Cancer Working Group. J Natl Cancer Inst 113, 808–819 (2020) [DOI] [PMC free article] [PubMed]
- 3.Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 34, 1134–1150 (2016) [DOI] [PMC free article] [PubMed]
- 4.Andre F, Ismaila N, Henry NL, Somerfield MR, Bast RC, Barlow W, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: ASCO Clinical Practice Guideline Update-Integration of Results From TAILORx. J Clin Oncol 37, 1956–1964 (2019) [DOI] [PubMed]
- 5.Acs B, Fredriksson I, Rönnlund C, Hagerling C, Ehinger A, Kovács A, et al. Variability in breast cancer biomarker assessment and the effect on oncological treatment decisions: a nationwide 5-year population-based study. Cancers (Basel) 13, 1166 (2021) [DOI] [PMC free article] [PubMed]
- 6.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of ki67 in breast cancer: recommendations from the international ki67 in breast cancer working group. J Natl Cancer Inst 103, 1656–1664 (2011) [DOI] [PMC free article] [PubMed]
- 7.Polley MY, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, et al. An international Ki67 reproducibility study. J Natl Cancer Inst 105, 1897–1906 (2013) [DOI] [PMC free article] [PubMed]
- 8.Polley MY, Leung SC, Gao D, Mastropasqua MG, Zabaglo LA, Bartlett JM, et al. An international study to increase concordance in Ki67 scoring. Mod Pathol 28, 778–786 (2015) [DOI] [PubMed]
- 9.Leung SCY, Nielsen TO, Zabaglo L, Arun I, Badve SS, Bane AL, et al. Analytical validation of a standardized scoring protocol for Ki67: phase 3 of an international multicenter collaboration. NPJ Breast Cancer 2, 16014 (2016) [DOI] [PMC free article] [PubMed]
- 10.Leung SCY, Nielsen TO, Zabaglo LA, Arun I, Badve SS, Bane AL, et al. Analytical validation of a standardised scoring protocol for Ki67 immunohistochemistry on breast cancer excision whole sections: an international multicentre collaboration. Histopathology 75, 225–235 (2019) [DOI] [PubMed]
- 11.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017) [DOI] [PMC free article] [PubMed]
- 12.Acs B, Pelekanou V, Bai Y, Martinez-Morilla S, Toki M, Leung SCY, et al. Ki67 reproducibility using digital image analysis: an inter-platform and inter-operator study. Lab Invest 99, 107–117 (2019) [DOI] [PubMed]
- 13.Aung TN, Acs B, Warrell J, Bai Y, Gaule P, Martinez-Morilla S, et al. A new tool for technical standardization of the Ki67 immunohistochemical assay. Mod Pathol 34, 1261–1270 (2021) [DOI] [PMC free article] [PubMed]
- 14.Malpica N, de Solorzano CO, Vaquero JJ, Santos A, Vallcorba I, Garcia-Sagredo JM, et al. Applying watershed algorithms to the segmentation of clustered nuclei. Cytometry 28, 289–297 (1997) [DOI] [PubMed]
- 15.Kos Z, Dabbs DJ. Biomarker assessment and molecular testing for prognostication in breast cancer. Histopathology 68, 70–85 (2016) [DOI] [PubMed]
- 16.Kayser K, Gortler J, Borkenfeld S, Kayser G. How to measure diagnosis-associated information in virtual slides. Diagn Pathol 6, Suppl 1 S9 (2011) [DOI] [PMC free article] [PubMed]
- 17.Robertson S, Azizpour H, Smith K, Hartman J. Digital image analysis in breast pathology-from image processing techniques to artificial intelligence. Transl Res 194, 19–35 (2018) [DOI] [PubMed]
- 18.Wienert S, Heim D, Kotani M, Lindequist B, Stenzinger A, Ishii M, et al. CognitionMaster: an object-based image analysis framework. Diagn Pathol 8, 34 (2013) [DOI] [PMC free article] [PubMed]
- 19.Laurinavicius A, Plancoulaine B, Laurinaviciene A, Herlin P, Meskauskas R, Baltrusaityte I, et al. A methodology to ensure and improve accuracy of Ki67 labelling index estimation by automated digital image analysis in breast cancer tissue. Breast Cancer Res 16, R35 (2014) [DOI] [PMC free article] [PubMed]
- 20.Klauschen F, Wienert S, Schmitt WD, Loibl S, Gerber B, Blohmer JU, et al. Standardized Ki67 diagnostics using automated scoring-clinical validation in the gepartrio breast cancer study. Clin Cancer Res 21, 3651–3657 (2015) [DOI] [PubMed]
- 21.Stalhammar G, Fuentes Martinez N, Lippert M, Tobin N,P Molholm I, Kis L, et al. Digital image analysis outperforms manual biomarker assessment in breast cancer. Mod Pathol 29, 318–329 (2016) [DOI] [PubMed]
- 22.Acs B, Madaras L, Kovacs KA, Micsik T, Tokes AM, Gyorffy B, et al. Reproducibility and prognostic potential of Ki-67 proliferation index when comparing digital-image analysis with standard semi-quantitative evaluation in breast cancer. Pathol Oncol Res 24, 115–127 (2018) [DOI] [PubMed]
- 23.Zhong F, Bi R, Yu B, Yang F, Yang W, Shui R. A comparison of visual assessment and automated digital image analysis of Ki67 labeling index in breast cancer. PLoS One 11, e0150505 (2016) [DOI] [PMC free article] [PubMed]
- 24.Stalhammar G, Robertson S, Wedlund L, Lippert M, Rantalainen M, Bergh J, et al. Digital image analysis of Ki67 in hot spots is superior to both manual Ki67 and mitotic counts in breast cancer. Histopathology 72, 974–989 (2018) [DOI] [PubMed]
- 25.Rimm DL, Leung SCY, McShane LM, Bai Y, Bane AL, Bartlett JMS, et al. An international multicenter study to evaluate reproducibility of automated scoring for assessment of Ki67 in breast cancer. Mod Pathol 32, 59–69 (2019) [DOI] [PubMed]
- 26.Robertson S, Acs B, Lippert M, Hartman J. Prognostic potential of automated Ki67 evaluation in breast cancer: different hot spot definitions versus true global score. Breast Cancer Res Treat 183, 161–175 (2020) [DOI] [PMC free article] [PubMed]
- 27.Koopman T, Buikema HJ, Hollema H, de Bock GH, van der Vegt B. Digital image analysis of Ki67 proliferation index in breast cancer using virtual dual staining on whole tissue sections: clinical validation and inter-platform agreement. Breast Cancer Res Treat 169, 33–42 (2018) [DOI] [PMC free article] [PubMed]
- 28.Plancoulaine B, Laurinaviciene A, Herlin P, Besusparis J, Meskauskas R, Baltrusaityte I, et al. A methodology for comprehensive breast cancer Ki67 labeling index with intra-tumor heterogeneity appraisal based on hexagonal tiling of digital image analysis data. Virchows Arch 467, 711–722 (2015) [DOI] [PubMed]
- 29.Arima N, Nishimura R, Osako T, Nishiyama Y, Fujisue M, Okumura Y, et al. The importance of tissue handling of surgically removed breast cancer for an accurate assessment of the Ki-67 index. J Clin Pathol 69, 255–259 (2016) [DOI] [PMC free article] [PubMed]
- 30.Mengel M, von Wasielewski R, Wiese B, Rüdiger T, Müller-Hermelink HK, Kreipe H. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J Pathol 198, 292–299 (2002) [DOI] [PubMed]
- 31.Benini E, Rao S, Daidone MG, Pilotti S, Silvestrini R. Immunoreactivity to MIB-1 in breast cancer: methodological assessment and comparison with other proliferation indices. Cell Prolif 30, 107–115 (1997) [DOI] [PMC free article] [PubMed]
- 32.Kalvala J, Parks RM, Green AR, Cheung KL. Concordance between core needle biopsy and surgical excision specimens for Ki-67 in breast cancer - a systematic review of the literature. Histopathology 80, 468–484 (2022) [DOI] [PubMed]
- 33.Janeva S, Parris TZ, Nasic S, De Lara S, Larsson K, Audisio RA, et al. Comparison of breast cancer surrogate subtyping using a closed-system RT-qPCR breast cancer assay and immunohistochemistry on 100 core needle biopsies with matching surgical specimens. BMC Cancer 21, 439 (2021). [DOI] [PMC free article] [PubMed]
- 34.Greer LT, Rosman M, Mylander WC, Hooke J, Kovatich A, Sawyer K, et al. Does breast tumor heterogeneity necessitate further immunohistochemical staining on surgical specimens? J Am Coll Surg 216, 239–251 (2013) [DOI] [PubMed]
- 35.Chen X, Sun L, Mao Y, Zhu S, Wu J, Huang O, et al. Preoperative core needle biopsy is accurate in determining molecular subtypes in invasive breast cancer. BMC Cancer 13, 390 (2013) [DOI] [PMC free article] [PubMed]
- 36.Chen X, Zhu S, Fei X, Garfield DH, Wu J, Huang O, et al. Surgery time interval and molecular subtype may influence Ki67 change after core needle biopsy in breast cancer patients. BMC Cancer 15, 822 (2015) [DOI] [PMC free article] [PubMed]
- 37.Kalkman S, Bulte JP, Halilovic, A Bult P, van Diest PJ. Brief fixation does not hamper the reliability of Ki67 analysis in breast cancer core-needle biopsies: a double-centre study. Histopathology 66, 380–387 (2015) [DOI] [PubMed]
- 38.Al Nemer A. The performance of Ki-67 labeling index in different specimen categories of invasive ductal carcinoma of the breast using 2 scoring methods. Appl Immunohistochem Mol Morphol 25, 86–90 (2017) [DOI] [PubMed]
- 39.Pölcher M, Braun M, Tischitz M, Hamann M, Szeterlak N, Kriegmair A, et al. Concordance of the molecular subtype classification between core needle biopsy and surgical specimen in primary breast cancer. Arch Gynecol Obstet 304, 783–790 (2021) [DOI] [PubMed]
- 40.Liu M, Tang SX, Tsang JYS, Shi YJ, Ni YB, Law BKB, et al. Core needle biopsy as an alternative to whole section in IHC4 score assessment for breast cancer prognostication. J Clin Pathol 71, 1084–1089 (2018) [DOI] [PubMed]
- 41.You K, Park S, Ryu JM, Kim I, Lee SK, Yu J, et al. Comparison of core needle biopsy and surgical specimens in determining intrinsic biological subtypes of breast cancer with immunohistochemistry. J Breast Cancer 20, 297–303 (2017) [DOI] [PMC free article] [PubMed]
- 42.Chen J, Wang Z, Lv Q, Du Z, Tan Q, Zhang D, et al. Comparison of core needle biopsy and excision specimens for the accurate evaluation of breast cancer molecular markers: a report of 1003 cases. Pathol Oncol Res 23, 769-775 (2017) [DOI] [PubMed]
- 43.Focke CM, Decker T, van Diest PJ. Reliability of the Ki67-labelling index in core needle biopsies of luminal breast cancers is unaffected by biopsy volume. Ann Surg Oncol 24, 1251–1257 (2017) [DOI] [PubMed]
- 44.Meattini I, Bicchierai G, Saieva C, De Benedetto D, Desideri I, Becherini C, et al. Impact of molecular subtypes classification concordance between preoperative core needle biopsy and surgical specimen on early breast cancer management: Single-institution experience and review of published literature. Eur J Surg Oncol 43, 642–648 (2017) [DOI] [PubMed]
- 45.Robertson S, Rönnlund C, de Boniface J, Hartman J. Re-testing of predictive biomarkers on surgical breast cancer specimens is clinically relevant. Breast Cancer Res Treat 174, 795–805 (2019) [DOI] [PMC free article] [PubMed]
- 46.Clark BZ, Onisko A, Assylbekova B, Li X, Bhargava R, Dabbs DJ. Breast cancer global tumor biomarkers: a quality assurance study of intratumoral heterogeneity. Mod Pathol 32, 354–366 (2019) [DOI] [PubMed]
- 47.Ekholm M, Grabau D, Bendahl PO, Bergh J, Elmberger G, Olsson H, et al. Highly reproducible results of breast cancer biomarkers when analysed in accordance with national guidelines - a Swedish survey with central re-assessment. Acta Oncol 54, 1040–1048 (2015) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.