Abstract
Background
Inflammatory Bowel Diseases with its complexity and heterogeneity could benefit from the increased application of Artificial Intelligence in clinical management.
Aim
To accurately predict adverse outcomes in patients with IBD using advanced computational models in a nationally representative dataset for potential use in clinical practice.
Methods
We built a training model cohort and validated our result in a separate cohort. We used LASSO and Ridge regressions, Support Vector Machines, Random Forests and Neural Networks to balance between complexity and interpretability and analyzed their relative performances and reported the strongest predictors to the respective models. The participants in our study were patients with IBD selected from The OptumLabs® Data Warehouse (OLDW), a longitudinal, real-world data asset with de-identified administrative claims and electronic health record (EHR) data.
Results
We included 72,178 and 69,165 patients in the training and validation set, respectively. In total, 4.1% of patients in the validation set were hospitalized, 2.9% needed IBD-related surgeries, 17% used long-term steroids and 13% of patients were initiated with biological therapy. Of the AI models we tested, the Random Forest and LASSO resulted in high accuracies (AUCs 0.70–0.92). Our artificial neural network performed similarly well in most of the models (AUCs 0.61–0.90).
Conclusions
This study demonstrates feasibility of accurately predicting adverse outcomes using complex and novel AI models on large longitudinal data sets of patients with IBD. These models could be applied for risk stratification and implementation of preemptive measures to avoid adverse outcomes in a clinical setting.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10620-022-07506-8.
Keywords: Artificial intelligence, Machine learning, Precision medicine, Big data, Inflammatory bowel diseases
Introduction
The burden of Inflammatory Bowel Disease (IBD) on patients as well as society is large. IBD is a progressive disease with a destructive character and is associated with substantial healthcare costs [1, 2]. Prevention of flares is key to preventing disease progression [3–5]. However, the disease course is unpredictable and reliable risk factors for flares are difficult to identify [5]. Finding an approach that identifies patients at risk for disease progression would help to better fine-tune treatment strategies in order to prevent adverse outcomes such as hospitalizations, long-term steroid use, the initiation of expensive biologics and surgeries. This could help reduce the substantial costs associated with IBD care and improve long-term outcomes [6].
The development of healthcare technologies driven by Artificial Intelligence (AI) is expected to see a growth of over $10 billion in just the next 5 years [7]. With the explosive amount of Electronic Medical Records (EMRs), having doubled in size since 2005, studying patient data is easier now than in any previous era [8]. By taking full advantage of EMR data and, other forms of patient information (e.g., wearables, microbiome/genetic testing, e-health applications, imaging), data-driven treatment plans targeted at the disease and individual level could be introduced. The opportunities to construct new strategies and technologies that turn these data into actionable provider recommendations are expected to rapidly grow, as showcased by the immense amount of funding that is going into companies that use AI for healthcare [9].
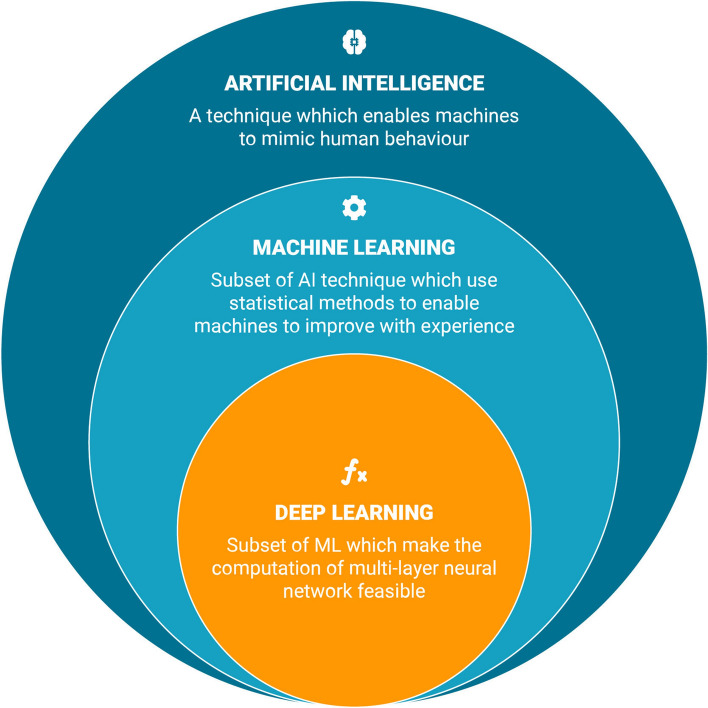
Recently, there have been multiple studies that were able to accurately and inexpensively use a subset of AI known as Machine Learning (ML) to predict a variety of outcomes and create distinct classifications for patients with IBD (Fig. 1) [10–18]. Han et al. created a gene-based ML classification model to better differentiate between patients with Crohn’s disease (CD) and ulcerative colitis (UC) [16]. Wei et al. were able to successfully create a genotype-based risk prediction model for IBD using a large sample of genetic data [14]. Beyond gene-based data, researchers have used AI models with insurance claims data to accurately predict IBD-related hospitalization or steroid use within a six-month period [10]. This ML approach outperformed more costly biomarker methods of predicting negative outcomes, such as testing for fecal calprotectin. These kind of AI approaches to healthcare have not been limited to IBD [19–23].
Fig. 1.
Context of the different models. AI is the broad umbrella term of techniques which enables machines to mimic human behavior, when talking about predictive models we usually refer to machine learning which is a subset of AI that uses statistical methods to improve the accuracy of their outcome with experience. Deep Learning is a subset that makes the computation of multi-layer neural networks feasible and thus improving the accuracy even further
However, studies using the most straightforward data resource, which are administrative databases due to the standardized format and accessibility, to build data-driven predictive models for patients with IBD were limited in their generalizability. The data came from public health insurance records, while the majority (67.2%) of US citizens use private insurance, and their samples have limited geographic spread [13, 24]. Additionally, these studies have not attempted to predict other costly negative outcomes such as IBD-related surgeries [10, 13]. To our knowledge, no other study has attempted to apply this ML approach to a larger set of private insurance claims data or use novel deep learning methods such as neural networks. Our goal is to assess the feasibility and performance of various ML models in early prediction of adverse outcomes for patients with IBD, including IBD-related surgeries, using a large private insurance claims dataset.
Methods
Study Objectives
The main objective of this study was to assess if variables extracted from insurance claims can accurately predict negative health outcomes in IBD. To achieve this, we assessed the performance of different Machine Learning and Deep Learning models to and compared the performances of the aforementioned models using different performance outcomes.
Data Collection
Deidentified medical, pharmacy and facility claims, were extracted from The OptumLabs® Data Warehouse (OLDW) includes claims from commercially insured individuals and Medicare Advantage beneficiaries (≥ 65 years old) who are representative of the US population with regards to geographical spread, age and race [25]. Patient-identifying data are removed from the OLDW by OptumLabs before access is granted to investigators. Therefore, this study is not considered human subjects research and is exempt from Institutional Review Board (IRB) regulation. Access to the data was granted as part of an academic collaboration between OLDW and UCLA.
We created two datasets: a training cohort and a validation cohort. The training cohort contained all patients that were continuously enrolled in their insurance plan between January 1, 2015 and December 31, 2016. The validation cohort includes patients who were continuously enrolled between January 1, 2016 and December 31, 2017. In each cohort, we aimed to predict outcomes in the second year (follow-up) using claims data available in the first year (baseline). We chose for a 1-year follow-up because enrollment cycles are annual and because IBD typically flares up in time frames of multiple months so we aim to pick up negative outcomes in this period.
Population
Patients with IBD were identified using a combination of inpatient and outpatient claims. Patients were included if they had at least two medical claim with diagnosis codes for IBD (International Classification of Diseases, Ninth Revision, Clinical Modification [ICD-9] 555.x or 556.x) OR one IBD-related medical claim and one pharmacy claim for IBD-related medication (Supplementary Table 4) in the first year of data.
To ensure enrollees had a specified period of continuous enrollment and the inability to identify an outcome was not due to missing claims data (e.g., enrollee claim was administered by another payor), a continuous enrollment code provided by OLDW was used to make sure the cohorts were continuously enrolled with the respective payor.
Predictive Variables
We constructed 108 variables related to IBD-related care using the claims in the first year of each dataset. These variables were defined based on definitions previously described by Vaughn et al. [13].The variables include the number of IBD-related claims, hospitalizations, emergency department (ED) visits, office visits, procedures, laboratory and imaging tests, medication use, relapse rate, and comorbidities (for a complete list, see Supplementary Table 1) [13]. Since the OptumLabs Data Warehouse is a curated database of claims, missing data are not an issue when constructing these variables.
Model Development
In our models, we aimed to predict IBD-related hospitalizations, initiation of biologics, long-term steroid use, and IBD-related surgery in the second year of the data (follow-up) using the 108 utilization-events that occurred in the prior year (baseline). There is consensus in the literature that these are negative outcomes for IBD that should be avoided [5, 6]. IBD-related hospitalizations were defined as the presence of any claim for an IBD-related inpatient hospital stay [13]. Initiation of biologics was defined as a pharmacy or medical claim for adalimumab, certolizumab pegol, infliximab or natalizumab in the second year, with no claim for that medicine in the first year. Long-term steroid use was defined as the use of hydrocortisone, prednisolone, dexamethasone, prednisone and/or methylprednisolone during a consecutive period longer than 90 days based on pharmacy and medical claims. IBD-related surgery was defined as any claim with a Current Procedural Terminology (CPT) code specific to an IBD related surgery (See supplementary Table 2 for a full overview).
Logistic and Machine Learning Models
After these datasets were constructed for both cohorts of patients, we trained several logistic regression and machine learning models: a Ridge regression, a LASSO regression, a Support Vector Machine, a Random Forest model, and a Neural Network [26] (See Table 1). Each of these models was trained to predict the probability of a patient incurring a specific negative health outcome in the next year, using the 108 variables from the previous year. We trained five models on the training set of patients and tested them on the validation set.
Table 1.
Introduction and description of different models
| Model | Explanation | Method | Advantages | Disadvantages |
|---|---|---|---|---|
| Ridge Logistic | This method creates a model that is not perfectly fit, or overfit, to the data in a given training set. In doing so, it reduces variance and makes the model a better predictor of data points outside of the training set | Regression |
Can reduce overfitting Shrinks effects towards 0 Fast/easy to implement |
Simplistic representation may be far from reality Assumptions may be difficult to justify with many predictors |
| LASSO Logistic | This method attempts to do the same thing as Ridge Regression but uses slightly different mathematical formulas that make it better in certain situations | Regression |
Can reduce overfitting Performs variable selection Fast/easy to implement |
Simplistic representation may be far from reality Variable selection is not robust to multicollinearity |
| Support Vector Machine | Attempts to find the largest separation between two groups. Sometimes the space of observations has to be transformed to find a clear separation | Machine learning |
Works well with many predictors Makes prediction easy by clearly segmenting population |
Lack of a clear separation can lead to poor performance Requires long training times for big data |
| Random Forest | Random forest is a collection of decision trees trained on different subsets of the data. Each decision tree decides the best places to cut so that observations from the same class fall on the same side of the cut | Machine learning |
Performs variable selection Good performance for linear and nonlinear relationships Fast/easy to implement |
Difficult to interpret Prone to overfitting |
| Neural Network | Neural networks consists of layers of nested linear models (neurons) with a nonlinear transformation (activation) after each layer. The output is often the probability that a given observation is a success | Deep learning |
Captures complex nonlinear relationships Fully utilizes big data |
Difficult to implement Requires many small decisions that can greatly affect performance |
A explanation of the different models used in our analysis is displayed below highlighting the advantages and disadvantages
Ridge regression and LASSO are regression techniques that place a penalty on the model coefficients to ensure that we do not overfit to the training data, we chose these two to avoid overfitting and to aim for simpler models with interpretable coefficients. Support Vector Machines attempt to separate the patients in the training set who did experience the negative health outcome from those who did not with the largest margin possible. After experimenting with various kernels, we decided on the Gaussian radial basis function. A Random Forest model generates a collection of decision trees, in which each decision tree attempts to find a cut point for each predictor that best separates patients who experienced the negative outcome from those that did not. The cut that achieves the best separation is added to the tree, and this process is repeated for each of the two resulting slices of the data, and so on until some minimum number of patients are left in each slice. To capture the nuances in the data, each tree is trained and evaluated on random subsets of the data drawn with replacement. To avoid having too many correlated trees that choose the same best predictors, at each split in the tree only a fraction of the predictors is considered.
Lastly, Neural Networks can identify complex nonlinear patterns in the data. These models consist of several imbedded linear functions, known as hidden layers, wrapped in nonlinear “activation” functions. These nonlinearities in the model work to capture the complicated relationships between the predictors and the probability that a patient will experience the negative outcome. The choice of activation function at each layer plays a big role in determining how well this relationship will be captured by the resulting model. After experimenting with several options, we found that a mix of standard and parametric Rectified Linear Units (ReLUs) performs the best. The last hidden layer is followed by a sigmoid activation function, which outputs a normalized score that we can interpret as the probability that the patient will experience the outcome.
Model Selection Rationale
We trained a battery of machine learning models to discriminate between patients who experienced negative outcomes and those that did not while emphasizing the clinical insights and practical significance that could be understood from the result. To choose the set of base models, on which we would improve with regularization and hyperparameter tuning, we considered the current gap between an algorithm’s complexity/performance and its explainability. We chose several simple linear models with different regularization penalties as they are easy to interpret and align with existing clinical knowledge but often miss complex associations between the variables. We also explored a variety of neural network architectures and tuning procedures to understand the extent to which nonlinear relationships in the data could be exploited to improve performance. These models are infamously difficult to understand, as theoretical notions such as statistical significance are difficult to define. With these two extremes covered the SVM and random forest models, we considered attempt to strike a balance between performance and interpretability by blending simple structures with complex training procedures. By comparing across models that cover this spectrum, we can find complicated relationships that lead to solid predictions and warrant prospective validation as well as simpler associations that are easy to validate through expert knowledge. We avoided training many complex models for the sake of isolating clear, practical relationships.
Performance of the Models
For each model, we obtain a prediction for each patient in the validation set. A series of cutoffs were then considered, and predictions above the cutoff were labeled as predicted true cases. With these labels, the true positive (sensitivity) and true negative (specificity) rates of the model were calculated based on which receiver operating curves (ROC) were constructed. The area under the ROC curve (AUC) for a specific model quantifies the overall certainty with which the model can predict outcomes at different cut-offs. The single cutoff with the highest geometric average of sensitivity and specificity was selected for each model and specificity, and sensitivity values were reported [27]. We defined a result < 0.60 as poor, scores between 0.6 and 0.8 as fair and scores above 0.8 as good.
Additionally, we calculated the Brier Score which measures the correctness of a model’s predictions by summing the differences between the predicted probability of an observation belonging to a class and its actual class label. A low Brier score indicates that the model on average confidently places observations into the correct class. While the AUC quantifies the accuracy of the model, the Brier score quantifies the certainty of the model. For example, if a model assigns a score of 0.51 to every at-risk patient and 0.49 to all other patients, then a cutoff of 0.5 will correctly classify every patient in the validation set and produce a good AUC, but it does not give us a sense of how certain we are about the predictions. The Brier score solves this by measuring the difference between the scores the model predicts (e.g., 0.51) and the true labels (e.g., 1). If all scores are closer to the true label, then the Brier score will be close to 0. In this way, the Brier score can be used to select the best model from a set with high AUC when the goal is to give not only accurate, but also strong predictions. This is relevant when extrapolating these results to potential meaningful use in a clinical setting.
Feature Importance (Except SVM)
The relative importance of the predictive variables in the different models was calculated. For the LASSO and Ridge regression, we looked at the magnitude of coefficients and their respective p values and present the odds ratio. For the Random Forest, we measured the importance of each variable by quantifying the change in accuracy of the final predictions after the variable is added to a tree. Larger values indicate the variable is more important. Since the Support Vector Machines did not result in accurate predictions, we did not investigate the relative importance of the predictors. For the neural network, we randomly shuffled the observations of a particular variable in the validation set and measured the change in the model’s AUC. Variables that create the largest negative change in AUC are defined as the most important.
TRIPOD Statement
Our methodology and research objectives were subject to the TRIPOD (Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis) statement which includes a 22-item checklist, which aims to improve the reporting of studies developing, validating, or updating a prediction model, whether for diagnostic or prognostic purposes [28]. See supplementary Table 5 for a full overview.
Tools and Software
Statistical analyses were performed using statistical package program R 3.4.0 and Python.
Results
Population
We included 72,178 patients in our training set and 69,165 patients in our validation set. For both sets, the claims from the baseline year (first) were used to generate the 108 predictive features, and the follow-up year (second) was used to create our four main outcomes.
Demographics
The mean age of the populations was around 48 years (SD 16.8) for both cohorts, and gender was distributed fairly evenly with approximately 52% being female. Both cohorts were predominantly non-Hispanic whites (66% in the training cohort, and 64% in the validation cohort). Looking at medications, biologics use was around 13% for both cohorts in the baseline year, and steroid use was around 27% for both cohorts. We found that 3% of patients in both cohorts had an IBD-related surgery in the baseline year and 6% had an IBD-related hospitalization (Table 2). For a complete overview of the extracted variables during the baseline years of both cohorts, including the average number of hospitalizations, emergency department (ED) visits, insurance coverage, office visits, procedures, laboratory and imaging tests, and medication use, see Supplementary Table 1.
Table 2.
Baseline demographics and variables of training and validation cohorts in the baseline year
| Variable | Training Set Baseline (2015) N = 72,178 |
Validation Set Baseline (2016) N = 69,165 |
|---|---|---|
| Age, mean (SD) | 48.5 years (16.8) | 47.9 years (16.5) |
| Female Gender, n (%) | 38,254 (53%) | 35,966 (52%) |
| Race, n (%) | ||
| White | 47,710 (66.1%) | 44,473 (64.3%) |
| Unknown | 12,776 (17.7%) | 12,381 (17.9%) |
| Black | 5052 (7%) | 5672 (8.2%) |
| Hispanic | 4692 (6.5%) | 4219 (6.1%) |
| Asian | 1949 (2.7%) | 2490 (3.6%) |
| Hospitalizations and ER visits in baseline year, n (%) | ||
| Any ER Visit (#103) | 10,827 (15%) | 11,066 (16%) |
| Any Hospitalization (#97) | 4331 (6%) | 4150 (6%) |
| Any IBD-related Hospitalization (#100) | 3609 (5%) | 3458 (5%) |
| Any IBD-related ER Visit (#105) | 2887 (4%) | 2767 (4%) |
| Any IBD-related surgery (#64) | 2165 (3%) | 2075 (3%) |
| Medication use during baseline year, n (%) | ||
| Any IBD Medication use (#1) | 28,149 (39%) | 15,908 (23%) |
| Any Aminosalicylate use (#2&6) | 12,270 (17%) | 11,758 (17%) |
| Any Antibiotic use (#8) | 7218 (10%) | 6917 (10%) |
| Any Corticosteroid use (#11,14,17) | 18,766 (26%) | 18,675 (27%) |
| Any Immunomodulator use (#21, 24, 27) | 5774 (8%) | 5533 (8%) |
| Any Biologics use (#42) | 8661 (12%) | 8991 (13%) |
| Adverse outcomes follow-up year | Follow-up year (2016) | Follow-up year (2017) |
|---|---|---|
| IBD-related hospitalizations | 3392 (4.70%) | 2863 (4.14%) |
| Initiation of biologics | 8661 (12%) | 9199 (13.3%) |
| Long-term steroid Use | 11,332 (15.7%) | 11,758 (17%) |
| IBD-related surgery | 2454 (3.4%) | 2006 (2.9%) |
# Refers to the corresponding feature in Supplementary Table 1
In the training cohort, 3392 (4.7%) patients had an IBD-related hospitalization, 2454 (3.4%) had IBD-related surgery, 11,332 (15.7%) used long-term steroids, and 8661 (12.0%) patients started biological therapy during the one year of follow-up (Table 2). In the validation cohort, 2863 (4.1%) patients had an IBD-related hospitalization, 2006 (2.9%) had an IBD-related surgery, 11,758 (17.0%) used long-term steroids, and 9199 (13.3%) of patients started biological therapy during the one year of follow-up (Table 2).
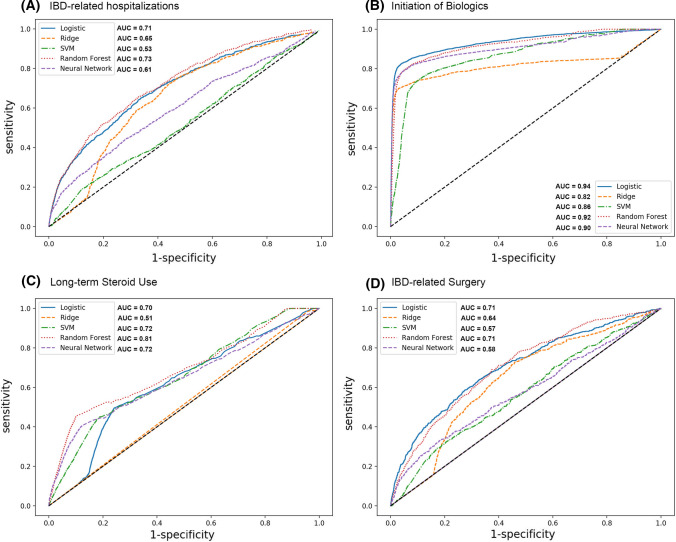
Performance the Validation Model
We included 72,178 patients in our training set (data from 2015 to 2016) and 69,165 patients in our validation set (date from 2016 to 2017). For the prediction of IBD-related hospitalizations, the Random Forest model performed most optimally with an AUC of 0.73 (66% sensitivity, 67% specificity) and a Brier score of 0.21 (See Table 3 and Fig. 2). For the prediction of Initiation of biologics, the LASSO regression performed best with an AUC of 0.94 (83% sensitivity, 96% specificity) and a Brier Score of 0.05, followed by the Random Forest with an AUC 0.92 (82% Sensitivity, 92% Specificity) and Brier Score of 0.10. Similarly, the Random Forest performed best for the prediction of Long-term steroid use with an AUC of 0.81 (48% Sensitivity, 86% Specificity) and Brier score of 0.15. For the prediction of IBD-related surgery, the LASSO Regression and Random Forest had the highest AUC, 0.71 and Brier scores of 0.22 and 0.21, respectively.
Table 3.
Performance of the different models for the four main outcomes
| Sensitivity | Specificity | AUC | Brier Score* | |
|---|---|---|---|---|
| IBD-related Hospitalizations | ||||
| Ridge Logistic | 72% | 56% | 0.65 | 0.95 |
| LASSO Logistic | 65% | 66% | 0.71 | 0.17 |
| Support Vector Machine | 54% | 48% | 0.53 | 0.04 |
| Random Forest | 66% | 67% | 0.73 | 0.21 |
| Neural Network | 57% | 58% | 0.61 | 0.04 |
| Initiation of Biologics | ||||
| Ridge Logistic | 70% | 97% | 0.82 | 0.07 |
| LASSO Logistic | 83% | 96% | 0.94 | 0.05 |
| Support Vector Machine | 75% | 89% | 0.86 | 0.10 |
| Random Forest | 82% | 92% | 0.92 | 0.10 |
| Neural Network | 81% | 93% | 0.90 | 0.05 |
| Long-term Steroid Use | ||||
| Ridge Logistic | 99% | 4% | 0.51 | 0.83 |
| LASSO Logistic | 52% | 74% | 0.70 | 0.83 |
| Support Vector Machine | 50% | 74% | 0.72 | 0.13 |
| Random Forest | 48% | 86% | 0.81 | 0.15 |
| Neural Network | 50% | 74% | 0.72 | 0.16 |
| IBD-related surgery | ||||
| Ridge Logistic | 72% | 55% | 0.64 | 0.97 |
| LASSO Logistic | 64% | 67% | 0.71 | 0.22 |
| Support Vector Machine | 54% | 55% | 0.57 | 0.03 |
| Random Forest | 69% | 63% | 0.71 | 0.21 |
| Neural Network | 50% | 63% | 0.58 | 0.03 |
*The Brier score measures the correctness of a model’s predictions by summing the differences between the predicted probability of an observation belonging to a class and its actual class label. A low Brier score indicates that the model on average confidently places observations into the correct class
Fig. 2.
Overview of the performance of the different models for the four main outcomes
Overall, the Random Forest resulted in high AUCs for all outcomes, as did the LASSO regression. The Neural Network performed well for some outcomes, but not others. The Support Vector Machine and Ridge regressions, on the other hand, consistently had lower performance than other models. Of the four outcomes included, the models were able to predict the initiation of biologics with the highest accuracy, while IBD-related surgery was the most challenging to predict.
Feature Importance
The relative importance of the predictive variables (Supplementary Table 1) in the different models was calculated except the SVM because of its poor performance. To predict IBD-related hospitalizations, long-term steroid use and IBD-related surgeries were strong predictors in both the LASSO and Ridge Regressions. Interestingly, the intensity of healthcare utilization as measured by the number of claims or office visits was the strongest predictors in the Random Forest model, which resulted in similar accuracy compared to the regression models. In the Neural Network, on the other hand, medication use variables were the most important predictors, but with much lower accuracy, indicating that this model was unable to identify the strongest relationship with IBD-related hospitalizations (Table 3).
Regarding initiation of biologics, across all models the use of previous steroids was strongly predictive of a patient being initiated on biologics. The LASSO and Ridge Regressions also found previous CRP laboratory test and IBD surgeries as strong predictors as well. The random forest, which had the highest accuracy overall, found more heterogeneous predictors including ED visits, number of upper endoscopies and X-ray, whereas the neural network mostly found previous use of steroids as the strongest predictor.
Concerning long-term steroid use, the regression models again found previous episodes of IBD medication use to be the strongest predictors. The random forest had the highest accuracy and found medical procedures such as imaging and laboratory tests and ED visits amongst one of the most predictive features. Similar to initiation of biologics, the neural network found episodes and use of IBD medication, in this particular instance aminosalicylates as the strongest predictor.
Lastly, for our fourth outcome IBD-related surgery we found comparable patterns within the regression models showing similar results with episodes of long-term steroids, imaging studies, gastroenterology-related visits and severe disease being the greatest predictors. The random forest, which was again one of the best performing models, found infliximab use as the strongest predictor, followed by the total of numbers of IBD-related claims, indicating overall utilization was a strong predictor of IBD-related surgery. Interestingly, the neural net again found use of aminosalicylates as the most predictive feature.
Discussion
Important Findings
This study demonstrated that it was feasible to accurately predict adverse outcomes in complex computational models (machine and deep learning) on large (Big Data) and representative longitudinal claims data sets of patients with IBD. We analyzed traditional models including LASSO and Ridge regressions, machine learning methods such as Support Vector Machines and Random Forests but also included more novel methods like Neural Networks, and successfully compared their relative performance. Overall, the Random Forest made the best predictions across all outcomes, which might indicate that the relationships between the claim’s features are best captured by a Random Forest model and that this model framework might work best for claims predictions in general.
Regarding feature importance, it is worth noting that each model/outcome pair may have a different set of relevant features. The regression models overall had comparable findings, with the most predictive features of negative outcomes being largely related to medication use. The random forest had the highest accuracy overall but had more heterogeneous findings, being less limited to medication use as the most predictive feature but also including procedures such as imaging and laboratory tests as strong predictors. Lastly, the neural net had the most consistent findings across all outcomes, which were mostly medication use related. The difference in findings across the models would argue for the need to explore various models depending on the available data and the choice of outcomes. Based on the research objectives and available data, different models may be more or less appropriate to capture the relationships between the predictors and the outcomes. Furthermore, more novel methods such as neural networks should be further investigated and explored in order to increase accuracy and to examine if they can potentially expose correlations and nonlinear relationships that might not be found in more conventional methods.
Comparisons & Limitations
Several others have used claims data to predict IBD-related utilization events in specific IBD sub-populations. For instance, Waljee et al. applied their model to a set of Veteran’s Heath Administration data, which limited their sample to a 93% male and old (mean age 59 years) population [10]; furthermore, public insurance is only used by a minority of US population [24]. Other prior works that have used ML approaches on private insurance data have been limited by the geographic spread of their sample [13]. To our knowledge, this is the first study to utilize this ML-based prediction approach on a nationally representative IBD population. Additionally, different outcomes were used in some of these studies. Waljee et al. used a composite measure capturing both hospitalization and corticosteroid use, where we have split up these outcomes and checked for long-term steroid use. Their composite measure had an AUC of 0.85 and Brier score of 0.20. We found similar results in our Random Forest model with a AUC of 0.73 and Brier score of 0.21 for hospitalizations and 0.81 AUC and 0.15 Brier Score for long-term steroid use. Furthermore, to our knowledge, our study is the first to predict IBD-related surgery using claims data. Additionally, the use of novel deep learning methods such as Neural Networks has not been described previously in the IBD literature. These new methods should be further explored and reported on as they have the potential to unlock new opportunities for personalized management in IBD and also because of the fact that these models are now feasible to run because of the increased availability of Big Data and increased computational resources.
There are some limitations worth noting to this study. While a data-driven approach to healthcare has great potential to improve patient outcomes, there are some limitations to ML that are worth noting. For one, ML algorithms can only describe correlations between variables or features of interest, not necessarily causation [29]. Furthermore, assumptions are generally made about data sets when applying a given ML algorithm to it, which can narrow the scope of the model in real-world situations [29]. In our case, we pre-defined 108 variables to include in our model. Additionally, some outcomes may have a more complicated (i.e., nonlinear) relationship with the predictors, and the models we chose may not capture those relationships. Also, we did not include data from the EMR in our prediction model, and inclusion of clinical variables could improve the predictive accuracy. However, administrative databases are more readily accessible due to the standardized format and therefore remain a more straightforward source of data for these initiatives.
Applying Outcomes in the Daily Clinical Practice
There are several ways that our models can be impactful in daily clinical practice. First, the odds ratios provided by the linear models (ridge logistic and LASSO logistic) can be used to evaluate the risk of patients. For example, we found that risk of hospitalization is strongly linked to previous acute IBD surgeries. Specifically, all else being equal an acute IBD surgery increases the odds of a patient being hospitalized by a factor of more than 8.
Second, the complex models that pick up on detailed interactions between the features can be used to make precise risk assessments based on an individual patient’s data. As demonstrated by the accuracy of these models, these risk assessments can be used to flag patients that are likely to have a negative outcome with enough notice that providers have time to react and course correct. For example, if we consider a patient with a set of features similar to that of the average patient in the training dataset, we can use our models to find that the probability of this patient being hospitalized within the next year is approximately 0.41. This value can give us a sense of the risk assumed by the average patient with IBD. Patients whose risk far exceeds this value can be treated as high risk monitored more frequently for predictive markers like CRP of fecal calprotectin. Additionally, we found our accuracy to be comparable to established clinical monitoring markers. Studies have shown a pooled sensitivity and specificity of 78% and a sROC of 0.83 for fecal calprotectin to predict relapse of quiescent IBD [30] (Table 4).
Table 4.
Feature importance of the different models
| Ridge Logistic (AUC = 0.65; Brier score = 0.95) | OR | LASSO Logistic (AUC = 0.71; Brier score = 0.17) | OR | Random Forest (AUC = 0.73; Brier score = 0.21) | Neural Network (AUC = 0.61; Brier score = 0.04) | |
|---|---|---|---|---|---|---|
| IBD-related hospitalizations | ||||||
| 1 | #65 Number of acute IBD surgeries | 8.72 | #20 Episodes of long-term steroids | 1.96 | #44 Number of IBD claims | #102 Number of ED visits |
| 2 | #64 Any IBD surgeries | 2.74 | #88 Number of Clostridium difficile stool tests | 1.57 | #49 Number of office visits | #36 Any certolizumab used this year |
| 3 | #88 Number of Clostridium difficile stool tests | 2.24 | #65 Number of acute IBD surgeries | 1.52 | #47 Number of UC claims | #35 Episodes of infliximab |
| 4 | #20 Episodes of long-term steroids | 1.72 | #43 Number of episodes of biologics | 1.52 | #94 Total number of claims | #5 Any oral aminosalicylates used this year |
| 5 | #54 Any IBD-related GI visits | 1.61 | #84 Any MR scans this year | 1.51 | #96 Number of hospitalizations | #30 Any adalimumab used this year |
| Ridge Logistic (AUC = 0.82; Brier score = 0.07) | OR | LASSO Logistic (AUC = 0.94; Brier score = 0.05) | OR | Random Forest (AUC = 0.92; Brier score = 0.10) | Neural Network (AUC = 0.90; Brier score = 0.05) | |
|---|---|---|---|---|---|---|
| Initiation of biologics | ||||||
| 1 | #42 Any Biologics this year | 4.65 | #42 Any Biologics this year | 8.72 | #8 Any antibiotics used this year | #16 Episodes of rectal steroids |
| 2 | #13 Episodes of budesonide | 2.71 | #13 Episodes of budesonide | 2.74 | #103 Any ED visits this year | #17 Any systemic steroids used |
| 3 | #90 Any TB tested this year | 2.31 | #90 Any TB tested this year | 2.24 | #10 Episodes of antibiotics | #19 Episodes of systemic steroids |
| 4 | #64 Any IBD surgeries | 2.29 | #23 Episodes of thiopurines | 1.72 | #80 Any X-rays this year | #20 Episodes of long-term steroids |
| 5 | #23 Episodes of thiopurines | 2.14 | #67 Number of c-reactive protein tests | 1.61 | #59 Number of upper endoscopies | #21 Any thiopurines used this year |
| Ridge Logistic (AUC = 0.51; Brier score = 0.83) | OR | LASSO Logistic (AUC = 0.70; Brier score = 0.83) | OR | Random Forest (AUC = 0.81; Brier score = 0.15) | Neural Network (AUC = 0.72; Brier score = 0.16) | |
|---|---|---|---|---|---|---|
| Long-term steroid use | ||||||
| 1 | #20 Episodes of long-term steroids | 2.47 | #20 Episodes of long-term steroids | 2.52 | #91 Any influenza vaccine this year | #2 Any rectal aminosalicylates used this year |
| 2 | #23 Episodes of thiopurines | 2.01 | #1 Any IBD medication use | 1.61 | #103 Any ED visits this year | #7 Episodes of oral aminosalicylates |
| 3 | #38 Episodes of certolizumab | 1.89 | #8 Any antibiotics used this year | 1.49 | #81 Number of CT scans | #8 Any antibiotics used this year |
| 4 | #32 Episodes of adalimumab | 1.80 | #32 Episodes of adalimumab | 1.42 | #90 Any TB tested this year | #3 Number of days rectal aminosalicylates used |
| 5 | #1 Any IBD medication use | 1.58 | #78 Any hepatitis B vaccination this year | 1.32 | #69 Number of sedimentation rate tests | #4 Episodes of rectal aminosalicylates |
| Ridge Logistic (AUC = 0.64; Brier score = 0.97) | OR | LASSO Logistic (AUC = 0.71; Brier score = 0.22) | OR | Random Forest (AUC = 0.71; Brier score = 0.21) | Neural Network (AUC = 0.58; Brier score = 0.03) | |
|---|---|---|---|---|---|---|
| IBD-related surgery | ||||||
| 1 | #11 Any budesonide this year | 4.85 | #108 Any severe disease this year | 1.96 | #33 Any infliximab used this year | #3 Number of days rectal aminosalicylates used |
| 2 | #65 Number of acute IBD surgeries | 3.32 | #11 Any budesonide this year | 1.78 | #44 Number of IBD claims | #2 Any rectal aminosalicylates used this year |
| 3 | #54 Any IBD-related GI visits | 3.18 | #65 Number of acute IBD surgeries | 1.76 | #81 Number of CT scans | #5 Any oral aminosalicylates used this year |
| 4 | #84 Any MR scans this year | 2.48 | #84 Any MR scans this year | 1.68 | #82 Any CT scans this year | #17 Any systemic steroids used |
| 5 | #20 Episodes of long-term steroids | 2.48 | #20 Episodes of long-term steroids | 1.68 | #51 Number of IBD office visits | #16 Episodes of rectal steroids |
In this table, we showcase the features that were most predictive for our four main outcomes
Additionally the features are broken down by the different statistical models used. The performance of the Support Vector Machine was excluded because of its overall poor performance
Lastly, alongside general conclusions about the patient population and risk assessments, these models can be used to evaluate and rank clinical recommendations at the patient level. In this way, the models can be used in conjunction with clinical knowledge to motivate actionable, tailored recommendations that are aimed at de-escalating the patient to a lower risk category. Returning to our example of the average patient, we can consider changes to their features that reduce the risk of hospitalization. By examining each feature individually, the model finds that similar patients to this one benefit from a Clostridium difficile stool test. Specifically, our patient is forecasted to see a reduction in their probability of being hospitalized from 0.41 to approximately 0.29 as a result of this intervention. Between these three applications of our results to clinical practice, it is clear that the models we have found provide the foundation for a novel, targeted approach to data-driven IBD care.
Future Outlook
Looking ahead, the practical reality of AI is an enigma to many practitioners (See Fig. 1 and Table 1). With boundless publications discussing the new wealth of electronic databases and promises of “Big Data,” most never go into details about what exactly these new technologies are doing to, for example, “outperform cardiologists reading EKGs” [9]. Unlike the days of small data sets collected through calculated experiment and observation, these data cannot be studied with the standard methods of statistical analysis [9]. The computations that are generally feasible in experimental settings require vast computational resources when the data are on the order of millions of observations. Therefore, smarter algorithms were created to perform statistical analysis on large data sets. Many would refer to this jump as the development of Machine Learning (ML), but formally it is closer to the sub-field of Computational Statistics. The real jump to ML utilizes the vast amounts of data in a sophisticated way that emphasizes accurate predictions of outcomes over significance and interpretability [9]. With this mindset change, outcomes can be evaluated by experts and the entire process can be incorporated into decision support in daily clinical practice. Now, without much effort from the user, algorithms can make predictions given new data and automatically make a recommendation or perform some action, appearing to have Artificial Intelligence (AI) [9]. With the increase in computational power and abundance of longitudinal patient data, applying machine learning and its subset of Deep Learning in Big Data sets has become feasible. In this study, we provide the first steps in this direction. Kim et al. have already showcased transferability of these models to different institutions, alleviating a major concern [19]. The next step would be to integrate these models in a prospective setting to study their performance on reliability, patient outcomes and costs.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- AI
Artificial intelligence
- ML
Machine learning
- EMR
Electronic medical record
Author contributions
AZ contributed to study concept and design; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; statistical analysis; study supervision. ZS contributed to study concept and design; drafting of the manuscript; analysis and interpretation of data; statistical analysis. AS contributed to drafting of the manuscript; analysis and interpretation of data; WKD contributed to study concept and design; critical revision of the manuscript for important intellectual content. DH contributed to study concept and design; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; statistical analysis; study supervision.
Declarations
Conflict of interest
Aria Zand: No conflicts of interest or financial support to disclose. Zack Stokes: no conflicts of interest or financial support to disclose. Arjun Sharma: no conflicts of interest or financial support to disclose. Welmoed K. van Deen received speaker fees from Ferring Pharmaceuticals. Daniel Hommes: no conflicts of interest or financial support to disclose.
Footnotes
An editorial commenting on this article is available at 10.1007/s10620-022-07511-x.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pariente B, Cosnes J, Danese S, et al. Development of the Crohn’s disease digestive damage score, the Lémann score. Inflammatory Bowel Dis. 2011;17(6):1415–1422. doi: 10.1002/ibd.21506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kappelman MD, Rifas-Shiman SL, Porter CQ, et al. Direct Health Care Costs of Crohn’s disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135(6):1907–1913. doi: 10.1053/j.gastro.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Haens G, Baert F, van Assche G, et al. Early combined immunosuppression or conventional management in patients with newly diagnosed Crohn’s disease: an open randomised trial. The Lancet. 2008;371(9613):660–667. doi: 10.1016/S0140-6736(08)60304-9. [DOI] [PubMed] [Google Scholar]
- 4.Kang B, Choi SY, Kim HS, et al. Mucosal healing in paediatric patients with moderate-to-severe luminal Crohn’s disease under combined immunosuppression: Escalation versus early treatment. J Crohn’s Colitis. 2016;10(11):1279–1286. doi: 10.1093/ecco-jcc/jjw086. [DOI] [PubMed] [Google Scholar]
- 5.Olivera P, Danese S, Jay N, et al. Big data in IBD: a look into the future. Nat Reviews Gastroenterol Hepatol. 2019;16(5):312–321. doi: 10.1038/s41575-019-0102-5. [DOI] [PubMed] [Google Scholar]
- 6.van der Valk ME, Mangen MJJ, Severs M, et al. Evolution of costs of inflammatory bowel disease over two years of follow-up. PLoS ONE. 2016;11:e0142481. doi: 10.1371/journal.pone.0142481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Statista. Global AI software market size 2018–2025 | Statista. Tractica. Published 2019. Accessed July 19, 2020. https://www.statista.com/statistics/607716/worldwide-artificial-intelligence-market-revenues/
- 8.Office-based Physician Electronic Health Record Adoption. Accessed June 24, 2020. https://dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php
- 9.Derrington D. Artificial Intelligence for Health and Health Care.; 2017. Accessed June 23, 2020. https://pdfs.semanticscholar.org/4f32/7be94508a5c1f2a6f09917d7dcf57698af24.pdf
- 10.Waljee AK, Lipson R, Wiitala WL, et al. Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients using machine learning. Inflammatory Bowel Dis. 2018;24(1):45–53. doi: 10.1093/ibd/izx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waljee AK, Liu B, Sauder K, et al. Predicting corticosteroid-free biologic remission with Vedolizumab in Crohn’s Disease. Inflammatory Bowel Dis. 2018;24(6):1185–1192. doi: 10.1093/ibd/izy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waljee AK, Wallace BI, Cohen-Mekelburg S, et al. Development and validation of machine learning models in prediction of remission in patients with moderate to severe Crohn disease. JAMA Network Open. 2019;2(5):e193721. doi: 10.1001/jamanetworkopen.2019.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughn DA, van Deen WK, Kerr WT, et al. Using insurance claims to predict and improve hospitalizations and biologics use in members with inflammatory bowel diseases. J Biomed Inform. 2018;81:93–101. doi: 10.1016/j.jbi.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Wei Z, Wang W, Bradfield J, et al. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Human Genetics. 2013;92(6):1008–1012. doi: 10.1016/j.ajhg.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menti E, Lanera C, Lorenzoni G, et al. Bayesian Machine Learning Techniques for revealing complex interactions among genetic and clinical factors in association with extra-intestinal Manifestations in IBD patients. AMIA Annu Symp Proc. 2016;2016:884–893. [PMC free article] [PubMed] [Google Scholar]
- 16.Han L, Maciejewski M, Brockel C, et al. A probabilistic pathway score (PROPS) for classification with applications to inflammatory bowel disease. Bioinformatics. 2018;34(6):985–993. doi: 10.1093/bioinformatics/btx651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai T, Lin TC, Bond A, et al. The association between arthralgia and vedolizumab using natural language processing. Inflammatory Bowel Dis. 2018;24(10):2242–2246. doi: 10.1093/ibd/izy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou JK, Chang M, Nguyen T, et al. Automated identification of surveillance colonoscopy in inflammatory bowel disease using natural language processing. Digestive Dis Sci. 2013;58(4):936–941. doi: 10.1007/s10620-012-2433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim E, Caraballo PJ, Castro MR et al. Towards more Accessible Precision Medicine: Building a more Transferable Machine Learning Model to Support Prognostic Decisions for Micro- and Macrovascular Complications of Type 2 Diabetes Mellitus. J Med Syst. 2019;43(7). 10.1007/s10916-019-1321-6 [DOI] [PubMed]
- 20.Nori VS, Hane CA, Martin DC et al. Identifying incident dementia by applying machine learning to a very large administrative claims dataset. PLoS ONE. 2019;14(7). 10.1371/journal.pone.0203246 [DOI] [PMC free article] [PubMed]
- 21.Chen S, Bergman D, Miller K et al. Using applied machine learning to predict healthcare utilization based on socioeconomic determinants of care. Am J Managed Care. 2020;26(1):26–31. 10.37765/ajmc.2020.42142 [DOI] [PubMed]
- 22.Xiao J, Ding R, Xu X, et al. Comparison and development of machine learning tools in the prediction of chronic kidney disease progression. J Translational Medi. 2019;17(1). 10.1186/s12967-019-1860-0 [DOI] [PMC free article] [PubMed]
- 23.Chiu YC, Chen HIH, Zhang T, et al. Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Medical Genomics. 2019;12(Suppl 1). 10.1186/s12920-018-0460-9 [DOI] [PMC free article] [PubMed]
- 24.Kinney ED. Health Insurance Coverage in the United States. In: Protecting American Health Care Consumers; 2020:23–40. 10.2307/j.ctv11smv14.6
- 25.OptumLabs and OptumLabs Data Warehouse (OLDW) Descriptions and Citation. Cambridge, MA: n.p., May 2019. PDF. Reproduced with permission from OptumLabs.
- 26.Hastie T, Tibshirani R, Friedman J. Elements of Statistical Learning 2nd Ed.; 2009.
- 27.Understanding ROC AUC: Pros and Cons. Why is Bier Score a Great Supplement? | by TinaGongting | Medium. Accessed November 21, 2020. https://medium.com/@penggongting/understanding-roc-auc-pros-and-cons-why-is-bier-score-a-great-supplement-c7a0c976b679
- 28.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Ann Internal Med. 2015;162(1):55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 29.Stewart M. The Limitations of Machine Learning | by Matthew Stewart, PhD Researcher | Towards Data Science. Published 2019. Accessed July 26, 2020. https://towardsdatascience.com/the-limitations-of-machine-learning-a00e0c3040c6
- 30.Mao R, Xiao Y, Gao X, et al. Fecal calprotectin in predicting relapse of inflammatory bowel diseases: A meta-analysis of prospective studies. Inflammatory Bowel Dis. 2012;18(10):1894–1899. doi: 10.1002/ibd.22861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.