Abstract
Carbon catabolite protein A (CcpA) is a global regulator of carbon metabolism in gram-positive bacteria, repressing transcription of genes for the utilization of secondary carbon sources in the presence of a readily metabolized carbon source and activating transcription of genes, such as ackA and pta, that are required for carbon excretion. The promoter region of the Bacillus subtilis ackA gene contains two catabolite responsive elements (cre sites), of which only the site closest to the promoter (cre2) binds CcpA to activate transcription. A region immediately upstream of the cre2 site is also important for transcriptional activation. The required elements in this region were further defined by mutagenesis. CcpA binds to the ackA promoter region in gel shift assays even in the presence of mutations in the upstream element that block transcriptional activation, indicating that this region has a function other than promoting binding of CcpA.
Carbon catabolite repression in Bacillus subtilis and other gram-positive bacteria is controlled by a mechanism different from that in gram-negative bacteria. The key regulator is carbon catabolite protein A (CcpA), which represses the transcription of various genes encoding proteins involved with the utilization of secondary carbon sources (12). CcpA also activates the transcription of genes involved in carbon excretion. These genes include pta and ackA, which function together to convert acetyl coenzyme A to acetate for excretion into the growth medium (11, 22, 26).
The CcpA protein is a member of the LacI-GaIR family of transcriptional repressors (13). Members of this family contain an amino-terminal helix-turn-helix DNA binding domain and carboxy-terminal regions involved with effector recognition and oligomerization (28). The activity of CcpA is controlled by HPr or the HPr homologue Crh, both of which are phosphorylated by an ATP-dependent kinase during growth in glucose (8, 9, 23, 24). Mutations which block this signaling pathway cause loss of glucose repression of many target genes and loss of transcriptional activation of ackA and pta (1, 2, 22, 27). Activation of ackA expression during growth in glucose is dependent on the cre2 CcpA binding site (centered at −56.5 relative to the transcription start site) and sequences upstream of cre2 (11, 27). The molecular mechanism of transcriptional activation by CcpA is unknown, and the role of the region upstream of the CcpA binding site has not been characterized.
Deletion analysis of the ackA upstream region.
Deletions of the region upstream of cre2 were generated by PCR, and transcriptional fusions to lacZ were generated using the plasmid pFG328 (11) and inserted in single copy into the B. subtilis chromosome by recombination into an SPβ prophage. Expression of the ackA-lacZ fusions was monitored during growth in TSS medium (5) containing 1% Casamino Acids (Difco) in the presence or absence of glucose (1%). Deletion constructs up to and including ACKBAM11, which contains 28 bp of sequence upstream of cre2, exhibited normal induction of ackA-lacZ expression (Table 1). ACKBAM6, containing 23 bp upstream of cre2, exhibited a small decrease in activation. These results map the 5′ end of the sequence elements required for complete activation of the ackA-lacZ fusion in the presence of glucose to between 23 and 28 bp upstream of cre2.
TABLE 1.
Deletion analysis of the 5′ end of the ackA promoter region
| ackA-lacZ fusion construct | ackA DNA | β-Galactosidase activitya for BR151MA
|
|
|---|---|---|---|
| −Glucose | +Glucose | ||
| Wild type | cre2 + 987 bpb | 150 | 500 |
| ACKBAM7 | cre2 + 33 bp | 180 | 500 |
| ACKBAM11 | cre2 + 28 bp | 140 | 500 |
| ACKBAM6 | cre2 + 23 bp | 110 | 360 |
| ACKKPN2 | cre2 + 11 bp | 50 | 50 |
Cells were grown in TSS medium (5) containing 1% Casamino Acids or 1% Casamino Acids plus glucose (1%). β-Galactosidase activities are expressed in Miller units (20) and indicate activity at 30 min prior to the time of entry of the culture into stationary phase. Growth experiments were performed at least two times and showed less than 10% variation.
The wild-type fusion contains a 1.35-kb EcoRI fragment, including 987 bp upstream of cre2, inserted into the plasmid pFG328 (11). Deletion constructs represent a 5′ truncation of this fragment and were generated by PCR using the 1.35-kb fragment as a template. All constructs were verified by DNA sequencing.
Random mutagenesis.
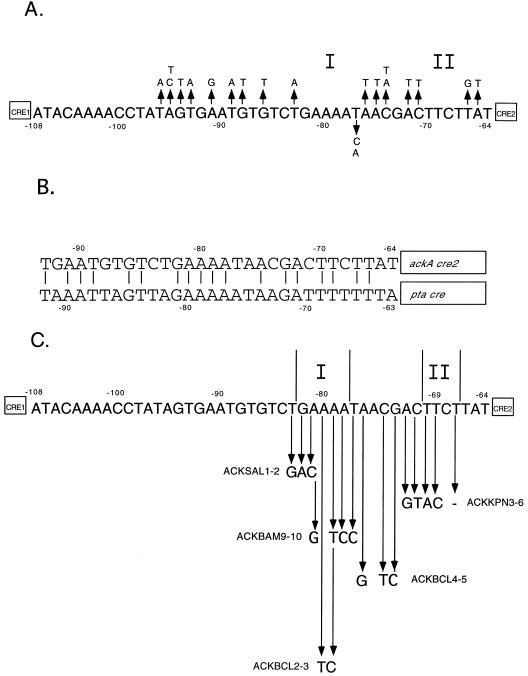
To identify the sequence elements required for activation, the 33 bp upstream of cre2 were randomly mutagenized by amplifying this region using an oligonucleotide containing 6% non-wild-type bases (94% wild-type) at each position in the region. This frequency of doping is predicted to create a pool of oligonucleotides with 1 to 3 mutations per individual oligonucleotide with a ≤1% probability of the wild-type sequence (15). The oligonucleotide included 13 bp of wild-type cre2 sequence at its 3′ end to ensure efficient annealing of primers containing mismatches at the 3′ end of the target sequence. The resulting pool of PCR fragments was inserted into the plasmid pFG328 to generate ackA-lacZ transcriptional fusions, and the plasmids were propagated as mixed pools and then introduced into the B. subtilis chromosome by recombination of the fusion into an SPβ prophage. Isolates that retained normal activation of ackA-lacZ transcription during growth on tryptose blood agar base (Difco) containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (40 μg/ml) and glucose (1%) were selected for further analysis, and the DNA sequence of the ackA upstream region was determined. Of the 34 candidates chosen, 17 were wild type, 2 had three mutations, 4 had two mutations, and 11 had one mutation. The results revealed two sequence elements in which no mutations appeared, the first between positions −82 and −77 (region I) and the second between positions −70 and −67 (region II) (Fig. 1A). These elements were presumed to be important for the activation of transcription.
FIG. 1.
Sequence of the ackA upstream region. Numbering is relative to the transcription start site. (A) Random mutagenesis of the region between nucleotides −96 and −64. Up arrows indicate mutations that allowed a significant degree of activation of the ackA promoter in the presence of glucose. Down arrows indicate mutations that did not allow significant activation. Two triple mutations were obtained: A(−95)C A(−91)G G(−86)T and G(−94)T A(−91)G T(−89)A. Four double mutations were obtained: A(−76)T A(−72)T, A(−95)T A(−72)T, A(−95)C C(−74)A, and T(−93)A C(−74)T. Eleven single mutations were obtained: A(−65)T, T(−66)G, C(−71)T, C(−74)A, A(−75)T, A(−76)T, T(−77)C, T(−77)A, T(−83)A, G(−88)T, and T(−96)A. (B) Alignment of sequences upstream of the ackA and pta cre sites. (C) Site-directed mutagenesis. Mutations were generated by PCR and introduced restriction sites at the positions shown. ACKKPN3-6 deleted a T residue at position −67. ACKBCL1-2T is the same as ACKBCL2-3 with the introduction of an additional T residue between positions −69 and −70.
The importance of these elements is also supported by conservation between the ackA and pta upstream regions (Fig. 1B), suggesting that transcriptional activation of ackA and that of pta operate by similar mechanisms. Interestingly, 7 of the 12 differences between the ackA and pta regions were found as single mutations in the random mutagenesis and therefore do not drastically affect expression of ackA. Four of the remaining five differences are at positions which were invariant in these experiments but are surrounded by positions which did change. The final difference between ackA and pta is within region II [C(−68)T], suggesting that T may be allowed at this position or that there are context-specific effects.
β-Galactosidase measurements for isolates with the mutations T(−96)A, T(−83)A, T(−77)A, C(−74)A, and T(−66)G, each of which contained only a single mutation, are shown in Table 2. The activation ratio ranged from 2.5- to 3.6-fold, relative to 3.4-fold for the wild-type fusion, and the expression level varied from half to twice that of the wild-type fusion. The C(−74)A mutant exhibited a basal level and glucose-activated transcriptional level two-fold over the wild-type level. This fusion was confirmed to be in single copy (data not shown). To determine if this increase in expression was dependent on CcpA, the mutant fusion was introduced into a ccpA mutant strain. While no activation in the presence of glucose was observed, the basal activity of the fusion was still increased over that of the wild-type fusion, indicating that CcpA is required for activation but that some other factor is leading to the increase in basal activity.
TABLE 2.
Effect of upstream region mutations on ackA-lacZ expression
| Mutation | β-Galactosidase activitya for:
|
|||
|---|---|---|---|---|
| BR151MA
|
BR151MACcp::Spcb
|
|||
| − Glucose | + Glucose | − Glucose | + Glucose | |
| None | 120 | 410 | 60 | 64 |
| T(−96)A | 140 | 400 | NTc | NT |
| T(−83)A | 180 | 430 | NT | NT |
| T(−77)A | 60 | 130 | NT | NT |
| C(−74)A | 240 | 860 | 160 | 160 |
| T(−66)G | 75 | 270 | NT | NT |
| ACKBAM9–10 | 45 | 80 | 32 | 32 |
| ACKKPN3–6 | 50 | 85 | NT | NT |
| ACKBCL1–2T | 50 | 98 | NT | NT |
| ACKBCL2–3 | 52 | 130 | 41 | 41 |
| ACKSAL1–2 | 35 | 75 | NT | NT |
| ACKBCL4–5 | 45 | 98 | NT | NT |
Cells were grown in TSS medium containing 1% Casamino Acids or 1% Casamino Acids plus glucose (1%). β-Galactosidase activities are expressed in Miller units (20) and indicate activity at 30 min prior to the time of entry of the culture into stationary phase. Growth experiments were performed at least two times and showed less than 10% variation.
BR151MACcp::Spc contains a null allele of ccpA (10).
NT, Not tested.
Site-directed mutagenesis of the region upstream of cre2.
Site-directed mutagenesis was performed on the sequence elements highlighted by the random mutagenesis (Fig. 1C). Each mutation drastically affected activation in the presence of glucose, confirming the importance of these sequence elements (Table 2). These constructs were generated in the context of the entire ackA upstream region, indicating that the presence of cre1 does not obviate the requirement for these elements. The ACKBAM9-10 and ACKBCL2-3 mutant fusions were also introduced into BR151MACcpA::Spc, and the residual activation was found to require CcpA.
Binding of CcpA to the ackA upstream region.
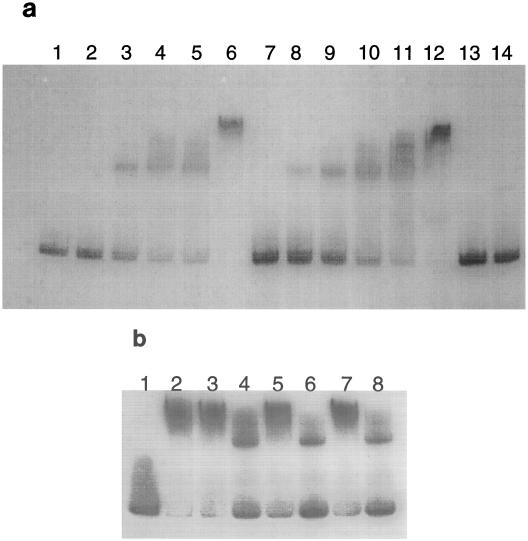
The sequence elements required for activation could function as a protein binding site, assist in the binding of CcpA to the cre2 site, or confer a DNA conformation required for activation of the ackA promoter. To test the hypothesis that the region upstream of ackA cre2 is important for CcpA binding, a gel mobility retardation assay was performed using wild-type and mutant DNA fragments. CcpA was purified to homogeneity by incorporating a six-histidine tag at the amino terminus followed by passage through a nickel-nitrilotriacetic acid (Ni-NTA) column, essentially as described for Bacillus megaterium CcpA (1). The His-tagged CcpA bound efficiently to a 163-bp wild-type ackA cre2-containing DNA fragment (Fig. 2a). It was previously shown that mutagenesis of cre2 conferred a loss of transcriptional activation from the ackA promoter, most likely due to the lower affinity of CcpA for the mutant cre2 site (27). It was determined that CcpA can still bind in vitro to the DNA fragment containing the mutant cre2, but with a significant reduction in affinity (data not shown). Since the sequence upstream of cre2 might affect CcpA binding to cre2, binding of CcpA to the mutant DNA fragments was tested. As represented by the site-directed mutant ACKKPN3-6, CcpA was still able to bind the fragments in each site-directed mutant (Fig. 2a and data not shown). In a competition assay (Fig. 2b), ACKSAL1-2 and ACKBAM9-10 DNA fragments were able to sequester CcpA from the labeled wild-type DNA as efficiently as did unlabeled wild-type DNA. It therefore appears that the upstream elements are not required for CcpA binding.
FIG. 2.
(a) Binding of CcpA-His to an ackA cre2-containing DNA fragment. The ackA DNA (163 bp; positions −139 to +24 relative to the transcription start site) was generated by PCR, labeled using T4 polynucleotide kinase and [γ-32P]ATP, and purified with the Qiagen QIAquick PCR purification kit. Mutant DNA was generated in parallel to the wild-type fragment. The nonspecific DNA probe (169 bp) contained a portion of the ccpA coding region. The intact ccpA coding region was isolated by PCR and inserted into the plasmid pQE30 (Qiagen) to generate the amino-terminal six-histidine tag, introduced into E. coli M15(pREP4) (Qiagen), and expressed upon addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 2 mM. The His-tagged protein was purified using nickel-nitrilotriacetic acid spin columns (Qiagen), and imidazole (250 mM) was used to elute the protein. Probe DNA (1 ng) was incubated with CcpA in a solution containing 10 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 1 mM EDTA, 50 mM KCl, 5% glycerol, 50 μg of bovine serum albumin/ml, and 0.05% igepal CA-630 (Sigma) in a 20-μl volume for 10 to 30 min at 37°C (18). Samples were mixed with nondenaturing sample buffer (40% sucrose, 0.25% bromphenol blue) and loaded directly onto a 5% polyacrylamide gel prepared in 6.7 mM Tris-HCl (pH 8.0)–1 mM EDTA–2.5% glycerol. Gels were run at 200 V for 1.5 h in a Protean II xi Cell (Bio-Rad) gel electrophoresis unit, dried, and exposed to X-ray film (Kodak) for autoradiography. Lanes 1 to 6, wild-type DNA (1 ng); lanes 7 to 12, ACKKPN3-6; lanes 13 and 14, CcpA9-10 (nonspecific sequence). Lanes 1, 7, and 13, no protein; lanes 2 and 8, 1 ng of CcpA-His (0.66 nM); lanes 3 and 9, 3 ng of CcpA-His (2.0 nM); lanes 4, 10, and 14, 7 ng of CcpA-His (4.6 nM); lanes 5 and 11, 10 ng of CcpA-His (6.6 nM); lanes 6 and 12, 20 ng of CcpA-His (13 nM). (b) Competition gel retardation assay. Unlabeled wild-type and mutant DNA was tested for the ability to sequester CcpA-His from the labeled wild-type probe (1 ng). The unlabeled DNA contained wild-type sequence (lanes 3 and 4), ACKSAL1-2 sequence (lanes 5 and 6), or ACKBAM9-10 sequence (lanes 7 and 8). Lanes 3, 5, and 7, 10 ng of unlabeled DNA; lanes 4, 6, and 8, 100 ng of unlabeled DNA; lanes 2 to 8, 50 ng of CcpA (33 nM); lane 1, no protein; lane 2, no competing DNA.
The requirement for these DNA elements for transcriptional activation of ackA suggests that additional factors may be involved. It is known that phosphorylated HPr or Crh is required for activation in vivo (27); however, in other systems these proteins have been shown to affect the efficiency of binding of CcpA to cre sites without directly contacting the DNA themselves (6, 7). Another unknown protein may be binding to the region upstream of the cre2 site. RNAP has been shown to respond to two bound activators to initiate transcription (14). For example, in an artificial promoter construct, λcI contacts the ς subunit of RNAP while cyclic AMP receptor protein (CRP) contacts the carboxy-terminal domain of the α subunit (α-CTD) of RNAP to stimulate transcriptional initiation (16). In the activation of the divergent malEp and malKp promoters, CRP effects a shift of the activator MalT to its functional sites in phase with the −10 region of the promoter to allow a favorable interaction with RNAP (25). For the proP P2 promoter, CRP (at −121.5) and Fis (at −41) act together to stimulate transcription initiation (19); each of these proteins individually makes contact with the α-CTD portion of RNAP. It is possible that some form of coactivation is occurring at the ackA promoter, which includes binding of CcpA to cre2 and binding of some other factor to the region upstream of cre2. Alternatively, the upstream region may interact directly with RNAP, perhaps functioning as an UP element to contact the α-CTD; if so, activation must be dependent on CcpA binding, perhaps to reposition the upstream region to permit contact with RNAP. Proteins such as integration host factor have been shown to bend the DNA to allow optimal contact between the subunits of RNAP and a DNA sequence or another protein binding upstream of the promoter (21). UP elements, which like the ackA upstream region contain A- and T-rich sequences, have been identified upstream of Escherichia coli and B. subtilis promoters and have been shown to interact with α-CTD (3). This interaction between the UP element and the α subunit of RNAP may assist the binding of RNAP to the promoter and further steps of transcription initiation (4). CcpA could also bend the DNA to facilitate an interaction between RNAP and region I and/or region II.
Further study will be required to determine the function of the upstream sequence elements and the molecular mechanism of transcriptional activation. Since these elements are conserved in pta, it is likely that activation is similar for the two genes; this would provide coordinate expression consistent with cotranscription of these genes in E. coli (17).
Acknowledgments
We thank Gregory Booton for technical assistance.
This work was supported by grant MCB-9723091 from the National Science Foundation.
REFERENCES
- 1.Deutscher J, Kuster E, Bergstedt U, Charrier U, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsHI gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estrem S T, Gaal T, Ross W, Gourse R L. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Estrem S T, Ross W, Gaal T, Chen Z W S, Niu W, Ebright R H, Gourse R L. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of RNA polymerase α subunit. Genes Dev. 1999;13:2134–2147. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher S H, Rosenkrantz M S, Sonenshein A L. Glutamine synthase gene of Bacillus subtilis. Gene. 1984;32:427–438. doi: 10.1016/0378-1119(84)90018-0. [DOI] [PubMed] [Google Scholar]
- 6.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 7.Galinier A, Deutscher J, Martin-Verstraete I. Phosphorylation of either Crh or HPr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. J Mol Biol. 1999;286:307–314. doi: 10.1006/jmbi.1998.2492. [DOI] [PubMed] [Google Scholar]
- 8.Galinier A, Haiech J, Kilhoffer M, Jaquinod M, Stulke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundy F J, Turinsky A J, Henkin T M. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundy F J, Waters D A, Allen S H G, Henkin T M. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J Bacteriol. 1993;175:7348–7355. doi: 10.1128/jb.175.22.7348-7355.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henkin T M. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 13.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 14.Hochschild A, Joung J K. Synergistic activation of transcription in Escherichia coli. In: Eckstein F, Lilley D M J, editors. Nucleic acids and molecular biology. Vol. 11. Berlin, Germany: Springer-Verlag; 1997. pp. 101–114. [Google Scholar]
- 15.Horwitz B H, DiMaio D. Saturation mutagenesis using mixed oligonucleotides and M13 templates containing uracil. Methods Enzymol. 1990;185:599–611. doi: 10.1016/0076-6879(90)85047-r. [DOI] [PubMed] [Google Scholar]
- 16.Joung J K, Koepp D M, Hochschild A. Synergistic activation of transcription by bacteriophage λcl protein and Escherichia coli cAMP receptor protein. Science. 1994;265:1863–1865. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- 17.Kakuda H, Hosono K, Shiroishi D, Ichihara S. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J Biochem. 1994;116:916–922. doi: 10.1093/oxfordjournals.jbchem.a124616. [DOI] [PubMed] [Google Scholar]
- 18.Kim J-H, Guvener Z T, Cho J Y, Chung K-C, Chambliss G H. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J Bacteriol. 1995;177:5129–5134. doi: 10.1128/jb.177.17.5129-5134.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLeod S M, Xu J, Johnson R C. Coactivation of the RpoS-dependent proP P2 promoter by Fis and cyclic AMP receptor protein. J Bacteriol. 2000;182:4180–4187. doi: 10.1128/jb.182.15.4180-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 21.Perez-Martin J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 22.Presecan-Siedel, Galinier E-A, Longin R, Deutscher J, Danchin A, Glaser P, Martin-Verstraete I. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J Bacteriol. 1999;181:6889–6897. doi: 10.1128/jb.181.22.6889-6897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reizer J, Hoischen C, Tigemeyer F, Rivolta C, Rabus R, Stulke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 24.Reizer J, Deutscher J, Saier M H., Jr Metabolite-sensitive, ATP-dependent, protein kinase catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system in Gram-positive bacteria. Biochimie. 1989;71:989–996. doi: 10.1016/0300-9084(89)90102-8. [DOI] [PubMed] [Google Scholar]
- 25.Richet E, Sogaard-Anderson L. CRP induces the repositioning of MalT at the Escherichia coli malKp promoter primarily through DNA bending. EMBO J. 1994;13:4558–4567. doi: 10.1002/j.1460-2075.1994.tb06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin B-S, Choi S-K, Park S-H. Regulation of the Bacillus subtilis phosphotransacetylase gene. J Biochem. 1999;126:333–339. doi: 10.1093/oxfordjournals.jbchem.a022454. [DOI] [PubMed] [Google Scholar]
- 27.Turinsky A J, Grundy F J, Kim J-H, Chambliss G H, Henkin T M. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J Bacteriol. 1998;180:5961–5967. doi: 10.1128/jb.180.22.5961-5967.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weickert M J, Adhya S. A family of bacterial regulators homologous to gal and lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]